Abstract
Polycomb group proteins are important epigenetic regulators for cell proliferation and differentiation, organ development, as well as initiation and progression of lethal diseases, including cancer. Upregulated Polycomb group proteins, including Enhancer of zeste homolog 2 (EZH2), promote proliferation, migration, invasion and metastasis of cancer cells, as well as self-renewal of cancer stem cells. In our study, we report that EZH2 and embryonic ectoderm development (EED) indicate respective direct interaction with androgen receptor (AR). In the context of AR-positive prostate cancer, EZH2 and EED regulate AR expression levels and AR downstream targets. More importantly, we demonstrate that targeting EZH2 with the small-molecule inhibitor astemizole in cancer significantly represses the EZH2 and AR expression as well as the neoplastic capacities. These results collectively suggest that pharmacologically targeting EZH2 might be a promising strategy for advanced prostate cancer.
Keywords: EZH2, EED, androgen receptor, astemizole, prostate cancer
Introduction
Prostate cancer (PCa) is a major health concern and the second most common cause of cancer-related mortality among men worldwide, especially in developed countries.1 After local therapy and hormone depletion therapy, most prostate cancer patients relapse and tumors become castration-resistant. For these castration-resistant prostate cancer (CRPC) patients, anti-AR or anti-androgen synthesis therapies, including enzalutamide (MDV3100), Apalutamide (ARN-509) and Zytiga (abiraterone acetate), are most commonly used.2 However, CRPC patients will soon develop drug-resistance to these therapies. Hence, there is a pressing need of new therapeutic targets and reagents for CRPC.
The Polycomb group proteins, which are considered paradigmatic epigenetic modulators, remodel the chromatin structure and subsequent transcriptional repression. Polycomb Repressive Complex 2 (PRC2), one of the two classes of Polycomb group proteins, catalyzes the trimethylation of lysine 27 on histone H3 (H3K27me3) on chromatin. The methylation requires physical interaction between EZH2 and EED, the two core catalytic subunits of PRC2.3,4
Recently, several EZH2 specific inhibitors that target the lysine methyltransferase activities of EZH2 have been developed, including the GSK126 by GSK, the EPZ5687 and EPZ6438 by Epizyme, and the EI1 by Novartis.5–8 Even though these EZH2 inhibitors successfully decrease the methylation marks on H3K27 at relatively low concentrations, they indicated limited utility to inhibit the progression of diffuse large B-cell lymphoma (DLBCL) that harbors the gain-of-function EZH2 mutations, and fail to slow down the growth of solid tumors without EZH2 or other mutations.9–11 Since these inhibitors do not alter EZH2 expression levels, new drugs that decrease EZH2 protein levels might be helpful to resolve this paradox.
It has been reported that EZH2 is a downstream target of AR in prostate cancer. AR directly binds to the upstream enhancer and promoter of EZH2 to activate EZH2 expression.12 EZH2 may also bind directly to AR in CRPC to regulate AR functions.13 However, the interaction between EZH2 and AR is not clearly understood. How EZH2/PRC2 regulates AR functions in CRPC also remains unknown. In our study, we elucidate how EZH2/PRC2 binds to AR to form a complex and alters AR functions by regulating AR expression levels. Furthermore, we newly discovered an EZH2 inhibitor, astemizole, an anti-histamine drug previously on the market as an allergy treatment. We demonstrated that degrading EZH2 with astemizole successfully decreases tumor progression, providing a new therapeutic strategy for advanced CRPC.
Materials and Methods
Cell lines
LNCaP, VCaP, 22Rv1 and HEK293T cells were purchased from ATCC. C4–2 was a gift from Dr. Leland W. Chang. All cell lines were cultured in DMEM (GenDEPOT) or RPMI-1640 (GenDEPOT) supplemented with 10% FBS (GenDEPOT) and used within 20 passages after receipt. The cells were cultured in a 37°C incubator and a humidified atmosphere with 5% CO2. All cell lines were authenticated by the University of Arizona Genetics Core using short tandem repeat (STR) profiling. Cell lines were mycoplasma negative as reported by routine lab tests.
Reagents and antibodies
GSK126 (406,228, MedKoo), EPZ5687 (S7004, Selleckchem), EPZ6438 (S7128, Selleckchem), EED226 (S8496, Selleckchem) and astemizole (3,489, Tocris) were dissolved in 100% ethanol or DMSO for cell treatment. Lipofectamine 3,000 (Thermo Fisher Scientific) was used to perform the transfection of EZH2 shRNA and EED shRNA (Sigma). The following antibodies were used: AR (06–680, Millipore), EZH2 (5,246, Cell Signaling), rabbit polyclonal anti-EED (09–774, Millipore), mouse monoclonal anti-EED (05–1,320, Millipore), normal rabbit IgG (12–370, Millipore), normal mouse IgG (12–371, Millipore), GST (sc-138, Santa Cruz), FLAG (14,793, Cell Signaling), PSA (A0562, Dako), GAPDH (sc-32,233, Santa Cruz), H3K27me3 (9,733, Cell Signaling), H3 (9,715, Cell signaling), β-Actin (A2228, Sigma), LC3-A/B (12,741, Cell Signaling).
Immunoprecipitation
Whole-cell lysate IP was performed by lysing cells in 1× NP-40 lysis buffer (2×) (GenDEPOT) or Pierce RIPA Buffer (Thermo Fisher Scientific) with protease and phosphatase inhibitor (Thermo Fisher Scientific). The lysate was kept on ice for 15 min and sonicated for 2 s on and 2 s off for 30 s and the insoluble pellet was removed after centrifugation. Lysates were pre-cleared using Dynabeads protein A (10002D, Invitrogen) or protein G (10004D, Invitrogen). Antibodies were added to lysates and incubated at 4°C for 2 hr. The immune complexes were then mixed with Dynabeads protein A (10002D, Invitrogen) or protein G (10004D, Invitrogen) at 4°C overnight, and beads were washed three times extensively with the corresponding lysis buffer.
