Abstract
As COVID-19 has reached pandemic status and the number of cases continues to grow, widespread availability of diagnostic testing is critical in helping identify and control the emergence of this rapidly spreading and serious illness. However, a lacking in making a quick reaction to the threat and starting early development of diagnostic sensing tools has had an important impact globally. In this regard, here we will review critically the current developed diagnostic tools in response to the COVID-19 pandemic and compare the different types through the discussion of their pros and cons such as nucleic acid detection tests (including PCR and CRISPR), antibody and protein-based diagnosis tests. In addition, potential technologies that are under development such as on-site diagnosis platforms, lateral flow, and portable PCR units are discussed. Data collection and epidemiological analysis could also be an interesting factor to incorporate with the emerging technologies especially with the wide access to smartphones. Lastly, a SWOT analysis and perspectives on how the development of novel sensory platforms should be treated by the different decision-makers are analyzed.
Keywords: COVID-19, SARS-CoV-2, Diagnostic kits, Sensors/biosensors, Response strategies
Graphical abstract
1. Introduction
The massive impact of COVID-19 on hospital systems resulted in aggressive policies initiated for alleviating or extenuating the propagation of the pandemic threat. These strategies have been mainly seen as desperate in many of the countries affected [1]. Regardless of the approaches arranged by the many decision-makers both nationally and locally, the most effective ones depend on laboratory testing and isolation of suspected patients. The World Health Organization highlighted the critical importance of laboratory testing for COVID-19 in understanding, tracking, case management, and transmission control of the virus spread [2]. Yet, the types of tests used by each country for the detection and epidemiological tracking are not easily available. Furthermore, the data of the currently used diagnostic kits and their characteristics are even less obtainable which in combination with the reports describing issues with false-negative results make the subject of high concern.
It has been reported that most of the deployed nucleic acid-based diagnostic tools for SARS-CoV-2 have shown important false-negative results especially for the swab samples assayed with reverse-transcription polymerase chain reaction (RT-PCR) [[3], [4], [5], [6]]. As a fact, a report describing a patient identified negatively for three consecutive times using the SARS-CoV-2 nucleic acid has been finally confirmed positive after chest computed tomography (CT) scan after which a fourth RT-PCR test gave a positive result [7]. As such, many groups are advocating the addition of CT scans with RT-PCR tests as a complete approach especially for highly suspicious cases [8,9]. Acquiring sample techniques from upper airway have been put under the spotlight for the high rate of false-negative results during testing [5,10]. Ye and colleagues have reviewed in an earlier work the lack of uniform protocols in sampling methods through the upper airway and demonstrated that nasopharyngeal aspirate gave more accurate results in addition to being the least risky to the staff performing the sampling [6]. On the other hand, some groups proposed sampling from the lower respiratory tract as a more accurate method [5,11].
Laboratory detection approaches for COVID-19 in biological samples demonstrate many pros and cons where the sequestration of the virus could be obtained via cell culture, quick antibody kits, blood samples, and other technologies such as CRISPR or biosensor based methodologies. They are all assays actively utilized in epidemiological studies and point of care applications [11,12]. Given the information above, it is obvious that there is a necessity to apply worldwide standard approaches for the prevention of such irregularity in data and facilitating a quicker and more effective response for eventual future COVID-19-like pandemics. It is necessary to combine the point of view of the decision-makers, healthcare providers, and researchers to make these strategies fast and efficient to any kind of threat. As such, knowing the methods being currently in use and identifying their advantages and disadvantages might be the first step while exploring technological development could provide more plausible solutions for next-generation diagnostic kits. This review is meant to critically examine the various types of diagnostic platforms used for COVID-19 and would explore the available technologies with great potential for implementation in the pandemic response. A SWOT analysis (strength, weakness, opportunities, and threats) of diagnostic kits is proposed. Lastly, what is the future of sensors development in this case, and how it can be perfected will be discussed.
2. What is SARS-COV-2 virus (COVID 19)?
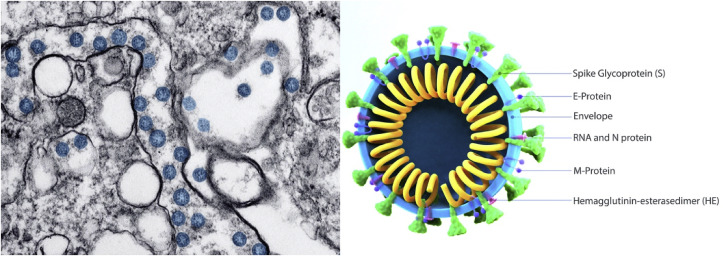
The coronavirus (SARS-CoV-2) was first identified in the region of Wuhan, China in December 2019 from which it got its COVID-19 name. It was identified in the bronchoalveolar lavages of patients after cell culture and transmission electron microscopy (Fig. 1 ). Observations showed a circular shape virus (60–140 nm) containing a genetic material and spike proteins on the surface. This structure was comparable to the viruses of the Coronaviridae family [13].
Fig. 1.
Morphology and structure of the SARS-CoV-2 virus observed under TEM [24].
The RNA of the virus encodes 27 proteins from which an RNA polymerase (RdRP) and 4 structural proteins could be found [14,15]. The RdRP functions with nonstructural proteins to conserve the genome fidelity. This RNA polymerase was found to share 96% similarity with bat coronavirus RaTG13 [15]. Between December to February, 104 strains were discovered sharing 99.9% sequence homology. However, variations in the genome have started to be discovered suggesting an important diversity [16].
SARS-CoV-2 contains 4 structural proteins on the surface (Spike glycoprotein (S), a small envelope protein (E), matrix protein (M), and nucleocapsid protein (N)) where the spike protein is the one responsible for the virus infection property [17]. Furthermore, the S protein facilitates receptor fixation and fusion to host cells which results in the transmission ability of the virus [18]. The gene responsible for S protein coding is less than 75% similar to other described SARS viruses while the other 3 proteins are more conserved [14,15]. The three structural proteins are responsible for wrapping the RNA, protein assembly, bourgeoning, and pathogenesis [[19], [20], [21]].
It has been shown that SARS-CoV-2 enters the cells through interaction with the angiotensin-converting enzyme 2 (ACE2) receptor [15]. ACE2 could be found in most human organs including epithelial cells (Lung alveolar and intestines cells) suggesting them being the primary region for infection onset while oral, nasal mucosa, and nasopharynx has been shown to lack ACE2 expression [22]. The virus diagnosis is generally isolated from oral swabs, lung fluids, and fecal matter [15,23]. It is important to understand the biological functions and features of the virus to allow researchers to develop effective diagnostic tools.
3. Diagnostic tests: what are they and what they sense?
Symptoms shown by infected patients are mostly non-specific and cannot be used as accurate diagnostic criteria for COVID-19 infection due to the similarities with many other respiratory infections such as influenza [25]. As such, molecular techniques and CT scans are seen as more appropriate for diagnosis and screening.
3.1. Nucleic acid detection via RT-PCR
According to the Center for Disease Control and Prevention (CDC), nucleic acid testing is the main approach for COVID-19 diagnostic [26]. RT-PCR technique is considered as the golden standard for diagnosing viral agents. It is based on the reverse transcription of the RNA into complementary DNA (cDNA) and amplifying a specific region [27]. The development of an adequate RT-PCR testing platform needs the selection and design of the right primer and the optimization of the assay conditions. Research has identified three regions on the viral genome with conserved sequences: RNA-dependent RNA polymerase gene (RdRP gene), envelope protein gene (E gene), and nucleocapsid protein gene (N gene) [28]. RdRP and E genes have demonstrated great analytical sensitivity for the detection compared to the N gene. Therefore, it is believed that the development of a two-target platform with a universal primer (all the coronaviruses strands known including SARS-CoV-2) and a specific SARS-CoV-2 primer would give more accurate identification.
