Abstract
Aims
With a large number of fatalities, coronavirus disease-2019 (COVID-19) has greatly affected human health worldwide. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus that causes COVID-19. The World Health Organization has declared a global pandemic of this contagious disease. Researchers across the world are collaborating in a quest for remedies to combat this deadly virus. It has recently been demonstrated that the spike glycoprotein (SGP) of SARS-CoV-2 is the mediator by which the virus enters host cells.
Main methods
Our group comprehensibly analyzed the SGP of SARS-CoV-2 through multiple sequence analysis and a phylogenetic analysis. We predicted the strongest immunogenic epitopes of the SGP for both B cells and T cells.
Key findings
We focused on predicting peptides that would bind major histocompatibility complex class I. Two optimal epitopes were identified, WTAGAAAYY and GAAAYYVGY. They interact with the HLA-B*15:01 allele, which was further validated by molecular docking simulation. This study also found that the selected epitopes are able to be recognized in a large percentage of the world's population. Furthermore, we predicted CD4+ T-cell epitopes and B-cell epitopes.
Significance
Our study provides a strong basis for designing vaccine candidates against SARS-CoV-2. However, laboratory work is required to validate our theoretical results, which would lay the foundation for the appropriate vaccine manufacturing and testing processes.
Keywords: COVID-19, SARS-CoV-2, Spike glycoprotein, Immunoinformatics, Epitope
Highlights
-
•
COVID-19 is regarded as an infectious disease, which caused by severe acute respiratory syndrome-coronavirus 2.
-
•
Our research group focused on vaccine design against SARS-CoV-2 utilizing various immunoinformatics tools.
-
•
For T-cell epitopes, our study mainly concentrated on the epitopes that bind with MHC class I molecules.
-
•
By utilizing the immunoinformatics database, we predicted three T-cell epitopes.
-
•
We determined the binding affinities of the epitopes with the HLA encoded by MHC through molecular docking studies.
-
•
Besides, our present study also predicted the B-cell epitopes, which presumably elicit a stronger immune response.
1. Introduction
Pandemics caused by severe life-threatening human pathogens have played a significant role in shaping human history. The world is currently battling a global pandemic rapidly developed in late December 2019. A cluster of pneumonia cases of unknown etiology was reported in Wuhan, the capital city of the Hubei Province in the People's Republic of China. Later, it was revealed that the causative agent of this outbreak was a novel coronavirus named SARS-CoV-2 (previously named 2019-nCoV). The clinical condition associated with this novel coronavirus is referred to as COVID-19 [[1], [2], [3]]. On March 11, 2020 the World Health Organization (WHO) categorized the current outbreak of COVID-19 as a pandemic. This viral infection appears to constitute a major global threat to humans and has had devastating effects worldwide. As of August 3, 2020, more than 17.9 million cases and over 685,000 deaths have been reported globally. Developed countries such as the USA, Italy, Spain, France, Germany, and the United Kingdom have experienced high mortality rates [4]. The number of COVID-19 cases has continued to escalate exponentially and is considered to be the largest outbreak of atypical pneumonia in recent times.
Tyrell and Bynoe first described the coronaviruses in 1966 [5]. Coronaviruses are pleomorphic or spherical particles with a positive single-strand RNA (+ssRNA). These viruses are very common among mammals and birds and can be transmitted to humans through pathogen spillover. They were named “coronavirus” because these virions consist of a core-shell and 9–12 nm-long crown-like surface spikes located on the outer surface of the virus, resembling a solar corona. Their genome size is the largest among all the RNA viruses, ranging from 27 to 32 kb in length, which encodes structural and nonstructural proteins of the coronavirus. Phenotypic and genotypic diversity allows them to adapt to new environments through mutation and recombination. Mutations that impact the surface proteins enable its sustainability and are challenging for the immune system, which makes SARS-CoV-2 infection unique compared to previous coronaviruses [6]. Among the four genera of coronaviruses (alpha, beta, gamma, and delta), the betacoronaviruses can cause severe disease and death in humans [7]. Including SARS-CoV-2, seven subtypes of coronaviruses have been identified in recent decades, all of which can infect humans.
SARS-CoV-2 differs from other betacoronaviruses in terms of its significantly higher infectivity and low mortality rate. It belongs to the B lineage of the betacoronavirus in the order Nidovirales, family Coronaviridae, and subfamily Orthocoronaviridae. After the two previously reported coronaviruses, SARS and MERS, this is the third zoonotic coronavirus outbreak of the 21st century and is closely related to its predecessors [8,9]. As it is a zoonotic virus, there is an intermediate host by which it was transmitted to humans. Preliminary investigations have predicted that SARS-CoV-2 underwent zoonotic transfer from bats to humans [9]. Primarily, environmental specimens taken from the Huanan wet market in Wuhan were found to be positive for SARS-CoV-2, but no specific association with any animal has yet been confirmed, according to a WHO report [3].
As this contagious disease is mainly a respiratory disease, it appears to affect only the lungs in most cases. The infection is transmitted between people during close contact, which occurs via the spraying of droplets from the infected individual when they cough or sneeze [10,11]. These droplets usually fall onto surfaces, and touching the contaminated surfaces followed by touching other parts of the face (such as one's eyes, nose, or mouth) may result in spreading the infection [12,13]. The symptoms of COVID-19 range from mild or moderate fever to severe pneumonia. Time from exposure to onset of symptoms ranges from 2 to 14 days, with an average of 5 days. It appears that viral spread can take place before symptoms appear. The clinical pathology largely resembles its two predecessors, with less severe upper respiratory and gastrointestinal symptoms. The most common symptoms or combination of symptoms include fever, fatigue, dry cough, dyspnea, alveolar edema, sore throat, new loss of taste or smell, and shortness of breath [14]. Older people with medical comorbidities or multi-organ failure, such as those with hypertension, cardiovascular disease, or diabetes, are more likely to become infected and suffer worse outcomes [14]. Severe cases can lead to cardiac injury, respiratory failure, acute respiratory syndrome, hepatic injury, neurological complications, or death [15].
The structure of SARS-CoV-2 includes a polyprotein (the open reading frame 1a and 1b, Orf1ab), four major structural proteins, the S protein (spike glycoprotein; SGP), E protein (envelope), M protein (membrane), and N protein (nucleocapsid), along with five accessory proteins, Orf3a, Orf6, Orf7a, Orf8 and Orf10 [16]. This novel coronavirus also encodes an additional glycoprotein that has acetyl esterase and hemagglutination (HE) attributes, and this glycoprotein does not occur in other coronaviruses [17]. Among the structural proteins, the S protein is a multifunctional molecular machine; a recent study by Walls et al. clarified the structure, function, and immunogenicity of the S protein [18]. The SGP can attach to a specific human host receptor, ACE2, on the surface of human cells. This receptor binding process mediates the entry of viral particles into the host cells with the aid of its protease, which cleaves the spike protein into the S1 and S2 subunits [19,20]. The SGP binds to ACE2, located on the surface of host cells, through the receptor binding domain (RBD), which is part of the S1 subunit. Subsequently, the viral and host membranes are fused by the S2 subunit. The viral genome RNA is released into the cytoplasm after this membrane fusion. Hence, developing neutralizing antibodies against the RBD of SARS-CoV-2 SGP might be an ideal vaccine strategy.
The identification of B-cell and T-cell epitopes for spike glycoproteins is critical for developing an effective vaccine. Although humans normally demonstrate an antibody response against viruses, only neutralizing antibodies can completely block the entry of viruses into human cells [21]. The location of antibody binding sites on a viral protein strongly affects the body's ability to produce neutralizing antibodies [22]. It is crucial to ascertain whether SARS-CoV-2 has any potential antibody binding sites (B-cell epitopes) in the RBD region, because this is the area where the virus interacts with its known human receptor, ACE2.
Apart from neutralizing antibodies, the human body also relies on cytotoxic CD8+ T cells and helper CD4+ T cells to eliminate viruses from the body. The presentation of a peptide will allow a B cell to receive stimulation from a helper T cell and become a plasma cell so that it can generate antibodies. T-cell epitopes in connection with MHC proteins are presented to T cells to elicit an antiviral T-cell response. Cytotoxic T cells recognize peptides that are received from the intracellular space presented by MHC class I molecules (CD8+ T-cell epitopes), while helper T cells recognize extracellular peptides presented by MHC class II molecules (CD4+ T-cell epitopes). The pMHC (peptide: MHC complex) interacts with the T-cell receptor and activates the cellular immune response. The inclusion of CD4+ T-cell epitopes plays a key role in vaccine design, as they provide cognate help and elicit vigorous humoral and cytotoxic CD8+ T cell responses and neutralizing antibodies [23]. T-cell epitope-based vaccines have been explored in recent years, as they can target conserved regions of the virus to elicit T cell responses and provide long-term protection against different strains of viruses [24].
Currently, no clinically proven vaccine grants immunity to SARS-CoV-2 infection. Researchers have examined different repurposed compounds from other viral infections to treat COVID-19, but the treatment benefit derived has been marginal or nonexistent in most cases [25]. However, with the rapid expansion of research in this area, it is hoped that a vaccine against SARS-CoV-2 could be developed in the not too distant future. To this end, several antibody development platforms have been explored, including DNA vaccines, RNA vaccines, protein subunit vaccines, virus-like particle (VLP)-based vaccines, and vector-based vaccines. However, further investigations are critical for developing a safe vaccine that is applicable in different age groups. For this purpose, extensive relevant research of the genomic and structural organization of SARS-CoV-2 is crucial. Recent studies have already reported the development of inactivated vaccine candidates against SARS-CoV-2; however, these candidates require further validation [26,27]. In addition, several institutes have isolated the SARS-CoV-2 strain for developing live-attenuated vaccine candidates; however, this type of vaccine requires extensive screening showing extremely low or no pathogenicity [28].
Many researchers have concentrated on making mRNA and DNA vaccines that eliminate the risk of any unwanted reactions. But these novel methods face many obstacles because of their experimental status and because they inherently carry very little antigenicity. Epitope-based vaccines offer a viable alternative since they can elicit potent immune responses without causing undesirable allergic reactions and have already been successfully implemented in combating other harmful pathogens. Designing vaccines using conventional methods is not only time consuming, it is also inefficient. Various immunoinformatics tools can help to significantly speed up this laborious process. Immunoinformatics tools, such as epitope prediction, can be used to construct vaccines in silico, and the results can then be experimentally validated [29]. Several in silico immunoinformatics-guided attributes have already been predicted as a basis for an epitope-based SARS-CoV-2 vaccine; these predicted epitopes have arisen from several of the pathogen's proteins, including the N and E proteins and the main protease (Mpro) [[30], [31], [32], [33]].
Effective promiscuous epitopes binding to a variety of human leukocyte antigen (HLA) alleles for wider dissemination with no human cross-reactivity are crucial because COVID-19 has affected populations worldwide. Our present study embarked upon the clear objective of designing an epitope-based peptide vaccine against SARS-CoV-2 infection using in silico methods by investigating its SGP. We targeted the epitopes of the SGP because they reportedly induce a longer-lasting immune response against SARS coronavirus [34]. We assessed the associated MHC alleles for the identified epitopes to determine those epitopes that would maximize population coverage across the world. As a result, using different computational tools, we designed an epitope-based peptide vaccine that would theoretically target the SARS-CoV-2 SGP with the expectation of subsequent wet laboratory validation.