For in vitro immunoprecipitation, AR-FL (346101–5,000 U, EMD Millipore), EZH2 (50,279, BPS Bioscience), EED (50,280, BPS Bioscience) and AR-NTD (ab82124, Abcam) were purchased from the vendor listed. RING1B was produced and fused with a GST tag. The proteins were mixed and added into chilled PBS (1 mL) with a protease and phosphatase inhibitor. 50 μL of the solution was aspirated as input. The remaining protein mixture was incubated with anti-AR antibody at 4°C for 2 hr. The immune complexes were then mixed with Dynabeads protein A (10002D, Invitrogen) at 4°C overnight, and beads were washed three times extensively with NP-40 lysis buffer.
The beads were eluted by 2× reducing SDS-sample buffer prepared by an equal volume of lysis buffer and 4× reducing SDS-sample buffer (BP-110R, Boston BioProducts) and heated to 95°C for 15 min.
Western blotting
To denature proteins, lysates were added to 1× reducing SDS-sample buffer prepared by lysis buffer and 4× reducing SDS-sample buffer (BP-110R, Boston BioProducts) and heated to 95°C for 10 min. Protein levels were assessed by standard SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes (162–0177, BIO-RAD). Images were captured using the ChemiDoc XRS+ Molecular Imager system (BIO-RAD). Primary antibodies used in western blot analyses are listed above. Blots were incubated overnight with primary antibodies at 4 C, followed by detection with Clean-Blot IP Detection Reagent (HRP) (21,230, Thermo Fisher Scientific), goat anti-mouse IgG (H+L)-HRP (SA001–500, GenDEPOT), or goat anti-rabbit IgG (H+L)-HRP (SA002–500, GenDEPOT) secondary antibody.
Mass spectrum analysis
The mass spectrum analysis was performed as previously described.14
Lentiviral constructs
Lentivirus was packaged by cotransfection of constructs with third-generation packaging plasmids pMD2.G, pRRE and pRSV/REV with Fugene HD (Roche) into 10-cm plates with HEK293T cells. The transfection mixture was replaced with growth medium 24 hr after transfection (2 μg of MDLG, 1 μg of VSVG, 1 μg of Rev, and 4 μg of target plasmid). The supernatant was collected at 72 and 96 hr after transfection and centrifuged to remove the cells. Lentiviral titers were determined by p24 assay, in addition to functional titration to determine the multiplicity of infection (MOI) of 1 for each initial batch of virus. Expression was verified by western blotting.
Fusion protein induction and purification
RING1B was cloned into pFN2K vector (Promega) in accordance to the manufacturer’s instructions. BL21 competent E. coli was used as bacterial host strain for the transformation. The transformed bacteria were added into 200 mL of LB medium containing 50 μg/mL of kanamycin. After shaking at 37°C for 2 hr, 100 ul of 0.1 M Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce the expression of fusion protein. The culture was collected by centrifugation after further incubation by shaking overnight at 16°C. The bacterial pellets were lysed using PBS supplemented with 1%Triton X-100 (GenDEPOT) and protease and phosphatase inhibitor (1,861,280, Thermo Fisher Scientific). For protein purification, the cell lysates were sonicated. The cleared supernatants were collected and incubated with Glutathione-Sepharose beads (17–0756–01, GE healthcare). The system was rotated at 4°C for 12 hr. The beads were washed for three times and the proteins were eluted from the beads with PBS supplemented with 0.1% NP-40 and 50 mM Glutathione (Sigma). The purified protein was collected and added with glycerol for preservation.
Reporter luciferase assays
The enhancer and promoter luciferase constructs were gifts from Dr. J. Chad Brenner and sequenced to confirm its precision. The promoters were cotransfected together with pRL-TK at a ratio of 10:1 into stable cell lines LNCaP and VCaP. Lentivirus packaged with EZH2 or EED shRNA was added 24 hr after cotransfection. Cells were lysed 24 hr later and conducted using the Dual-Luciferase Reporter Assay System (E1910, Promega). The bioluminescence was read on Synergy 2 Multi-Mode Reader (BioTek). PSA and TMPRSS2 promoter luciferase activity was normalized with Renilla luciferase activity. Each experiment was performed in quadruplicate.
RNA isolation and RT-qPCR
Total RNA was isolated from cells to generate cDNA using the RNA MiniPrep kit (Direct-zol, R2052, ZYMO Research) and amfiRivert cDNA Synthesis Platinum Master Mix (R5600–100, GenDEPOT). Each cDNA sample was amplified using iTaq Universal SYBR Green Supermix (172–5,124,BioRad) on the QuantStudio 6 Flex Real-time PCR System (403115082, GE Healthcare). Briefly, the reaction conditions consisted of 2 μL of cDNA and 0.2 μM primers in a 10 μL total volume of super mix. The whole system was hold at 95°C for 10 min to denature. Then each cycle consisted of denaturation at 95°C for 30 s and annealing/extension at 60°C for 30 s. GAPDH was used as an endogenous control to normalize each sample. The primers are listed in Supporting Information Table 1.
RNA-sequencing analysis
The RNA-seq reads were mapped to the human reference genome version hg19 using TopHat (version 2.0.12) default parameters.15 The human reference gene set (RefSeq gene) was downloaded from https://www.ncbi.nlm.nih.gov/refseq/rsg/. Cuffdiff (v2.0.12) was used to calculate gene expression level and the significance of differential expression based on the classic-FPKM using default parameters.16 We used p value<0.05 as a threshold to select differentially expressed genes. For clustering analysis, we used hierarchical clustering method with Spearman correlation distance to cluster samples based on the log scaled FPKM, and used MORPHEUS (https://software.broadinstitute.org/morpheus/) to plot the heat map. We used Fisher’s exact test to calculate p values for significance of overlapping between two groups of genes. Gene set enrichment analysis (GSEA) was applied to assess the significance of associations between AR target genes and genes affected by astemizole treatment or EZH2 knockdown.17 To compare the expression level of EZH1 and EZH2, gene expression data for metastatic prostate tumor was collected from the Gene Expression Omnibus (GEO) database (accession no. GSE35988). Raw expression data was downloaded as a SOFT formatted family file. The expression value is the log2 ratio of prostate tissue (test) / pooled benign prostate tissue (reference). To analyze differential expression in different prostate tumor stage, gene expression data for EZH1 and EZH2 and clinical data were obtained from The Cancer Genome Atlas (TCGA) database via cBioPortal (http://www.cbioportal.org). For gene expression data, the relative expression (z score) of an individual gene comparing to the gene expression distribution in a reference population was analyzed. The reference population was all tumors that are diploid for the gene in question. Two-tailed Wilcox test was used to access the significance for differential expression when two groups were compared.