Optimizing the assay conditions can involve many of the steps included in the RT-PCR method such as the chemicals used, temperatures, and incubation time. There are two approaches in conducting RT-PCR: (1) one-step assay which implies that transcription and amplification are realized at the same time. This approach is quick and highly reproducible however due to the simultaneous occurrence of transcription and amplification, the optimization of such a technique becomes really difficult and results in a low target amplicon generation; (2) the two-step assay on the other hand, is performed in a sequential order which gives higher sensitivity than the one-step assay. Furthermore, the two-step assay is time-consuming and requires additional parameters to be optimized [29]. It should be noted that these assays need special care in selecting controls because it could be a determining factor for the reliability of the assays and to avoid errors.
The number of developed nucleic-acid-based detection platforms that have been approved is quite important. The majority are applied to samples from different regions of the respiratory tract depending on the symptoms exhibited by patients. Furthermore, depending on the region targeted, different sampling methods are used. Nasopharyngeal/oropharyngeal swabs, washes, and nasal aspirates are seen when sampling from the upper respiratory tract while sputum and tracheal aspirates are used when from the lower respiratory tract. Depending on the onset of the infection date, the detectable levels of the virus will change. During the first 14 days of infection, sputum and nasal swabs are reliable for sampling whereas throat swabs were found to be irregular after 8 days of symptoms observation [30,31]. Negative results do not necessarily mean the absence of infection, it could be due to inadequate sampling, low virus quantities in the sampled region of the respiratory tract, or the presence of a mutation on the viral genome [8,32].
RT-PCR diagnosis though recommended by authorities, it still has many disadvantages. The probability of having false negative needs the addition of complementary tests such as CT scans. Due to the pandemic, the PCR reagents and test kits are not keeping up with the rate of consumption. Hospitals with PCR infrastructures are mainly focused on centralized cities and are less found in the outskirts and far places. Lastly, RT-PCR detection is based on the presence of the SARS-CoV-2 in the sample which means that the test will not identify asymptomatic patients who recovered from the infection, and thus spread prevention measures would not be applied.
3.2. CT scans
Given the high consumption of RT-PCR kits and the false-negative results observed, the Chinese medical authorities opted for CT scans as a complementary measure for COVID-19 diagnosis [33]. The procedure is non-invasive and consists of taking X-ray images of different sections of the chest area. The images are examined by expert radiologists to identify any abnormal characteristics related to the disease. These features are diverse and depend on the infection stage [34]. It has been found that during the early onset of the infection (first 2 days), 56% of the cases presented similar CT results while after 10 days, the lungs' implication was observed. The most commonly observed characteristics include lung consolidation and bilateral/peripheral opacities. The early days of infection showed ground-glace opacities while after the disease progresses further, paving patterns development and consolidation of the lungs are seen [35,36]. Given the scans observations in addition to earlier studies, CT scans showed a higher sensitivity (86–98%) in addition to less false-negative results compared to RT-PCR [8,25,37,38]. However, a huge disadvantage of CT scans is that many characteristics observed for COVID-19 are overlapping with other viral pneumonia and thus making the specificity of the test alarmingly low (25%) [8]. The main drawbacks of CT scans are the costly price, the necessity of highly experienced staff, and being not specific to COVID-19. As such, there a need for exploring other technologies to overcome the issues observed.
3.3. Rising test strategies for COVID-19 diagnosis
Given the urgency of the current pandemic and the need for a quick response, the WHO suggested that priority must be given to nucleic acid and protein test platforms for point of care (POC) applications in the short term [2]. As for the long run, the focus should be directed toward combinatorial applications and multiplex platform development. In addition, serological kits for protein detection should be added to enhance the effectiveness of surveillance and tracking. Unlike RT-PCR, these tests offer the benefit of post-infection/recovery detection which facilitates tracking convalesced patients and superior estimation of virus propagation by the scientists. In addition, on-site devices are cost-effective and easy to operate which lighten the burden on clinics and centralized laboratories [39].
3.3.1. Nucleic acid testing platforms
3.3.1.1. Isothermal amplification techniques
This technology is currently under development for COVID-19 detection. Isothermal amplification methods present great advantages due to the use of a single temperature during the process, easy to perform without the requirement of highly trained staff, and a similar level of sensitivity as PCR [40]. From the many techniques included in this technology, loop-mediated isothermal amplification (LAMP) takes the front. Research has developed reverse transcription LAMP (RT-LAMP) platforms specific for COVID-19 testing and has been clinically tested [[41], [42], [43], [44]]. The technique is based on the use of 4–6 primers and DNA polymerase which gives a higher specificity to LAMP [45]. LAMP-based diagnostic tests are detected via turbidity, colorimetric, or fluorescence [46]. The technique is simple to perform and visualize, has low background interference, does not prerequisite a thermocycler, and the reaction is quick (less than 1 h) [45]. The main disadvantage is the reaction and primers optimization difficulty thus more development is needed for an effective isothermal amplification technique for COVID-19 [46].
The potential of isothermal amplification techniques could be elevated into a multiplex platform either at the amplification or the visualization steps. Multiplexing can utilize specific optical polymeric units for barcoding which are selective to particular biomarkers thus giving the ability to detect various analytes (DNA or protein) from the same sample [47]. As such, multiplexing provides increased information from a single sample and enhances its clinical importance [48]. As of now, the multiplexed assays are still engineered for laboratory use only. However, many efforts are working on the development of on-site devices. The issue of this technology lies within the visualization unit. The presence of different barcode signals needs an intricate readout device to distinguish the different signals. In the aim of overcoming the issue, quantum dots have been proposed for barcoding due to the easiness of excitation (ex. Battery-operated device) and reading the emission signals (ex. Smartphone camera). For example, diagnosing patients with hepatitis B after isothermal reverse polymerase amplification using quantum dot barcodes showed a high level of specificity and sensitivity (>90%) [47]. Barcoded isothermal amplification techniques could be used in the design of diagnosis platforms for specific targets of various diseases such as respiratory viruses (including SARS-CoV-2) and sexually transmitted diseases. Barcodes have a great potential to be tableted which could facilitate the transportation and distribution of the different reagents [49].
3.3.1.2. SHERLOCK detection strategy
One of the emerging nucleic acid-based tests with the potential for SARS-CoV-2 detection is SHERLOCK. This detection strategy is based on the gene-editing tool CRISPR and uses a variant of Cas9 called Cas13a ribonuclease for RNA sensing [50]. Targets from the virus RNA is reverse transcribed to cDNA and followed by isothermal amplification. The resultant product is transformed back to RNA which interacts with Cas13a [51]. When in presence of the target molecule to be detected, Cas13a gets activated and cleaves the fluorescent probe yielding a signal. The advantage of this approach is the storability of the components (freeze-dried) and the high sensitivity shown in testing was demonstrated in a study over the Zika virus [52]. Concerning the COVID-19 situation, two studies have been published proposing a SHERLOCK protocol for SARS-CoV-2 detection [53].
3.3.2. Protein testing platforms
Another approach for diagnosing COVID-19 is the detection of viral spike proteins and the antibodies generated in the patients after infection. The level variation of detectable viral proteins during the infection period makes the detection process quite difficult. It has been shown that salivary levels of viral protein are high during the first days of infection after which, a progressive decline is observed [54]. On the other hand, antibodies present an indirect approach to SARS-CoV-2 sensing. Antibodies are mainly important in the tracing and supervision of the viral infection. The main advantage during the development of serological kits is the observed cross-reactivity between SARS-CoV-2 antibodies and other coronaviruses’ antibodies. It has been shown that the S protein from SARS-CoV-2 and SARS-CoV tested in the plasma sample demonstrated a high level of reactivity [55].