2. Materials and methods
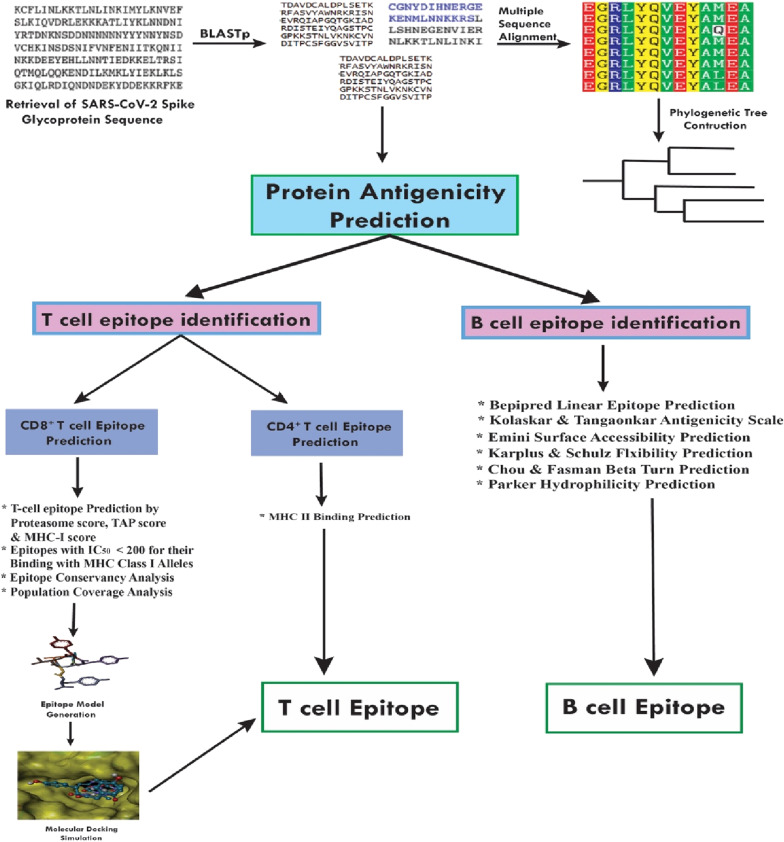
The methodologies used for peptide vaccine development for SARS-CoV-2 SGP are shown in Fig. 1 .
Fig. 1.
Workflow of the methodologies used in epitope-based vaccine design from SARS-CoV-2 Spike Glycoprotein.
2.1. Protein sequence retrieval and sequence analysis
The SARS-CoV-2 SGP sequence was extracted from the UniProt database (UniProt entry: P0DTC2) in FASTA format [35]. The features, function, structure, and evaluation of the sequence were mainly based on the process of sequence analysis, which subjects DNA, RNA, or peptide sequences to a wide range of analytical methods. We screened homologous sequences from the BLASTp database and selected those sequences that were most similar to that of SARS-CoV-2 SGP. We also performed multiple sequence alignment (MSA) using the ClustalW web server with default settings, and a phylogenetic tree was established using the Clustal tree format and the EMBL-EBI web server [36].
2.2. Protein antigenicity prediction
To determine the potent antigenic protein of the SARS-CoV-2 SGP, we used the online server VaxiJen v2.0, with a default threshold value [37]. All the antigenic proteins of SARS-CoV-2 SGP with their respective scores were obtained and then sorted in Notepad++.
2.3. T-cell epitope prediction
2.3.1. CD8+ T-cell epitope identification
The NetCTL 1.2 server was used to identify T-cell epitopes; a 0.95 threshold was applied to maintain a sensitivity and specificity of 0.90 and 0.95, respectively [38]. The tool expanded the prediction for 12 MHC-I supertypes and unified the prediction of peptide MHC-I binding, proteasomal C-terminal cleavage with TAP transport efficiency. These predictions were performed by an artificial neural network. A weighted TAP transport efficiency matrix and a combined algorithm for MHC-I binding and proteasomal cleavage efficiency were then used to determine the overall scores and translated into sensitivity/specificity. Based on this overall score, the ten best peptides (epitopes) were selected for further evaluation.
To predict peptides that bind to MHC-I, we used a tool from the Immune Epitope Database (IEDB) to calculate half-maximal inhibitory concentration (IC50) values for peptides binding to specific MHC-I molecules. For the binding analysis, all the frequently used alleles (http://tools.iedb.org/mhci/) with a word length of nine residues and a binding affinity < 200 nm were selected for further analysis [39]. The MHC-NP tool provided by the IEDB server was used to assess the probability that a given peptide was naturally processed and would bind to a given MHC molecule [40].
2.3.2. Epitope conservancy and immunogenicity prediction
The degree of similarity between the epitope and the target (i.e. given) sequence is elucidated by epitope conservancy. This property of an epitope indicates its availability in a range of different strains. The web-based tool from IEDB analysis resources was used to analyze the epitope conservancy [41]. Immunogenicity prediction can infer the degree to which a particular epitope will produce an immunogenic response. The T-cell class I pMHC immunogenicity predictor at IEDB, which uses amino acid properties as well as considering their position within the peptide, will predict the immunogenicity of a class I peptide MHC (pMHC) complex [42].
2.3.3. Allergenicity assessment
Allergenicity of the predicted epitope was calculated using AllerTop v2.0. This is an alignment-free server used for in silico-based allergenicity prediction of a protein based on its physicochemical properties [43].
2.3.4. Analysis of HLA–epitope interaction by molecular docking
2.3.4.1. Epitope model generation
A web-based server, PEP-FOLD, was used to predict the 3D structures of the selected epitopes [44]. For each sequence, the server predicted five probable structures. The energy of each structure was determined by SWISS-PDB VIEWER, and the structure with the lowest energy was chosen for further analysis [45].
2.3.4.2. Retrieval of HLA allele molecule
The 3D structure of the HLA-B*15:01 (PDB ID: 5TXS) was retrieved from the Protein Data Bank (RCSB-PDB) (https://www.rcsb.org/).
2.3.4.3. Molecular docking analysis
Molecular docking analysis was performed using AutoDock Vina in PyRx 0.8. The HLA-B*15:01 allele was considered as receptor protein and the identified epitopes were considered as ligand molecules [46]. First, we used the protein preparation wizard of UCSF Chimera (version 1.11.2) to prepare the protein for docking analysis by deleting the attached ligand and adding hydrogens and Gasteiger–Marsili charges. The file was then converted into pdbqt format using OpenBabel [47]. The energy form of the ligand was minimized and converted to pdbqt format by OpenBabel in PyRx 0.8. The parameters used for the docking simulation were set to default. The size of the grid box in AutoDock Vina was kept at 64.1887 × 51.9339 × 62.1079 Å for X, Y, and Z-axes, respectively. AutoDock Vina was implemented via the shell script offered by AutoDock Vina developers. The docking results were presented as negative scores in kcal/mol, as the binding affinity of a ligand with its receptor is calculated as a negative value [48].
2.4. CD4+ T-cell epitope identification
To predict MHC class II potential epitopes, we utilized the MHC II prediction tools from the IEDB database (http://tools.iedb.org/mhcii/) [49]. For MHC II binding prediction, we used the human MHC alleles from Greenbaum et al. [50]. The MHC class II groove can bind peptides of different lengths. All epitopes that were predicted to bind to the alleles with an IC50 score of less than 50 were selected for further analysis.
2.5. Prediction of population coverage
The population coverage calculation tools from IEDB were employed to determine the individual epitope and multi-epitope population coverage for the targeted potential MHC I and MHC II alleles [51]. Here, we used the allelic frequency of the interacting HLA alleles to predict the population coverage for the corresponding epitope, both individually and combined.
2.6. Linear B-cell epitope identification
The B-cell epitope prediction aimed to identify potential antigens that would give rise to humoral immunity. To detect B-cell epitopes, various tools from IEDB were used to identify B cell antigenicity, along with Emini surface accessibility prediction, Kolaskar and Tongaonkar antigenicity scale, Karplus and Schulz flexibility prediction, and BepiPred linear epitope prediction analysis [[52], [53], [54], [55]]. Because the antigenic regions of a protein are part of beta-turns, the Chou and Fasman beta-turn prediction tool was also used [56].
3. Results and discussion
3.1. Sequence retrieval and analysis
The protein sequence of the SARS-CoV-2 SGP was retrieved from the UniProt database and then BLASTp was performed. From the many identified homologs, we selected only 17 that had greater than 60% sequence identity. MSA was performed (Supplementary Data 1) and a phylogenetic tree was constructed (Fig. S1). The MSA and phylogenetic results indicated that the protein sequences have a close relationship.
3.2. Antigenic protein prediction
The most potent antigenic protein of SARS-CoV-2 SGP was predicted by VaxiJen v2.0, which is based on the auto-cross covariance transformation of protein sequences into uniform vectors of principal amino acid properties. The overall antigen prediction score was 0.4683 (probable antigen) at a threshold value of 0.4. A recent review of the genomic and proteomic analysis of SARS-CoV-2 found that certain of its proteins, including M, SGP, E, and N, had the ability to confer a protective immune response against SARS-CoV-2 [57]. Additionally, a recent study proposed that epitopes on the SGP and N proteins of SARS-CoV-2 elicited strong T-cell immune responses for longer periods of time; therefore, these might be ideal vaccine candidates [58]. Significantly, the receptor-binding domain (RBD) of the SGP of two other coronaviruses, SARS-CoV and MERS-CoV, provokes strongly potent neutralizing antibody responses and have potential to be developed as vaccines against SARS and MERS infection [59,60]. In alignment with previous analyses on other coronaviruses and on other recent evidence, the current study applied computational biology techniques and immunoinformatics tools to identify an epitope-based vaccine candidate utilizing the SARS-CoV-2 SGP. We predicted the antigenicity of SGP with the VaxiJen server. However, a recent study predicted the E protein of SARS-CoV-2 to be the best immunogenic target for designing a vaccine; however, another study that used the Vaxign-ML website proposed SGP as the best vaccine candidate [32,61]. The Vaxijen tool mainly incorporates the physicochemical properties of protein sequences for its classification, but the Vaxign-ML server encompasses biological data for its predictions of vaccine candidates [62]. Several studies have documented that SGP protrudes from the virion envelope and binds to the cellular receptors, playing a crucial role in host cell entry [63,64]. Other research has suggested that proteins involved in viral fusion, which depends on the envelope protein and glycoprotein region, elicit the most extensive immune response, along with a cytotoxic T lymphocytes (CTL) response [65,66]. In line with previous studies and considering the best immunogenicity, we selected the SGP of SARS-CoV-2 to predict an epitope-based vaccine, which may lead to further analysis of the SARS-CoV-2 vaccination strategy.
3.3. T-cell epitope identification
3.3.1. CD8+ T-cell epitope identification
On the basis of the highest combinatorial score and MHC class I binding, 23 epitopes were predicted by NetCTL 1.2 from the given protein sequence. The antigenicity of each selected peptide was predicted by VaxiJen v2.0, and we found that ten epitopes were antigenic (Table S1). The MHC-I binding prediction tool from the IEDB server was used to assess affinity; this tool is based on the stabilized matrix method. We chose the MHC-I alleles for which the epitopes showed the highest affinity (IC50 < 200 nm). Proteasomes play an important role during the conversion of protein into peptide; after proteasomal cleavage, peptide–MHC molecules are transported to the cell membrane where they are presented to T helper cells. The total score of each epitope-HLA interaction was taken into consideration; a higher score implied a higher processing efficiency. The epitope WTAGAAAYY was found to interact with 17 MHC class I alleles: HLA-A*29:02, HLA-A*26:01, HLA-A*68:01, HLA-C*12:03, HLA-B*15:25, HLA-B*35:01, HLA-C*03:02, HLA-A*30:02, HLA-A*01:01, HLA-B*15:01, HLA-B*15:02, HLA-C*16:01, HLA-A*25:01, HLA-C*02:09, HLA-C*02:02, HLA-C*12:02, and HLA-C*14:02 (Table 1 ). In addition, two other epitopes, namely GAAAYYVGY and STQDLFLPF, interacted with MHC I alleles. The former interacted with five HLAs and the latter interacted with nine. However, like WTAGAAAYY, both epitopes interacted with HLA-B*15:25 and HLA-B*15:01. The epitope GAAAYYVGY showed a stronger affinity for binding HLA-B*15:25, whereas WTAGAAAYY had the strongest immunogenicity predicted by the I-pMHC immunogenicity prediction analysis, and the binding affinity of WTAGAAAYY to HLA-B*15:01 was relatively higher (Table 1). Furthermore, all the predicted epitopes had a maximum identity for conservancy, and 100% maximum identity was found (Table 1). Therefore, we selected the aforementioned three epitopes for further analysis. In addition, MHC-NP found that these epitopes would naturally bind to the MHC class I alleles (Table S2).