Data accessibility
The GEO accession number for the RNA-seq data sets reported in this paper is GSE124268.
Cell growth assay
Cells were seeded in 96-well plates and treated at concentration gradients for 72 hr. Bioluminescence was measured to quantify cell viability using CellTiter-Glo® Luminescent Cell Viability Assay Kit (Promega) and was read on Synergy 2 Multi-Mode Reader (BioTek). The cell proliferation curve was drawn and fit by the bioluminescence to drug concentration. Half-maximum inhibitory concentration (IC50) was calculated with nonlinear fitting.
Wound healing assay
Cell migration capacities were detected using wound healing assay. C4–2 cells were plated with 80–90% confluence in 6-well plates. Wounds were created across the monolayer of cell culture using a bio-clean pipette tip. The cells were incubated in serum-free medium supplemented with 5 or 10 μM of astemizole after rinsed with PBS. Wound closure were captured at 0, 24 and 72 hr.
Boyden chamber invasion assay
Polycarbonate membrane cell culture inserts (CLS3422, Corning) were applied with Basement Membrane Matrix (Cultrex). After the matrix condensed at 37°C in cell incubator, the inserts were added with 1 × 105 of C4–2 cells in RPMI-1640 without FBS. The outside wells were added with RPMI-1640 with FBS. Astemizole or ethanol was added to keep the same concentration inside and outside of the inserts. The inserts were fixed with methanol and cells that permeated through the membrane were stained with 0.5% crystal violet. Images were captured and cell count was calculated.
Autophagy assay
C4–2 cells were seeded in 6-well plates and treated with astemizole at dose gradients for 72 hr. Cells were lysed for western blotting to detect LC3-A/B. Densitometry measurements of bands were quantitated and calculated in ImageJ. In another set of plates, autophagosome activity was detected with specificdye using an autophagy assay kit (MAK138, Sigma). The pictures were captured under fluorescence microscopy, and bioluminescence was read on Synergy 2 Multi-Mode Reader (BioTek).
Apoptosis assay
C4–2 cells were plated and treated with astemizole at dose gradients in 6 well plates for 72 hr. Apoptosis was detected using FITC annexin V apoptosis detection kit (556,547, BD Biosciences). The staining was analyzed by flow cytometry (LX200 Luminex Multiplexing Assay system).
Murine prostate tumor xenograft model
CB17SCID mice were purchased from Charles River. Animal care and conditions were followed in accordance with institutional and National Institutes of Health protocols and guidelines, and all studies were approved by Houston Methodist Institution Animal Care and Use Committee. Tumor xenograft model was induced as previously described.18 Mice were anesthetized using 2% isoflurane (inhalation), and 2 × 106 of VCaP prostate cancer cells suspended in 100 μL of PBS with 50% Basement Membrane Matrix (Cultrex) were implanted subcutaneously into the dorsal flank on the right side of each mouse. Tumor volumes were measured by length (a), width (b) and calculated as tumor volume = MIN(a)2 × MAX(b)/2. For VCaP castration-resistant prostate tumor model, VCaP tumor-bearing mice were castrated when tumors grew to approximately 200–300 mm3 in size (approximately 5 weeks after implantation of tumor cells) and once tumors started to relapse, mice were randomized and treated with vehicle or astemizole (50 mg kg−1) daily (5 days per week), and terminated 28 days later. A total of 20 mice were utilized, with 12 mice in vehicle-treated group and 8 mice in astemizole-treated group. Body weight of mice was also monitored during the course of the study. Kaplan–Meier analysis of tumor volume doubling time was performed as previously described.19,20
Immunohistochemistry
Mice were sacrificed for tumor tissues. Part of tumor tissues were fixed in 10% neutral-buffered formalin, processed and embedded in paraffin. EZH2(1:1000, AR(1:600) and PSA (1:2000) staining were developed using DAB (Vector Laboratories, Burlingame, CA) followed by Hematoxylin counterstaining (Sigma, St. Louis, MO). Detection was developed by Alexa 594 nm conjugated secondary antibodies (Molecular Probes, Eugene, OR) and visualized with microscopes (Daco). The slides were scanned and then quantitated using ImageJ to determine the proportion of stained cells. The results were normalized with the vehicle control group.
Statistical analysis
No statistical method was used to predetermine sample size. Mice were assigned at random to treatment groups and, where possible, mixed among cages. There were no inclusion or exclusion criteria. Whenever possible, the investigators were blinded to group allocation during the experiments and when assessing outcomes. Experiments were repeated two to three times. Data were analyzed using Prism 6.0 software (GraphPad) and presented as mean ± SEM. The p values were assessed using a two-tailed unpaired Student’s t-test or a two-way analysis of variance (ANOVA), with significance considered as follows: *p < 0.05; **p < 0.01; and ***p < 0.001. For tumor-free mice frequency, statistics were done with log-rank (Mantel–Cox) test.
Results
EZH2 and EED directly interact with AR in prostate cancer
Our previous mass spectrometry analysis14 indicated that Polycomb Group protein EED interacts with AR (Supporting Information Fig. 1). To confirm this finding, we performed immunoprecipitation with the anti-EED antibody, followed by immunoblot analysis using lysates from prostate cancer cell line VCaP. Two distinct anti-EED antibodies pulled down AR successfully (Fig. 1a). In addition, we used anti-EZH2 and anti-AR antibodies to perform immunoprecipitation in 22Rv1, C4–2, LNCaP and VCaP, and discovered that EZH2 and AR were able to pull down each other in all four AR-positive prostate cancer cell lines (Fig. 1b).