At this time, the development of serological tests for specific antibodies is still ongoing [[56], [57], [58]]. Since the start of the COVID-19 pandemics, many companies have taken the initiative to develop serological and antibody test and have applied for emergency use authorization (EUA) from the FDA (Table 1 ). Recently, a study proposing an enzyme-linked immunosorbent assay (ELISA) for the detection of specific immunoglobulin G and M (IgG and IgM) in human serum of COVID-19 positive patients (confirmed by RT-PCR) was proposed [58]. They utilized the Rp3 nucleocapsid protein from SARS-CoV-2 (which has 90% homology to other SARS viruses). Results showed that 50% and 80% of patients were positive for IgM and IgG, respectively at day 0 after which a further increase to 80% and 100% after 5 days was seen. The antibodies were detected in different matrices (respiratory, blood, and fecal matter samples). According to these studies, a possibility for other proteins or cellular components to be used as biomarkers is quite high. It has been shown that COVID-19 positive patients had an increased C-reactive protein level in addition to immune cells such as lymphocytes [25]. The issue in targeting the irregularities of these markers is that they are common with other diseases. Again, a multiplex approach including antibodies and other biomarkers could greatly enhance the specificity.
Table 1.
Serological tests in development by test kit manufacturers and commercial laboratories [59].
| Manufacturer | Diagnostic Test under EUA | Technology |
|---|---|---|
| Quidel Corporation | Sofia 2 SARS Antigen FIA | Antigen |
| Wadsworth Center, New York State Department of Health | New York SARS-CoV Microsphere Immunoassay for Antibody Detection | Serology Total Antibody |
| Bio-Rad Laboratories, Inc | Platelia SARS-CoV-2 Total Ab assay | Serology Total Antibody |
| Ortho Clinical Diagnostics, Inc. | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack | Serology Total Antibody |
| Autobio Diagnostics Co. Ltd. | Anti-SARS-CoV-2 Rapid Test | Serology IgM and IgG |
| Chembio Diagnostic System, Inc | DPP COVID-19 IgM/IgG System | Serology IgM and IgG |
| Cellex Inc. | qSARS-CoV-2 IgG/IgM Rapid Test | Serology IgM and IgG |
| Abbott Laboratories Inc. | SARS-CoV-2 IgG assay | Serology IgG only |
| DiaSorin Inc. | LIAISON SARS-CoV-2 S1/S2 IgG | Serology IgG only |
| Ortho-Clinical Diagnostics, Inc. | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack | Serology IgG only |
| EUROIMMUN US Inc. | Anti-SARS-CoV-2 ELISA (IgG) | Serology IgG |
| Mount Sinai Laboratory | COVID-19 ELISA IgG Antibody Test | Serology IgG |
| Roche Diagnostics | Elecsys Anti-SARS-CoV-2 | Serology Antibody |
| DiaSorin Inc. | LIAISON SARS-CoV-2 S1/S2 IgG | IgG, CLIA |
| Ortho-Clinical Diagnostics, Inc. | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack | IgG, CLIA |
| Babson Diagnostics, Inc. | Babson Diagnostics aC19G1 | IgG, CLIA |
| Abbott Laboratories Inc. | SARS-CoV-2 IgG assay | IgG, CMIA |
| Mount Sinai Laboratory | COVID-19 ELISA IgG Antibody Test | IgG, ELISA |
| EUROIMMUN US Inc. | Anti-SARS-CoV-2 ELISA (IgG) | IgG, ELISA |
| InBios International, Inc. | SCoV-2 Detect IgG ELISA | IgG, ELISA |
| Emory Medical Laboratories | SARS-CoV-2 RBD IgG test | IgG, ELISA |
| Healgen Scientific LLC | COVID-19 IgG/IgM Rapid Test Cassette (Whole Blood/Serum/Plasma) | IgM and IgG Lateral Flow |
| Hangzhou Biotest Biotech Co., Ltd. | RightSign COVID-19 IgG/IgM Rapid Test Cassette | IgM and IgG Lateral Flow |
| Biohit Healthcare (Hefei) Co. Ltd. | Biohit SARS-CoV-2 IgM/IgG Antibody Test Kit | IgM and IgG Lateral Flow |
| Hangzhou Laihe Biotech Co., Ltd. | LYHER Novel Coronavirus (2019-nCoV) IgM/IgG Antibody Combo Test Kit (Colloidal Gold) | IgM and IgG Lateral Flow |
| Vibrant America Clinical Labs | Vibrant COVID-19 Ab Assay | IgM and IgG, CLIA |
| Cellex Inc. | qSARS-CoV-2 IgG/IgM Rapid Test | IgM and IgG, Lateral Flow |
| Autobio Diagnostics Co. Ltd. | Anti-SARS-CoV-2 Rapid Test | IgM and IgG, Lateral Flow |
| Ortho Clinical Diagnostics, Inc. | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack | Total Antibody, CLIA |
| Siemens Healthcare Diagnostics Inc. | Atellica IM SARS-CoV-2 Total (COV2T) | Total Antibody, CLIA |
| Siemens Healthcare Diagnostics Inc. | ADVIA Centaur SARS-CoV-2 Total (COV2T) | Total Antibody, CLIA |
| Siemens Healthcare Diagnostics Inc. | Dimension Vista SARS-CoV-2 Total antibody assay (COV2T) | Total Antibody, CLIA |
| Siemens Healthcare Diagnostics Inc. | Dimension EXL SARS-CoV-2 Total antibody assay (CV2T) | Total Antibody, CLIA |
| Roche Diagnostics | Elecsys Anti-SARS-CoV-2 | Total Antibody, ECLIA |
| Bio-Rad Laboratories, Inc. | Platelia SARS-CoV-2 Total Ab assay | Total Antibody, ELISA |
| Wadsworth Center, New York State Department of Health | New York SARS-CoV Microsphere Immunoassay for Antibody Detection | Total Antibody, FMIA |
3.3.3. On-site testing platforms
On-site tests are an important tool for point-of-care diagnosis. Their main advantage resides in obtaining results without sending samples to laboratories. This proves very useful for communities lacking advanced infrastructures.
3.3.3.1. Lateral flow assays
One of the approaches that are under development is the lateral flow assay for COVID-19 diagnosis [57]. It is a paper strip coated with gold nanoparticles functionalized with antibodies. The test is qualitative and results can be observed by a simple color change in the lines due to gold nanoparticles clustering via plasmon banding. Commercial tests for either IgG and IgM have shown a 100% specificity while their sensitivity and accuracy ranges between 60 and 90%. In addition, a lateral flow test for both IgG and IgM demonstrated a sensitivity reaching 80% [57]. Nucleic acids could also be integrated into lateral flow assays. A research team has reported the combination of RT-LAMP tests with the lateral flow method for the detection of MERS-CoV [60]. The challenge observed with on-site tools is their single-use application and lower sensitivity compared to RT-PCR. However, to improve the assays, many researchers have been working on developing various approaches for amplifying the signal readout [61].
3.3.3.2. Microfluidic devices
Microfluidic devices are another option for point-of-care diagnostic. They comprise a small chip (made of paper, glass, or polydimethyl sulfoxide) containing micro-sized channels and reaction spaces. The device processes the sample in different ways (ex. Vacuum, capillarity, electro-kinetics). The advantages proposed by microfluidics consist of the small size of the device and the sample volume, and the quick time for detection [62]. A microfluidics-based smartphone dongle for antibody detection of sexually transmitted infections has been proposed. The device showed a high sensitivity level (between 90 and 100%) [63]. Though no attempts to adapt this technology to SARS-CoV-2 detection, the technology stands as a viable option for COVID-19's nucleic acid or specific proteins.
3.3.3.3. Portable RT-PCR devices
Given the strength of RT-PCR described earlier, its main challenge resides in the need of equipped infrastructures, skilled and highly trained staff, and at least 6–24 h of waiting. As such, this technology is not efficient in the rapid-screening of a huge number of individuals in crowded places. A very interesting instrument that has been developed and widely used in plant pathogenesis could be adapted to SARS-CoV-2 detection. The device is a portable DNA detection unit that in addition to targeting viral RNA, can analyze host biomarkers which represent a complementary approach for precise detection and avoiding false-positives. As the virus structure and components have been studied, it was shown that the S and N proteins are predominantly present and long-lasting by T-cell responses [64]. This suggests the use of S, N, E, and M proteins’ nucleic acids as targets to be tested via the device.