Table 1.
The potential CD8+ T-cell epitopes along with their interacting MHC class I alleles and total processing score, epitopes conservancy hits and pMHC-I immunogenicity score.
| Epitopes | NetCTL Combined score |
Epitope_ Conservancy_Hit (MAX. Identity %) |
MCH-I interaction with an affinity of IC50 and the total score (proteasome score, TAP score, MHC-I score, processing score) |
pMHC-I immunogenicity score |
|---|---|---|---|---|
| WTAGAAAYY | 3.1128 | 100 | HLA-A*29:02 (1.51), HLA-A*26:01 (1.43), HLA-A*68:01 (1.12), HLA-C*12:03 (0.99), HLA-B*15:25 (0.97), HLA-B*35:01 (0.96), HLA-C*03:02 (0.91), HLA-A*30:02 (0.90), HLA-A*01:01 (0.89), HLA-B*15:01 (0.78), HLA-B*15:02 (0.68), HLA-C*16:01 (0.62), HLA-A*25:01 (0.56), HLA-C*02:09 (0.53), HLA-C*02:02 (0.53), HLA-C*12:02 (0.52), HLA-C*14:02 (0.24) | 0.15259 |
| CNDPFLGVY | 1.3355 | 100 | HLA-A*01:01 (0.36) | 0.15232 |
| GAAAYYVGY | 1.2194 | 100 | HLA-B*15:25 (1.03), HLA-A*29:02 (0.81), HLA-B*15:01 (0.69), HLA-A*30:02 (0.56), HLA-B*15:02 (0.39) | 0.09963 |
| ITDAVDCAL | 1.1680 | 100 | HLA-C*05:01 (0.62), HLA-C*08:02 (0.13), HLA-C*08:01 (0.08), HLA-C*03:04 (−0.41), HLA-C*03:03 (−0.41), HLA-C*16:01 (−0.43) | 0.08501 |
| STQDLFLPF | 1.0468 | 100 | HLA-B*15:25 (0.77), HLA-B*15:01 (0.60), HLA-B*15:02 (0.50), HLA-A*32:01 (0.48), HLA-C*16:01 (0.42), HLA-C*03:02 (0.35), HLA-B*35:01 (0.02), HLA-C*12:03 (−0.03), HLA-A*29:02 (−0.11) | 0.06828 |
| TSNQVAVLY | 3.0758 | 100 | HLA-A*01:01 (0.76), HLA-A*29:02 (0.68), HLA-A*30:02 (0.54), HLA-B*58:01 (0.33) | −0.01327 |
| KTSVDCTMY | 2.3795 | 100 | HLA-A*30:02 (1.04), HLA-B*58:01 (0.47) | −0.11115 |
| MTSCCSCLK | 1.0963 | 100 | HLA-A*68:01 (0.26), HLA-A*11:01 (−0.07), HLA-A*03:01 (−0.74), HLA-A*30:01 (−0.92), HLA-A*31:01 (−0.93), HLA-A*33:03 (−1.19) | −0.36816 |
| STECSNLLL | 2.3492 | 100 | HLA-C*05:01 (−0.27) | −0.20478 |
| GAEHVNNSY | 1.9960 | 100 | – | −0.00296 |
Notes: MHC-I alleles that have an interacting affinity lower than 200 nm are represented, and total processing scores are shown as enclosed numbers.
By concentrating on potential MHC class I peptide epitopes, we predicted not only T-cell but also B-cell epitopes. Both of these epitope types are capable of showing immune responses in several ways. Many criteria must be met when identifying a protein sequence-based epitope as a potential vaccine candidate, whereby allergenicity is regarded as one of the most important factors. We input the SARS-CoV-2 SGP sequence into the VaxiJen server and predicted the antigenicity of the protein sequence. Ten potent epitopes were predicted by the NetCTL 1.2 server, and the epitopes were further used in the progressive analysis that also showed MHC class I interaction. Subsequently, all peptides except GAEHVNNSY were shown to interact with the MHC class I alleles. WTAGAAAYY interacted with the most MHC class I alleles. Among them, the allele HLA-A*29:02 had the highest binding score (1.51). Interestingly, three of the epitopes, WTAGAAAYY, GAAAYYVGY, and STQDLFLPF, showed binding interaction with the same MHA class I allele (HLA-B*15:25), of which GAAAYYVGY had the highest affinity. Additionally, the aforementioned epitopes also interacted with HLA-B*15:01, and epitope WTAGAAAYY showed the highest affinity for it. In addition, the epitope conservancy was predicted by the IEDB conservancy analysis tool, and all of our predicted epitopes had a maximum identity of 100%. Hence, these three epitopes were selected for further analysis.
3.3.2. Allergenicity assessment
The AllerTop server assesses the allergic reactions caused by a vaccine candidate in an individual that might be harmful or life-threatening. The allergenicity of the selected epitopes was calculated using the AllerTop tool, and they were predicted as probable non-allergens.
Allergenicity is regarded as a notable obstacle during vaccine development. CD4+ T cells are the primary actors responsible for provoking an allergic reaction, but immunoglobulin E and type 2 T helper cells can also stimulate allergic reactions [67]. We evaluated the allergenicity using AllerTop 2.0. This tool is well-known for its high sensitivity to identify new allergens in relation to known allergens [43]. AllerTop predicted our selected epitopes to be non-allergenic.
3.3.3. Molecular docking analysis of the HLA–epitope interaction
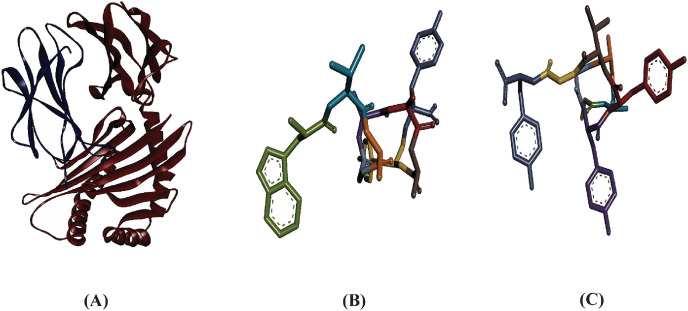
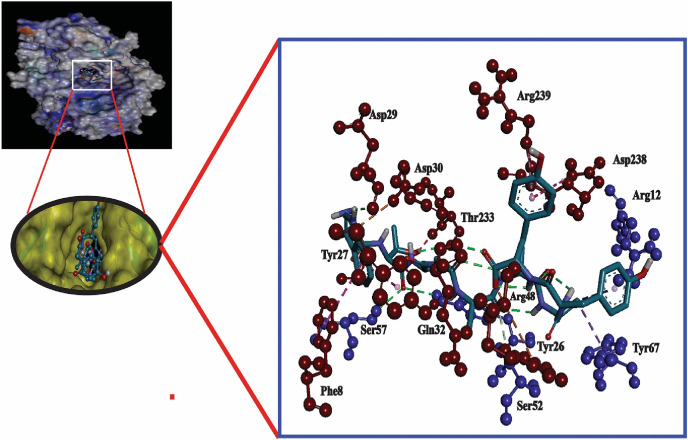
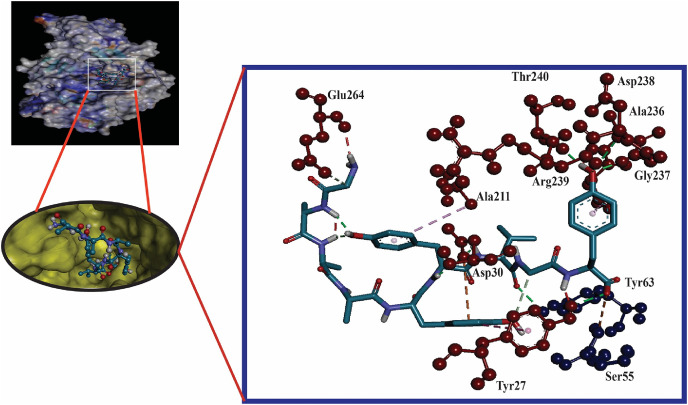
Here, the interaction between the HLA molecules and our predicted potential epitopes was validated by molecular docking simulation using AutoDock Vina in PyRx 0.8. We previously found that alleles HLA-B*15:25 and HLA-B*15:01 interacted with the three predicted epitopes. However, the 3D structure of HLA-B*15:25 was not available in the Protein Data Bank (PDB). Although the binding affinities of the selected epitopes were greater for HLA-B*15:25, we selected HLA-B*15:01 for study because it was available from the PDB database. The structures of HLA-B*15:01 and the selected epitopes are shown in Fig. 2 . For the docking analysis, we considered the HLA molecule as the receptor and the selected three epitopes as ligand molecules. The docking experiments showed that the epitope WTAGAAAYY interacted with HLA-B*15:01 with the highest binding affinity, which was calculated as −10.0 kcal/mol; the binding affinity of GAAAYYVGY was almost equal to that of GAAAYYVGY (−9.4 kcal/mol) (Table 2 )). By visualizing the docking r. However, the binding affinity of the epitope STQDLFLPF was found to be only −8.3 kcal/mol (Table 2). By visualizing the docking results, it was clear that WTAGAAAYY formed hydrogen bonds (H-bonds) with six amino acid residues (Gln32, Thr233, Asp29 from the A-chain; Arg12, Tyr26, Ser7 from the B-chain) and pi-pi stacking with Asp238, Tyr27, and Phe8 from the A-chain (Fig. 3 ). No unfavorable bonds were visualized for the epitope WTAGAAAYY, albeit two unfavorable donor-donor bonds were found for GAAAYYVGY. In addition, more H-bonds were found for GAAAYYVGY, including six from A-chain residues (Asp238, Gly237, Thr240, Arg239, Asp30, Tyr27) and one from Tyr63 in the B-chain. There were also favorable interactions with Ser55 from the B-chain, and pi-pi stacking was visualized for Tyr27 from the A-chain, which also formed a H-bond with the HLA molecule (Fig. 4 ).
Fig. 2.
Three dimensional representation of (A) HLA-B*15:01 allele, (B) Epitope WTAGAAAYY, and (C) Epitope GAAAYYVGY.
Table 2.
Binding affinities of the selected epitopes with HLA-B*15:01.
| Epitope | Binding affinity (kcal/mol) |
|---|---|
| WTAGAAAYY | −10.0 |
| GAAAYYVGY | −9.4 |
| STQDLFLPF | −8.3 |
Fig. 3.
Molecular docking analysis of epitope WTAGAAAYY with HLA-B*15:01 allele. The interacting A-chain residues are displayed as red ball and stick, the interacting B-chain residues are displayed in blue ball and sick, conventional hydrogen bonds are displayed as green line, pi-pi/pi-alkyl stacking are displayed as pink lines, unfavorable bumps are displayed as red lines.
Fig. 4.
Molecular docking analysis of epitope GAAAYYVGY with HLA-B*15:01 allele. The interacting A-chain residues are displayed as red ball and stick, the interacting B-chain residues are displayed in blue ball and sick, conventional hydrogen bonds are displayed as green line, pi-pi/pi-alkyl stacking are displayed as pink lines, unfavorable bumps are displayed as red lines.
