6.1. Background
Cancer treatment across multiple solid and hematologic malignancies has been revolutionized with the approval of checkpoint inhibitor therapy (CIT) and adoptive cell therapy (ACT). However, clinical benefit is derived in only a subset of patients. The next frontier in cancer immunotherapy is to unveil the innate and/or adaptive mechanisms of immune resistance that limit the efficacy of therapy in the majority of cancer patients [1]. A prevailing phenomenon observed in immune resistant cancers is an absent dialog between the cancer and immune cells [2]. This occurs in the context of “immune silent” [3, 4] and, even perhaps, “immune-excluded” tumors [2, 5, 6]. The latter are characterized by T cells restricted to the periphery of cancer nests. Since the activated T cells do not come in direct contact with the cancer cells, CIT is rendered immaterial unless the tumor microenvironment (TME) can be reprogrammed to facilitate T cell homing and infiltration [7].
Similarly, ACT may be affected by immune exclusion processes that limit direct T cell contact with targeted antigen-expressing cancer cells. More than two decades ago, Pockaj et al. [8] observed that in a great proportion of cases, adoptively transferred tumor infiltrating lymphocytes (TIL) labeled with radioactive 111Indium, which facilitates their tracking in vivo, did not localize to the targeted metastatic tumor sites. None of the patients in whom TILs did not traffic to the metastatic lesions experienced tumor regression suggesting that TIL localization to the tumor is an absolute requirement for the success of ACT. Furthermore, even successful homing of TILs to the tumor site was not entirely sufficient as only a proportion of patients responded to therapy even under this circumstance underlining the principle that cancer immune resistance results from distinct biologic processes [9]. We observed in patients with melanoma that response to TIL therapy is associated with the local expression of CXCR3 and CCR5-ligand chemokines capable of recruiting activated T cells to the metastatic site [10]. These observations highlight the multifactorial requirements needed to achieve an effective anti-tumor immune response [8, 10]. One may hypothesize that in the case of successful trafficking of TILs to the target lesions without accompanying tumor regression, immune exclusion mechanisms may have prevented the appropriately homed T cells from infiltrating the tumor nest and employing their full effector functions. Such hypothesis, however, has never been tested.
While it may be intuitive to attribute the silent immune landscape to a lack of tumor cell immunogenicity due to low tumor mutational burden [11, 12] and/or loss of the antigen processing and presentation machinery [13], immune exclusion may represent a more complicated phenomenon. The presence of T cells at the periphery of tumor nests suggests that chemo-attraction recruits the T cells to the tumor microenvironment and an antigenic stimulus promotes their persistence. However, a barrier prevents the T cells from infiltrating the tumor nests and to engage in direct cytotoxic cancer cell killing. Various hypotheses suggesting mechanical and/or functional barriers (Table 6.1) have been raised but no definitive consensus has been reached about the clinical relevance that each of them may play in human cancers.
Table 6.1.
Hypotheses raised to explain immune exclusion (N.B. mechanisms may not be mutually exclusive)
| Mechanical barriers | Physical impediment to a direct contact between T cells and cancer cells | |
|---|---|---|
| Stromal fibrosis | Filaggrin and desmosomal proteins | [27] |
| Endothelin B receptor | [53, 66, 67] | |
| Defects in physical interaction between immune cells and TME | [62] | |
| Transforming growth factor (TGF)-β-induced fibrosis | [26, 63, 64] | |
| Epithelial mesenchymal transition | [57–61] | |
| Vascular access | Vascular endothelial growth factor (VEGF) | [68, 69] |
| Functional Barriers | Biological or metabolic interactions between cancer, stromal and immune cells limiting migration, function, and/or survival of T cells | |
| Metabolic barriers | Nutrient depletion by cancer cells | [125–127] |
| Warburg and reverse Warburg effect (lacto-genesis) | [73, 76, 78, 79, 127] | |
| Metabolic reprogramming of immune cells | [48, 128] | |
| Altered lipid metabolism | [129] | |
| Hypoxia | [80–85] | |
| K+ levels | [74, 75] | |
| Glutaminase-dependent metabolism | [130] | |
| Metabolic inhibitors | [125, 131–134] | |
| Cyclooxygenase and prostaglandin metabolism | [135–137] | |
| Soluble factors | Cytokine/chemokine gradients | [28, 81, 82, 100, 101] |
| VEGF-α-mediated immune suppression | [68, 69, 87, 89, 92] | |
| TGF-β | [26, 63, 93, 94] | |
| Tumor-associated immune and stromal infiltrate suppressive mechanisms | [76, 101, 138–144] | |
| Danger sensing | Tolerogenic cell death/absent immunogenic cell death | [97–99] |
| Adenosine signaling | [101–103, 105] | |
| TAM receptor tyrosine kinases | [6, 106–108, 145] | |
| “Don’t eat me” signals (CD47/signal regulatory protein (SIRP)-α axis) | [70–72] | |
| Tumor cell-intrinsic signaling | Tumor cell-intrinsic β-catenin/signaling | [54–56, 121, 122] |
| Extended PI3K pathway signaling | [115–120] | |
| Tumor cell-intrinsic STAT-3 activation | [111–114] | |
| Tumor cell-intrinsic MAPK signaling | [16, 45] | |
| Dynamic Barriers | Interactions between cancer and T cells resulting in limited function | |
| Checkpoint receptor/ligand interactions | [46–48] | |
6.2. Immune Infiltration and the Continuum of Immune Surveillance
Immunohistochemical (IHC) documentation of CD8+ T cells infiltration of solid cancers is a favorable prognostic biomarker [14–16], whereas a high density of CD163+ tumor associated macrophages (TAMs) is associated with poor survival [17]. Moreover, high ratios of CD8+ T cells over CD68+ myeloid cells or CD163+ TAMs bear favorable predictive and prognostic implications in patients with various cancers [18, 19]. However, an in-depth understanding of the contribution of the presence of distinct immune cell subsets within the TME can be better interpreted when morphological description is combined with functional characterization [3, 20, 21]. Representative transcriptional patterns such as the immunologic constant of rejection (ICR) [20, 22] and the tumor inflammation signature (TIS) [3] include evidence of the activation of interferon (IFN)-γ signaling, immune effector mechanisms and the expression of CCR5 and CXCR3-ligand chemokines. These immune effector signatures have been confirmed as independent prognostic biomarkers and predictors of response to various types of IO agents. The ICR has been shown to predict prolonged survival in several independent data sets of breast cancer [16, 23, 24] and other cancers (Roeland’ et al. manuscript submitted). Thus, the ICR has been applied to sub-classify cancers according to degree of immune activation in association with a favorable clinical connotation [7]. Moreover, the ICR was observed to be a predictor of immune responsiveness to the systemic administration of human recombinant interleukin (IL)-2 [21] and to ACT with TILs [10]. The expression of the ICR signature is consistently accompanied by the expression of genes associated with immune regulatory function that have been implied to determine immune resistance [7]. These include transcripts representative of regulatory T cell and myeloid suppressor cell presence, and activation of the IL-23-Th17 axis, the phosphatidyl-inositol-4,5-bisphosphate 3-kinase (PI3K)-γ pathway, the checkpoint cluster, and the IDO/NOS metabolic immune suppressors [7]. In addition, a transcriptional pattern representative of ICD was found to be strongly associated with the ICR suggesting a leading role for ICD as a determinant of immune activation. The congregation of immune effector and immune regulatory mechanisms within the same cancer landscape suggests that an evolutionary balance is required for the survival of antigenic tumors in the immune-competent host that depends on offsetting immunogenicity with compensatory mechanisms of tolerance [7]. In this case, resistance of tumors to IO agents and in particular CIT, maybe due to the multiple immune suppressive forces that operate within the TME; this may explain why targeting a single pathway with monotherapy by CIT may not suffice to induce tumor rejection [7, 25]. Further, discussion about the biology of immune active cancers and related compensatory immune resistance is beyond the scope of this chapter as it was discussed elsewhere [1, 7] and is the subject of another chapter in this book (Bedognetti et al., The biology of immune active cancers and their regulatory mechanisms).