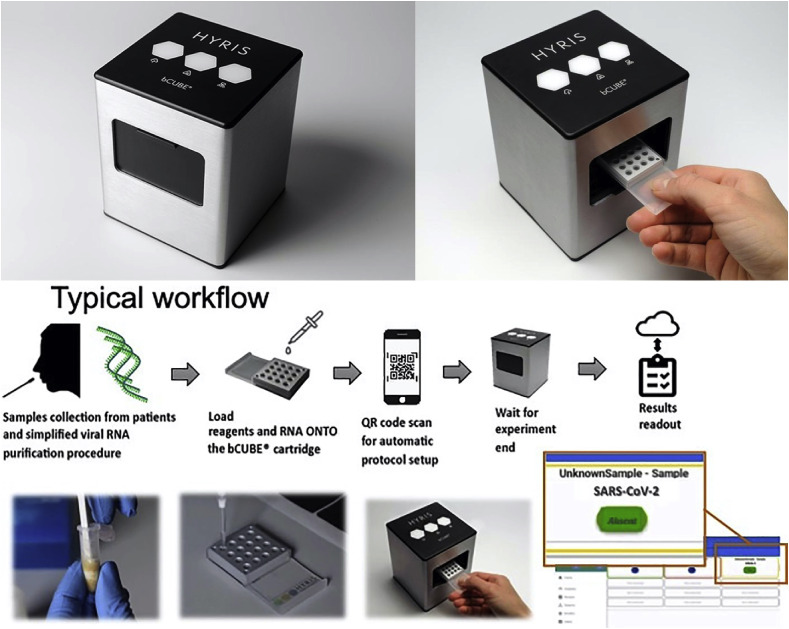
The proposed device is commercially known under the name of bCUBE®, and it is developed by Hyris Ltd. The device is a small unit (10 × 10 × 12 cm) able to perform both temperature cycles and isothermal analysis which allows the screening of a large number of nucleic acids. It can run custom thermal cycle protocols and linked to a centralized database. The handling and results analysis are controlled via any mobile device such as smartphones. The device can be multiplexed using customizable cartridges handling up to 36 samples with a simple workflow (Fig. 2 ). Furthermore, the platform has already been certified to both European and North American standards. The analysis of RNA by the instrument takes less than 2 h on-site without the need for laboratory or highly trained individuals. The gene expression analysis could be performed on oral or nasal swab samples with simplified and quick RNA extraction methods [65].
Fig. 2.
Portable PCR device bCUBE® and how it is used. Reproduced with permission from Ref. [65]. Copyright 2020 John Wiley and Sons.
It should be noted that the technologies described above present high feasibility in the fight against COVID-19. Many emerging technologies-based platforms are under development that could be adapted for SARS-CoV-2 diagnosis. Table 2 presents a more comprehensive review of these options. For example, electrochemical sensors, paper-based systems, and surface-enhanced Raman scattering based systems. These strategies are still in early development and cannot be fully utilized for COVID-19 diagnosis.
Table 2.
Emerging Diagnostic tools with potential SARS-CoV-2 application.
| Diagnostic Platform | On-site application | Mode of action | Matrices (# of samples) | Reference |
|---|---|---|---|---|
| 1. Nucleic acid detection | ||||
| CRISPR | Yes | PCR, perform CRISPR/Ca9-mediated lateral flow nucleic assay (CASLFA) | Serum (110) | [66] |
| CRISPR | Yes | RPA, SHERLOCK multiplexed signal detection via fluorescence | Nasopharyngeal swabs (384) | [50] |
| LAMP | No | Isothermal DNA synthesis using self-recurring strand displacement reactions; positive detection leads to increased sample turbidity | Throat swabs (53) | [67] |
| RPA | No | Forward and reverse primars blind to DNA and amplify strands at 37 °C | Fecal and nasal swabs (30) | [68] |
| NASBA | No | Transcription-based amplification for RNA targets | Nasal swabs (138) | [69] |
| RCA | No | DNA polymerase used to extend a circular primer and repeatedly replicate the sequence | Serum (7) | [70] |
| RT-LAMP | No | Reverse transcriptase LAMP reaction for RNA targets | Nasopharyngeal aspirates (59) | [71] |
| Quantum dot barcode | Yes | Multiplexed quantum beads capture viral DNA for RPA detection | Serum (72) | [47] |
| Magnetic bead | No | Magnetic beads isolate bacteria for PCR detection | Stool (17) | [72] |
| 2. Protein/antibody detection | ||||
| Smartphone dongle | Yes | Microfluidics-based cassette operating an ELISA | Blood (96) | [63] |
| Paramagnetic bead | No | Magnetic separation of protein targets | Serum (12) | [73] |
| Magnetic bead isolation | No | Magnetic isolation of bacteria | Synovia (12) | [74] |
| ELISA | No | Enzymatic reaction to produce colored product in presence of target | Serum (30) | [75] |
| SIMOA | No | Digital readout of colored product by enzymatic reaction in presence of target | Serum (30) | [76] |
| Biobarcode assay | No | Protein signal is indirectly detected by amplifying DNA conjugated to gold nanoparticle | Serum (18) | [77] |
| Rapid antigen test | Yes | Gold-coated antibodies produce colorimetric signal on paper in presence of target | Serum (117) | [78] |
| 3. Whole pathogene detection | ||||
| Magnetic bead isolation | No | Magnetic isolation of bacteria | Synovia (12) | [74] |
4. How are biosensors and bioelectronics beneficial for virus detection?
There is no doubt that biosensing technology has a bright future ahead. This fact is further highlighted by the constant increase of published papers and patents reporting the subjects of viruses and biosensors. This represents a healthy research activity with a 16% growth rate annually [79]. Biosensors should offer quick and efficient detection of viral diseases with high levels of specificity and sensitivity [80]. These criteria are crucial in the success or failure of the detection technology. As such, the choice of the targets of any given pathogen can be a deciding factor. There are two strategies followed: viral nucleic acid or specific proteins or biomarkers. Nanotechnology-based biosensors are known for their promising results in addition to their advantage of being highly customizable through labeling and biofunctionalization.
4.1. Detection methods
Advances in virus sensing have been achieved in electrochemical, fluorescence, light scattering, surface-enhanced Raman scattering (SERS), and SPR sensors. All approaches focus on three main targets: nucleic acids, antibodies, and protein affinity. The objective of biosensor development is to propose an effective alternative tool for diagnosis in a miniaturized shape, time-effective, and ease to manipulate without high-level facilities. The question on the importance of biosensors and bioelectronics is not recent, researchers have been exploring and progressing in viral biosensing in preparation of any new outbreak. From the many reviews present on the subject, three reports have been of great interest given the details given on the role of biosensors and their potential in future pandemic outbreaks [79,81,82]. Biosensors are now able to exhibit high sensitivity with ultra-low LODs. While gold electrodes are the most frequently used working electrodes, many new materials such as polymers, nanoparticles, and composites have been in constant emergence.
To avoid redundancy and repetition, it should be noted that many of the examples described across the manuscript fall under the biosensors category. Indeed, the important potential of biosensor and bioelectronics in the fight against any COVID-19-like outbreaks could be seen.