MHC class I and class II molecules play a pivotal role by presenting peptides on the cell surface that can then be identified by T cells. MHC I class molecules present shorter molecules (generally 8–11 amino acids long), and MHC class II molecules present longer peptides (typically 13–17 amino acid residues) [68]. In the current study, we determined the binding affinity of the predicted peptide sequences with HLA-B*15:01 because the structure of this allele was available in the PDB database. The findings from the molecular docking analysis revealed that epitopes WTAGAAAYY and GAAAYYVGY had the highest affinity interactions with the HLA-B*15:01 allele, as a more negative result implies a stronger interaction [69]. Furthermore, these two epitopes were able to interact with the allele through H-bonds and hydrophobic (pi-pi stacking, pi-alkyl) interactions, and attractive charges also contributed to binding in the case of GAAAYYVGY. Conversely, the epitope STQDLFLPF had a less negative docking score; hence, its binding interaction was weaker in comparison with the other two epitopes.
3.4. Predication of T helper cell epitopes and MHC class II interaction analysis
In this experiment, potential CD4+ T-cell peptide epitopes were identified by the MHC class II prediction tool from the IEDB website, which provided 56 core epitopes for the selected HLA-DRB alleles with an IC50 < 50 nm. The peptides with the core epitopes IVNNATNVV, LKYNENGTI, FLVLLPLVS, FELLHAPAT, FTISVTTEI, FCTQLNRAL, FNGLTGTGV, LALHRSYLT, FQTLLALHR, and YFKIYSKHT interacted with most of the MHC class II alleles. These results are shown in Table 3 .
Table 3.
List of epitopes that had the binding affinity with the MHC Class II alleles. Notes: MHC-II alleles that have an interacting affinity lower than 50 nm are represented.
| Core epitope | Allele | Percentile rank | Peptide | Start | End | SMM IC50 |
|---|---|---|---|---|---|---|
| LIVNNATNV | HLA-DRB1*13:02 | 0.01 | KTQSLLIVNNATNVV | 113 | 127 | 3.00 |
| HLA-DRB1*01:01 | 9.50 | TQSLLIVNNATNVVI | 114 | 128 | 49.00 | |
| IVNNATNVV | HLA-DRB1*13:02 | 0.01 | TQSLLIVNNATNVVI | 114 | 128 | 3.00 |
| HLA-DRB1*13:02 | 0.01 | QSLLIVNNATNVVIK | 115 | 129 | 3.00 | |
| HLA-DRB1*13:02 | 0.01 | SLLIVNNATNVVIKV | 116 | 130 | 3.00 | |
| HLA-DRB1*13:02 | 0.01 | LLIVNNATNVVIKVC | 117 | 131 | 4.00 | |
| HLA-DRB1*13:02 | 0.03 | LIVNNATNVVIKVCE | 118 | 132 | 9.00 | |
| HLA-DRB1*13:02 | 0.09 | IVNNATNVVIKVCEF | 119 | 133 | 15.00 | |
| SKTQSLLIV | HLA-DRB1*13:02 | 0.03 | SKTQSLLIVNNATNV | 112 | 126 | 8.00 |
| QPRTFLLKY | HLA-DRB1*13:02 | 0.06 | QPRTFLLKYNENGTI | 271 | 285 | 11.00 |
| LKYNENGTI | HLA-DRB1*13:02 | 0.06 | PRTFLLKYNENGTIT | 272 | 286 | 11.00 |
| HLA-DRB1*13:02 | 0.06 | RTFLLKYNENGTITD | 273 | 287 | 11.00 | |
| HLA-DRB1*13:02 | 0.06 | TFLLKYNENGTITDA | 274 | 288 | 11.00 | |
| HLA-DRB1*13:02 | 0.06 | FLLKYNENGTITDAV | 275 | 289 | 11.00 | |
| HLA-DRB1*13:02 | 0.36 | LLKYNENGTITDAVD | 276 | 290 | 32.00 | |
| HLA-DRB1*13:02 | 0.36 | LKYNENGTITDAVDC | 277 | 291 | 32.00 | |
| YNYLYRLFR | HLA-DRB1*11:01 | 0.09 | GGNYNYLYRLFRKSN | 446 | 460 | 7.00 |
| HLA-DRB1*11:01 | 0.09 | GNYNYLYRLFRKSNL | 447 | 461 | 7.00 | |
| HLA-DRB1*11:01 | 0.29 | KVGGNYNYLYRLFRK | 444 | 458 | 10.00 | |
| HLA-DRB1*11:01 | 0.29 | VGGNYNYLYRLFRKS | 445 | 459 | 10.00 | |
| FNFSQILPD | HLA-DRB1*04:05 | 0.21 | DFGGFNFSQILPDPS | 796 | 810 | 26.00 |
| FSQILPDPS | HLA-DRB1*04:05 | 0.21 | FGGFNFSQILPDPSK | 797 | 811 | 26.00 |
| HLA-DRB1*04:05 | 0.23 | GGFNFSQILPDPSKP | 798 | 812 | 27.00 | |
| HLA-DRB1*04:05 | 0.42 | GFNFSQILPDPSKPS | 799 | 813 | 38.00 | |
| HLA-DRB1*04:05 | 0.42 | FNFSQILPDPSKPSK | 800 | 814 | 38.00 | |
| FLVLLPLVS | HLA-DRB1*01:01 | 0.24 | MFVFLVLLPLVSSQC | 1 | 15 | 5.00 |
| HLA-DRB1*01:01 | 0.24 | FVFLVLLPLVSSQCV | 2 | 16 | 5.00 | |
| HLA-DRB1*04:05 | 0.49 | MFVFLVLLPLVSSQC | 1 | 15 | 41.00 | |
| HLA-DRB1*04:05 | 0.52 | FVFLVLLPLVSSQCV | 2 | 16 | 43.00 | |
| HLA-DRB1*11:01 | 0.59 | MFVFLVLLPLVSSQC | 1 | 15 | 18.00 | |
| HLA-DRB1*11:01 | 0.59 | FVFLVLLPLVSSQCV | 2 | 16 | 18.00 | |
| HLA-DRB1*01:01 | 1.30 | VFLVLLPLVSSQCVN | 3 | 17 | 11.00 | |
| HLA-DRB1*01:01 | 1.80 | FLVLLPLVSSQCVNL | 4 | 18 | 13.00 | |
| HLA-DRB1*11:01 | 2.10 | VFLVLLPLVSSQCVN | 3 | 17 | 45.00 | |
| VLSFELLHA | HLA-DRB1*01:01 | 0.24 | RVVVLSFELLHAPAT | 509 | 523 | 5.00 |
| FELLHAPAT | HLA-DRB1*01:01 | 0.24 | VVVLSFELLHAPATV | 510 | 524 | 5.00 |
| HLA-DRB1*01:01 | 0.24 | VVLSFELLHAPATVC | 511 | 525 | 5.00 | |
| HLA-DRB1*01:01 | 0.24 | VLSFELLHAPATVCG | 512 | 526 | 5.00 | |
| HLA-DRB1*01:01 | 0.24 | LSFELLHAPATVCGP | 513 | 527 | 5.00 | |
| HLA-DRB1*01:01 | 2.30 | SFELLHAPATVCGPK | 514 | 528 | 16.00 | |
| HLA-DRB1*01:01 | 2.30 | FELLHAPATVCGPKK | 515 | 529 | 16.00 | |
| SKVGGNYNY | HLA-DRB1*11:01 | 0.29 | SKVGGNYNYLYRLFR | 443 | 457 | 10.00 |
| FGAISSVLN | HLA-DRB1*01:01 | 4.20 | SSNFGAISSVLNDIL | 967 | 981 | 25.00 |
| HLA-DRB1*01:01 | 4.20 | SNFGAISSVLNDILS | 968 | 982 | 25.00 | |
| HLA-DRB1*01:01 | 4.90 | LSSNFGAISSVLNDI | 966 | 980 | 28.00 | |
| HLA-DRB1*01:01 | 5.60 | QLSSNFGAISSVLND | 965 | 979 | 31.00 | |
| WTFGAGAAL | HLA-DRB1*09:01 | 0.33 | ITSGWTFGAGAALQI | 882 | 896 | 42.00 |
| HLA-DRB1*01:01 | 8.90 | ITSGWTFGAGAALQI | 882 | 896 | 46.00 | |
| HLA-DRB1*01:01 | 9.50 | TSGWTFGAGAALQIP | 883 | 897 | 49.00 | |
| FGAGAALQI | HLA-DRB1*09:01 | 0.33 | SGWTFGAGAALQIPF | 884 | 898 | 42.00 |
| HLA-DRB1*09:01 | 0.34 | TSGWTFGAGAALQIP | 883 | 897 | 43.00 | |
| HLA-DRB1*09:01 | 0.35 | GWTFGAGAALQIPFA | 885 | 899 | 44.00 | |
| HLA-DRB1*09:01 | 0.39 | WTFGAGAALQIPFAM | 886 | 900 | 46.00 | |
| FLPFFSNVT | HLA-DRB1*15:01 | 0.33 | TQDLFLPFFSNVTWF | 51 | 65 | 34.00 |
| HLA-DRB1*15:01 | 0.33 | QDLFLPFFSNVTWFH | 52 | 66 | 34.00 | |
| HLA-DRB1*15:01 | 0.37 | DLFLPFFSNVTWFHA | 53 | 67 | 35.00 | |
| HLA-DRB1*15:01 | 0.37 | STQDLFLPFFSNVTW | 50 | 64 | 36.00 | |
| FTISVTTEI | HLA-DRB1*07:01 | 0.40 | AIPTNFTISVTTEIL | 713 | 727 | 13.00 |
| HLA-DRB1*07:01 | 0.40 | PTNFTISVTTEILPV | 715 | 729 | 13.00 | |
| HLA-DRB1*07:01 | 0.40 | TNFTISVTTEILPVS | 716 | 730 | 13.00 | |
| HLA-DRB1*07:01 | 0.47 | IPTNFTISVTTEILP | 714 | 728 | 14.00 | |
| HLA-DRB1*07:01 | 2.50 | NFTISVTTEILPVSM | 717 | 731 | 38.00 | |
| HLA-DRB1*07:01 | 2.60 | FTISVTTEILPVSMT | 718 | 732 | 39.00 | |
| YGSFCTQLN | HLA-DRB1*01:01 | 4.90 | LLQYGSFCTQLNRAL | 753 | 767 | 28.00 |
| HLA-DRB1*04:05 | 0.73 | LLQYGSFCTQLNRAL | 753 | 767 | 49.00 | |
| FCTQLNRAL | HLA-DRB1*01:01 | 3.70 | GSFCTQLNRALTGIA | 757 | 771 | 23.00 |
| HLA-DRB1*01:01 | 3.90 | YGSFCTQLNRALTGI | 756 | 770 | 24.00 | |
| HLA-DRB1*01:01 | 4.40 | LQYGSFCTQLNRALT | 754 | 768 | 26.00 | |
| HLA-DRB1*01:01 | 4.40 | QYGSFCTQLNRALTG | 755 | 769 | 26.00 | |
| HLA-DRB1*01:01 | 8.00 | SFCTQLNRALTGIAV | 758 | 772 | 42.00 | |
| HLA-DRB1*01:01 | 8.20 | FCTQLNRALTGIAVE | 759 | 773 | 43.00 | |
| LYRLFRKSN | HLA-DRB1*11:01 | 0.42 | NYNYLYRLFRKSNLK | 448 | 462 | 13.00 |
| HLA-DRB1*11:01 | 0.42 | YNYLYRLFRKSNLKP | 449 | 463 | 13.00 | |
| HLA-DRB1*11:01 | 0.52 | NYLYRLFRKSNLKPF | 450 | 464 | 16.00 | |
| HLA-DRB1*11:01 | 2.10 | LYRLFRKSNLKPFER | 452 | 466 | 46.00 | |
| LFLPFFSNV | HLA-DRB1*15:01 | 0.46 | HSTQDLFLPFFSNVT | 49 | 63 | 40.00 |
| LLALHRSYL | HLA-DRB1*15:01 | 0.46 | TRFQTLLALHRSYLT | 236 | 250 | 40.00 |
| HLA-DRB1*11:01 | 2.10 | RFQTLLALHRSYLTP | 237 | 251 | 44.00 | |
| HLA-DRB1*11:01 | 2.10 | FQTLLALHRSYLTPG | 238 | 252 | 44.00 | |
| TNFTISVTT | HLA-DRB1*07:01 | 0.47 | IAIPTNFTISVTTEI | 712 | 726 | 14.00 |
| WLGFIAGLI | HLA-DRB1*01:01 | 4.60 | WYIWLGFIAGLIAIV | 1214 | 1228 | 27.00 |
