Abstract
Background & Aims
The miR-34a gene is a direct target of p53 and is commonly silenced in colorectal cancer (CRC). Here we identified the receptor tyrosine kinase CSF1R as a direct miR-34a target and characterized CSF1R as an effector of p53/miR-34a-mediated CRC suppression.
Methods
Analyses of TCGA-COAD and three other CRC cohorts for association of mRNA expression and signatures with patient survival and molecular subtypes. Bioinformatics identification and experimental validation of miRNA and transcription factor targets. Functional analysis of factors/pathways in the regulation of epithelial-mesenchymal transition (EMT), invasion, migration, acquired chemo-resistance and metastasis. Analyses of protein expression and CpG methylation within primary human colon cancer samples.
Results
In primary CRCs increased CSF1R, CSF1 and IL34 expression was associated with poor patient survival and a mesenchymal-like subtype. CSF1R displayed an inverse correlation with miR-34a expression. This was explained by direct inhibition of CSF1R by miR-34a. Furthermore, p53 repressed CSF1R via inducing miR-34a, whereas SNAIL induced CSF1R both directly and indirectly via repressing miR-34a in a coherent feed-forward loop. Activation of CSF1R induced EMT, migration, invasion and metastasis of CRC cells via STAT3-mediated down-regulation of miR-34a. 5-FU resistance of CRC cells was mediated by CpG-methylation of miR-34a and the resulting elevated expression of CSF1R. In primary CRCs elevated expression of CSF1R was detected at the tumor invasion front and was associated with CpG methylation of the miR-34a promoter as well as distant metastasis.
Conclusions
The reciprocal inhibition between miR-34a and CSF1R and its loss in tumor cells may be relevant for therapeutic and prognostic approaches towards CRC management.
Keywords: EMT, Metastasis, Chemoresistance, 5-FU, Tumor Progression, MicroRNAs
Abbreviations used in this paper: 5-aza, 5-aza-2’-deoxycytidine; 5-FU, 5-fluorouracil; cDNA, complementary DNA; CMS, consensus molecular subtype; COAD, colon adenocarcinoma; CRC, colorectal cancer; CRIS, colorectal cancer intrinsic subtype; CSF1R, colony-stimulating factor 1 receptor; Dox, doxycycline; EMT, epithelial-mesenchymal transition; EMT-TF, epithelial-mesenchymal transition–inducing transcription factor; GSEA, gene set enrichment analysis; IL, interleukin; miRNA, microRNA; mRNA, messenger RNA; MSP, methylation-specific polymerase chain reaction; qPCR, quantitative real-time polymerase chain reaction; PDX, patient-derived xenograft; RTK, receptor tyrosine kinase; siRNA, small interfering RNA; TAM, tumor-associated macrophage; TCGA, The Cancer Genome Atlas; TSA, trichostatin A; VIM, vimentin
Graphical abstract
Summary.
We show that the receptor tyrosine kinase colony-stimulating factor 1 receptor is under negative control by the p53-inducible microRNA miR-34a. In primary colorectal cancers, this regulation is alleviated, and elevated colony-stimulating factor 1 receptor levels contribute to critical features of tumor cells, such as invasiveness, chemoresistance, and metastasis.
Most cancer patients die as a consequence of metastatic spread.1 More than 50% of colorectal cancer (CRC) patients will develop liver metastases, leading to ineffective chemotherapy and increased mortality.2,3 Epithelial-mesenchymal transition (EMT) of primary tumor cells is one of the first steps of the metastatic cascade.4 Several signals that emanate from cells in the tumor microenvironment promote EMT. Among these are cytokines that activate receptor tyrosine kinases (RTKs).5 Together with the PDGFR and c-Kit receptors, the colony-stimulating factor 1 receptor (CSF1R), which is encoded by the c-fms proto-oncogene, belongs to the group of type III RTKs.6, 7, 8 Transforming potential has been assigned to the viral homolog (v-fms) and c-fms.9,10 Binding of its ligand CSF1 or of the more recently identified ligand, interleukin 34 (IL34), induces homodimerization and activation of CSF1R.11,12 Subsequently, the Ras/Raf/MAPK, PI3K/AKT, and JAK/STAT pathways are activated.12, 13, 14 CSF1R has important functions in macrophages, and inhibition of CSF1R in tumor-associated macrophages (TAMs) represents an attractive therapeutic strategy.15 However, the significance of CSF1R expression in tumor cells of epithelial origin is less well characterized. Interestingly, elevated expression of CSF1 and CSF1R has been associated with metastases and progression of breast cancer.16 Notably, colorectal cancer patients with a more advanced tumor stage display elevated serum levels of CSF1, implying that CSF1R signaling may be involved in CRC progression.17
The p53 protein functions as a transcription factor that suppresses a variety of malignant processes.18 Besides inducing protein-coding genes, p53 also induces the expression of microRNA (miRNA)-encoding genes, which have been shown to represent important mediators of p53 functions.19 Among these microRNAs, miR-34a often shows the most pronounced induction by p53. MiR-34a inhibits the expression of multiple targets that have been implicated in the progression of CRC, such as SNAIL, ZNF281, IL6R, INH3, and PAI-1.20, 21, 22, 23, 24 The miR-34a gene is silenced by CpG methylation of its promoter in ∼75% of all CRCs.25,26 Furthermore, the downregulation of miR-34a by CpG methylation or p53 mutation has been associated with distant metastasis in CRCs.24,27
Here, we show that elevated CSF1R expression is associated with poor survival of CRC patients and inversely correlates with miR-34a expression. Furthermore, we demonstrate that CSF1R represents a direct miR-34a target. Our results imply that deregulation of the p53/miR-34a/CSF1R/STAT3 pathway, which we characterize here, may contribute to initiation, progression, and chemoresistance of CRCs.