Figure 1.
PRC2 protein EED and EZH2 interact with AR. (a) Immunoprecipitation of VCaP cell lysates with the indicated mouse monoclonal anti-EED antibody (05–1,320, Millipore), rabbit polyclonal anti-EED antibody (09–774, Millipore), control IgG and anti-AR antibody was followed by immunoblot analysis. Representative graph from at least three independent experiments is shown. (b) Immunoprecipitation of 22Rv1,C4–2, LNCaP and VCaP cell lysates with anti-EZH2, anti-AR antibody and control IgG was followed by immunoblot analysis. (c) HEK293T cells transfected with Halo-AR (full length), Halo-DBD, Halo-LBD, Halo-NTD plasmids and empty vector were lysed and subjected to pull-down assay using HaloLink resin (Promega), followed by immunoblot analysis. (d) Purified EZH2 and EED were respectively mixed with AR (full length) and pulled down with anti-AR antibody and protein A beads. RING1B served as a negative control. (e) Purified EED was mixed with AR N-terminal fragment and pulled down with anti-AR antibody and protein A beads. RING1B served as a negative control.
AR has three functional domains: N-Terminal Domain (NTD), DNA Binding Domain (DBD) and Ligand Binding Domain (LBD) (Supporting Information Fig. 2). To determine which AR domain is involved in AR-EED interaction, we first overexpressed halo-tagged full-length AR (AR-FL), AR-NTD, AR-DBD and AR-LBD in HEK293T cells. Pulldown assays demonstrated that EED interacted with AR-NTD as well as AR-FL, whereas EZH2 interacted with AR-DBD and AR-FL (Fig. 1c).
Next, we performed in vitro interaction assay using the purified proteins. As expected, EED was detected to directly interact with AR and AR-NTD, and EZH2 was also detected to interact with AR-FL (Figs. 1d and 1e). Our discoveries collectively reveal the intense interaction between PRC2 and AR, which implies the significant function of PRC2 in the progression of prostate cancer.
PRC2 regulates AR and AR pathway
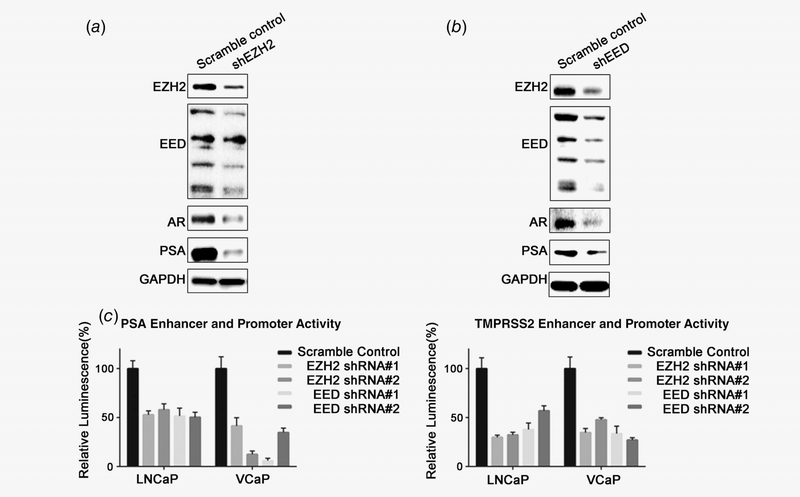
When we knocked down EED by EED specific shRNA packaged in lentivirus, AR and PSA were significantly decreased along with EED (Fig. 2a). Similarly, knocking down EZH2 by shRNA also decreased AR and PSA levels (Fig. 2b). To confirm that EED and EZH2 regulate AR signaling, we transfected firefly luciferase reporters, which have PSA and TMPRSS2 upstream region promoter and enhancer regions, containing AR binding sites, into EED and EZH2 stable knockdown cells. As shown in Figure 2c, in both LNCaP and VCaP cells, PSA and TMPRSS2 promoter activities were significantly decreased by knocking down EED or EZH2. These results collectively confirmed that PRC2 regulates the AR pathway in prostate cancer.
Figure 2.
EZH2 and EED knockdown decreases AR and downstream targets. (a) EZH2 was depleted by shRNA in C4–2 cells. After 48 hr, cells were lysed and blotted by EZH2, EED (rabbit polyclonal anti-EED antibody, 09–774, Millipore), AR, PSA and GAPDH. (b) EED was depleted by shRNA in C4–2 cells. After 48 hr, cells were lysed and blotted by EZH2, EED (rabbit polyclonal anti-EED antibody, 09–774, Millipore), AR, PSA and GAPDH. (c) LNCaP and VCaP cells were subjected to cotransfection of PSA or TMPRSS2 firefly luciferase reporter constructs and pRL-TK (Renilla luciferase). Lentivirus packaged with two distinct shRNAs of EZH2 or EED were added 24 hr after the cotransfection to knockdown EZH2 or EED. The luciferase activity was normalized using Renilla bioluminescence.
Astemizole, a newly identified PRC2 inhibitor, represses both EZH2 and AR
Since EZH2 and EED are found to be profoundly involved in the epigenetic aberrations of prostate cancer progression, tremendous efforts have been made to develop PRC2 inhibitors. GSK126, EPZ5687 and EPZ6438, which suppress EZH2 methyltransferase activities, were successfully developed. EED226 is a newly discovered PRC2 inhibitor, which targets the H3K27me3 binding site of EED.21 Intriguingly, these inhibitors failed to alter AR and AR downstream targets (Fig. 3a). However, astemizole22, a newly identified PRC2 inhibitor which can disrupt the EZH2-EED interaction and then induce the degradation of EED and EZH2 proteins, successfully decreased EZH2, AR and PSA levels (Fig. 3a). The expression levels of EZH1, a paralog of EZH2 in mammals and the other known H3K27 methyltransferase, are very weak or not detectable in prostate cancer tissues, and much lower than the levels of EZH2 in prostate cancer (Supporting Information Fig. 3a and b). Because EZH1 expression levels are not altered in prostate cancer, while EZH2 is significantly upregulated in high-grade prostate cancer tissues (Supporting Information Fig. 3c–f), we focused on investigating the effect of astemizole on EZH2 in our study. We tested astemizole in different AR-positive prostate cell lines, and similar results were observed (Figs. 3b–3d). The data suggest that EZH2 regulates AR independently of its methyltransferase activity. Moreover, astemizole is a PRC2 inhibitor with promising inhibitory effects targeting both EZH2 and AR.