| HLA-DRB1*01:01 | 8.40 | WPWYIWLGFIAGLIA | 1212 | 1226 | 44.00 | |
| HLA-DRB1*01:01 | 8.40 | PWYIWLGFIAGLIAI | 1213 | 1227 | 44.00 | |
| FNGLTGTGV | HLA-DRB1*01:01 | 0.56 | CVNFNFNGLTGTGVL | 538 | 552 | 7.00 |
| HLA-DRB1*01:01 | 0.56 | VNFNFNGLTGTGVLT | 539 | 553 | 7.00 | |
| HLA-DRB1*01:01 | 0.56 | NFNFNGLTGTGVLTE | 540 | 554 | 7.00 | |
| HLA-DRB1*01:01 | 0.56 | FNFNGLTGTGVLTES | 541 | 555 | 7.00 | |
| HLA-DRB1*01:01 | 3.60 | NFNGLTGTGVLTESN | 542 | 556 | 22.00 | |
| HLA-DRB1*01:01 | 3.60 | FNGLTGTGVLTESNK | 543 | 557 | 22.00 | |
| LALHRSYLT | HLA-DRB1*15:01 | 0.58 | RFQTLLALHRSYLTP | 237 | 251 | 45.00 |
| HLA-DRB1*15:01 | 0.60 | FQTLLALHRSYLTPG | 238 | 252 | 46.00 | |
| HLA-DRB1*15:01 | 0.72 | QTLLALHRSYLTPGD | 239 | 253 | 49.00 | |
| HLA-DRB1*01:01 | 5.80 | QTLLALHRSYLTPGD | 239 | 253 | 32.00 | |
| HLA-DRB1*01:01 | 8.40 | TLLALHRSYLTPGDS | 240 | 254 | 44.00 | |
| LLQYGSFCT | HLA-DRB1*15:01 | 0.58 | CSNLLLQYGSFCTQL | 749 | 763 | 45.00 |
| HLA-DRB1*15:01 | 0.60 | SNLLLQYGSFCTQLN | 750 | 764 | 46.00 | |
| HLA-DRB1*15:01 | 0.72 | ECSNLLLQYGSFCTQ | 748 | 762 | 49.00 | |
| ITRFQTLLA | HLA-DRB1*01:01 | 1.30 | GINITRFQTLLALHR | 232 | 246 | 11.00 |
| HLA-DRB1*11:01 | 1.90 | GINITRFQTLLALHR | 232 | 246 | 41.00 | |
| FIAGLIAIV | HLA-DRB1*01:01 | 2.60 | YIWLGFIAGLIAIVM | 1215 | 1229 | 18.00 |
| SVYAWNRKR | HLA-DRB1*11:01 | 0.63 | TRFASVYAWNRKRIS | 345 | 359 | 19.00 |
| YAWNRKRIS | HLA-DRB1*11:01 | 0.63 | RFASVYAWNRKRISN | 346 | 360 | 19.00 |
| HLA-DRB1*11:01 | 0.63 | FASVYAWNRKRISNC | 347 | 361 | 19.00 | |
| HLA-DRB1*11:01 | 0.63 | ASVYAWNRKRISNCV | 348 | 362 | 19.00 | |
| HLA-DRB1*11:01 | 0.63 | SVYAWNRKRISNCVA | 349 | 363 | 19.00 | |
| KCVNFNFNG | HLA-DRB1*01:01 | 0.67 | KCVNFNFNGLTGTGV | 537 | 551 | 8.00 |
| VIGIVNNTV | HLA-DRB1*13:02 | 0.70 | CDVVIGIVNNTVYDP | 1126 | 1140 | 49.00 |
| HLA-DRB1*13:02 | 0.70 | DVVIGIVNNTVYDPL | 1127 | 1141 | 49.00 | |
| LLLQYGSFC | HLA-DRB1*15:01 | 0.72 | TECSNLLLQYGSFCT | 747 | 761 | 49.00 |
| FQTLLALHR | HLA-DRB1*01:01 | 0.91 | ITRFQTLLALHRSYL | 235 | 249 | 9.00 |
| HLA-DRB1*01:01 | 0.91 | TRFQTLLALHRSYLT | 236 | 250 | 9.00 | |
| HLA-DRB1*11:01 | 1.20 | ITRFQTLLALHRSYL | 235 | 249 | 27.00 | |
| HLA-DRB1*11:01 | 1.20 | TRFQTLLALHRSYLT | 236 | 250 | 27.00 | |
| HLA-DRB1*01:01 | 1.30 | INITRFQTLLALHRS | 233 | 247 | 11.00 | |
| HLA-DRB1*01:01 | 1.30 | NITRFQTLLALHRSY | 234 | 248 | 11.00 | |
| HLA-DRB1*11:01 | 1.80 | INITRFQTLLALHRS | 233 | 247 | 40.00 | |
| HLA-DRB1*11:01 | 1.80 | NITRFQTLLALHRSY | 234 | 248 | 40.00 | |
| HLA-DRB1*01:01 | 3.10 | RFQTLLALHRSYLTP | 237 | 251 | 20.00 | |
| HLA-DRB1*01:01 | 3.10 | FQTLLALHRSYLTPG | 238 | 252 | 20.00 | |
| LVKQLSSNF | HLA-DRB1*01:01 | 8.20 | QALNTLVKQLSSNFG | 957 | 971 | 43.00 |
| YRLFRKSNL | HLA-DRB1*11:01 | 1.20 | YLYRLFRKSNLKPFE | 451 | 465 | 27.00 |
| MIAQYTSAL | HLA-DRB1*01:01 | 6.00 | LLTDEMIAQYTSALL | 864 | 878 | 33.00 |
| DYSVLYNSA | HLA-DRB1*01:01 | 1.30 | SNCVADYSVLYNSAS | 359 | 373 | 20.00 |
| YSVLYNSAS | HLA-DRB1*01:01 | 2.60 | CVADYSVLYNSASFS | 361 | 375 | 18.00 |
| HLA-DRB1*01:01 | 2.60 | ADYSVLYNSASFSTF | 363 | 377 | 18.00 | |
| HLA-DRB1*01:01 | 2.90 | NCVADYSVLYNSASF | 360 | 374 | 19.00 | |
| HLA-DRB1*01:01 | 8.00 | DYSVLYNSASFSTFK | 364 | 378 | 42.00 | |
| HLA-DRB1*01:01 | 8.00 | YSVLYNSASFSTFKC | 365 | 379 | 42.00 | |
| IRASANLAA | HLA-DRB1*01:01 | 9.30 | AEIRASANLAATKMS | 1016 | 1030 | 48.00 |
| YFKIYSKHT | HLA-DRB1*11:01 | 1.30 | NIDGYFKIYSKHTPI | 196 | 210 | 28.00 |
| HLA-DRB1*11:01 | 1.30 | IDGYFKIYSKHTPIN | 197 | 211 | 28.00 | |
| HLA-DRB1*11:01 | 1.30 | KNIDGYFKIYSKHTP | 195 | 209 | 29.00 | |
| HLA-DRB1*11:01 | 1.30 | DGYFKIYSKHTPINL | 198 | 212 | 29.00 | |
| HLA-DRB1*01:01 | 7.70 | NIDGYFKIYSKHTPI | 196 | 210 | 41.00 | |
| HLA-DRB1*01:01, | 7.70 | IDGYFKIYSKHTPIN | 197 | 211 | 41.00 | |
| HLA-DRB1*01:01 | 7.70 | DGYFKIYSKHTPINL | 198 | 212 | 41.00 | |
| HLA-DRB1*01:01 | 8.40 | KNIDGYFKIYSKHTP | 195 | 209 | 44.00 | |
| LTVLPPLLT | HLA-DRB1*01:01 | 1.30 | FNGLTVLPPLLTDEM | 855 | 869 | 11.00 |
| HLA-DRB1*01:01 | 1.60 | QKFNGLTVLPPLLTD | 853 | 867 | 12.00 | |
| HLA-DRB1*01:01 | 1.60 | KFNGLTVLPPLLTDE | 854 | 868 | 12.00 | |
| HLA-DRB1*01:01 | 1.60 | NGLTVLPPLLTDEMI | 856 | 870 | 12.00 | |
| HLA-DRB1*01:01 | 6.30 | GLTVLPPLLTDEMIA | 857 | 871 | 34.00 | |
| HLA-DRB1*01:01 | 6.80 | LTVLPPLLTDEMIAQ | 858 | 872 | 37.00 | |
| IDGYFKIYS | HLA-DRB1*11:01 | 1.30 | FKNIDGYFKIYSKHT | 194 | 208 | 31.00 |
| FKNIDGYFK | HLA-DRB1*01:01 | 7.70 | FKNIDGYFKIYSKHT | 194 | 208 | 41.00 |
| YTSALLAGT | HLA-DRB1*01:01 | 7.50 | MIAQYTSALLAGTIT | 869 | 883 | 40.00 |
| HLA-DRB1*01:01 | 8.20 | IAQYTSALLAGTITS | 870 | 884 | 43.00 | |
| HLA-DRB1*01:01 | 9.10 | AQYTSALLAGTITSG | 871 | 885 | 47.00 | |
| IAGLIAIVM | HLA-DRB1*01:01 | 2.50 | IWLGFIAGLIAIVMV | 1216 | 1230 | 17.00 |
| HLA-DRB1*01:01 | 2.50 | LGFIAGLIAIVMVTI | 1218 | 1232 | 17.00 | |
| HLA-DRB1*01:01 | 2.60 | WLGFIAGLIAIVMVT | 1217 | 1231 | 18.00 | |
| HLA-DRB1*01:01 | 3.60 | GFIAGLIAIVMVTIM | 1219 | 1233 | 22.00 | |
| HLA-DRB1*01:01 | 8.90 | FIAGLIAIVMVTIML | 1220 | 1234 | 46.00 | |
| LSSNFGAIS | HLA-DRB1*01:01 | 5.10 | KQLSSNFGAISSVLN | 964 | 978 | 29.00 |
| VKQLSSNFG | HLA-DRB1*01:01 | 7.70 | NTLVKQLSSNFGAIS | 960 | 974 | 41.00 |
| HLA-DRB1*01:01 | 8.00 | LNTLVKQLSSNFGAI | 959 | 973 | 42.00 | |
| HLA-DRB1*01:01 | 8.90 | ALNTLVKQLSSNFGA | 958 | 972 | 46.00 | |
| HLA-DRB1*01:01 | 8.90 | TLVKQLSSNFGAISS | 961 | 975 | 46.00 | |
| DSKTQSLLI | HLA-DRB1*07:01 | 3.30 | FGTTLDSKTQSLLIV | 106 | 120 | 48.00 |
| HLA-DRB1*07:01 | 3.30 | GTTLDSKTQSLLIVN | 107 | 121 | 48.00 | |
| HLA-DRB1*07:01 | 3.30 | TTLDSKTQSLLIVNN | 108 | 122 | 49.00 | |
| FAMQMAYRF | HLA-DRB1*01:01 | 3.60 | QIPFAMQMAYRFNGI | 895 | 909 | 22.00 |
| HLA-DRB1*01:01 | 3.70 | IPFAMQMAYRFNGIG | 896 | 910 | 23.00 | |
| HLA-DRB1*01:01 | 3.90 | ALQIPFAMQMAYRFN | 893 | 907 | 24.00 | |
| HLA-DRB1*01:01 | 3.90 | LQIPFAMQMAYRFNG | 894 | 908 | 24.00 | |
| IAQYTSALL | HLA-DRB1*01:01 | 5.80 | DEMIAQYTSALLAGT | 867 | 881 | 32.00 |
| HLA-DRB1*01:01 | 6.30 | TDEMIAQYTSALLAG | 866 | 880 | 34.00 | |
| HLA-DRB1*01:01 | 6.30 | EMIAQYTSALLAGTI | 868 | 882 | 34.00 | |
| HLA-DRB1*01:01 | 6.50 | LTDEMIAQYTSALLA | 865 | 879 | 35.00 | |
| YLQPRTFLL | HLA-DRB1*01:01 | 4.40 | AYYVGYLQPRTFLLK | 264 | 278 | 26.00 |
| HLA-DRB1*01:01 | 4.90 | YYVGYLQPRTFLLKY | 265 | 279 | 28.00 | |
| HLA-DRB1*01:01 | 5.60 | YVGYLQPRTFLLKYN | 266 | 280 | 31.00 | |
| HLA-DRB1*01:01 | 5.80 | VGYLQPRTFLLKYNE | 267 | 281 | 32.00 | |
| ISGINASVV | HLA-DRB1*01:01 | 4.40 | LGDISGINASVVNIQ | 1166 | 1180 | 26.00 |
| HLA-DRB1*01:01 | 4.60 | DLGDISGINASVVNI | 1165 | 1179 | 27.00 | |
| HLA-DRB1*01:01 | 4.90 | GDISGINASVVNIQK | 1167 | 1181 | 28.00 | |
| HLA-DRB1*01:01 | 5.10 | VDLGDISGINASVVN | 1164 | 1178 | 29.00 | |
| QIPFAMQMA | HLA-DRB1*01:01 | 5.10 | AALQIPFAMQMAYRF | 892 | 906 | 29.00 |
| ITGRLQSLQ | HLA-DRB1*01:01 | 5.80 | RLITGRLQSLQTYVT | 995 | 1009 | 32.00 |