6.3. Prevalence of the Immune Excluded Phenotype
The phenomenon of immune exclusion has been described by several investigators [2, 5, 6, 26–32]. Galon et al. [14] observed that the infiltration of CD8 T lymphocytes within tumor nests combined with their peri-tumoral presence is the principal parameter that prognosticates improved survival of patients with colorectal cancer with higher accuracy than the classical TNM staging. This observation was recently validated by an international, multi-institutional study [15]. In contrast, the limited presence of T cells at the periphery of tumor nests without intra-tumoral infiltration, which corresponds to what we define as “immune exclusion,” did not bear any significant impact on survival [14, 15, 33, 34]. This observation powerfully suggests that immune exclusion represents a functionally distinct biological entity with distinct clinical implication.
It is well accepted that in most cases, individual cancers segregate according to three distinct landscapes: immune active, immune silent, and immune excluded [28] although occasionally a combination of these landscapes can be observed within the same tumor [35]. The prevalence of each landscape in cancers of different histology remains undetermined. This information may be critical for the stratification of patients during clinical trials [30, 36–40]. A recent study systematically measured by IHC the pattern of immune infiltration (“topography”) of 965 histological tissue slides from 177 patients bearing cancers of different histology [28]. The tissue specimens were categorized according to the number of cells per mm2 in three spatial compartments: outer invasive margin (0–500 μm outside the tumor invasion front), inner invasive margin (0–500 μm inside the tumor invasion front), and in the tumor core (>500 μm inside the invasion front). Preliminary analysis demonstrated that there was a strong correlation between the infiltration of the tumor core and the inner invasive margin. Thus, these two categories were combined to define immune active or “hot” tumors. Whereas, tumors with high immune cell density in the outer invasive margin and low density in the core were defined as immune “excluded” tumors (parenthetically, this is the most precise definition of immune exclusion that we have encountered so far). Low density in all compartments characterized “cold” tumors. Distinct cutoff values were used for different immune cell types including CD3+ (363 cells/mm2), CD8+ (295 cells/mm2), Foxp3+ (62 cells/mm2), PD1+ (6 cells/mm2), CD68+ (310 cells/mm2), and CD163+ cells (559 cells/mm2). Distinct tumor types displayed remarkably different patterns of immune infiltration. Cancers deemed sensitive to IO agents and in particular CIT, such as melanoma, lung adenocarcinoma, lung squamous cell carcinoma, and head and neck squamous cell carcinoma (HNSSC), displayed a high frequency of simultaneously CD3-hot, CD8-hot, and PD1-hot tumors whereas colorectal cancers presented most frequently with an immune excluded phenotype. With bivariate immune cell analysis, several reciprocally exclusive behaviors were observed: while CD3 and CD8-expressing T cell infiltrates were generally concordant, inverse correlations were observed between CD8 and Foxp3 T cells and CD68 (TAMs) infiltrates. The critical observation of this study suggested that bivariate classifications including several immune cellular components are most likely to provide clinically relevant information. An interesting example was provided by Tsujikawa et al. [30] wherein HNSSC were segregated according to the presence of oncogenic human papilloma virus (HPV) known to be associated with an immunogenic gene signature [41, 42]. The HPV positive cases were sub-classified into tumors that were predominantly expressing only lymphoid signatures and tumors that included a mixture of lymphoid and myeloid cell signatures. Interestingly, in the case of HPV-associated cancers, the worst prognostic connotation was associated with the presence of a strong myeloid infiltrate that was independent of the lymphoid infiltrate [30]. This study was performed on tissue microarrays that did not allow spatial discrimination of invasive margins versus tumor core assessment. Thus, it remains unknown whether lymphoid to myeloid ratios are also affected by spatial distribution according to the “geocentric” description of cancer immune exclusion presented in this chapter. A critical relevant observation of this study was the realization that immune excluded cancers are much more prevalent across cancer histologies than generally perceived. This observation is important because it may suggest that an opportunity for the design of next-generation IO agents is to determine the prevalent mechanisms of immune exclusion and address them to expand the effectiveness of IO approaches to cancer in which immunity is relevant but literally plays only a “peripheral” role.
We propose that the biology of immune excluded tumors is partially similar to that of the immune active as compared to the immune silent phenotype. This hypothesis is based on the premise that chemo-attraction can recruit the T cells to the tumor periphery and that some immunogenic stimulus can preserve their persistence. The question remains, however, whether additional mechanisms are required to stimulate immune infiltration that are present in hot tumors but not in immune excluded (active recruitment). Conversely, immune exclusion could be determined by chemo-repulsive mechanisms that are not present in hot tumors.
Limited observations in HNSCC suggest that immune excluded cancers are transcriptionally indistinguishable from the immune active ones. We applied the hallmark ICR signature to segregate HNSCC samples and observed that both immune excluded and immune infiltrated tumors demonstrated a full display of an immune effector Th1-like polarization of T cells. Consistent with previous observations [16, 24, 43–45], the Th1 polarization was combined with the presence of immune-suppressive mechanisms (Pai SI. et al., ASCO Abstract # 6052). Thus, the distinction between the immune excluded versus the immune active phenotypes is primarily the spatial resolution and localization of immune cell populations and these two immune phenotypes cannot be distinguished solely based on functional signatures [28]. The ICR signature includes Th1 polarization markers such as IFN-γ-related transcripts, granzyme, and perforin that are detectable upon cognate activation of T cells by antigen exposure. Thus, it appears that in the immune excluded HNSCCs studied, CD8 T cells come into contact with and recognize cancer cells at the border of the tumor nest but some functional barrier prevents their infiltration into the tumor. However, this conclusion may not apply to all cancers. In melanoma and ovarian cancer, we observed that the expression of genes regulating physical barriers was in some cases inversely correlated with the ICR signature [27]. This observation suggests that when mechanical barriers prevent T cell infiltration, no direct contact occurs between cancer cells and T cells and, therefore, no activation of immune effector gene signatures can be observed. Since this study did not include extensive histological examination of the samples, it remains unknown if the inverse correlation relates to immune silent or immune excluded cancers.
Functional barriers may not be present in baseline conditions but they may be induced only when a contact occurs between T cells and cancer cells preventing subsequent infiltration into tumor nests [46–48]. As discussed later, we refer to this concept as dynamic barriers.
Thus, we propose that mechanisms potentially responsible for immune exclusion could be segregated into three functional categories according to the relevance of (1) mechanical barriers that pose a physical impediment which prevents the contact between T cells and cancer cells, (2) functional barriers that consist of steady-state biological and/or metabolic interactions between cancer, stromal, and immune cells which limit the migration, function and/or survival of T cells, and, lastly, (3) dynamic barriers that are induced upon the interaction between cancer and immune cells but then prevents further T cell recruitment, migration, and/or survival.
Distinguishing these three mechanisms of immune exclusion has important implications in guiding novel IO therapeutics aimed at overcoming immune resistance. For example, a lack of chemo-attraction of T cells to the tumor site may not be the rate-limiting factor in achieving a successful anti-tumor immune response in immune excluded tumors as compared to immune silent cancers. Thus, immune excluded tumors may require a completely different therapeutic approach compared to immune silent cancers as suggested by the previously described 111Indium-labeled TIL study [8]. Furthermore, understanding these mechanisms of immune exclusion has critical therapeutic implications in the successful application of ACT of solid tumors.
In the case of functional and dynamic barriers, the implication that some albeit aborted interactions occur between cancer and T cells at the periphery of the tumor nests presents the optimistic opportunity that interference with functional barriers by genetically reprogramming T cell function may overcome these immune exclusion mechanisms [49–51]. An example of such therapeutic opportunity is represented by the regulation of PD1 expression on T cells by genetic knockout or conditional knockdown to overcome the interaction with PDL1 molecules expressed either by tumor cells or TAMs at the periphery of tumor nests [46] or the constitutive or conditional expression of factors aimed at counteracting immune suppressive signals [52].
6.4. Mechanisms of Immune Exclusion
The biology establishing immune exclusion, defined as the presence of T cell lining the outer margins of tumor nests [28], remains unknown. In particular, it is unknown whether a prominent mechanism is responsible for most cases rather than this well-defined phenotype resulting from the convergence of multiple inconsistently and unpredictably occurring biological disruptions that in distinct ways hamper T cell infiltration within the tumors nests. Here, we separate potential mechanisms according to main categories (Table 6.1).