Results
Association of CSF1R, CSF1, and IL34 Expression With Clinical Parameters in CRCs
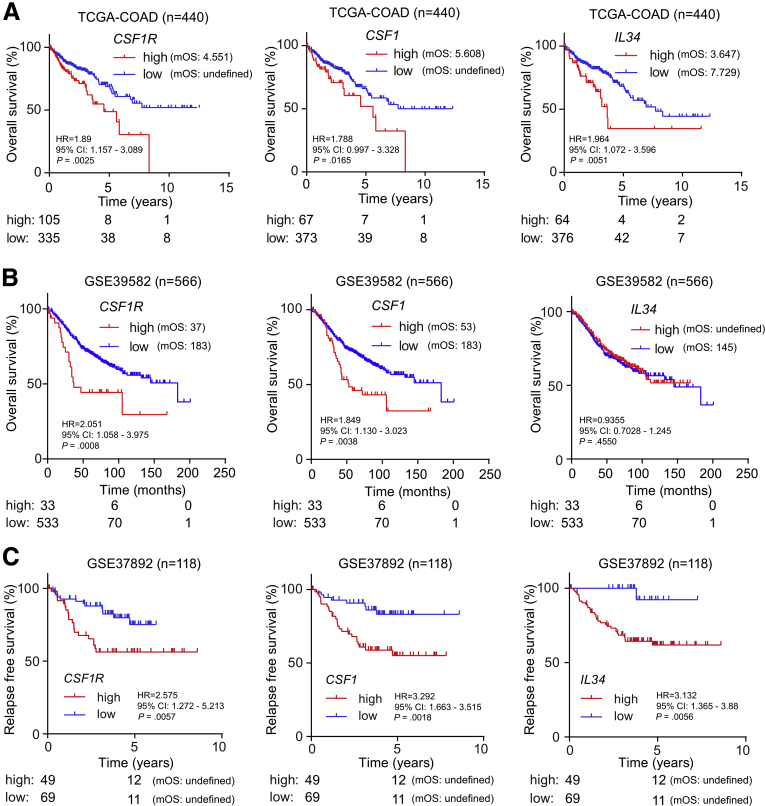
In order to determine the potential clinical relevance of CSF1R and its ligands in CRC, we analyzed their expression in 440 primary CRC samples represented within The Cancer Genome Atlas (TCGA) database.28 Thereby, we found that increased expression of CSF1R, as well as CSF1 and IL-34 messenger RNAs (mRNAs) in primary CRCs was significantly associated with decreased survival of patients (Figure 1A). In another cohort of 566 CRC patient samples,29 elevated CSF1R and CSF1, but not IL34 mRNA expression was associated with poor overall survival (Figure 1B). Moreover, elevated CSF1R, CSF1, and IL34 mRNA expression was also associated with decreased relapse-free survival in an independent cohort comprising 118 patients (Figure 1C).30 Therefore, we could confirm the findings obtained within the TCGA colon adenocarcinoma (TCGA-COAD) cohort in 2 independent CRC cohorts.
Figure 1.
Association of elevated CSF1R, CSF1, and IL34 expression with survival in primary CRCs. Kaplan-Meier analyses of survival with data from the (A) TCGA database, the (B) GSE39582, and the (C) GSE37892 cohorts using log-rank tests. Below the graphs, the numbers of patients with high or low expression of the indicated mRNA at the respective time point is provided. CI, confidence interval; HR, hazard ratio; mOS, median overall survival.