Vaccination strategies that only depend on CD8+ T-cell immunity might be inadequate, because although CD8+ T cells stimulate extremely early responses, they might be unable to elicit long-term protective responses [70]. CD4+ T cells play a pivotal role in the adaptive immune response by maintaining and promoting the expansion of cytotoxic T cells, maintaining the memory of CD8+ T cells, and communicating with the innate immune cells [23]. In addition, CD4+ cells help CD8+ cells to generate strong primary and memory responses, as well as protective immunity against many types of bacterial and viral infections [71,72]. Previously, the development of vaccines was rudimentarily based on B cell immunity, whereas more recently it has been well established that T-cell epitopes provide a more long-lasting immune response, which is primarily mediated by CD4+ T cells as a result of antigenic drift [73]. The prediction of CD4+ T-cell epitopes mainly focuses on the peptide binding to the MHC class II proteins [74]. In the current study, we predicted several CD4+ T-cell epitopes by applying the criterion of IC50 < 50 nm. This threshold is considered to indicate more favorable immunogenic properties of the predicted T-cell epitopes, where a lower IC50 value represents a higher binding affinity [75]. The current research work is more specific than several previous studies, for instance, Alam et al. used IC50 < 100 nm for predicting CD4+ T-cell epitopes [76]. Moreover, our predicted T-cell epitopes were identical to the epitopes predicted in previous studies. The peptides with the core epitopes LIVNNATNV, IVNNATNVV, SKTQSLLIV, YNYLYRLFR, FNFSQILPD, SKVGGNYNY, YGSFCTQLN, LYRLFRKSN, LLALHRSYL, WLGFIAGLI, LALHRSYLT, LLQYGSFCT, FIAGLIAIV, FQTLLALHR, LVKQLSSNF, YRLFRKSNL, IRASANLAA, IAGLIAIVM, and ITGRLQSLQ were identical to those found in the previous study by Grifoni et al., which mapped the T-cell epitopes of SARS-CoV and SARS-CoV-2 [77]. Further, our predicted peptide AALQIPFAMQMAYRF (region 892–906) interacts with HLA-DRB1*01:01, which is in line with the previous study by Ahmed et al. [16].
3.5. Population coverage analysis
We first analyzed the individual epitopes that interacted with MHC class I alleles. West Africa had the highest coverage region for epitopes WTAGAAAYY and STQDLFLPF, at 76.33% and 58.93%, respectively (Table S3). South Asia had the highest coverage region for the epitope GAAAYYVGY, at 32.67% (Table S3). Next, we performed a population coverage analysis combining multiple epitopes. The cumulative population coverage was obtained for the three predicted epitopes, WTAGAAAYY, GAAAYYVGY, and STQDLFLPF. For the population coverage analysis of the multiple epitopes, the results were in agreement with those of the individual predicted epitopes, showing 77.98% coverage for West Africa, which was the highest coverage region. Europe had the second-highest coverage, at 77.91% (Table 4 ). The population coverage results for MHC class I alleles are shown in Fig. S2-S5.
Table 4.
Analysis of the population coverage using potential MHC I and MHC II interacted alleles for the proposed multi-epitope vaccine against SARS-CoV-2.
| Population/Area | MHC I Combined |
MHC II Combined |
||||
|---|---|---|---|---|---|---|
| Coverage (%)a | Average hitb | PC90c | Coverage (%)a | Average hitb | PC90c | |
| Central Africa | 63.97 | 1.56 | 0.28 | 46.24 | 1.12 | 0.19 |
| Central America | 2.19 | 0.04 | 0.10 | 23.76 | 0.43 | 0.13 |
| East Africa | 60.78 | 1.40 | 0.25 | 51.00 | 1.51 | 0.20 |
| East Asia | 62.39 | 1.49 | 0.27 | 57.95 | 1.65 | 0.24 |
| Europe | 77.91 | 1.86 | 0.45 | 65.71 | 2.16 | 0.29 |
| North Africa | 72.00 | 1.77 | 0.36 | 57.03 | 1.15 | 0.23 |
| North America | 65.50 | 1.59 | 0.29 | 63.77 | 2.01 | 0.28 |
| Northeast Asia | 56.06 | 1.54 | 0.23 | 36.94 | 0.77 | 0.16 |
| Oceania | 35.97 | 0.67 | 0.16 | 43.67 | 0.94 | 0.18 |
| South Africa | 70.08 | 1.71 | 0.33 | 5.91 | 0.12 | 0.21 |
| South America | 46.65 | 0.91 | 0.19 | 26.99 | 0.71 | 0.14 |
| South Asia | 69.11 | 1.53 | 0.32 | 60.68 | 1.63 | 0.25 |
| Southeast Asia | 44.14 | 1.08 | 0.18 | 35.62 | 0.67 | 0.16 |
| Southwest Asia | 60.13 | 1.14 | 0.25 | 31.97 | 0.76 | 0.15 |
| West Africa | 77.98 | 2.17 | 0.45 | 47.72 | 1.25 | 0.19 |
| West Indies | 56.56 | 1.22 | 0.23 | 54.15 | 1.72 | 0.22 |
Projected population coverage.
Average number of epitope hits/HLA combinations recognized by the population.
Minimum number of epitope hits/HLA combinations recognized by 90% of the population.
We next conducted a population coverage analysis for MHC class II alleles. As in the MHC class I analysis, we first individually predicted the population coverage for the MHC II alleles to find the maximum MHC II interaction. The core epitope LALHRSYLT showed the maximum coverage, which was 34.63% for the North American region (Table S4). Three core epitopes, namely YFKIYSKHT, FLVLLPLVS, and FQTLLALHR, exhibited greater than 25% coverage for Europe (Table S4). Epitope FLVLLPLVS showed 33.74% and 34.08% coverage for East Africa and East Asia, respectively (Table S4). The cumulative coverage of the targeted epitopes interacting with MHC class II alleles was then analyzed. The predicted epitopes were found to have a coverage of greater than 60% for three regions, Europe, North America, and South Asia, of which the maximum coverage was for Europe (65.71%) (Table 4).
Population coverage is important to understand during vaccine development because HLA varies by ethnicity and geographical region. We utilized the population coverage tool from IEDB to conduct the population coverage analysis for the predicted three epitopes, both individually and combined, for MHC class I and II alleles. Individually, for MHC class I alleles, West Africa had the highest percentage of population coverage for epitope WTAGAAAYY, followed by STQDLFLPF. The individual population coverage analysis for MHC class I alleles showed that the coverage for epitope WTAGAAAYY was greater than 50% for most regions, whereas GAAAYYVGY covered less than 40% of all regions. The selected three epitopes combined covered almost all available regions worldwide, and the highest coverage was observed in West Africa. Surprisingly, of the regions where the most cases of the infection were reported (namely, Europe and North America), the coverage was greater than 65%; thus, the coverage of Europe was nearly identical to that of West Africa. Interestingly, the epitopes showed 62.39% coverage in East Asia, where the first COVID-19 case was reported. The results of our predictions agree with those of Ahmed et al., who found that a multi-epitope vaccine has a larger population coverage [16]. However, like a multi-epitope vaccine, WTAGAAAYY individually covered most of the world's population. In addition, the predicted epitopes that interacted with the MHC class II alleles showed results similar to those of the MHC class I alleles, where a multi-epitope vaccine would cover a larger percentage of the population than individual epitopes could.
3.6. Linear B-cell epitope prediction
In the current study, linear B-cell epitopes were identified by utilizing an amino acid-based method.
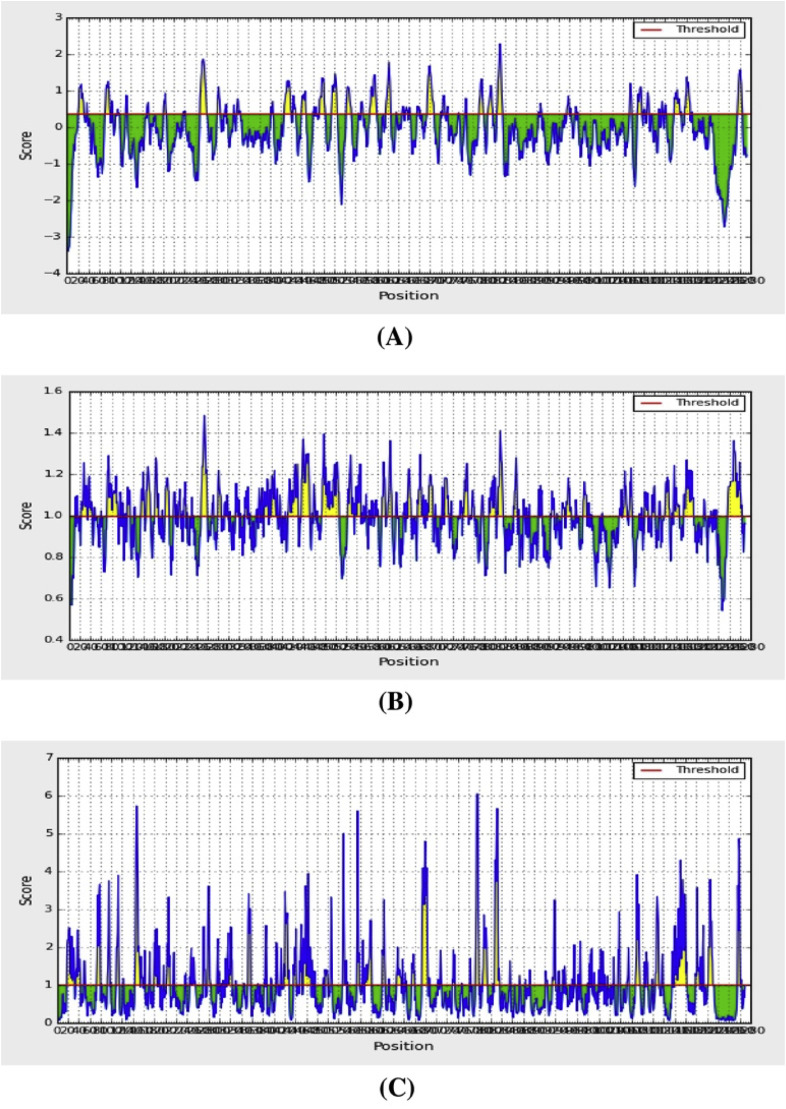
The BepiPred linear epitope prediction tool was applied, which is regarded as the best method for predicting linear B-cell epitopes. This tool uses a hidden Markov model. A total of 68 linear epitopes were predicted by BepiPred. By selecting only those with lengths ≥7 amino acid residues, 19 epitopes were finally included for further analysis (Table 5 ). The β-turns were predicted by the Chaus and Fasman β-turn prediction method. Previous research has shown that the antigenic part of the protein mostly remains as β-sheet [78]. The region containing residues 807–813 was predicted to be a β-turn region, with a score of 1.413; this was almost the highest score (Table S5). The region of residues 499–505 had a score of 1.257, and the 1140–1146 region had a score of 1.057; these were all greater than the average score (Fig. 5 ).