Figure 3.
Astemizole functions as a better PRC2 inhibitor by degrading PRC2 and AR. (a)C4–2 cells were treated with GSK126 (2 μM), EPZ5687 (5 and 10 μM), EPZ6438 (20 and 40 μM), EED226 (10 and 20 μM), astemizole (10 μM) as well as vehicle and lysed for immunoblot analysis 72 hr after drug treatment. (b-d)C4–2, LNCaP and VCaP cells treated with astemizole at dose gradients and lysed for immunoblot analysis 72 hr after treatment.
Astemizole has EZH2 and AR inhibitory effects similar to EZH2 shRNA
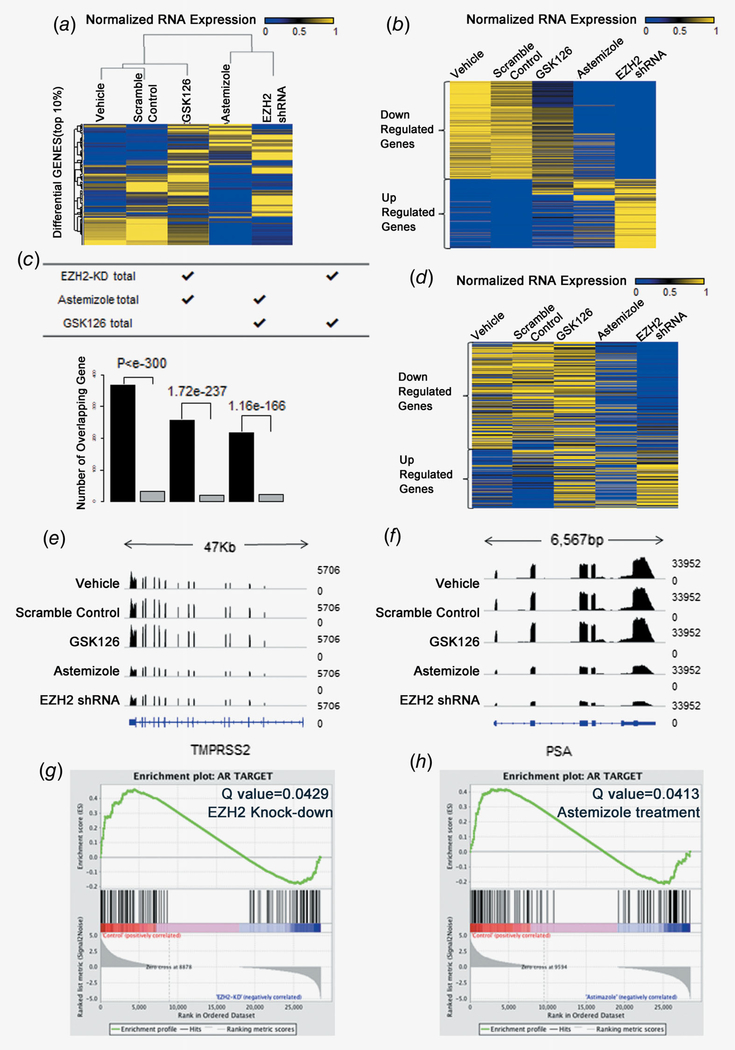
To investigate EZH2 inhibitory effects, we performed RNA-seq for C4–2 cells treated with astemizole, GSK126 and EZH2 shRNA. We retrieved 1,571 (top 10%) genes that display the largest expression variation across these samples, and clustered the samples based on the expression values for these genes. This unbiased comparison revealed that astemizole-treated samples were the closest to the shRNA-treated samples, whereas the GSK126-treated samples had a larger distance to the shRNA-treated sample (Fig. 4a). We further defined genes that were up or down regulated after shRNA treatment, and found that the expression profile of these genes was closer between astemizole-treated and shRNA-treated samples than between GSK126-treated and shRNA-treated samples (Fig. 4b). We observed a 67.89% overlap of downregulated genes from the astemizole-treated and shRNA-treated C4–2 cells. Notably, the number of overlapped differential genes was significantly larger than the number of genes overlapped by chance (Fig. 4c), and the overlap was even more significant between astemizole-treated and shRNA-treated samples (Fisher’s exact test p < 1e-300) than between GSK126-treated and shRNA-treated samples (p = 1.16e-166).
Figure 4.
EZH2 knockdown and astemizole treatment demonstrate similar inhibition patterns of AR signaling blockage.(a) EZH2 knockdown and astemizole-treated samples cluster together based on log expression of 1,571 (top 10%) high variation genes. (b) Heat maps for the expression level of genes down- or up-regulated by EZH2 knockdown, GSK126 and astemizole treatment. (c) The number of overlapped differential genes in each paired group is significantly larger than the number of genes overlapped by chance. (d) 426 AR-induced genes were compared and the expression is similar between EZH2 knockdown and astemizole-treated samples. (e) Comparison of PSA gene track between groups. (f) Comparison of PSA gene track between groups. (g) GSEA shows that AR target genes are significantly enriched (Q value = 0.0429) in downregulated genes due to EZH2 knockdown. (h) GSEA shows that AR target genes are significantly enriched (Q value = 0.0413) in downregulated genes due to astemizole treatment.