6.4.1. Mechanical Barriers
Physical impediment preventing contact between T cells and cancer cells
In two independent cohorts that included 114 metastatic melanoma and 186 ovarian cancer samples, we identified eight genes that encode for proteins with mechanical barrier function. The expression of those genes was inversely correlated with that of the Th1-like ICR immune signature. Their expression was also associated with worse prognosis in patients with melanoma [27]. Among them, the expression of the desmosomal protein dystonin (DST) marked cancers lacking in absolute the expression of the Th1 immune signature suggesting that no interactions were present in these tumors between T cells and cancer cells. The other seven genes demarcated a set of cancers characterized by reduced and variable expression of the ICR signature suggesting that in this case some functional interactions were occurring. Importantly, expression of the latter barrier molecules occurred independently of the expression of endothelin receptor B and, in a mutually exclusive manner, with the activation of the β-catenin signaling pathway both reported to interfere with T cell infiltration into human cancers [53–56]. These observations suggest that several mechanisms of immune exclusion may shape the cancer immune landscapes and that such mechanisms may be at times mutually exclusive.
Interestingly, DST expression identified a subset of melanoma cases in which absolute absence of CD8+ gene signatures was observed but this was not associated with decreased patient survival. This supports the concept that the absolute presence of T cells within a given TME is not a singular prognostic determinant and possibly the ratio between lymphoid over myeloid or other cellular infiltrate may better define prognostic significance [18, 19, 30]. This hypothesis, however, was not explored by this study. Limited IHC analyses performed on those samples validated the expression of the barrier molecules at the protein level. In addition, these limited IHC analyses suggested that the expression of protein that could function as physical barriers was predominantly observed in cancer cells. The expression of these markers was associated morphologically with the cold or immune silent TME and this observation is also consistent with the exclusion of transcriptional ICR signatures suggesting that mechanical barriers are more likely to be relevant to the immune silent than the immune excluded landscape. Conversely, the true immune excluded landscape is more likely due to functional rather than physical barriers. Clearly more extensive analyses combining histological with functional methods will be needed to test this hypothesis.
Various other mechanisms are likely to play a role in influencing T cell migration. For instance, epithelial to mesenchymal transition altering the stromal composition has been accounted for in the development of a mechanical barrier [27, 57–59]. This in turn is largely mediated by transforming growth factor (TGF)-β activity [60, 61].
Epithelial to mesenchymal transition is not the only way in which TGF activity may lead to fibrosis mediated in cancer [26, 62, 63]. This pleiotropic cytokine is indeed essential for the induction of fibrotic responses through a cross talk with extracellular matrix components during organic tissue regeneration. Moreover, TGF-β is a critical component of the TME mediating resistance to immune surveillance mechanism through several direct and indirect mechanisms that restricts T cell trafficking and function by acting on the modulation of various cell populations contributing to the development of mesenchymal stromal cells [26, 64] and cancer-associated fibroblasts (CAF) [63]. The latter bear several immune regulatory properties beyond the generation of fibrous material contributing simultaneously to the development of both physical and functional barriers. For this reason, TGF-β is considered a promising therapeutic target particular for immune silent cancers. Its role in determining specifically immune exclusion remains, however, indeterminate [65].
Another mechanism of physical exclusion of T cells could be related to limited vascularization; it can be presumed that less vascularized tumors may be less prone to immune infiltration. In addition, various endothelial receptors may play either facilitator or inhibitor roles by mediating immune cell translocation into the extravascular space [53, 66, 67]. Independent of specific receptors mediating T cell transposition across the vascular structures, vascular endothelia growth factors (VEGF) seems to play a prominent role by mediating not only the access of immune cells into tumors but also their function [68, 69]. The latter mechanism probably plays a major role in determining immune resistance to IO agents and, therefore, similarly to TGF mediates the activation of both functional as well as mechanical barriers.
6.4.2. Functional Barriers
Pre-existing biological and/or metabolic interactions between cancer, stromal, and immune cells limit the migration, function, and/or survival of T cells
The mechanisms limiting immune infiltration through adaptive or innate interactions are extensive. Moreover, other mechanisms may be indirectly determining immune exclusion by dampening the production of pro-inflammatory and/or chemo-attractive signals. For instance, innate immune cells may less efficiently process and present apoptotic cancer cells for immune activation because of the presence of “don’t eat me signals” [70–72] and/or other tolerogenic mechanisms in which cancer cells may die, as discussed later.
It is important to emphasize that, to our knowledge, there is no integrated view about how various mechanisms may relate to each other in determining immune exclusion in human cancers either from a causative standpoint or through converging biologic routes. Moreover, the prevalence of distinct mechanisms and their weight in shaping the TME of human cancer is unclear and it is unknown whether this plethora reflects an equal distribution of various ways that immune exclusion may be randomly distributed across different cancer types or a predominant mechanism is responsible for most cases while others may only occasionally play a significant role. In other words, the longstanding debate of whether cancer is one disease or rather a mosaic of diverse diseases is best exemplified by the chaotic models used to describe immune exclusion. For instance, Schwartz et al. [73] suggested that most of the hallmarks of cancer can be attributed to the Warburg effect according to which, even in aerobic conditions, cancer cells favor glycolysis over oxidative phosphorylation. According to this view, the metabolic impairment of oxidative phosphorylation triggers a cascade of events that include the fractal shape of tumors, the secretion of collagen by CAFs, the production of lactic acid that elicits an acidic TME with direct and indirect effects on innate and adaptive immune functions. Furthermore, this can induce increased intracellular alkalosis in cancer cells and alteration of mitochondrial function leading to dysregulation of the ion concentration in the TME with its increasingly recognized effects on T cell function [74, 75]. Whether this hypothesis or modification around it reverses the Warburg effect whereby cancer cells induce aerobic glycolysis in neighboring CAFs [76] is debatable as discussed by Xu et al. [77]. However, its weight is not as important as the general concept suggested by the authors that efforts should be placed to identify convergent views regarding this otherwise chaotic portrait of immune exclusion.
An integrated approach to resolve the enigma of immune exclusion should include the simultaneous analysis of biomarkers representative of distinct mechanisms of immune exclusion in the same tumor samples. This approach can identify associations that may have similar upstream mechanistic determinants. The analysis may prove even more effective if a topographical analysis will be performed surveying the centripetal gradients within individual tumor nests. To our knowledge, such comprehensive analysis has never been done nor is currently being entertained by any investigation on human cancer samples and we encourage such efforts as part of the systematic and integrated approach to the solution of the quandary of cancer immune resistance [1]. A better understanding of the dominant mechanisms leading to immune exclusion and their primary causation may lead to a better rationale for drug development and for the selection of sound combination therapies.
A comprehensive discussion about individual theories proposed to explain a functional basis for immune exclusion is beyond the purpose of this chapter and we refer to the content arranged into sub-categories in Table 6.1. Here, we are limiting the discussion to principles that reflect similar biologic facets. We propose that functional barriers could be categorized into those determined by metabolic alterations specific to the TME as exemplified by the Warburg effect [73, 78, 79] as discussed previously.
A separate role may be played by hypoxia [80]. In fact, most cancers thrive in hypoxic conditions due to suboptimal vascularization. This in turn, has been shown to lead to profound depression of T cell function that is sufficient on its own to lead to immune exclusion [81–85]. Related to hypoxia is the complex and pleiotropic role that VEGF family members play in the TME by regulating lymph angiogenesis. It is likely that hypoxia is at least a cause of increased VEGF family members including VEGF-A, -C, and -D presence in the TME [86]. Although intuitively VEGF should increase lymphocyte infiltration in tumors due to increased angiogenesis, most observations refute this concept demonstrating that indeed anti-angiogenic therapies enhance immune infiltration of T cells [69, 87–89]. Indeed VEGF has been shown to mediate direct and indirect immune suppression through a series of distinct mechanisms [68] once again pointing out that the border between physical and functional barriers is not always well demarcated and that often, the same pathways may lead to contrasting effects due to the pleiotropic properties of the cells involved and the factors that they secrete. Thus, like for the Warburg effect, hypoxia may represent a principal component of immune exclusion mediating directly and indirectly immune infiltration and function. To complicate things, it has been shown that VEGF family members can be overexpressed constitutively by cancer cells independent of hypoxic conditions that stimulate their production [90–92].