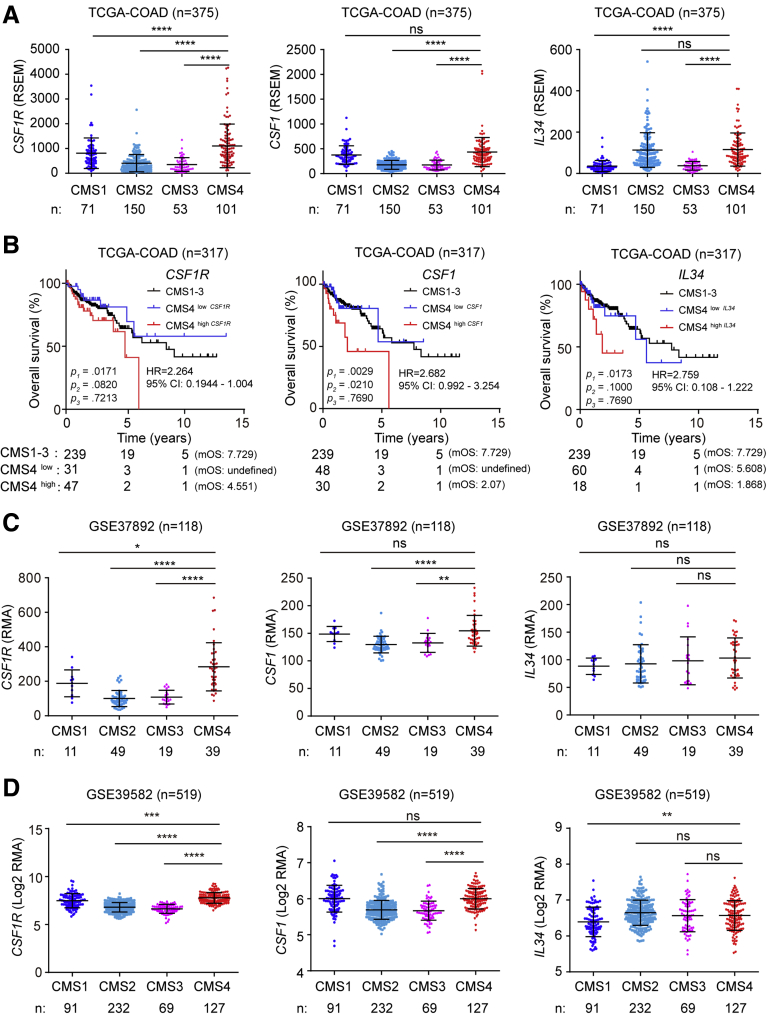
The consensus molecular subtype (CMS) classification is one of the most robust classification systems for CRCs and is based on comprehensive gene expression profiles.31 CRCs belonging to the CMS4, which displays a mesenchymal signature and the worst prognosis among the 4 different CMSs,31 showed the highest expression of CSF1R, CSF1, and IL34 (Figure 2A). Next, we stratified CMS4 tumors into 2 subgroups with either elevated or low expression of CSF1R, CSF1, and IL34 mRNAs (Figure 2B). Patients with CMS4 CRCs that displayed either high CSF1R, CSF1, or IL34 expression had a significantly shorter overall survival than patients with CRCs classified as CMS1–3 or CMS4 with low CSF1R, CSF1 or IL34 expression. Furthermore, also in the 2 other cohorts, expression levels of CSF1R and CSF1 were elevated in CMS4 tumors (Figure 2C and D).
Figure 2.
Association of CSF1R, CSF1, and IL34 expression with CMS subtypes. (A) CSF1R, CSF1, and IL34 mRNA expression in CRCs belonging to the indicated consensus molecular subtypes (CMS). (B) Kaplan-Meier analysis of overall survival of patients with primary CRCs classified as CMS1–3 or CMS4 with either high CSF1R/CSF1/IL34 or low CSF1R/CSF1/IL34 expression levels. P1: CMS4high vs CMS4low; P2: CMS4high vs CMS1-3; P3: CMS4low vs CMS1–3. The number of patients in each group was listed below the graph. CSF1R, CSF1, and IL34 mRNA expression in CRC patient samples from the (C) GSE37892 and (D) GSE39582 datasets classified according to the indicated consensus molecular subtypes (CMS). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. ns, not significant; RSEM, RNA sequencing by expectation maximization.
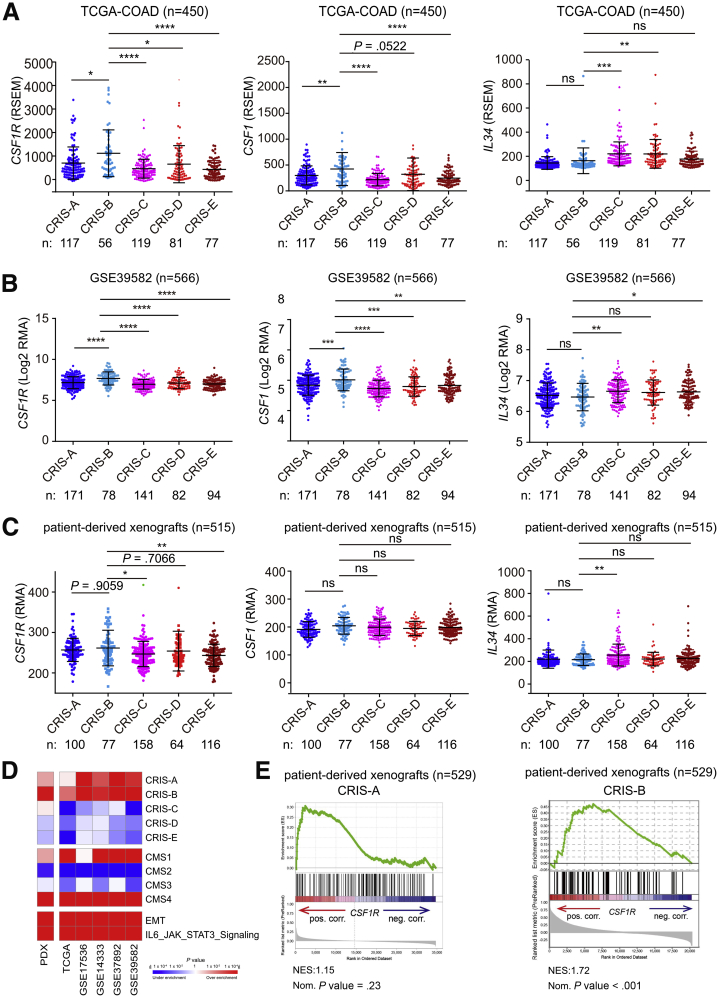
However, mRNAs displaying elevated expression in CMS4-type tumors may originate from stromal cells and therefore confound the gene expression profiles of CRCs.32, 33, 34 To overcome this caveat, patient-derived xenografts (PDXs) have been used to generate mRNA expression signatures by microarray analyses, in which the contribution of (murine) stromal mRNAs to whole-tumor mRNA expression patterns was selectively eliminated by the use of human-specific probe sets.34 Thereby, 5 different colorectal cancer intrinsic subtypes (CRIS) were defined. Apart from classifying PDX-derived tumors, these were used to reclassify previously established publicly available CRC patient cohorts into CRIS subtypes, such as the TCGA-COAD cohort.34 Notably, CSF1R mRNA expression within the TCGA-COAD cohort was elevated in the CRIS-B subtype (Figure 3A), which is characterized by TGF-β pathway activity, epithelial-mesenchymal transition and poor prognosis.34 Moreover, expression of the CSF1 ligand, but not of IL34, was elevated in the CRIS-B subtype of tumors within the TCGA-COAD cohort. We validated these findings in the additional cohort comprising 566 cases. Again, expression of both CSF1R and its ligand CSF1, but not IL34, was elevated in CRIS-B tumors (Figure 3B).