Expression changes of AR target genes in response to astemizole treatment also had a pattern analogous to that in response to EZH2 knockdown. Upon analyzing 426 AR-induced genes,23 we found that the expression patterns of both AR upregulated and downregulated genes were similar between astemizole-treatment and EZH2-knockdown samples (Fig. 4d). Further, 113 AR target genes defined by another independent data resource19 also showed similar patterns (Supporting Information Fig. 4). Manual inspection on PSA and TMPRSS2, two known downstream targets of AR, revealed that astemizole and EZH2 shRNA induced the same pattern of RNA expression change (Figs. 4e and 4f). Gene set enrichment analysis (GSEA) also confirmed that AR target genes were significantly enriched in genes downregulated by EZH2 knockdown as well as astemizole treatment. In conclusion, astemizole is a promising inhibitor of EZH2 and AR pathway (Figs. 4g and 4h).
Astemizole inhibits prostate cancer tumor growth
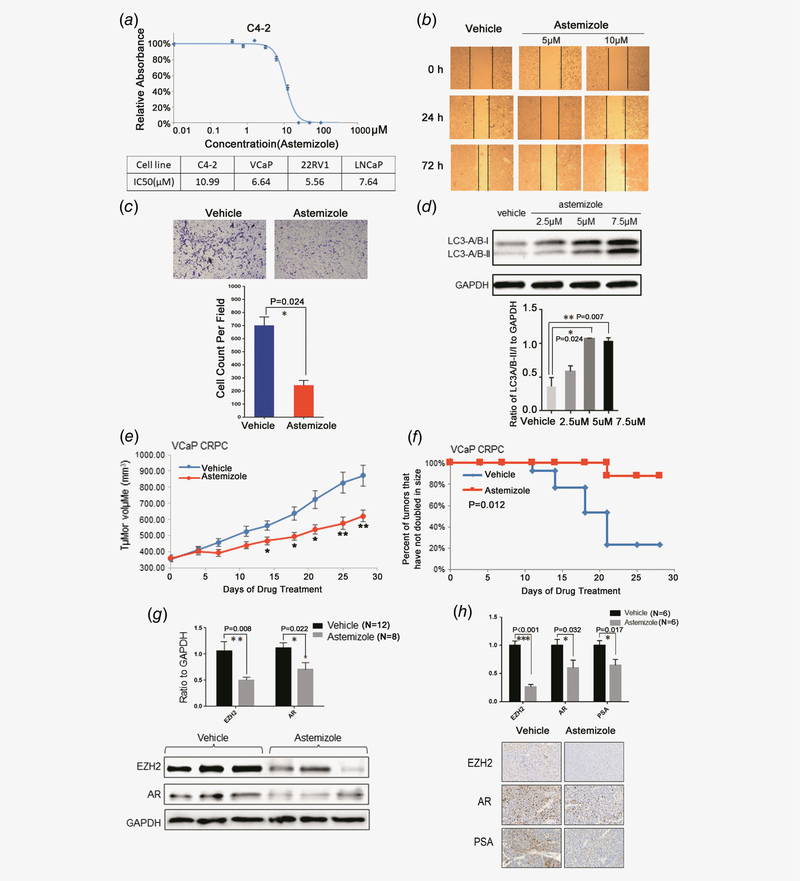
To further investigate if inhibiting EZH2 and AR by astemizole has any effect on the phenotypes of prostate cancer cells, we first performed cell growth assay for AR-positive prostate cancer cell lines LNCaP, VCaP, C4–2 and 22Rv1 (Fig. 5a, Supporting Information Fig. 5), and analyzed the antiproliferative effect of astemizole treatment at different doses. As expected, astemizole was effective on inhibiting the proliferation of each cell line at low dose (Fig. 5a). Next, we performed the wound healing assay for C4–2 cells and demonstrated that astemizole impaired migration capacities (Fig. 5b). Furthermore, we detected thwarted invasive abilities after astemizole treatment in C4–2 cells by performing Boyden Chamber invasion assay (Fig. 5c).
Figure 5.
Astemizole has potent therapeutic effects on prostate cancer. (a) Astemizole critically thwarts cell proliferation in C4–2 and other AR-positive prostate cancer cell lines. (b) The wound healing assay indicates that astemizole compromises the migration of C4–2 cells. (c) Astemizole decreases the invasive abilities of C4–2 cells compared to vehicle treatment. Cell count was analyzed and the difference was statistically significant. (d)C4–2 cells were treated with 2.5, 5 and 7.5 μM of astemizole. Cells were lysed 48 hr after treatment and blotted with anti-LC3-A/B antibody. The ratio of LC3-A/B-II/I to GAPDH was elevated as dose increased, which indicates that astemizole induces autophagy in prostate cancer cells. (e) Castration-resistant VCaP xenograft mouse models were generated. Castrated mice bearing CPRC xenografts received vehicle or astemizole treatment (50 mg kg−1) daily (5 days per week). Caliper measurements were taken every 4 days to determine tumor volume. Mean tumor volume SEM, *p < 0.05, **p < 0.01 vs. vehicle was marked. (f) Kaplan–Meier survival plot compares progression-free survival. (g) Upper panel: Proteins were blotted and quantitated to compare the protein levels of EZH2 and AR in astemizole-treated group (n = 8) compared to vehicle-treated group (n = 12). Lower panel: The expression of EZH2 and AR was decreased in response to astemizole treatment. (h) The proportion of the cells stained with EZH2/AR/PSA in astemizole-treated group (n = 6) were significantly lower than that in vehicle-treated group (n = 6).
Recently, a study reported that EZH2 regulates autophagy via the mTOR signaling pathway and EZH2 knockdown would significantly induce autophagy.24 We observed that the ratio of LC3-A/B-II to LC3-A/B-I was significantly elevated due to astemizole treatment (Fig. 5d). We also detected induced autophagy by staining a proprietary fluorescent autophagosome marker and observed that astemizole treatment significantly promoted the formation of autophagosome (Supporting Information Fig. 6). These results suggest that astemizole functions as a potent EZH2 inhibitor and induces autophagy in prostate cancer cells. However, astemizole treatment did not alter induced apoptosis (Supporting Information Fig. 7), which implies autophagy might be a major phenotypic alteration as a result of astemizole treatment.