An increasing gradient of repulsing signals is best exemplified by TGF-β. As previously mentioned, this extremely pleiotropic factor exercises effects on the TME way beyond the stimulation of the production of fibrotic material [26, 63]. Beyond its effects on stromal cells including CAFs, TGF-β acts directly on immune cells and in particular CD4+ T cells [93, 94].
Chemo-attraction (and lack thereof) is a factor that regulates immune infiltration. While several chemokines have been implicated in the recruitment of T cells and other immune effector cells to the TME, it appears that CXCR-3 and CCR-5 ligand chemokines play a dominant role in immune surveillance and they are most significantly correlated with CD8 T cell infiltration as part of the ICR signature [10, 20, 22, 95]. The central role that these two families of cytokines play in immune surveillance has been clearly established for both immune active and immune silent cancer as they are tightly correlated to these two phenotypes [7]. The question remains, however, about the role that they may play in the context of immune excluded tumors. Intuitively, these or other T cell attracting chemokines should be present to attract T cells at the periphery of tumor nests, although this presumption has never been tested. It is, therefore, possible that for unknown reasons an abrupt reduction in the gradient of chemo-attraction from the center to the periphery, such as expression of CXCR3 and CCR5 associated chemokines, may reduce the chemo-attractive propulsion that occurs at the periphery. For instance, it was recently suggested that in melanoma chemokine expression is modulated by cancer cell-intrinsic pathways, such as microphthalmia-associated transcription factor, whose expression is associated with that of CXCL-10 [96]. Alternatively, while the expression of these chemokines is stably maintained within the concentric circles of the tumor nest, additional overpowering repulsive mechanisms emanated from the center may counterbalance the attractive signals gradually limiting the progression of T cells.
Evolving patterns of cell death from the germinal center to the periphery may create a gradient of increasing chemo-attraction. Possibly, cancer cells may rapidly dedifferentiate from a stem cell-like core to a degenerate progeny at the periphery prone to Immunogenic Cell Death (ICD) [97–99]. This could be tested by surface expression of calreticulin or other ICD markers [100]. Alternatively, cancer cell death occurs only at the periphery while the “germinal” centers continue to actively proliferate (this could be tested using proliferation markers). Thus, Damage Associated Molecular Patterns (DAMPs) may be present only at the periphery. To our knowledge, this possibility has never been entertained and remains only speculative. A more likely possibility is that various mechanisms affecting the processing of stressed or dying cells may play a significant role by dampening pro-inflammatory signals. Several of these mechanisms include adenosine signaling [101–105], TAM receptor tyrosine kinases [6, 106–108] and the “don’t eat me” signals through the CD47/signal regulatory protein (SIRP)-α interactions [70–72]. Extracellular adenosine is an almost ubiquitous component of tissues exposed to stress. It is generated in response to pro-inflammatory conditions and in particular hypoxia [105] when stressed cells release ATP. The latter is then degraded by CD39 into AMP and subsequently turned into adenosine by CD73. Adenosine in turn interacts with several G-protein-coupled receptors almost ubiquitously expressed on the surface of most immune cells. These receptors promote anti-inflammatory responses as described comprehensively by Young A et al. [104]. Similarly, TAM receptors (Tyro3, AXL and Mertk) are tyrosine kinases that orchestrated a prominent regulatory role particular on innate immune cell functions by promoting the phagocytosis of apoptotic cells and inhibiting inflammation [109]. Through this process, TAM receptors inhibit natural dendritic cell and macrophage maturation signals [109]. Consequently, their targeting promotes the activation of immune responses against cancer as well exemplified by the potentiation of radiation effects by the inhibition of Mertk [107]. Similarly, to TAM receptors, other cancer cell/immune checkpoint interactions may occur as well exemplified by the CD47/SIPR-α interactions [70–72, 110]. Although CD47 interacts with its ligand expressed by cells of the myeloid lineage by inhibiting phagocytosis and clearance of dying cells, it appears that it also plays a direct anti-inflammatory role by regulating the activation of innate immune effector cells [70]. Thus, it could be considered a cell surface checkpoint molecule target for immunotherapy [71, 72].
A final category of potential mechanisms of immune exclusion is represented by the direct effect that cancer-intrinsic signaling pathways can play in modulating chemo-attractive signals and immune modulatory responses. Among them, the role played by tumor cell-intrinsic signal transducer and activator of Transcription (STAT)-3 [111–114] and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) [115–120] signaling have been extensively described. In addition, the contribution of alterations in β-catenin [54–56, 121, 122] and MAPK [16, 45] signaling have been described. Interestingly, both PI3K and STAT-3 signaling are prominent features of myeloid cell activation [113, 114, 123, 124] and, therefore, their role in immune modulation is often confused with the genuine alteration of intrinsic cancer pathways. Interestingly, most studies referring to dysregulation of the PI3K pathway refer to mutations of genes involved in cancer cell signaling. However, the same pathway is critical in the activation of myeloid suppressor dendritic cell function downstream of TAM receptor signaling [107, 108]. This functional signature is totally independent of the cancer cells mutational status and it is purely related to immune cell functions determined by other signaling mechanisms. For instance, we noticed that transcriptional signatures associated to activation of the PI3K pathway were most frequently observed in immune active cancers [7, 25] and this activation was most likely due to the myeloid cells, while in those tumors no specific somatic mutations relevant to cancer cell biology were noted particularly related to STAT3 and PI3K. Conversely, β-catenin and MAPK activation have been associated with mutational status of genes within the respective pathways and are, therefore, to be considered purely related to activation of cancer cell-intrinsic pathways [43, 44]. We believe, however, that both mechanisms are not likely to be relevant to immune exclusion as both are tightly associated with the immune silent cancer phenotype and the absolute lack of expression of the ICR signature [7]. We recently validated this observation in a set of 9,282 tumor samples in a Pan-cancer analysis of cancer of 31 different histologies (Roelands J. et al., submitted). Yet, no specific analysis attempting to match morphologic features with transcriptional activation have been so far entertained to verify this assumption.
6.4.3. Dynamic Barriers
Biological interactions between cancer and T cells that result in limited function
As previously described, the regulatory interactions that limit T cell infiltration into tumor nests may not be present in baseline conditions but develop as a consequence of the first encounter between T cells and cancer cells. This is well exemplified by the inducible activation of PDL-1 in response to IFN-γ signaling. It has been observed in the context of head and neck squamous cell carcinoma that programmed cell death protein-1 (PD-1) is observable in functionally anergic PD-1 expressing TILs at the periphery of tumor nests. The latter are flanked by tumors and/or CD68+ TAMs that express programmed death ligand-1 (PDL-1). This observation suggests that a functional cross-check occurs dynamically at the initial encounter between T cell and cancer cells possibly triggered by the production of IFN-γ by T cells upon exposure to cognate stimulation. This in turn, induces expression of PDL1 by either cancer or TAMs that serves as a negative feedback loop to limit the function of CD4+ and CD8+ T cells [46] (Fig. 6.1). Similarly, close dynamic interactions between PD-1+ CD8+ T cells and PD-L1 expression cancer cells, and/or CD68+ TAMs were reported in human papillomavirus-associated HNSSC [47]. Finally, a cross talk between T cells and dendritic cells expressing PDL1 in response to IFN-γ and IL-12 stimulation plays a critical role in modulating immune responsiveness [48]. Whether these dynamic interactions may be at the basis of immune exclusion has been sufficiently investigated but we believe that future studies should include extensive analyses integrating transcriptional with morphological description of cancer immune phenotypes.