Figure 3.
Association of CSF1R, CSF1, and IL34 expression with CRIS subtypes. The indicated mRNA expression in CRC patient samples from the (A) TCGA-COAD, (B) GSE39582, and (C) PDX cohorts were classified according to the indicated CRIS. (D) GSEA results for CSF1R expression from PDX, TCGA, and indicated Gene Expression Omnibus datasets. Genes were preranked by expression correlation coefficient (Pearson’s r) with CSF1R for each dataset. (E) CSF1 and IL34 mRNA expression in CRC patient samples from the GSE76402 cohort classified according to the indicated CRIS. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. ns, not significant.
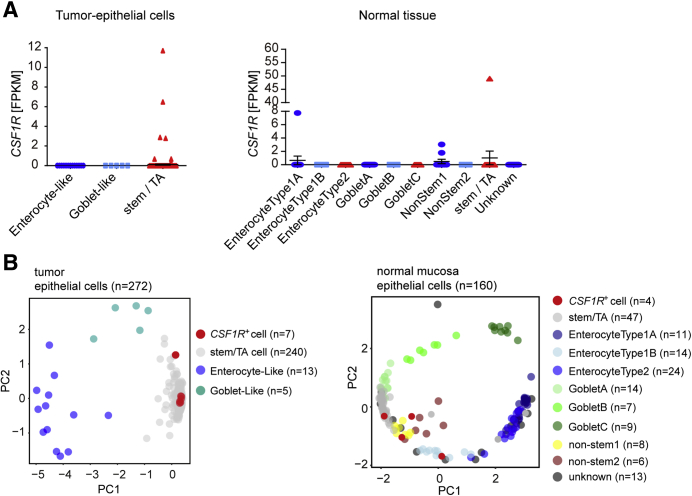
To further validate our findings, we also analyzed CSF1R expression in CRIS subtypes of PDX samples. CSF1R expression was elevated in the CRIS-B subtype when compared with the other subtypes, albeit without statistical significance in case of the CRIS-A and CRIS-D subtypes (Figure 3C). Gene set enrichment analyses (GSEAs) showed a strong positive correlation of CSF1R mRNA expression with CRIS-B and CMS4 gene signatures, whereas either negative or nonsignificant correlations of CSF1R mRNA with signatures from all other CRIS or CMS subclasses in PDX samples were observed (Figure 3D and E), indicating that tumor intrinsic CSF1R expression is associated with a mesenchymal tumor phenotype. In addition, elevated CSF1R expression was associated with CRIS-B/CMS4 signatures, such as EMT and IL6/JAK/STAT3 signaling. Moreover, mRNA expression data from whole tumors showed strong positive correlations of CSF1R with CRIS-A and CRIS-B and CMS1 and CMS4, as well as EMT and IL6/JAK/STAT3 signaling–associated signatures in the majority of analyzed patient cohorts. Furthermore, analysis of published single-cell RNA sequencing results.35 obtained from primary colorectal tumors and matched normal mucosa revealed that CSF1R is specifically expressed in colonic epithelial and tumor cells with stem/TA-like features (Figure 4A and B).
Figure 4.
Single-cell sequencing based analysis of CSF1R expression. (A) CSF1R mRNA expression derived from single-cell sequencing data of normal colonic (n = 160) and CRC tissue (n = 271) (GSE81861). (B) Principal component analysis plot based on the reference component analysis single cell RNA sequencing clustering algorithm showing the clustering of different cell types in tumor epithelial cells and cells from normal mucosa. CSF1R-expressing cells are indicated in red. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. PC, principal component.