To evaluate the therapeutic efficacy of astemizole in CRPC, we utilized castration-resistant mouse xenograft models with implanted VCaP cells. We observed that astemizole significantly inhibited tumor growth when compared to vehicle treatment (Fig. 5e and Supporting Information Fig. 8a). The survival analysis also revealed the anti-tumor effects of astemizole (Fig. 5f) without effect on body weight in the mice xenograft models (Supporting Information Fig. 8b). Furthermore, we extracted protein and RNA from the harvested tumor tissue from mice xenograft models. By immunoblot analysis with anti-EZH2 and anti-AR, we found that EZH2 and AR were decreased in astemizole-treated tumors compared to vehicle-treated tumors (Fig. 5g). We also utilized the formalin-fixed tumor tissues assayed for immunochemistry. We observed that astemizole inhibited the expression of EZH2 and AR, which further validated that astemizole had therapeutic effects by targeting EZH2 and AR in tumor (Fig. 5h). Furthermore, real-time qPCR analysis showed that the well-known AR targets were also decreased by astemizole treatment in xenograft tumors (Supporting Information Fig. 9).
Collectively, our results suggest that astemizole may be repurposed as a feasible treatment for castration-resistant prostate cancer with less adverse effects.
Discussion
EZH2 and other PRC2 components are well-known transcriptional repressors that methylate H3K27 and condense chromatin conformation. We previously reported that EZH2 directly represses multiple downstream targets, including ADRB2, CDH1, rap1GAP, SLIT2 and miRNAs (miR-203, miR-200 family and miR-181 family) by binding to their promoter regions.12,25–28 Other groups also reported that tumor suppressors, such as DAB2IP and miRNAs let-7 family, are EZH2 and PRC2 downstream targets.29,30 The majority of previous reports suggest that EZH2 and PRC2 perform their oncogenic function by repressing these tumor suppressors. However, several groups have recently reported that EZH2 might perform its oncogenic functions by interacting with novel binding partners, such as RELA and RELB, and regulating the NF-κB pathway, and these functions are independent of its lysine methyltransferase activities.31 Besides histone H3, EZH2 can methylate several nonhistone proteins, including GATA4, STAT3, RORa and JARID2, and also regulates the transcriptional activities of these transcriptional factors.32–35 In our study, we discovered that, in prostate cancer, EZH2 and EED directly interact with AR. Intriguingly, knocking down EZH2 remarkably decreased AR at both transcript and protein levels, and then reduced the expression levels of AR activated genes, such as PSA and TMPRSS2. RNA-Seq also supported the activation of AR targets via EZH2. Our data support the idea that in addition to functioning as a transcriptional repressor, EZH2 is also an AR coactivator and activates many downstream targets in prostate cancer.
Targeting EZH2 for advanced cancer patients has been proposed for many years. DZNep was the first discovered EZH2 inhibitor that decreases protein levels but not transcript levels.36 However, it was demonstrated that DZNep is a paninhibitor for several histone lysine methyltransferases.37 In addition, many reports showed that DZNep lacks therapeutic specificity. Since 2012, several other EZH2 inhibitors targeting its enzymatic activities have been developed by pharmaceutical companies. These EZH2 specific inhibitors successfully remove the methyl-groups from histone H3K27 at low concentrations. However, many studies revealed that targeting enzymatic activities of EZH2 failed to inhibit tumor progression of most solid tumors, except in those harboring EGFR or ARID1A mutants.10,11 Recently, a study reported a new EZH2 inhibitor, astemizole, which could disrupt the interaction between EZH2 and EED, to degrade EZH2 proteins.22 In our study, we compared the effects of astemizole and GSK126 in treating prostate cancer. We demonstrated that astemizole treatment could mimic the effect of knocking down EZH2 by siRNAs. Our RNA-Seq analyses revealed that most dysregulated genes by EZH2 siRNAs were also altered by astemizole treatment, but not altered by GSK126. In comparison to the enzymatic inhibitors of EZH2, only astemizole decreased EZH2 protein levels, AR and AR signaling (Fig. 4a). More importantly, we demonstrated that astemizole significantly inhibits the cell growth and tumor growth of CRPC, even though these tumors do not harbor previously known mutations. The EZH2 protein itself is more important than its enzymatic activity for cancer initiation and progression. Therefore, EZH2 degraders, rather than EZH2 enzyme inhibitors, are more potent for advanced cancers. Furthermore, our murine xenografts provide a rationale for repurposing the previously approved anti-histamine drug for treating CRPC patients, for which there is a pressing need to develop more treatment options. For CRPC patients resistant to available anti-AR and/or anti-androgen drugs, astemizole could be one of the available last options.
Supplementary Material
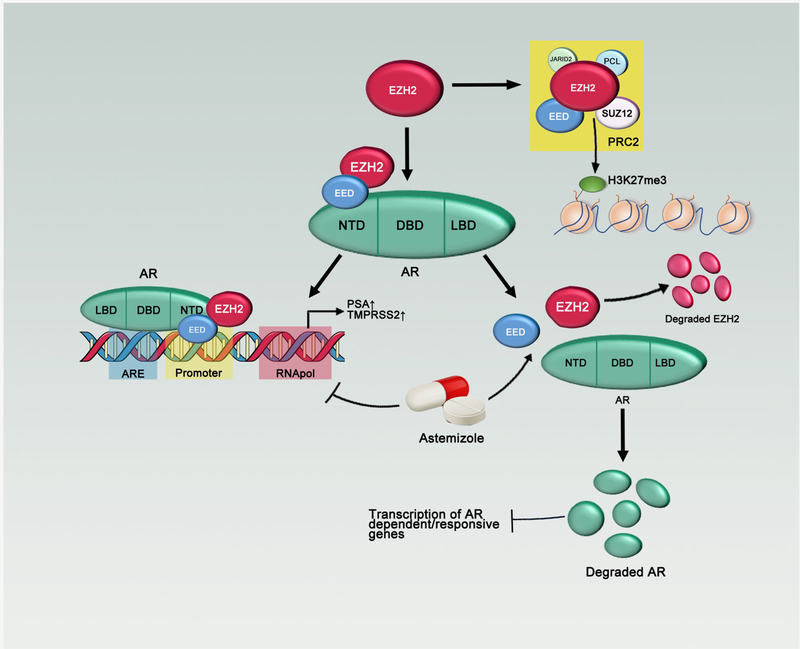
Figure 6.