Fig. 6.1. The Paradox of Immune Exclusion—
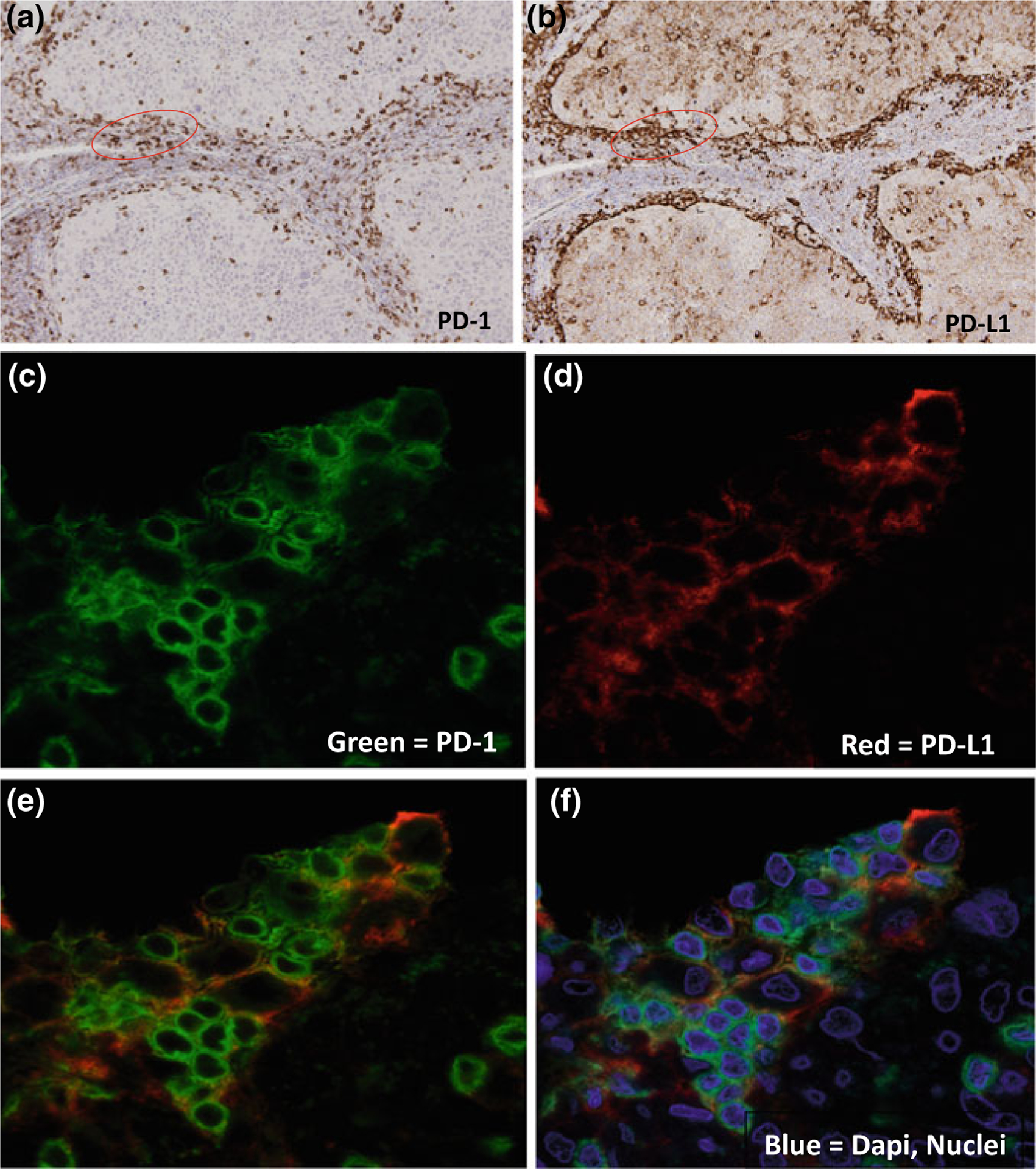
Example of distinct theories that may explain the confined presence of T cells at the periphery of immune excluded cancer nests. a. Single immunohistochemical staining of PD-1 receptor on T cells in HNSCCs. Although there is some T cell infiltration into the tumor nests, the majority of the activated T cells remain in the tumor stroma and in the periphery of the tumor nest depicting immune exclusion. b Single immunohistochemical staining of PD-L1 on the tumor and/or immune cells on the same HNSCCs cut in serial section. There is strong PD-L1 expression in the periphery of the tumor nests as well as on tumor infiltrating TAMs. c Single immunofluorescent staining of PD-1 receptor. This is a high magnification of the area circled in red in Panel A. d Single immunofluorescent staining of PD-L1. This is a high magnification of the area circled in Panel B. e Dual immunofluorescent staining of PD-1 and PD-L1 with areas of overlap depicted as orange. f Dual immunofluorescent staining of PD-1 and PD-L1 with blue DAPI staining of single cell nuclei
6.5. Clinical Implications
Improved understanding of the mechanisms that drive immune exclusion has important clinical implications in the development of novel therapeutic strategies aimed to overcome immune resistance against IO agents, including ACT. It remains to be clarified whether a predominant biology is at the basis of most immune excluded cases. Future studies may reveal that a complex combination of mechanical, functional, and dynamic barriers may shape the immune biology of individual tumors attesting for the need for more in-depth precision medicine-based molecular and histological characterization to be implemented for the selection of appropriate monotherapy or combination therapeutics.
Abbreviations
- ACT
Adoptive Cell Therapy
- CAF
Cancer-Associated Fibroblast
- CIT
Checkpoint Inhibitor Therapy
- CSC
Cancer Stem Cells
- DST
Dystonin
- GC
Germinal Center
- HNSCC
Head and Neck Squamous Cell Carcinoma
- HPV
Human Papilloma Virus
- ICD
Immunogenic Cell Death
- ICR
Immunologic Constant of Rejection
- IFN
Interferon
- IHC
Immunohistochemistry
- IL
Interleukin
- IO
Immune Oncology
- PD1
Programmed cell Death protein-1
- PDL1
Programmed Death Ligand-1
- PI3K
Phosphatidyl-Inositol-4,5-bisphosphate 3-Kkinase
- SIRP-α
Signal Regulatory Protein-α
- STAT
Signal Transducer and Activator of Transcription
- TAMs
Tumor-Associated Macrophages
- TGF
Transforming Growth Factor
- TIL
Tumor Infiltrating Lymphocytes
- TIS
Tumor Inflammation Signature
- TME
Tumor Microenvironment
- VEGF
Vascular Endothelial Growth Factor
Contributor Information
Sara I. Pai, Massachusetts General Hospital, Harvard University, Boston, MA, USA
Alessandra Cesano, ESSA Pharmaceuticals, Inc., South San Francisco, CA, USA.