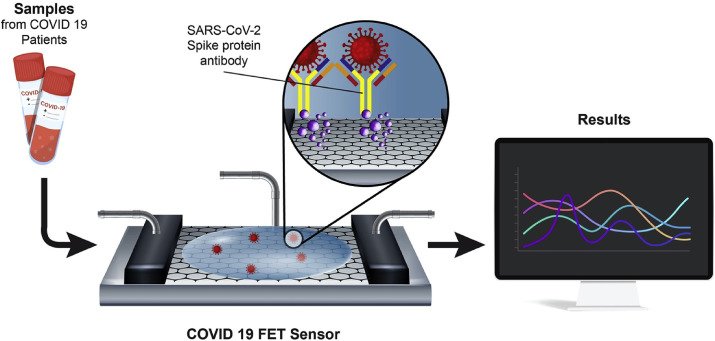
A recent addition to the use of biosensors in COVID-19 detection was provided by a team from Korea. They developed a fast SARS-CoV-2 detection kit from a nasopharyngeal swab sample using a field-effect transistor (FET)-Based Biosensor [83]. The sensor was constructed by conjugating the graphene of the FET with an antibody against the spike protein of the COVID-19 via 1-pyrenebutyric acid N-hydroxysuccinimide ester, which is an interfacing molecule as a probe linker (Fig. 3 ). The platform was able to detect the S protein as low as 1.0 fg/mL in PBS while in clinical transport medium it reached 100 fg/mL. Furthermore, the FET sensor successfully detected SARS-CoV-2 in culture medium (LOD = 1.6 × 101 pfu/mL) and clinical samples (LOD = 2.42 × 102 copies/mL).
Fig. 3.
Schematic diagram of COVID-19 FET sensor operation procedure [83].
5. Opinions towards diagnostic kits development
When a global scale emergency with public health implications happens, many actions demand urgent consideration and funding. In the wake of the recent COVID-19 outbreak, there has been quite a debate on which strategy should be followed by the different law and decision-makers. Though the WHO have been communicating their recommendations, there was a reaction latency in some countries and downplaying the threat. Given the fast spreading of the virus and the experience from past epidemics (such as H1N1, Ebola, and the others), the experts' response was to “flatten the curve” and that's by implementing many strategies for social distancing, lockdown, and testing people at risk for disease surveillance.
While the development of specific treatment and vaccine is still in progress, rapid diagnostic strategies are the main tool at hand to limit the propagation via case identification, isolation, and affiliation tracking. Diagnostic tools development can be divided into 4 steps: (1) Conceptualization and design step is very crucial where researchers use experiences from the past and the current technologies to come up with innovative and efficient approaches. 2) Initial testing on small cohorts. (3) Selecting and implementing clinical testing with a larger group of patients. (4) Validation of the technology and proceeding for manufacturing and commercialization.
Governments have initiated a series of major emergency research programs on virus genomics, antivirals, clinical trials, vaccines, diagnostics, and animal models in order to fight the viral threat. WHO has given a list of research areas that need to be prioritized in the immediate vs. the long-term (Table 3 ) [84]. However, even with all the measures planned or the safety measures put in motions, some requirements are still not fulfilled. According to doctors and general healthcare providers, the most important issue during the COVID-19 outbreak is manpower and equipment [85]. Furthermore, researchers advocate that communication is necessary to both health-care workers and the general population in order to implement infection prevention and control measures that are based on sound scientific principles [86].
Table 3.
Priority research areas with immediate, intermediate, and longer-term goals [84].
| Immediate Goals | Intermediate Goals | Long-term Goals | |
|---|---|---|---|
| Diagnostics | RNA assays, antibody & antigen assays, point of care detection | Multiplex diagnostic platforms | Prognostic markers |
| Therapeutics | Remdesivir, favipiravir, chloroquine, plasma, TCM | intravenous immunoglobulin (IVIg) | Innovative approaches (CRISPR-CAS; RNAi; Cell-based; positive hits from library screening) |
| Vaccines | Development of animal models | mRNA candidates and candidate viral vectors | inactivated candidates and subunit candidates |
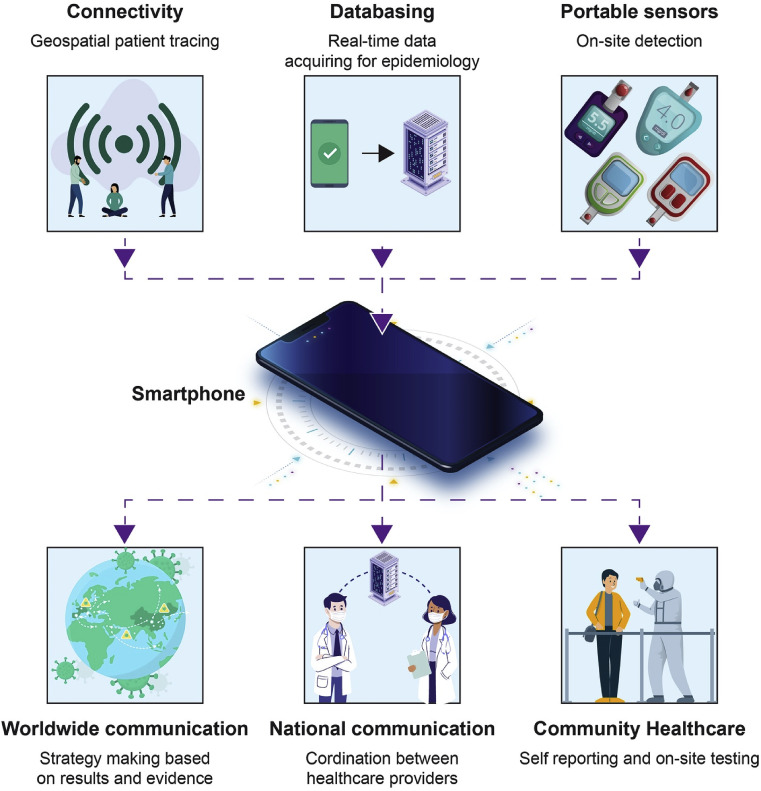
5.1. Smartphones for disease management
Having the spread under control necessitates important surveillance, disseminating data, and infected people tracking [87,88]. Authorities need quick and easy tools of communication in order to manage the propagation. Mobile phones represent an adequate means to fulfill remote reporting, collecting data, and on-site testing owing to their widespread use, connectivity, hardware, and computational power (Fig. 4 ) [89,90]. Smartphone use is a constant rise around the world even is Saharan regions which makes the technology a suitable tool for coordination during COVID-19-like pandemics [90]. In the case of COVID-19, an important insufficiency in communication and reporting has been seen [91,92]. Without having good communication networks between the different administrations and healthcare agencies, the propagation rate of the disease may be greatly influenced [93]. Smartphones could be used to deposit epidemiological data onto public health databases which could be a good starting point for disease control and planning strategies for outbreak response. Smartphones have the ability to be linked with diagnostic tools (dongles) which provide on-site results and geospatial data that helps national and global authorities to implement coordinated strategies. This approach has been positively applied in tracing various infectious diseases (HIV, Ebola, Tuberculosis) [[94], [95], [96]]. Mobile phone applications connect people and doctors during quarantine and help alleviate any mental or other fears that may occur [97]. Furthermore, self-reported symptoms through apps are a great help for epidemiologists and public health agencies to collect information related to transmission behavior [98].
Fig. 4.
Role of smartphones in diagnostics and data acquisition [99].
6. Strengths, weaknesses, opportunities, and threats (SWOT) analysis of diagnostic platforms
The biosensors field has been expanding for the last years from the research of new applications to markets [100]. Businesses target applications in developing countries and low-cost consumables. Indeed, new approaches are mostly orphans and need more work to fulfill the criteria to advance for production. Urgent situations where infrastructures are lacking or insufficient in addition to the need for decentralized tests as we are seeing during the COVID 19 pandemic are crucial factors that can make use of developed technology to facilitate on-site testing. To understand whether current biosensor technology is appropriate for the healthcare sector during pandemic times, technical characteristics, cost, lack of manpower, as well as the finance and policies should be taken into account.
R&D has been working hard for overcoming all the challenges described by creating user-friendly portable biosensors that have no or low necessity for reagents. This is made possible through the miniaturization of the devices, use of nanotechnologies such as nanoparticles and nanowires [101,102], microspotting [103] in addition to linking with smartphones to enable on-site analysis [104]. Another factor of biosensors is their flexibility and versatility where multiplexing is seen as a great solution for accurate results and data analysis [105].