Model for the regulation of EZH2/EED on androgen receptor. EZH2 and EED directly bind to AR and regulate its downstream targets. Astemizole is a newly identified PRC2 disruptor, which degrades EZH2 and AR.
What’s new?
Polycomb group proteins are epigenetic regulators with important roles in cancer initiation and progression. Among them, EZH2 is a downstream target of androgen receptor (AR) in prostate cancer. How EZH2 regulates AR functions in castration-resistant prostate cancer (CRPC) however remains unclear. This study reveals that EZH2 and EED--the two core catalytic subunits of the PRC2 class of Polycomb group proteins--play a critical role related to the AR pathway in prostate cancer. Moreover, astemizole was a potent PRC2 disruptor that significantly represses EZH2 and AR expression in prostate cancer cells, thus representing a potential medication for castration-resistant prostate cancer.
Acknowledgements
We appreciate the strong support from Dr. Anastasia K. Yocum, Dr. Leland W. Chang and Dr. J. Chad Brenner. We thank Johnique T. Atkins for comments and editing our study. We appreciate the support from Dr. Bella Shmaltsuyeva and Dr. Shanshan Zhang from Pathology Core Facility of Northwestern University.
Houston Methodist Research Institute, Prostate Cancer Foundation (13YOUN007 to Q.C.), U.S. Department of Defense (W81XWH-15-1-0639 and W81XWH-17-1-0357 to Q.C.), American Cancer Society (TBE-128382 to Q.C.), and NIH/NCI (R01CA208257 to Q.C.); K.C. is supported in part by grants from NIH/NHLBI (R01CA208257, HL100397 and HL099997) and Department of Defense (W81XWH-17-1-0357);
Grant sponsor: NIH/NHLBI; Grant numbers: HL099997, HL100397; Grant sponsor: NIH/NCI; Grant number: R01CA208257; Grant sponsor: American Cancer Society; Grant number: TBE-128382; Grant sponsor: U.S. Department of Defense; Grant numbers: W81XWH-15-1-0639, W81XWH-17-1-0357; Grant sponsor: Prostate Cancer Foundation; Grant number: 13YOUN007; Grant sponsor: Houston Methodist Research Institute; Grant sponsor: Northwestern University
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Rathkopf D, Scher HI. Androgen receptor antagonists in castration-resistant prostate cancer. Cancer J 2013; 19:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002; 111:185–96. [DOI] [PubMed] [Google Scholar]
- 4.Montgomery ND, Yee D, Chen A, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 2005;15:942–7. [DOI] [PubMed] [Google Scholar]
- 5.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012;492:108–12. [DOI] [PubMed] [Google Scholar]
- 6.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 2012;8:890–6. [DOI] [PubMed] [Google Scholar]
- 7.Knutson SK, Warholic NM, Wigle TJ, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci USA 2013;110:7922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci USA 2012;109:21360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap DB, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011;117:2451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fillmore CM,Xu C,Desai PT,et al. EZH2inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 2015;520:239–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 2015;21:231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Yu J, Mani RS, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010;17:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012;338: 1465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Q, Wang X, Zhao M, et al. The central role of EED in the orchestration of polycomb group complexes. Nat Commun 2014;5:3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YC, Morley M, Brandimarto J, et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics 2015;105:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asangani I A, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014; 510:278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S, Zhao D, Yan L, et al. BMI1 regulates androgen receptor in prostate cancer independently of the polycomb repressive complex 1. Nat Commun 2018;9:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speers C, Zhao SG, Kothari V, et al. Maternal embryonic Leucine zipper kinase (MELK) as a novel mediator and biomarker of Radioresistance in human breast cancer. Clin Cancer Res 2016;22: 5864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi W, Zhao K, Gu J, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol 2017;13:381–8. [DOI] [PubMed] [Google Scholar]
- 22.Kong X, Chen L, Jiao L, et al. Astemizole arrests the proliferation of cancer cells by disrupting the EZH2-EED interaction of polycomb repressive complex 2. J Med Chem 2014;57:9512–21. [DOI] [PubMed] [Google Scholar]
- 23.Zhao JC, Yu J, Runkle C, et al. Cooperation between Poly comb and androgen receptor during oncogenic transformation. Genome Res 2012;22: 322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei FZ, Cao Z, Wang X, et al. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy 2015;11:2309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Q, Yu J, Dhanasekaran SM, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008;27:7274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Cao Q, Yu J, et al. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene 2010;29:5370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Q, Mani RS, Ateeq B, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 2011;20: 187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee R, Mani RS, Russo N, et al. The tumor suppressor gene rap 1 GAP is silenced by miR101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene 2011;30: 4339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem 2005;280: 22437–44. [DOI] [PubMed] [Google Scholar]
- 30.Kong D, Heath E, Chen W, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One 2012;7: e33729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee ST, Li Z, Wu Z, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell 2011;43: 798–810. [DOI] [PubMed] [Google Scholar]
- 32.He A, Shen X, Ma Q, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev 2012;26:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013;23: 839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JM, Lee JS, Kim H, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell 2012;48:572–86. [DOI] [PubMed] [Google Scholar]
- 35.Sanulli S, Justin N, Teissandier A, et al. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol Cell 2015;57:769–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007;21: 1050–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miranda TB, Cortez CC, Yoo CB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther 2009;8:1579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for the RNA-seq data sets reported in this paper is GSE124268.