Francesco M. Marincola, Refuge Biotechnologies, Inc., Menlo Park, CA, USA
References
- 1.Bedognetti D, Ceccarelli M, Galluzzi L, Lu R, Palucka K, Samayoa J et al. (2019) Toward a comprehensive view of cancer immune responsiveness: a synopsis from the SITC workshop. J Immunother Cancer 7(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen DS, Mellman I (2017) Elements of cancer immunity and the cancer-immune set point. Nature 541(7637):321–330 [DOI] [PubMed] [Google Scholar]
- 3.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR et al. (2017) IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127(8):2930–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS et al. (2014) Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515(7528):563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joyce JA, Fearon DT (2015) T cell exclusion, immune privilege, and the tumor microenvironment. Science 348(6230):74–80 [DOI] [PubMed] [Google Scholar]
- 6.Aguilera TA, Giaccia AJ (2017) Molecular pathways: oncologic pathways and their role in T-cell exclusion and immune evasion-A new role for the AXL receptor tyrosine kinase. Clin Cancer Res 23(12):2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turan T, Kannan D, Patel M, Matthew Barnes J, Tanlimco SG, Lu R et al. (2018) Immune oncology, immune responsiveness and the theory of everything. J Immunother Cancer 6(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pockaj BA, Sherry RM, Wei JP, Yannelli JR, Carter CS, Leitman SF et al. (1994) Localization of 111indium-labeled tumor infiltrating lymphocytes to tumor in patients receiving adoptive immunotherapy. Augmentation with cyclophosphamide and correlation with response. Cancer 73(6):1731–1737 [DOI] [PubMed] [Google Scholar]
- 9.Wang E, Uccellini L, Marincola FM (2012) A genetic inference on cancer immune responsiveness. Oncoimmunology. 1(4):520–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedognetti D, Spivey TL, Zhao Y, Uccellini L, Tomei S, Dudley ME et al. (2013) CXCR3/CCR5 pathways in metastatic melanoma patients treated with adoptive therapy and interleukin-2. Br J Cancer 109(9):2412–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF et al. (2015) Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350 (6266):1387–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z et al. (2018) Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 24(6):724–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S (2000) Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol 74:181–273 [DOI] [PubMed] [Google Scholar]
- 14.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C et al. (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313(5795):1960–1964 [DOI] [PubMed] [Google Scholar]
- 15.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C et al. (2018) International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391(10135):2128–2139 [DOI] [PubMed] [Google Scholar]
- 16.Hendrickx W, Simeone I, Anjum S, Mokrab Y, Bertucci F, Finetti P et al. (2017) Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology. 6(2):e1253654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fridman WH, Pages F, Sautes-Fridman C, Galon J (2012) The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12(4):298–306 [DOI] [PubMed] [Google Scholar]
- 18.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF et al. (2011) Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov 1(1):54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruffell B, Coussens LM (2015) Macrophages and therapeutic resistance in cancer. Cancer Cell 27(4):462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galon J, Angell HK, Bedognetti D, Marincola FM (2013) The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 39(1):11–26 [DOI] [PubMed] [Google Scholar]
- 21.Weiss GR, Grosh WW, Chianese-Bullock KA, Zhao Y, Liu H, Slingluff CL Jr et al. (2011) Molecular insights on the peripheral and intratumoral effects of systemic high-dose rIL-2 (aldesleukin) administration for the treatment of metastatic melanoma. Clin Cancer Res 17(23):7440–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang E, Worschech A, Marincola FM (2008) The immunologic constant of rejection. Trends Immunol 29(6):256–262 [DOI] [PubMed] [Google Scholar]
- 23.Miller LD, Chou JA, Black MA, Print C, Chifman J, Alistar A et al. (2016) Immunogenic subtypes of breast cancer delineated by gene classifiers of immune responsiveness. Cancer Immunol Res 4(7):600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertucci F, Finetti P, Simeone I, Hendrickx W, Wang E, Marincola FM et al. (2018) The immunologic constant of rejection classification refines the prognostic value of conventional prognostic signatures in breast cancer. Br J Cancer 119(11):1383–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu R, Turan T, Samayoa J, Marincola FM (2017) Cancer immune resistance: can theories converge? Emerg Top Life Sci 1(5):411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y et al. (2018) TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554(7693):544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salerno EP, Bedognetti D, Mauldin IS, Deacon DH, Shea SM, Pinczewski J et al. (2016) Human melanomas and ovarian cancers overexpressing mechanical barrier molecule genes lack immune signatures and have increased patient mortality risk. Oncoimmunology 5(12): e1240857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kather JN, Suarez-Carmona M, Charoentong P, Weis CA, Hirsch D, Bankhead P et al. (2018) Topography of cancer-associated immune cells in human solid tumors. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenplate AR, Johnson DB, Ferrell PB Jr, Irish JM (2016) Systems immune monitoring in cancer therapy. Eur J Cancer 61:77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH et al. (2017) Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep 19(1):203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D et al. (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18(1):248–262 [DOI] [PubMed] [Google Scholar]
- 32.van der Woude LL, Gorris MAJ, Halilovic A, Figdor CG, de Vries IJM (2017) Migrating into the tumor: a roadmap for T Cells. Trends Cancer 3(11):797–808 [DOI] [PubMed] [Google Scholar]
- 33.Fridman WH, Dieu-Nosjean MC, Pages F, Cremer I, Damotte D, Sautes-Fridman C et al. (2013) The immune microenvironment of human tumors: general significance and clinical impact. Cancer Microenviron 6(2):117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bindea G, Mlecnik B, Angell HK, Galon J (2014) The immune landscape of human tumors: implications for cancer immunotherapy. Oncoimmunology 3(1):e27456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jimenez-Sanchez A, Memon D, Pourpe S, Veeraraghavan H, Li Y, Vargas HA et al. (2017) heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell 170(5):927–938.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kather JN, Halama N, Jaeger D (2018) Genomics and emerging biomarkers for immunotherapy of colorectal cancer. Semin Cancer Biol 52(Pt 2):189–197 [DOI] [PubMed] [Google Scholar]
- 37.Emens LA, Ascierto PA, Darcy PK, Demaria S, Eggermont AMM, Redmond WL et al. (2017) Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. Eur J Cancer 81:116–129 [DOI] [PubMed] [Google Scholar]
- 38.Masucci GV, Cesano A, Hawtin R, Janetzki S, Zhang J, Kirsch I et al. (2016) Validation of biomarkers to predict response to immunotherapy in cancer: Volume I—pre-analytical and analytical validation. J Immunother Cancer 4:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butterfield LH, Disis ML, Fox BA, Kaufman DR, Khleif SN, Wang E et al. (2018) SITC 2018 workshop report: Immuno-Oncology Biomarkers: State of the Art. J Immunother Cancer 6(1):138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haris M, Bagga P, Hariharan H, McGettigan-Croce B, Johnson LA, Reddy R (2017) Molecular imaging biomarkers for cell-based immunotherapies. J Transl Med 15(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M et al. (2015) Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res 21(4):870–881 [DOI] [PubMed] [Google Scholar]
- 42.Thurlow JK, Pena Murillo CL, Hunter KD, Buffa FM, Patiar S, Betts G et al. (2010) Spectral clustering of microarray data elucidates the roles of microenvironment remodeling and immune responses in survival of head and neck squamous cell carcinoma. J Clin Oncol 28(17):2881–2888 [DOI] [PubMed] [Google Scholar]
- 43.Roelands J, Decock J, Boughorbel S, Rinchai D, Maccalli C, Ceccarelli M et al. (2017) A collection of annotated and harmonized human breast cancer transcriptome datasets, including immunologic classification. F1000Res 6:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roelands J, Kuppen PJK, Vermeulen L, Maccalli C, Decock J, Wang E et al. (2017) Immunogenomic classification of colorectal cancer and therapeutic implications. Int J Mol Sci 18(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH et al. (2018) The immune landscape of cancer. Immunity 48(4):812–830.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattox AK, Lee J, Westra WH, Pierce RH, Ghossein R, Faquin WC et al. (2017) PD-1 expression in head and neck squamous cell carcinomas derives primarily from functionally anergic CD4(+) TILs in the presence of PD-L1(+) TAMs. Cancer Res 77(22):6365–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B et al. (2013) Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res 73(6):1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C et al. (2018) Successful Anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-gamma and IL-12. Immunity. 49(6):1148–61 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y (2017) Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res 23(9):2255–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salas-Mckee J, Kong W, Gladney WL, Jadlowsky JK, Plesa G, Davis MM et al. (2019) CRISPR/Cas9-based genome editing in the era of CAR T cell immunotherapy. Hum Vaccin Immunother 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simeonov DR, Marson A (2019) CRISPR-Based Tools in Immunity. Annu Rev Immunol 37:571–597 [DOI] [PubMed] [Google Scholar]