Table 5.
List of predicted B cell epitopes from BepiPred linear epitope prediction analysis.
| No. | Start | End | Peptide | Length |
|---|---|---|---|---|
| 1 | 21 | 31 | RTQLPPAYTNS | 11 |
| 2 | 71 | 81 | SGTNGTKRFDN | 11 |
| 3 | 249 | 261 | LTPGDSSSGWTAG | 12 |
| 4 | 318 | 324 | FRVQPTE | 7 |
| 5 | 407 | 420 | VRQIAPGQTGKIAD | 14 |
| 6 | 439 | 447 | NNLDSKVGG | 9 |
| 7 | 473 | 483 | YQAGSTPCNGV | 11 |
| 8 | 495 | 506 | YGFQPTNGVGYQ | 12 |
| 9 | 523 | 532 | TVCGPKKSTN | 10 |
| 10 | 567 | 580 | RDIADTTDAVRDPQ | 14 |
| 11 | 597 | 606 | VITPGTNTSN | 10 |
| 12 | 675 | 687 | QTQTNSPRRARSV | 13 |
| 13 | 772 | 780 | VEQDKNTQE | 9 |
| 14 | 788 | 797 | IYKTPPIKDF | 10 |
| 15 | 805 | 816 | ILPDPSKPSKRS | 12 |
| 16 | 1069 | 1077 | PAQEKNFTT | 9 |
| 17 | 1137 | 1148 | VYDPLQPELDSF | 12 |
| 18 | 1157 | 1167 | KNHTSPDVDLG | 11 |
| 19 | 1256 | 1265 | FDEDDSEPVL | 10 |
Fig. 5.
Combined B-cell linear epitope prediction using (A) Bepipred linear epitope prediction, (B) Chou & Fasman beta-turn prediction, (C) Emini surface accessibility prediction methods.
The Emini surface accessibility prediction method was also employed in this study. The average surface accessibility was 1.0, with a minimum of 0.042. In accordance with the above B-cell epitope results, the peptide region of 810–815 was predicted to have the best surface accessibility, with a score of 5.662 (Fig. 5).
The Kolaskar and Tongaonkar method was employed to predict antigenicity. This method evaluates antigenicity based on the physicochemical properties of amino acids and their abundances in experimentally known epitopes. A total of 46 B-cell epitopes were predicted by this method (Table 6 ). The average antigenic propensity of the SARS-CoV-2 SGP epitopes was 1.041, with a maximum of 1.261. The highest score was found for amino acids 5–11. The regions of 490–496 and 491–497 had equal scores of 1.067 (Fig. 6 ). The scores for the amino acid regions 558–564, 703–709, and 1140–1146 were 1.065, 1.026, and 1.051, respectively, and were all greater than the average value (Fig. 6).
Table 6.
List of predicted B-cell epitopes from Kolaskar and Tongaonkar antigenicity prediction method.
| Start | End | Peptide | Length |
|---|---|---|---|
| 4 | 18 | FLVLLPLVSSQCVNL | 15 |
| 34 | 41 | RGVYYPDK | 8 |
| 44 | 51 | RSSVLHST | 8 |
| 53 | 60 | DLFLPFFS | 8 |
| 81 | 87 | NPVLPFN | 7 |
| 115 | 121 | QSLLIVN | 7 |
| 125 | 134 | NVVIKVCEFQ | 10 |
| 136 | 146 | CNDPFLGVYYH | 11 |
| 168 | 174 | FEYVSQP | 7 |
| 210 | 216 | INLVRDL | 7 |
| 223 | 230 | LEPLVDLP | 8 |
| 239 | 248 | QTLLALHRSY | 10 |
| 263 | 270 | AAYYVGYL | 8 |
| 272 | 278 | PRTFLLK | 7 |
| 288 | 295 | AVDCALDP | 8 |
| 333 | 339 | TNLCPFG | 7 |
| 359 | 371 | SNCVADYSVLYNS | 13 |
| 376 | 385 | TFKCYGVSPT | 10 |
| 488 | 495 | CYFPLQSY | 8 |
| 505 | 527 | YQPYRVVVLSFELLHAPATVCGP | 23 |
| 592 | 599 | FGGVSVIT | 8 |
| 607 | 615 | QVAVLYQDV | 9 |
| 617 | 627 | CTEVPVAIHAD | 11 |
| 647 | 653 | AGCLIGA | 7 |
| 667 | 674 | GAGICASY | 8 |
| 687 | 693 | VASQSII | 7 |
| 723 | 730 | TTEILPVS | 8 |
| 735 | 741 | SVDCTMY | 7 |
| 750 | 763 | SNLLLQYGSFCTQL | 14 |
| 781 | 788 | VFAQVKQI | 8 |
| 837 | 843 | YGDCLGD | 7 |
| 847 | 853 | RDLICAQ | 7 |
| 858 | 864 | LTVLPPL | 7 |
| 873 | 880 | YTSALLAG | 8 |
| 959 | 966 | LNTLVKQL | 8 |
| 973 | 979 | ISSVLND | 7 |
| 1003 | 1011 | SLQTYVTQQ | 9 |
| 1030 | 1037 | SECVLGQS | 8 |
| 1057 | 1070 | PHGVVFLHVTYVPA | 14 |
| 1079 | 1085 | PAICHDG | 7 |
| 1123 | 1132 | SGNCDVVIGI | 10 |
| 1174 | 1179 | ASVVNI | 12 |
| 1221 | 1256 | IAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKF | 36 |
Fig. 6.
Combined B-cell linear epitope prediction using (D) Karplus & Schulz flexibility prediction, (E) Kolaskar & Tongaonkar antigenicity, (F) Parker hydrophilicity prediction methods.
The Karplus and Schulz flexibility prediction method found an average flexibility of 0.993 and a minimum of 0.866. The residue region 809–815 was found to be flexible, with a score of 1.101; this result was nearly the highest score (1.125). The next highest score was for residues 810–816, with a score of 1.1 (Fig. 6). The amino acid regions from 700 to 706, 910–916, and 1140–1146 had very similar scores of 1.029, 1.031, and 1.028, respectively (Fig. 6).
The Parker hydrophilicity prediction tool found an average hydrophilicity score of 1.238 for the SARS-CoV-2 SGP. The highest score was for the amino acid region 1257–1263 (Fig. 6).
Many vaccines are comprised of attenuated or killed pathogens. However, peptide vaccines that are capable of eliciting immune responses against specific pathogens contain peptides that are linear B cell epitopes [79]. The B-cell epitopes are antigenic determinants and are specifically recognized by the immune system because they represent specific pieces of antigens that bind to B lymphocytes; these epitopes are crucial for vaccine design [80]. A previous study demonstrated that linear B-cell epitopes can induce immunity against Pseudomonas aeruginosa in mice [81]. Synthetic peptides that contain linear B-cell epitopes are pivotal for producing antibodies against a specific protein; these antibodies can be used in various applications, such as in diagnostic tools or screening assays [82]. Importantly, B-cell epitopes elicit strong immune responses and do not cause any side effects. Here, we also predicted B-cell epitopes utilizing the IEDB database. The recent study by Grifoni et al. predicted B-cell epitopes of SARS-CoV-2 and SARS-CoV; the authors found that the SGP of SARS-CoV-2 contained the largest number of B-cell epitopes [77]. Moreover, they also predicted seven epitope regions of the SARS-CoV-2 SGP, which were the amino acid residues 491–505, 558–562, 703–704, 793–794, 810, 914, and 1140–1146. In the current study, we utilized several tools from the IEDB database to predict linear B-cell epitopes. As a result, we identified several B-cell epitopes from the SARS-CoV-2 SGP that are in agreement with those identified by Grifoni et al. (Table S6) [77]. In addition, the Grifoni study found that the predicted epitopes had a sequence identity greater than 65% [77]. The predicted B-cell epitopes were also in agreement with another previous study that had predicted several SARS-CoV-2 SGP B-cell epitopes (Table S6) [83].
Our study is not exhaustive, and in silico work has several pros and cons. The vaccine development process is tedious, and it can take years to develop a vaccine for use in humans, especially when the technologies are new and have not been extensively used before. Although institutes such as Curevac, Inovio, and Applied DNA Sciences are working on vaccines focusing on the SGP of SARS-CoV-2, it is not possible to predict how long it will take to obtain a successful vaccine. Realistically, an animal model is a prerequisite for testing the protectiveness of a vaccine, but developing an animal model for SARS-CoV-2 has been difficult. Vaccines also must be tested for toxicity in various animals, for example, rabbits, which typically takes several months to complete. Human vaccines must be produced in accordance with current good manufacturing practices (cGMPs), which requires trained personnel, exclusive facilities, appropriate documentation, and quality raw materials. As a result, vaccine candidates that are in the preclinical phase might require specialized processes that need to be developed from scratch. Realistically, it will require more than 1 year to license any vaccine for SARS-CoV-2.
4. Conclusions
Improvements in the field of immunoinformatics have been an important factor in advancing the prediction of peptide-based vaccines. Viruses can activate both T-cell and humoral immunity. The epitopes predicted in this study might improve immunity against SARS-CoV-2. Our work is based on the fundamental principles of immunity, whereby the recognition of foreign bodies by the host immune cells provokes an immune response in which relevant information is transferred to both T cells and B cells. Here, utilizing in silico simulations, our predicted epitopes were able to interact with CD8+ cells when the epitopes were presented as antigens. However, the present study is only a preliminary approach for predicting an epitope-based vaccine against SARS-CoV-2. We hope that these predicted epitopes will lay the foundation for the further experimental design of vaccine candidates against SARS-CoV-2.
Declaration of competing interest
The author declares that there is no competing interest in this work.
Acknowledgments
The authors would like to thank Cambridge Proofreading & Editing LLC. (https://proofreading.org/) for editing a draft of this manuscript. Prof. T. Ben Hadda acknowledges the offer of the bioinformatics facilities by Prof Y. Zarhloule even in this critical situation of COVID-19, and the financially support by the Deanship of Scientific Research at Umm Al-Qura University to Prof. F.A. Almalki (18-MED-1-02-0003).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.compbiomed.2020.103967.
Data availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author contributions
Study concept and design: A.R., S.A.S., T.B.H., F.A.A., and T.B.E. Acquisition of data: A.R., S.A.S., A.S., A.P. F.A.A. and T.B.H. Analyses and interpretation of data: A.R., S.A.S., A.M.T., N.U.E., N.J.M., M.M.C., T.A.E., S.C., S.S., F.N. and T.B.E. Drafting the manuscript: A.R., S.A.S., S.J.M., F.A.A., T.B.H., and T.B.E. Critical revision of the manuscript for important intellectual content: S.J.M., F.A.A., T.B.H., and T.B.E. Technical or material support: A.R. Study supervision: F.A.A., T.B.H., and T.B.E.