Next, we also evaluated association of CSF1R expression with other clinical or pathological variables in both the TCGA-COAD and the GSE39582 cohorts in order to exclude potentially confounding factors that might affect patient survival. CSF1R expression did not display a significant association with age, gender, and tumor stage (UICC/Union for International Cancer Control). However, elevated CSF1R expression in CRCs was associated with mismatch repair–deficient/microsatellite instability, as well as with the CMS4 or CRIS-B molecular subtypes of CRCs (Table 1). A Cox multiple regression analysis demonstrated prognostic power of high CSF1R expression independent from age, gender, microsatellite instability status, and tumor stage (Table 2). The GSE37892 cohort was not included here, because the necessary patient data were incomplete.
Table 1.
Clinical Data and CSF1R mRNA Expression in Colon Cancer Cases From 2 Independent Patient Cohorts
| Characteristic | TCGA-COAD |
GSE39582 |
||||||
|---|---|---|---|---|---|---|---|---|
| Total |
CSF1R |
P | Total |
CSF1R |
P | |||
| Low | High | Low | High | |||||
| All patients | 440 (100.0) | 335 (76.1) | 105 (23.9) | 566 (100.0) | 532 (76.1) | 34 (23.9) | ||
| Age | ||||||||
| <median | 206 (46.9) | 164 (79.6) | 42 (20.4) | .109 | 283 (51.2) | 272 (96.1) | 11 (3.9) | .033a |
| ≥median | 234 (53.1) | 171 (73.1) | 63 (26.9) | 282 (48.8) | 259 (91.8) | 23 (8.2) | ||
| Sex | ||||||||
| Male | 235 (53.4) | 182 (77.4) | 53 (22.6) | .490 | 310 (54.8) | 290 (93.5) | 20 (6.5) | .624 |
| Female | 205 (46.6) | 153 (74.6) | 52 (25.4) | 256 (45.2) | 242 (94.5) | 14 (5.5) | ||
| UICC stage | ||||||||
| I | 73 (17.0) | 58 (79.5) | 15 (20.5) | .378 | 33 (5.9) | 33 (100) | 0 (0) | .497 |
| II | 169 (39.4) | 122 (72.2) | 47 (27.8) | 264 (47.0) | 248 (93.9) | 16 (6.1) | ||
| III | 126 (29.4) | 97 (77.0) | 29 (23.0) | 205 (36.5) | 191 (93.2) | 14 (6.8) | ||
| IV | 61 (14.2) | 50 (82.0) | 11 (18.0) | 60 (10.7) | 56 (93.3) | 4 (6.7) | ||
| MSI status | ||||||||
| MSS/MSI-low | 323 (81.2) | 253 (78.3) | 70 (21.7) | .005a | 444 (85.5) | 424 (95.5) | 20 (4.5) | .029 |
| MSI-high | 75 (18.8) | 47 (62.7) | 28 (37.3) | 75 (14.5) | 67 (89.3) | 8 (10.7) | ||
| CMS subtype | ||||||||
| CMS1–3 | 253 (73.3) | 219 (86.6) | 34 (13.4) | <.0001a | 360 (73.9) | 351 (97.5) | 9 (2.5) | <.0001a |
| CMS4 | 92 (26.7) | 41 (44.6) | 51 (55.4) | 127 (26.1) | 107 (84.3) | 20 (15.7) | ||
| CRIS subtype | ||||||||
| CRIS-A/C–E | 283 (87.9) | 219 (77.4) | 64 (22.6) | .002a | 488 (86.2) | 468 (95.9) | 20 (4.1) | <.0001a |
| CRIS-B | 39 (12.1) | 21 (53.8) | 18 (46.2) | 78 (13.8) | 64 (82.1) | 14 (17.9) | ||
Values are n (%).
Percentage values are given in parentheses. Association of CSF1R expression with clinical parameters was analyzed using chi-square tests. CSF1R low or high status was defined according to Figures 1A and 1B, respectively.
CMS, consensus molecular subtype; COAD, colon adenocarcinoma; CRIS, colorectal cancer intrinsic subtypes; MSI, microsatellite instability; UICC, Union for International Cancer Control; TCGA, The Cancer Genome Atlas.
P < .05.
Table 2.
Multiple Regression Analysis of Overall Survival in Colon Cancer Cases From 2 Independent Cohorts
| Variable | TCGA-COAD |
GSE395823 |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age ≥median | 2.02 | 1.26–3.24 | .003 | 2.11 | 1.55–2.88 | .001 |
| Male vs female | 1.08 | 0.69–1.68 | .745 | 0.71 | 0.52–0.96 | .025 |
| UICC stage | 2.27 | 1.74–2.96 | <.0001 | 2.14 | 1.67–2.58 | <.0001 |
| MSI status | 1.01 | 0.55–1.82 | .985 | 0.82 | 0.49–1.32 | .392 |
| CSF1R high | 1.80 | 1.10–2.93 | .018a | 1.96 | 1.18–3.26 | .009a |
Cox proportional hazards models were used for multiple regression analyses.
CI, confidence interval; HR, hazard ratio; MSI, microsatellite instability; UICC, Union for International Cancer Control.
P < .05.