- 52.Rosewell Shaw A, Porter CE, Watanabe N, Tanoue K, Sikora A, Gottschalk S et al. (2017) Adenovirotherapy delivering cytokine and checkpoint inhibitor augments CAR T cells against metastatic head and neck cancer. Mol Ther 25(11):2440–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kandalaft LE, Facciabene A, Buckanovich RJ, Coukos G (2009) Endothelin B receptor, a new target in cancer immune therapy. Clin Cancer Res 15(14):4521–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF (2019) WNT/beta-catenin pathway activation correlates with immune exclusion across human cancers. Clin Cancer Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spranger S, Bao R, Gajewski TF (2015) Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523(7559):231–235 [DOI] [PubMed] [Google Scholar]
- 56.Dangaj D, Barras D, Coukos G (2019) Tumor landscapes: beta-catenin drives immune desertification. Clin Cancer Res [DOI] [PubMed] [Google Scholar]
- 57.Fintha A, Gasparics A, Rosivall L, Sebe A (2019) Therapeutic targeting of fibrotic epithelial-mesenchymal transition-an outstanding challenge. Front Pharmacol 10:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Runyan RB, Savagner P (2018) Epithelial-mesenchymal transition and plasticity in the developmental basis of cancer and fibrosis. Dev Dyn 247(3):330–331 [DOI] [PubMed] [Google Scholar]
- 59.Kurata T, Fushida S, Kinoshita J, Oyama K, Yamaguchi T, Okazaki M et al. (2018) Low-dose eribulin mesylate exerts antitumor effects in gastric cancer by inhibiting fibrosis via the suppression of epithelial-mesenchymal transition and acts synergistically with 5-fluorouracil. Cancer Manag Res 10:2729–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katsuno Y, Lamouille S, Derynck R (2013) TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 25(1):76–84 [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez DM, Medici D (2014) Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal 7(344):re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreau HD, Piel M, Voituriez R, Lennon-Dumenil AM (2018) Integrating physical and molecular insights on immune cell migration. Trends Immunol 39(8):632–643 [DOI] [PubMed] [Google Scholar]
- 63.Caja L, Dituri F, Mancarella S, Caballero-Diaz D, Moustakas A, Giannelli G et al. (2018) TGF-beta and the tissue microenvironment: relevance in fibrosis and cancer. Int J Mol Sci 19(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Araujo FV, Carrillo-Galvez AB, Martin F, Anderson P (2018) TGF-beta and mesenchymal stromal cells in regenerative medicine, autoimmunity and cancer. Cytokine Growth Factor Rev 43:25–37 [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Di C, Zhang X, Wang J, Wang F, Yan JF et al. (2019) Transforming growth factor beta signaling pathway: a promising therapeutic target for cancer. J Cell Physiol [DOI] [PubMed] [Google Scholar]
- 66.Buckanovich RJ, Facciabene A, Kim S, Benencia F, Sasaroli D, Balint K et al. (2008) Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat Med 14(1):28–36 [DOI] [PubMed] [Google Scholar]
- 67.Coffman L, Mooney C, Lim J, Bai S, Silva I, Gong Y et al. (2013) Endothelin receptor-A is required for the recruitment of antitumor T cells and modulates chemotherapy induction of cancer stem cells. Cancer Biol Ther 14(2):184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li YL, Zhao H, Ren XB (2016) Relationship of VEGF/VEGFR with immune and cancer cells: staggering or forward? Cancer Biol Med 13(2):206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Yan J, Liu B (2018) Targeting VEGF/VEGFR to modulate antitumor immunity. Front Immunol 9:978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matlung HL, Szilagyi K, Barclay NA, van den Berg TK (2017) The CD47-SIRPalpha signaling axis as an innate immune checkpoint in cancer. Immunol Rev 276(1):145–164 [DOI] [PubMed] [Google Scholar]
- 71.Ring NG, Herndler-Brandstetter D, Weiskopf K, Shan L, Volkmer JP, George BM et al. (2017) Anti-SIRPalpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity. Proc Natl Acad Sci U S A. 114(49):E10578–E10585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiskopf K (2017) Cancer immunotherapy targeting the CD47/SIRPalpha axis. Eur J Cancer 76:100–109 [DOI] [PubMed] [Google Scholar]
- 73.Schwartz L, Supuran CT, Alfarouk KO (2017) The warburg effect and the hallmarks of cancer. Anticancer Agents Med Chem 17(2):164–170 [DOI] [PubMed] [Google Scholar]
- 74.Gurusamy D, Clever D, Eil R, Restifo NP (2017) Novel “Elements” of immune suppression within the tumor microenvironment. Cancer Immunol Res 5(6):426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH et al. (2019) T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363 (6434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG et al. (2009) The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 8(23):3984–4001 [DOI] [PubMed] [Google Scholar]
- 77.Xu XD, Shao SX, Jiang HP, Cao YW, Wang YH, Yang XC et al. (2015) Warburg effect or reverse Warburg effect? a review of cancer metabolism. Oncol Res Treat 38(3):117–122 [DOI] [PubMed] [Google Scholar]
- 78.Morrot A, da Fonseca LM, Salustiano EJ, Gentile LB, Conde L, Filardy AA et al. (2018) Metabolic symbiosis and immunomodulation: how tumor cell-derived lactate may disturb innate and adaptive immune responses. Front Oncol 8:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A et al. (2016) LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab 24(5):657–671 [DOI] [PubMed] [Google Scholar]
- 80.Bartrons R, Caro J (2007) Hypoxia, glucose metabolism and the Warburg’s effect. J Bioenerg Biomembr 39(3):223–229 [DOI] [PubMed] [Google Scholar]
- 81.Hatfield SM, Kjaergaard J, Lukashev D, Belikoff B, Schreiber TH, Sethumadhavan S et al. (2014) Systemic oxygenation weakens the hypoxia and hypoxia inducible factor 1alpha-dependent and extracellular adenosine-mediated tumor protection. J Mol Med (Berl) 92(12):1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hatfield SM, Kjaergaard J, Lukashev D, Schreiber TH, Belikoff B, Abbott R et al. (2015) Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med 7(277):277ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hatfield SM, Sitkovsky M (2016) A2A adenosine receptor antagonists to weaken the hypoxia-HIF-1alpha driven immunosuppression and improve immunotherapies of cancer. Curr Opin Pharmacol 29:90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daniel SK, Sullivan KM, Labadie KP, Pillarisetty VG (2019) Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin Transl Med 8(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatfield S, Veszeleiova K, Steingold J, Sethuraman J, Sitkovsky M (2019) Mechanistic justifications of systemic therapeutic oxygenation of tumors to weaken the hypoxia inducible factor 1alpha-mediated immunosuppression. Adv Exp Med Biol 1136:113–121 [DOI] [PubMed] [Google Scholar]
- 86.Morfoisse F, Renaud E, Hantelys F, Prats AC, Garmy-Susini B (2015) Role of hypoxia and vascular endothelial growth factors in lymphangiogenesis. Mol Cell Oncol 2(4):e1024821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lapeyre-Prost A, Terme M, Pernot S, Pointet AL, Voron T, Tartour E et al. (2017) Immunomodulatory activity of VEGF in cancer. Int Rev Cell Mol Biol 330:295–342 [DOI] [PubMed] [Google Scholar]
- 88.Veitenhansl M, Stegner K, Hierl FX, Dieterle C, Feldmeier H, Gutt B et al. (2004) 40(th) EASD annual meeting of the european association for the study of diabetes, Munich, Germany, 5–9 Sept 2004. Diabetologia 47(Suppl 1):A1–A464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsukita Y, Okazaki T, Ebihara S, Komatsu R, Nihei M, Kobayashi M et al. (2019) Beneficial effects of sunitinib on tumor microenvironment and immunotherapy targeting death receptor5. Oncoimmunology 8(2):e1543526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo P, Fang Q, Tao HQ, Schafer CA, Fenton BM, Ding I et al. (2003) Overexpression of vascular endothelial growth factor by MCF-7 breast cancer cells promotes estrogen-independent tumor growth in vivo. Cancer Res 63(15):4684–4691 [PubMed] [Google Scholar]
- 91.Petit AM, Rak J, Hung MC, Rockwell P, Goldstein N, Fendly B et al. (1997) Neutralizing antibodies against epidermal growth factor and ErbB-2/neu receptor tyrosine kinases down-regulate vascular endothelial growth factor production by tumor cells in vitro and in vivo: angiogenic implications for signal transduction therapy of solid tumors. Am J Pathol 151(6):1523–1530 [PMC free article] [PubMed] [Google Scholar]
- 92.Gabrilovich DI, Chen HL, Girgis KR, Cunningham HT, Meny GM, Nadaf S et al. (1996) Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat Med 2(10):1096–1103 [DOI] [PubMed] [Google Scholar]
- 93.Fu S, Zhang N, Yopp AC, Chen D, Mao M, Chen D et al. (2004) TGF-beta induces Foxp3+ T-regulatory cells from CD4+ CD25− precursors. Am J Transplant 4(10):1614–1627 [DOI] [PubMed] [Google Scholar]