Funding
This work is conducted with the individual funding of all authors.
Ethical approval
Not required.
ORCID identification
-
•
A. Rakib: https://orcid.org/0000-0003-3335-0368
-
•
F. Nainu: https://orcid.org/0000-0003-0989-4023
- •
-
•
A. Shahriar: https://orcid.org/0000-0001-5529-8893
-
•
A.M. Tareq: https://orcid.org/0000-0003-2704-7610
-
•
N.U. Emon: https://orcid.org/0000-0001-7567-4796
-
•
S. Chakraborty: https://orcid.org/0000-0002-0178-4869
-
•
T. Ben Hadda: https://orcid.org/0000-0002-5633-6203.
-
•
F.A. Almalki: https://orcid.org/0000-0003-4048-1526
-
•
T. Bin Emran: https://orcid.org/0000-0003-3188-2272
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus--infected pneumonia in Wuhan, China. JAMA. 2020;323(3):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus--infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Organization W.H. vol. 72. 2020. (Coronavirus Disease 2019 (COVID-19): Situation Report). [Google Scholar]
- 5.Tyrrell D.A.J., Bynoe M.L. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966:76–77. doi: 10.1016/s0140-6736(66)92364-6. [DOI] [PubMed] [Google Scholar]
- 6.Christie J.M., Chapel H., Chapman R.W., Rosenberg W.M. Immune selection and genetic sequence variation in core and envelope regions of hepatitis C virus. Hepatology. 1999;30:1037–1044. doi: 10.1002/hep.510300403. [DOI] [PubMed] [Google Scholar]
- 7.Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong C., Jiang L., Chen Y., Jiang Q. Biorxiv; 2020. Evolution and Variation of 2019-novel Coronavirus. [Google Scholar]
- 9.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanne J.P. 2020. Chest CT Findings in 2019 Novel Coronavirus (2019-nCoV) Infections from Wuhan, China: Key Points for the Radiologist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Sanche, Y.T. Lin, C. Xu, E. Romero-Severson, N. Hengartner, R. Ke, Early Release-High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus vol. 2, (n.d.). [DOI] [PMC free article] [PubMed]
- 12.W.H. Organization, et al., Q&A on Coronaviruses (COVID-19), Retrieved April 6th. (2020).
- 13.Organization W.H. 29 March 2020. Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations: scientific brief; p. 2020. [Google Scholar]
- 14.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed S.F., Quadeer A.A., McKay M.R. 2020. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Hu Y., Song Z.-G., Tao Z.-W., Tian J.-H., Pei Y.-Y. BioRxiv; China: 2020. Complete Genome Characterisation of a Novel Coronavirus Associated with Severe Human Respiratory Disease in Wuhan. [Google Scholar]
- 18.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. J. Thorac. Dis. 2013;5:S142. doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suarez D.L., Schultz-Cherry S. Immunology of avian influenza virus: a review. Dev. Comp. Immunol. 2000;24:269–283. doi: 10.1016/s0145-305x(99)00078-6. [DOI] [PubMed] [Google Scholar]
- 22.Briney B., Sok D., Jardine J.G., Kulp D.W., Skog P., Menis S., Jacak R., Kalyuzhniy O., De Val N., Sesterhenn F. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell. 2016;166:1459–1470. doi: 10.1016/j.cell.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosa D.S., Ribeiro S.P., Cunha-Neto E. CD4+ T cell epitope discovery and rational vaccine design. Arch. Immunol. Ther. Exp. (Warsz). 2010;58:121–130. doi: 10.1007/s00005-010-0067-0. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Zhang S., Tan S., Zheng B., Gao G.F. Revival of the identification of cytotoxic T-lymphocyte epitopes for immunological diagnosis, therapy and vaccine development. Exp. Biol. Med. 2011;236:253–267. doi: 10.1258/ebm.2010.010278. [DOI] [PubMed] [Google Scholar]
- 25.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir--ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. (80–. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J. Cell; 2020. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020;8:153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poland G.A., Ovsyannikova I.G., Kennedy R.B., Haralambieva I.H., Jacobson R.M. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS A J. Integr. Biol. 2011;15:625–636. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakib A., Sami S.A., Islam M.A., Ahmed S., Faiz F.B., Khanam B.H., Marma K.K.S., Rahman M., Uddin M.M.N., Bin Emran T. 2020. Epitope-Based Immunoinformatics Approach on Nucleocapsid Protein of Severe Acute Respiratory Syndrome-Coronavirus-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peele K.A., Srihansa T., Krupanidhi S., Sai A.V., Venkateswarulu T.C. Design of multi-epitope vaccine candidate against SARS-CoV-2: a in-silico study. J. Biomol. Struct. Dyn. 2020:1. doi: 10.1080/07391102.2020.1770127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong E., Wong M.U., Huffman A., He Y. BioRxiv; 2020. COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C.H., Koohy H. 2020. (In Silico Identification of Vaccine Targets for 2019-nCoV). F1000Research. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du L., Zhao G., Li L., He Y., Zhou Y., Zheng B.-J., Jiang S. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem. Biophys. Res. Commun. 2009;384:486–490. doi: 10.1016/j.bbrc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.UniProt The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugembe D.L., Ekii A.O., Ndembi N., Serwanga J., Kaleebu P., Pala P. Computational MHC-I epitope predictor identifies 95% of experimentally mapped HIV-1 clade A and D epitopes in a Ugandan cohort. BMC Infect. Dis. 2020;20:1–16. doi: 10.1186/s12879-020-4876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giguère S., Drouin A., Lacoste A., Marchand M., Corbeil J., Laviolette F. MHC-NP: predicting peptides naturally processed by the MHC. J. Immunol. Methods. 2013;400:30–36. doi: 10.1016/j.jim.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Bui H.-H., Sidney J., Li W., Fusseder N., Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinf. 2007;8:361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.-H., Grey H., Sette A. A consensus epitope prediction approach identifies the breadth of murine T CD8+-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 43.Dimitrov I., Flower D.R., Doytchinova I. AllerTOP-a server for in silico prediction of allergens. BMC Bioinf. 2013:S4. doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maupetit J., Derreumaux P., Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37:W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 46.Dallakyan S. Scripps Res. Inst; 2008. PyRx-python Prescription V. 0.8; p. 2010. [Google Scholar]
- 47.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009 doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen M., Lundegaard C., Worning P., Lauemøller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bui H.-H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf. 2006;7:1–5. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emini E.A., V Hughes J., Perlow D., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 54.Karplus P.A., Schulz G.E. Prediction of chain flexibility in proteins. Naturwissenschaften. 1985;72:212–213. [Google Scholar]
- 55.Larsen J.E.P., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou P.Y., Fasman G.D. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 57.S.K. Patel, M. Pathak, R. Tiwari, M.I. Yatoo, Y.S. Malik, R. Sah, A.A. Rabaan, K. Sharun, K. Dhama, D.K. Bonilla-Aldana, et al., A Vaccine Is Not Too Far for COVID-19, (n.d.). [DOI] [PubMed]
- 58.Ramaiah A., Arumugaswami V. 2020. Insights into Cross-Species Evolution of Novel Human Coronavirus 2019-nCoV and Defining Immune Determinants for Vaccine Development, BioRxiv. [Google Scholar]
- 59.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.-T.K., Zhou Y. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PloS One. 2013;8 doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W., Makhawi A.M. Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: an immunoinformatics approach. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/2683286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalsass M., Brozzi A., Medini D., Rappuoli R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019;10:113. doi: 10.3389/fimmu.2019.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinha S.K., Shakya A., Prasad S.K., Singh S., Gurav N.S., Prasad R.S., Gurav S.S. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 66.Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 67.Kallinich T., Beier K.C., Wahn U., Stock P., Hamelmann E. T-cell co-stimulatory molecules: their role in allergic immune reactions. Eur. Respir. J. 2007;29:1246–1255. doi: 10.1183/09031936.00094306. [DOI] [PubMed] [Google Scholar]
- 68.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Peter Walter P. Garland Science.[Google Scholar]; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- 69.Ahmed S., Rakib A., Islam M.A., Khanam B.H., Faiz F.B., Paul A., Chy M.N.U., Bhuiya N.M.M.A., Uddin M.M.N., Ullah S.M.A. In vivo and in vitro pharmacological activities of Tacca integrifolia rhizome and investigation of possible lead compounds against breast cancer through in silico approaches. Clin. Phytosci. 2019;5:36. [Google Scholar]
- 70.Khanolkar A., Badovinac V.P., Harty J.T. CD8 T cell memory development: CD4 T cell help is appreciated. Immunol. Res. 2007;39:94–104. doi: 10.1007/s12026-007-0081-4. [DOI] [PubMed] [Google Scholar]
- 71.Bevan M.J. Helping the CD8+ T-cell response. Nat. Rev. Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 72.Novy P., Quigley M., Huang X., Yang Y. CD4 T cells are required for CD8 T cell survival during both primary and memory recall responses. J. Immunol. 2007;179:8243–8251. doi: 10.4049/jimmunol.179.12.8243. [DOI] [PubMed] [Google Scholar]
- 73.Chiou S.-S., Fan Y.-C., Crill W.D., Chang R.-Y., Chang G.-J.J. Mutation analysis of the cross-reactive epitopes of Japanese encephalitis virus envelope glycoprotein. J. Gen. Virol. 2012;93:1185–1192. doi: 10.1099/vir.0.040238-0. [DOI] [PubMed] [Google Scholar]
- 74.Oyarzún P., Ellis J.J., Bodén M., Kobe B. PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity. BMC Bioinf. 2013;14:1–11. doi: 10.1186/1471-2105-14-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khan M.K., Zaman S., Chakraborty S., Chakravorty R., Alam M.M., Bhuiyan T.R., Rahman M.J., Fernández C., Qadri F., Seraj Z.I. In silico predicted mycobacterial epitope elicits in vitro T-cell responses. Mol. Immunol. 2014;61:16–22. doi: 10.1016/j.molimm.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 76.Alam A., Ali S., Ahamad S., Malik M.Z., Ishrat R. From ZikV genome to vaccine: in silico approach for the epitope-based peptide vaccine against Zika virus envelope glycoprotein. Immunology. 2016;149:386–399. doi: 10.1111/imm.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. 2020. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2, Cell Host Microbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hasan A., Hossain M., Alam J. A computational assay to design an epitope-based Peptide vaccine against Saint Louis encephalitis virus. Bioinf. Biol. Insights. 2013;7 doi: 10.4137/BBI.S13402. BBI--S13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nardin E.H., Calvo-Calle J.M., Oliveira G.A., Nussenzweig R.S., Schneider M., Tiercy J.-M., Loutan L., Hochstrasser D., Rose K. A totally synthetic polyoxime malaria vaccine containing Plasmodium falciparum B cell and universal T cell epitopes elicits immune responses in volunteers of diverse HLA types. J. Immunol. 2001;166:481–489. doi: 10.4049/jimmunol.166.1.481. [DOI] [PubMed] [Google Scholar]
- 80.Shey R.A., Ghogomu S.M., Esoh K.K., Nebangwa N.D., Shintouo C.M., Nongley N.F., Asa B.F., Ngale F.N., Vanhamme L., Souopgui J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci. Rep. 2019;9:1–18. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hughes E.E., Gilleland H.E., Jr. Ability of synthetic peptides representing epitopes of outer membrane protein F of Pseudomonas aeruginosa to afford protection against P. aeruginosa infection in a murine acute pneumonia model. Vaccine. 1995;13:1750–1753. doi: 10.1016/0264-410x(95)00166-x. [DOI] [PubMed] [Google Scholar]
- 82.Schellekens G.A., Visser H., De Jong B.A.W., Van Den Hoogen F.H.J., Hazes J.M.W., Breedveld F.C., Van Venrooij W.J. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2000;43:155–163. doi: 10.1002/1529-0131(200001)43:1<155::AID-ANR20>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 83.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S.-S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.