CSF1R Represents a Direct Target of miR-34a
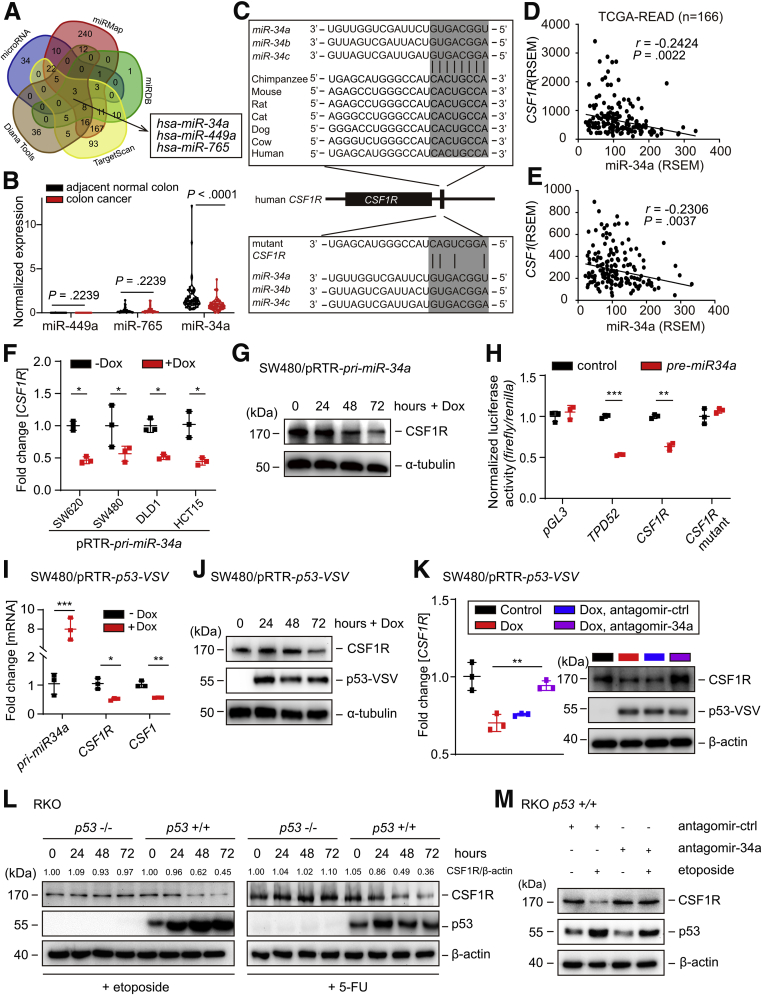
In order to determine whether the upregulation of CSF1R expression in CRCs may be due to the downregulation of microRNAs that negatively control the CSF1R mRNA, we examined the 3′-UTR of CSF1R for the presence of potential seed-matching sites. Only 3 different microRNAs were identified by all 5 algorithms used here (Figure 5A): miR-34a and miR-449a share the same, whereas miR-765 has a different seed sequence. Analysis of miRNA expression data obtained from 61 paired colon cancer and adjacent normal colon samples revealed that only miR-34a showed significant downregulation in primary CRCs when compared with normal colonic tissue, whereas miR-449a and miR-765 expression did not display significant downregulation in the primary CRCs (Figure 5B). Therefore, we decided to focus miR-34a. Notably, the miR-34a seed-matching sequence within the CSF1R 3′-UTR is highly conserved in other species (Figure 5C). In line with these observations, expression of miR-34a showed an inverse correlation with CSF1R and CSF1 in primary CRCs (Figure 5D and E). However, we did not detect a miR-34a seed-matching site in the CSF1 mRNA (data not shown). Ectopic expression of an doxycycline (Dox)-inducible pri-miR-34a allele resulted in a significant downregulation of CSF1R mRNA levels in 4 different human CRC lines (Figure 5F). Furthermore, ectopic expression of pri-miR-34a in mesenchymal-like SW480 cells, which display low expression of endogenous miR-34a, also resulted in downregulation of CSF1R protein expression (Figure 5G). In a dual-reporter assay, ectopic miR-34a significantly repressed the activity of a wild-type CSF1R 3′-UTR reporter and also repressed a TPD52 (a known miR-34a target) reporter. However, a CSF1R 3′-UTR reporter with mutations in the miR-34a seed-matching sequence was refractory to miR-34a (Figure 5H). Therefore, miR-34a directly represses CSF1R expression via a conserved miR-34a seed-matching sequence.
Figure 5.