Even when a biosensor is matured enough to obtain satisfactory analytical results, it seems that another challenge is observed during the implementation and deployment of the devices. This challenge is situated generally upstream and downstream of the sensor during the workflow. Sample collection (upstream side) is often underestimated (ex. Blood collection) while in the downstream side, in the absence of a doctor or an expert, no further steps could be done further. Indeed, some diagnostics cannot be made with just a biosensor especially with the risk of positives/negatives readings. As such, authorities need to make adequate preparation with competent staff and centralized data collection to make an appropriate decision and plan the following steps. Without taking these precautions, there is a risk for the biosensors to become just “gadgets” resulting in misdiagnosing, wasting resources, and pollution. Even though these are not the responsibility of researchers and companies, a collective organization with governments, decision-makers, and financers needs to be put in place [100].
Biosensors struggle to exit laboratories to reach industrialization even in developed countries. Genome sequencing is an example where during the last decade, the costs were cut down from 100 M$/subject to 1.0 k$/subject [106,107]. This shows how much the companies contributed to lowering the costs to be able for use in low-income places or extreme situations such as the COVID-19. Creating cost-effective technologies could be reached through the use of inexpensive materials [108], open-source programs [109], and free solutions for optical detection and image processing [110,111]. These are tools able to greatly reduce the cost, however, many companies are not prone to open-source and prefer protecting their own developed tools and platforms.
Other than the financial restriction, regulations coming from strict international healthcare standards are another barrier to be addressed especially with the requirement for certification and licensing resulting in increased competition between companies. Besides, even when researchers develop promising diagnostic tools, it is quite difficult and time-consuming to reach a final product that complies with regulation especially when unforeseen and uncontrollable factors are implicated such as delays and irregularities [100].
From the many examples of biosensors developed for the use in the COVID-19 pandemic, we chose the paper-based flow lateral test as an example for a Strengths-Weaknesses-Opportunities-Threats (SWOT) analysis. This analysis could be applicable to many of the developed on-site platforms (Table 4 ). The main strength is the cost-effectiveness and facile use. The straightforward results allow doctors to interpret and make decisions quickly and plan further actions. Sensors provide good opportunities due to the versatility of their application. The main disadvantage is the tests are qualitative and fragile. As for the threat, the main point is that most public and private investors focus more on drug and vaccine development instead of diagnostic.
Table 4.
SWOT analysis of on-site diagnostic devices. Reused from Ref. [100]. Copyright 2018 by the authors. Licensee MDPI, Basel, Switzerland.
| Strengths—Internal Positive Factors |
Weaknesses—Internal Negative Factors |
| •Ultra-low-cost (less than 1 $) | •Not quantitative |
| •Power-supply-free | •Fragile |
| •Safe disposable | •Single-use |
| •Portable | •Very basic sample processing only |
| •No need for specialized operator | |
| •Equipment-free | |
| •Multiplexing capability | |
| Opportunities—External Positive Factors | Threats—External Negative Factors |
| •Many addressable needs (e.g., infectious diseases diagnostics, vaccination optimization, nutritional monitoring). | •Regulatory agencies may delay/oppose clinical validation |
| •Reimbursement from the state may motivate hospitals/patients to make use of them. | •Much fewer investments than in drug discovery and vaccine development |
| •Slow adoption by physicians and mistrust of results | |
| •Low demand because of low revenue of the patients |
7. Perspectives and conclusions
The presence of diagnosis technologies from earlier works provided a head-start during the COVID 19 outbreak. These technologies are the fruits of years of development and optimization which made them important players in the fight against COVID-19. Past experiences from various pandemics have paved a road for COVID-19 identification and detection development. Identification of the virus through microscopy and gene analysis greatly helped the design of adequate PCR primers and markers in a record time of 3 weeks compared to SARS-CoV identification that took around 5 months [112]. The approaches resulting in the design of RT-PCR diagnosis tests are considered as a first-line defense against the pandemic. Afterward, efforts are made for the development of serologic platforms and portable devices that could complement the PCR findings. As of now, a need for efficient on-site and multiplex platforms is in urgent need. Technologies still in the second and third development phase (Phase 2 and 3) should receive further focused development in order to produce high-quality on-site tools that can be deployed quickly. The combination of detection platforms with mobile phones should not be neglected and should be more implemented to create a clear communication network.
In conclusion, diagnostics play a pivotal role during COVID-19-like outbreaks enabling authorities access to information that facilitates logistic adjustment and provides help to where it is needed, and eventually, these strategies can limit the virus spread and lower mortality rates. COVID-19 data is in constant enrichment due to the efforts of the different parties implicated in the fight and therefore many of the information described in this report may change. Lastly, it should be emphasized that neither diagnostics nor medication and vaccine development can be done without the fundamental studies contributed by the research society.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. Authors thank Dr. Korhan Ege for the fruitful discussions.
References
- 1.Cohen J., Kupferschmidt K. Countries test tactics in 'war' against COVID-19. Science. 2020;367(6484):1287–1288. doi: 10.1126/science.367.6484.1287. [DOI] [PubMed] [Google Scholar]
- 2.WHO . 2020. Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans. who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance. [Google Scholar]
- 3.Han H., Luo Q., Mo F., Long L., Zheng W. The Lancet Infectious Diseases; 2020. SARS-CoV-2 RNA More Readily Detected in Induced Sputum than in Throat Swabs of Convalescent COVID-19 Patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li D., Wang D., Dong J., Wang N., Huang H., Xu H., Xia C. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J. Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Kang H., Liu X., Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020;92(6):538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye G., Li Y., Lu M., Chen S., Luo Y., Wang S., Wang Y., Wang X. Experience of different upper respiratory tract sampling strategies for detection of COVID-19. J. Hosp. Infect. 2020;105(1):1–2. doi: 10.1016/j.jhin.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao W., Li M. Travel Med Infect Dis; 2020. Clinical Diagnostic Value of CT Imaging in COVID-19 with Multiple Negative RT-PCR Testing; p. 101627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei P., Fan B., Mao J., Wang P. Multiple parameters required for diagnosis of COVID-19 in clinical practice. J. Infect. 2020;S0163–4453(20) doi: 10.1016/j.jinf.2020.03.016. 30142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong X., Cao Y.Y., Lu X.X., Zhang J.J., Du H., Yan Y.Q., Akdis C.A., Gao Y.D. Eleven faces of coronavirus disease. Allergy. 2019;2020 doi: 10.1111/all.14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microb. Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen K.S., Ye Z.W., Fung S.Y., Chan C.P., Jin D.Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J., Cui J., Qian Z., Wang Y., Zhang H., Duan Y., Wu X., Yao X., Song Y., Li X., Wu C., Tang X. National Science Review; 2020. On the Origin and Continuing Evolution of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State -nCo V.C.I.T. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullock H., Tamin A. A digitally-enhanced microscopic image shows a coronavirus infection in blue of the first case discovered in the United States. 2020. https://phil.cdc.gov/Details.aspx?pid=23354