- 94.Dahmani A, Delisle JS (2018) TGF-beta in t cell biology: implications for cancer immunotherapy. Cancers (Basel) 10(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S et al. (2018) CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—a target for novel cancer therapy. Cancer Treat Rev 63:40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wiedemann GM, Aithal C, Kraechan A, Heise C, Cadilha BL, Zhang J et al. (2019) Microphthalmia-associated transcription factor (MITF) regulates immune cell migration into melanoma. Transl Oncol 12(2):350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Green DR, Ferguson T, Zitvogel L, Kroemer G (2009) Immunogenic and tolerogenic cell death. Nat Rev Immunol 9(5):353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G (2017) Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 17(2):97–111 [DOI] [PubMed] [Google Scholar]
- 99.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P et al. (2018) Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ 25(3):486–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kepp O, Senovilla L, Vitale I, Vacchelli E, Adjemian S, Agostinis P et al. (2014) Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 3(9):e955691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gorzalczany Y, Sagi-Eisenberg R (2019) Role of mast cell-derived adenosine in cancer. Int J Mol Sci 20(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Antonioli L, Blandizzi C, Pacher P, Hasko G (2013) Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer 13(12):842–857 [DOI] [PubMed] [Google Scholar]
- 103.Muller-Haegele S, Muller L, Whiteside TL (2014) Immunoregulatory activity of adenosine and its role in human cancer progression. Expert Rev Clin Immunol 10(7):897–914 [DOI] [PubMed] [Google Scholar]
- 104.Young A, Mittal D, Stagg J, Smyth MJ (2014) Targeting cancer-derived adenosine: new therapeutic approaches. Cancer Discov 4(8):879–888 [DOI] [PubMed] [Google Scholar]
- 105.Stagg J, Smyth MJ (2010) Extracellular adenosine triphosphate and adenosine in cancer. Oncogene 29(39):5346–5358 [DOI] [PubMed] [Google Scholar]
- 106.Akalu YT, Rothlin CV, Ghosh S (2017) TAM receptor tyrosine kinases as emerging targets of innate immune checkpoint blockade for cancer therapy. Immunol Rev 276(1):165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crittenden MR, Baird J, Friedman D, Savage T, Uhde L, Alice A et al. (2016) Mertk on tumor macrophages is a therapeutic target to prevent tumor recurrence following radiation therapy. Oncotarget 7(48):78653–78666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang B, Fang L, Wu HM, Ding PS, Xu K, Liu RY (2016) Mer receptor tyrosine kinase negatively regulates lipoteichoic acid-induced inflammatory response via PI3K/Akt and SOCS3. Mol Immunol 76:98–107 [DOI] [PubMed] [Google Scholar]
- 109.Lemke G, Rothlin CV (2008) Immunobiology of the TAM receptors. Nat Rev Immunol 8(5):327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barclay AN, Van den Berg TK (2014) The interaction between signal regulatory protein alpha (SIRPalpha) and CD47: structure, function, and therapeutic target. Annu Rev Immunol 32:25–50 [DOI] [PubMed] [Google Scholar]
- 111.Ahn R, Sabourin V, Bolt AM, Hebert S, Totten S, De Jay N et al. (2017) The Shc1 adaptor simultaneously balances Stat1 and Stat3 activity to promote breast cancer immune suppression. Nat Commun 8:14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su YL, Banerjee S, White SV, Kortylewski M (2018) STAT3 in tumor-associated myeloid cells: multitasking to disrupt immunity. Int J Mol Sci 19(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y, Shen Y, Wang S, Shen Q, Zhou X (2018) The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett 415:117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu H, Lee H, Herrmann A, Buettner R, Jove R (2014) Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 14(11):736–746 [DOI] [PubMed] [Google Scholar]
- 115.Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S et al. (2016) PI3Kgamma is a molecular switch that controls immune suppression. Nature 539 (7629):437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daragmeh J, Barriah W, Saad B, Zaid H (2016) Analysis of PI3K pathway components in human cancers. Oncol Lett 11(4):2913–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu P, Cheng H, Roberts TM, Zhao JJ (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 8(8):627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Di Cristofano A (2017) SGK1: the dark side of PI3K signaling. Curr Top Dev Biol 123: 49–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Orlacchio A, Ranieri M, Brave M, Arciuch VA, Forde T, De Martino D et al. (2017) SGK1 is a critical component of an AKT-independent pathway essential for pi3k-mediated tumor development and maintenance. Cancer Res 77(24):6914–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiaobo Y, Qiang L, Xiong Q, Zheng R, Jianhua Z, Zhifeng L et al. (2016) Serum and glucocorticoid kinase 1 promoted the growth and migration of non-small cell lung cancer cells. Gene 576(1 Pt 2):339–346 [DOI] [PubMed] [Google Scholar]
- 121.Spranger S, Gajewski TF (2016) Tumor-intrinsic oncogene pathways mediating immune avoidance. Oncoimmunology 5(3):e1086862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Spranger S, Gajewski TF (2018) Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer 18(3):139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH et al. (2016) Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 539(7629):443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kumar V, Cheng P, Condamine T, Mony S, Languino LR, McCaffrey JC et al. (2016) CD45 phosphatase inhibits STAT3 transcription factor activity in myeloid cells and promotes tumor-associated macrophage differentiation. Immunity 44(2):303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Platten M, Wick W, Van den Eynde BJ (2012) Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res 72(21):5435–5440 [DOI] [PubMed] [Google Scholar]
- 126.Fultang L, Vardon A, De Santo C, Mussai F (2016) Molecular basis and current strategies of therapeutic arginine depletion for cancer. Int J Cancer 139(3):501–509 [DOI] [PubMed] [Google Scholar]
- 127.Kremer JC, Prudner BC, Lange SES, Bean GR, Schultze MB, Brashears CB et al. (2017) Arginine deprivation inhibits the warburg effect and upregulates glutamine anaplerosis and serine biosynthesis in ASS1-deficient cancers. Cell Rep 18(4):991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cascone T, McKenzie JA, Mbofung RM, Punt S, Wang Z, Xu C et al. (2018) Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab 27(5):977–87.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Iwamoto H, Abe M, Yang Y, Cui D, Seki T, Nakamura M et al. (2018) Cancer lipid metabolism confers antiangiogenic drug resistance. Cell Metab 28(1):104–17 e5 [DOI] [PubMed] [Google Scholar]
- 130.Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC et al. (2018) Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175(7):1780–95 e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Q, Tomei S, Ascierto ML, De Giorgi V, Bedognetti D, Dai C et al. (2014) Melanoma NOS1 expression promotes dysfunctional IFN signaling. J Clin Invest 124(5):2147–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Munn DH, Bronte V (2016) Immune suppressive mechanisms in the tumor microenvironment. Curr Opin Immunol 39:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mondanelli G, Ugel S, Grohmann U, Bronte V (2017) The immune regulation in cancer by the amino acid metabolizing enzymes ARG and IDO. Curr Opin Pharmacol 35:30–39 [DOI] [PubMed] [Google Scholar]
- 134.Zhai L, Spranger S, Binder DC, Gritsina G, Lauing KL, Giles FJ et al. (2015) Molecular Pathways: Targeting IDO1 and Other Tryptophan Dioxygenases for Cancer Immunotherapy. Clin Cancer Res 21(24):5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE et al. (2015) Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 162(6):1257–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang D, DuBois RN (2018) Role of prostanoids in gastrointestinal cancer. J Clin Invest 128(7):2732–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang L, Yamagata N, Yadav R, Brandon S, Courtney RL, Morrow JD et al. (2003) Cancer-associated immunodeficiency and dendritic cell abnormalities mediated by the prostaglandin EP2 receptor. J Clin Invest 111(5):727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaipainen A, Kieran MW, Huang S, Butterfield C, Bielenberg D, Mostoslavsky G et al. (2007) PPARalpha deficiency in inflammatory cells suppresses tumor growth. PLoS ONE 2(2):e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ma X, Aoki T, Tsuruyama T, Narumiya S (2015) Definition of Prostaglandin E2-EP2 Signals in the Colon Tumor Microenvironment That Amplify Inflammation and Tumor Growth. Cancer Res 75(14):2822–2832 [DOI] [PubMed] [Google Scholar]
- 140.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS et al. (2013) Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA 110(50):20212–20217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M et al. (2017) Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 214(3):579–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Arnold JN, Magiera L, Kraman M, Fearon DT (2014) Tumoral immune suppression by macrophages expressing fibroblast activation protein-alpha and heme oxygenase-1. Cancer Immunol Res 2(2):121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ohlund D, Elyada E, Tuveson D (2014) Fibroblast heterogeneity in the cancer wound. J Exp Med 211(8):1503–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saleh SMI, Bertos N, Gruosso T, Gigoux M, Souleimanova M, Zhao H et al. (2017) Identification of Interacting Stromal Axes in Triple-Negative Breast Cancer. Cancer Res 77(17):4673–4683 [DOI] [PubMed] [Google Scholar]
- 145.Grabiec AM, Hussell T (2016) The role of airway macrophages in apoptotic cell clearance following acute and chronic lung inflammation. Semin Immunopathol 38(4):409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]