Characterization of CSF1R as a miR-34a target. (A) Bioinformatics prediction of matching seed sequences in the CSF1R 3′-UTR using 5 different algorithms. (B) Expression of the indicated mature miRNAs in paired samples of colon cancer and adjacent normal colon (n = 61). Data is derived from the cohort GSE4826736 and was subjected to a paired t test. (C) Scheme of the miR-34a seed, the seed-matching sequences and its targeted mutation in the 3′-UTR of the CSF1R mRNA. The seed and seed-matching sequences are high-lighted in gray. Black vertical bars indicate complementarity between the miR-34a seed and the CSF1R seed-matching sequence. (D, E) Correlative analysis between miR-34a and the indicated mRNAs in the samples of the TCGA collection of rectal adenocarcinomas (READs) (n = 166) using the Pearson coefficient. (F) qPCR analysis of CSF1R expression in 3 different colorectal cancer cell lines 72 hours after induction of pri-miR-34a expression by addition of Dox. (G) Western blot analysis of CSF1R expression after induction of pri-miR-34a in SW480 cells by addition of Dox for the indicated periods. (H) Dual-reporter assay after transfection with the indicated pre-miR-34a oligonucleotides and human CSF1R 3′-UTR reporter constructs. (I) qPCR analysis of the indicated mRNAs and (J) Western blot analysis of CSF1R expression 72 hours after induction of ectopic p53 by addition of Dox to SW480/pRTR-p53-VSV cells. (K) qPCR (left) and Western blot (right) analysis of SW480/pRTR-p53-VSV cells transfected with antago-miR-34a or control oligonucleotides for 24 hours and/or subsequently treated with Dox for 48 hours. (L) Western blot analysis of CSF1R expression in RKO p53+/+ and RKO p53–/– cells after addition of etoposide (20 μM) or 5-FU (25 μg/mL) for the indicated periods. (M) Western blot analysis of CSF1R proteins in RKO p53+/+ cells transfected with antagomir-miR-34a or antagomir control oligonucleotides for 24 hours and subsequent exposure to etoposide (20 μM) for 48 hours or DMSO. In panels F, H, I, and K, mean values ± SD are provided. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
p53 Represses CSF1R via Inducing miR-34a
Because miR-34a is directly induced by p53, we determined whether p53 activation would also repress CSF1R. Indeed, ectopic expression of p53 repressed CSF1R mRNA and protein expression in SW480 cells (Figure 5I and J). In addition, p53 activation suppressed CSF1 expression (Figure 5I). Furthermore, the repression of CSF1R by p53 was alleviated by inactivation of miR-34a via treatment with miR-34a–specific antagomirs, demonstrating that miR-34a mediates the repression of CSF1R by ectopic p53 (Figure 5K). In addition, treatment with the DNA-damaging agents etoposide or 5-fluorouracil (5-FU) caused the downregulation of CSF1R protein expression in p53+/+ but not in isogenic p53–/– RKO cells (Figure 5L). Moreover, the repression of CSF1R by activation of endogenous p53 by treatment with etoposide was prevented by miR-34a–specific antagomirs (Figure 5M). Taken together, these results show that p53 activation leads to a miR-34a–mediated repression of CSF1R expression.
Coherent Feed-Forward Regulation of CSF1R by SNAIL and miR-34a
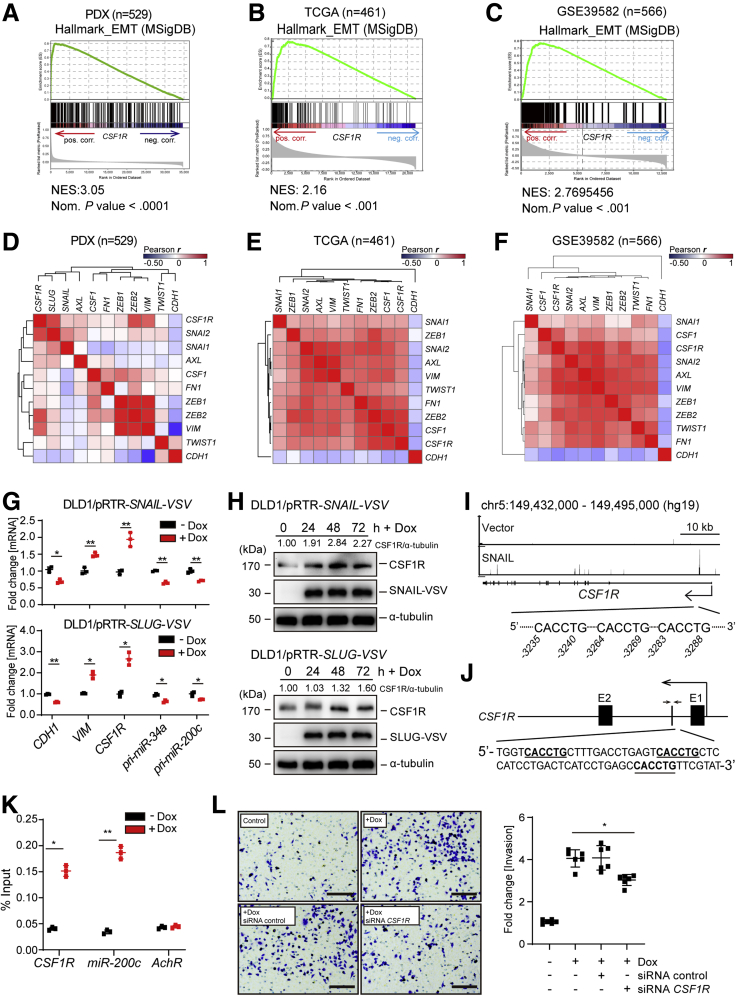
Because CSF1R expression was elevated in CRCs classified as CRIS-B subtype, which is characterized by a mesenchymal expression profile, we determined whether CSF1R expression is associated with EMT-specific gene expression profiles. GSEAs showed that CSF1R mRNA expression is strongly associated with the expression of EMT-specific signature mRNAs represented by the EMT hallmark gene set in PDX samples (Figure 6A). A similar correlation was found in the TCGA-COAD cohort and an additional cohort containing 566 CRC samples (Figure 6B and C). More specifically, CSF1R mRNA expression was positively associated with the expression of “canonical” EMT-TFs, such as SNAIL and SLUG, and mesenchymal markers such as vimentin (VIM), whereas it displayed an inverse correlation with E-cadherin (CDH1) (Figure 6D–F).