- 25.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment C. Expert group for, clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CDC Novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2020. https://www.fda.gov/media/134922/download 2019. [DOI] [PMC free article] [PubMed]
- 27.Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B., Takeda N., Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41(4):1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corman V., Bleicker T., Brünink S., Zambon M. WHO; Geneva: 2020. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RT-PCR. [Google Scholar]
- 29.Wong M.L., Medrano J.F. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39(1):75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 30.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J., Peng L., Wong G., Zheng H., Liao M., Feng K., Li J., Yang Q., Zhao J., Zhang Z., Liu L., Liu Y. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020:2020. 02.11.20021493. [Google Scholar]
- 32.Winichakoon P., Chaiwarith R., Liwsrisakun C., Salee P., Goonna A., Limsukon A., Kaewpoowat Q. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00297-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W., Yan F. Patients with RT-PCR-confirmed COVID-19 and normal chest CT. Radiology. 2020;295(2):E3. doi: 10.1148/radiol.2020200702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E.Y.P., Ng M.Y., Khong P.L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K., Li S., Shan H., Jacobi A., Chung M. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi T., de la Guardia M., Baradaran B. Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. Trac. Trends Anal. Chem. 2020;125:115842. [Google Scholar]
- 40.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 41.Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. medRxiv; 2020. Rapid Detection of Novel Coronavirus (COVID-19) by Reverse Transcription-Loop-Mediated Isothermal Amplification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W., Dang X., Wang Q., Xu M., Zhao Q., Zhou Y., Zhao H., Wang L., Xu Y., Wang J., Han S., Wang M., Pei F., Wang Y. medRxiv; 2020. Rapid Detection of SARS-CoV-2 Using Reverse Transcription RT-LAMP Method. [Google Scholar]
- 43.Yu L., Wu S., Hao X., Li X., Liu X., Ye S., Han H., Dong X., Li X., Li J., Liu J., Liu N., Zhang W., Pelechano V., Chen W.-H., Yin X. iLACO, medRxiv; 2020. Rapid Colorimetric Detection of COVID-19 Coronavirus Using a Reverse Tran-Scriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Plat-form. [Google Scholar]
- 44.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. medRxiv; 2020. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. [Google Scholar]
- 45.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 47.Kim J., Biondi M.J., Feld J.J., Chan W.C. Clinical validation of quantum dot barcode diagnostic technology. ACS Nano. 2016;10(4):4742–4753. doi: 10.1021/acsnano.6b01254. [DOI] [PubMed] [Google Scholar]
- 48.Elnifro E.M., Ashshi A.M., Cooper R.J., Klapper P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000;13(4):559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udugama B., Kadhiresan P., Samarakoon A., Chan W.C.W. Simplifying assays by tableting reagents. J. Am. Chem. Soc. 2017;139(48):17341–17349. doi: 10.1021/jacs.7b07055. [DOI] [PubMed] [Google Scholar]
- 50.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Connell M.R. Molecular mechanisms of RNA targeting by cas13-containing type VI CRISPR-cas systems. J. Mol. Biol. 2019;431(1):66–87. doi: 10.1016/j.jmb.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 52.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Wang J., Gu B., Xu T. medRxiv; 2020. Development and Evaluation of A CRISPR-Based Diagnostic for 2019-novel Coronavirus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chun Chan J.M., Yip G.K., Hong Chik T.S., Wang Y., Chung Choi C.Y., Lin Y., Ng W.W., Zhao J., Poon L.L.M., Malik Peiris J.S., Wilson I.A., Mok C.K.P. bioRxiv; 2020. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. 03.15.993097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai X., Chen J., Hu J., Long Q., Deng H., Fan K., Liao P., Liu B., Wu G., Chen Y., Li Z., Wang K., Zhang X., Tian W., Xiang J., Du H., Wang J., Hu Y., Tang N., Lin Y., Ren J., Huang L., Wei J., Gan C., Chen Y., Gao Q., Chen A., He C., Wang D., Hu P., Zhou F., Huang A., Liu P., Wang D. Cold Spring Harbor Laboratory; 2020. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Corona Virus Disease 2019 (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. medRxiv; 2020. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold- Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19) [Google Scholar]
- 58.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.FDA List of commercial serologic and molecular tests under Emergency Use Authorization (EUA) 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#covid19ivd
- 60.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J., Yu B., Yan F., Hu X., Wu F., Jiao C., Hou P., Xu S., Zhao Y., Feng N., Wang J., Sun W., Wang T., Gao Y., Yang S., Xia X. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spengler M., Adler M., Niemeyer C.M. Highly sensitive ligand-binding assays in pre-clinical and clinical applications: immuno-PCR and other emerging techniques. Analyst. 2015;140(18):6175–6194. doi: 10.1039/c5an00822k. [DOI] [PubMed] [Google Scholar]
- 62.Foudeh A.M., Fatanat Didar T., Veres T., Tabrizian M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip. 2012;12(18):3249–3266. doi: 10.1039/c2lc40630f. [DOI] [PubMed] [Google Scholar]
- 63.Laksanasopin T., Guo T.W., Nayak S., Sridhara A.A., Xie S., Olowookere O.O., Cadinu P., Meng F., Chee N.H., Kim J., Chin C.D., Munyazesa E., Mugwaneza P., Rai A.J., Mugisha V., Castro A.R., Steinmiller D., Linder V., Justman J.E., Nsanzimana S., Sia S.K. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015;7(273):273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 64.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88(19):11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinelli F., Perrone A., Della Noce I., Colombo L., Lo Priore S., Romano S. Immunol Rev; 2020. Application of a Portable Instrument for Rapid and Reliable Detection of SARS-CoV-2 Infection in Any Environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H., Zhang G., Sun J., Zhou X. Clustered regularly interspaced short palindromic repeats/cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14(2):2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 67.Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Van Tu P., Tien N.T., Tashiro M., Odagiri T. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J. Virol Methods. 2007;141(2):173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 68.Amer H.M., Abd El Wahed A., Shalaby M.A., Almajhdi F.N., Hufert F.T., Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol Methods. 2013;193(2):337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wat D., Gelder C., Hibbitts S., Cafferty F., Bowler I., Pierrepoint M., Evans R., Doull I. The role of respiratory viruses in cystic fibrosis. J. Cyst. Fibros. 2008;7(4):320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martel N., Gomes S.A., Chemin I., Trepo C., Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J. Virol Methods. 2013;193(2):653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 71.Shirato K., Nishimura H., Saijo M., Okamoto M., Noda M., Tashiro M., Taguchi F. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J. Virol Methods. 2007;139(1):78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nilsson H.O., Aleljung P., Nilsson I., Tyszkiewicz T., Wadstrom T. Immunomagnetic bead enrichment and PCR for detection of Helicobacter pylori in human stools. J. Microbiol. Methods. 1996;27(1):73–79. [Google Scholar]
- 73.Aytur T., Foley J., Anwar M., Boser B., Harris E., Beatty P.R. A novel magnetic bead bioassay platform using a microchip-based sensor for infectious disease diagnosis. J. Immunol. Methods. 2006;314(1–2):21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 74.Bicart-See A., Rottman M., Cartwright M., Seiler B., Gamini N., Rodas M., Penary M., Giordano G., Oswald E., Super M., Ingber D.E. Rapid isolation of Staphylococcus aureus pathogens from infected clinical samples using magnetic beads coated with fc-mannose binding lectin. PloS One. 2016;11(6) doi: 10.1371/journal.pone.0156287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rowe T., Abernathy R.A., Hu-Primmer J., Thompson W.W., Lu X.H., Lim W., Fukuda K., Cox N.J., Katz J.M. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J., Ferrell E.P., Randall J.D., Provuncher G.K., Walt D.R., Duffy D.C. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thaxton C.S., Elghanian R., Thomas A.D., Stoeva S.I., Lee J.S., Smith N.D., Schaeffer A.J., Klocker H., Horninger W., Bartsch G., Mirkin C.A. Nanoparticle-based bio-barcode assay redefines "undetectable" PSA and biochemical recurrence after radical prostatectomy. Proc. Natl. Acad. Sci. U. S. A. 2009;106(44):18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bosch I., de Puig H., Hiley M., Carre-Camps M., Perdomo-Celis F., Narvaez C.F., Salgado D.M., Senthoor D., O'Grady M., Phillips E., Durbin A., Fandos D., Miyazaki H., Yen C.W., Gelvez-Ramirez M., Warke R.V., Ribeiro L.S., Teixeira M.M., Almeida R.P., Munoz-Medina J.E., Ludert J.E., Nogueira M.L., Colombo T.E., Terzian A.C.B., Bozza P.T., Calheiros A.S., Vieira Y.R., Barbosa-Lima G., Vizzoni A., Cerbino-Neto J., Bozza F.A., Souza T.M.L., Trugilho M.R.O., de Filippis A.M.B., de Sequeira P.C., Marques E.T.A., Magalhaes T., Diaz F.J., Restrepo B.N., Marin K., Mattar S., Olson D., Asturias E.J., Lucera M., Singla M., Medigeshi G.R., de Bosch N., Tam J., Gomez-Marquez J., Clavet C., Villar L., Hamad-Schifferli K., Gehrke L. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017;9(409) doi: 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kizek R., Krejcova L., Michalek P., Merlos Rodrigo M., Heger Z., Krizkova S., Vaculovicova M., Hynek D., Adam V. 2015. Nanoscale Virus Biosensors: State of the Art, Nanobiosensors in Disease Diagnosis; p. 47. [Google Scholar]
- 80.Solanki P.R., Patel M.K., Kaushik A., Pandey M.K., Kotnala R.K., Malhotra B.D. Sol-gel derived nanostructured metal oxide platform for bacterial detection. Electroanalysis. 2011;23(11):2699–2708. [Google Scholar]
- 81.Cesewski E., Johnson B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020;159:112214. doi: 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pejcic B., De Marco R., Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst. 2006;131(10):1079–1090. doi: 10.1039/b603402k. [DOI] [PubMed] [Google Scholar]
- 83.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 84.WHO Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19) 2020. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
- 85.Goh Y., Chua W., Lee J.K.T., Ang B.W.L., Liang C.R., Tan C.A., Choong D.A.W., Hoon H.X., Ong M.K.L., Quek S.T. Operational strategies to prevent coronavirus disease 2019 (COVID-19) spread in radiology: experience from a Singapore radiology department after severe acute respiratory syndrome. J. Am. Coll. Radiol. 2020;S1546–1440(20):30306–30309. doi: 10.1016/j.jacr.2020.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao Y., Torok M.E. Taking the right measures to control COVID-19. Lancet Infect. Dis. 2020;20(5):523–524. doi: 10.1016/S1473-3099(20)30152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gates B. Responding to covid-19 - a once-in-a-century pandemic? N. Engl. J. Med. 2020;382(18):1677–1679. doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 88.Smith R.D. Responding to global infectious disease outbreaks: lessons from SARS on the role of risk perception, communication and management. Soc. Sci. Med. 2006;63(12):3113–3123. doi: 10.1016/j.socscimed.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nayak S., Blumenfeld N.R., Laksanasopin T., Sia S.K. Point-of-Care diagnostics: recent developments in a connected age. Anal. Chem. 2017;89(1):102–123. doi: 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wood C.S., Thomas M.R., Budd J., Mashamba-Thompson T.P., Herbst K., Pillay D., Peeling R.W., Johnson A.M., McKendry R.A., Stevens M.M. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature. 2019;566(7745):467–474. doi: 10.1038/s41586-019-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun H., Dickens B.L., Chen M., Cook A.R., Clapham H.E. medRxiv; 2020. Estimating Number of Global Importations of COVID-19 from Wuhan, Risk of Transmission outside Mainland China and COVID-19 Introduction Index between Countries outside Mainland China. [Google Scholar]
- 92.Zhang R., Liu H., Li F., Zhang B., Liu Q., Li X., Luo L. medRxiv; 2020. Transmission and Epidemiological Characteristics of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infected Pneumonia (COVID-19): Preliminary Evidence Obtained in Comparison with 2003-SARS. [Google Scholar]
- 93.Skowronski D.M., Petric M., Daly P., Parker R.A., Bryce E., Doyle P.W., Noble M.A., Roscoe D.L., Tomblin J., Yang T.C., Krajden M., Patrick D.M., Pourbohloul B., Goh S.H., Bowie W.R., Booth T.F., Tweed S.A., Perry T.L., McGeer A., Brunham R.C. Coordinated response to SARS, vancouver, Canada. Emerg. Infect. Dis. 2006;12(1):155–158. doi: 10.3201/eid1201.050327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brangel P., Sobarzo A., Parolo C., Miller B.S., Howes P.D., Gelkop S., Lutwama J.J., Dye J.M., McKendry R.A., Lobel L., Stevens M.M. A serological point-of-care test for the detection of IgG antibodies against Ebola virus in human survivors. ACS Nano. 2018;12(1):63–73. doi: 10.1021/acsnano.7b07021. [DOI] [PubMed] [Google Scholar]
- 95.Danquah L.O., Hasham N., MacFarlane M., Conteh F.E., Momoh F., Tedesco A.A., Jambai A., Ross D.A., Weiss H.A. Use of a mobile application for Ebola contact tracing and monitoring in northern Sierra Leone: a proof-of-concept study. BMC Infect. Dis. 2019;19(1):810. doi: 10.1186/s12879-019-4354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iribarren S.J., Schnall R., Stone P.W., Carballo-Dieguez A. Smartphone applications to support Tuberculosis prevention and treatment: review and evaluation. JMIR Mhealth Uhealth. 2016;4(2):e25. doi: 10.2196/mhealth.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiang Y.T., Yang Y., Li W., Zhang L., Zhang Q., Cheung T., Ng C.H. Timely mental health care for the 2019 novel coronavirus outbreak is urgently needed. Lanc. Psychiatr. 2020;7(3):228–229. doi: 10.1016/S2215-0366(20)30046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karimuribo E.D., Mutagahywa E., Sindato C., Mboera L., Mwabukusi M., Kariuki Njenga M., Teesdale S., Olsen J., Rweyemamu M. A smartphone app (AfyaData) for innovative one health disease surveillance from community to national levels in africa: intervention in disease surveillance. JMIR Public Health Surveill. 2017;3(4):e94. doi: 10.2196/publichealth.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 100.Migliozzi D., Guibentif T. Assessing the potential deployment of biosensors for point-of-care diagnostics in developing countries: technological, economic and regulatory aspects. Biosensors. 2018;8(4):119. doi: 10.3390/bios8040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He B., Morrow T.J., Keating C.D. Nanowire sensors for multiplexed detection of biomolecules. Curr. Opin. Chem. Biol. 2008;12(5):522–528. doi: 10.1016/j.cbpa.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pingarron J.M., Yanez-Sedeno P., Gonzalez-Cortes A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta. 2008;53(19):5848–5866. [Google Scholar]
- 103.Shumaker-Parry J.S., Zareie M.H., Aebersold R., Campbell C.T. Microspotting streptavidin and double-stranded DNA arrays on gold for high-throughput studies of protein-DNA interactions by surface plasmon resonance microscopy. Anal. Chem. 2004;76(4):918–929. doi: 10.1021/ac034964v. [DOI] [PubMed] [Google Scholar]
- 104.Martinez A.W., Phillips S.T., Carrilho E., Thomas S.W., 3rd, Sindi H., Whitesides G.M. Simple telemedicine for developing regions: camera phones and paper-based microfluidic devices for real-time, off-site diagnosis. Anal. Chem. 2008;80(10):3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Volpetti F., Garcia-Cordero J., Maerkl S.J. A microfluidic platform for high-throughput multiplexed protein quantitation. PloS One. 2015;10(2) doi: 10.1371/journal.pone.0117744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mardis E.R. A decade's perspective on DNA sequencing technology. Nature. 2011;470(7333):198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 107.Wetterstrand K.A. GSP); 2019. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program.https://www.genome.gov/sequencingcostsdata/ [Google Scholar]
- 108.Rolland J.P., Mourey D.A. Paper as a novel material platform for devices. MRS Bull. 2013;38(4):299–305. [Google Scholar]
- 109.Pearce J.M. Maximizing returns for public funding of medical research with open-source hardware. Health Pol. Technol. 2017;6(4):381–382. [Google Scholar]
- 110.Pitrone P.G., Schindelin J., Stuyvenberg L., Preibisch S., Weber M., Eliceiri K.W., Huisken J., Tomancak P. OpenSPIM: an open-access light-sheet microscopy platform. Nat. Methods. 2013;10(7):598–599. doi: 10.1038/nmeth.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. J. Am. Med. Assoc. 2020;323(13):1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]