Figure 6.
Regulation by SNAIL and SLUG links CSF1R to EMT and invasion. Genes were preranked by expression correlation coefficient (Pearson r) with CSF1R in descending order from left (positive correlation) to right (negative correlation) based on RNA expression data obtained from (A) GSE76402, (B) TCGA-COAD, and (C) GSE39582 and association of hallmark EMT genes with CSF1R expression was subsequently analyzed by GSEA. (D–F) Heatmap depicting a hierarchically clustered correlation matrix of pairwise expression correlation coefficients (Pearson r) between previously described direct miR-34a target genes, EMT markers, and CSF1R mRNA. (G) qPCR analysis of the indicated mRNAs 72 hours after addition of Dox to DLD1/pRTR-SNAIL-VSV cells (top) and DLD1/pRTR-SLUG-VSV cells (bottom). (H) Western blot analysis of CSF1R expression after addition of Dox for the indicated periods in DLD1/pRTR-SNAIL-VSV cells (top) and DLD1/pRTR-SLUG-VSV cells (bottom). (I) SNAIL-VSV–derived chromatin immunoprecipitation sequencing (ChIP-Seq) results were obtained after induction of ectopic SNAIL in DLD1 cells and displayed using the UCSC genome browser. (J) Scheme of the first intron of human CSF1R. Putative SNAIL binding sites are indicated as bold letters in the DNA sequence. Small arrows indicate the amplicon used in panel K for quantitative ChIP analysis. (K) ChIP analysis of SNAIL occupancy at the first intron of CSF1R and promoter of miR-200c 24 hours after addition of Dox or cells left untreated using anti-VSV and anti-rabbit-IgG antibodies. AchR served as negative control. (L) Boyden chamber invasion assay of DLD1/pRTR-SNAIL-VSV cells after the indicated treatments. In panels G, K, and L, mean values ± SD are provided. ∗P < .05 and ∗∗P < .01. NES, normalized enrichment score.
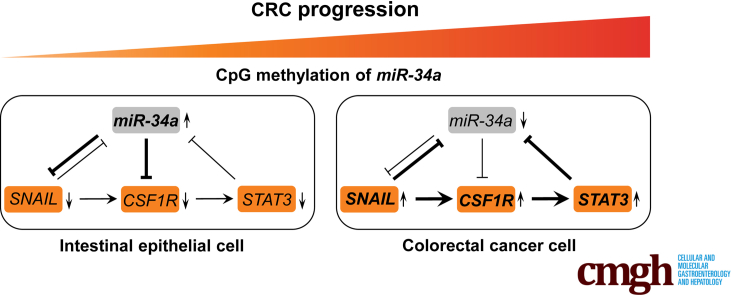
We have previously shown that the EMT-TFs SNAIL and SLUG negatively regulate miR-34a expression by directly binding to its promoter.20 Because CSF1R is repressed by miR-34a, a downregulation of miR-34a by SNAIL should presumably lead to induction of CSF1R. Therefore, we determined CSF1R expression levels after ectopic expression of SNAIL or SLUG in epithelial-like DLD1 cell pools harboring Dox-inducible expression vectors encoding either SNAIL or SLUG. Indeed, CSF1R mRNA showed robust induction concomitantly with repression of pri-miR-34a transcription after ectopic expression of SNAIL or SLUG in DLD1 cells (Figure 6G). Consistent with a previous report,20 pri-miR-200c was also repressed by SNAIL or SLUG. The upregulation of CSF1R mRNA after activation of SNAIL or SLUG was accompanied by an increase in CSF1R protein levels (Figure 6H). Although repression of miR-34a by SNAIL is presumably a critical component in the regulation of CSF1R, we asked whether direct activation by these EMT-TFs may also contribute to the induction of CSF1R expression. Indeed, we detected SNAIL occupancy in the first intron of CSF1R in a genome-wide chromatin immunoprecipitation-sequencing analysis of DLD1 cells (Figure 6I) (H. Hermeking et al, 2019, unpublished data), suggesting that CSF1R is also directly regulated by SNAIL. Accordingly, we identified a cluster of 3 closely spaced SNAIL binding sites with the sequence 5′-[CACCTG]-3′ within the first intron of the CSF1R gene (Figure 6J). By quantitative chromatin immunoprecipitation of this region, SNAIL occupancy at the first intron of the CSF1R gene was confirmed (Figure 6K). Moreover, small interfering RNA (siRNA)–mediated suppression of CSF1R in SNAIL-expressing DLD1 cells resulted in a decrease in invasion (Figure 6L). Taken together, these results demonstrate a coherent feed-forward regulation of CSF1R expression by SNAIL and miR-34a. In addition, CSF1R may be a critical downstream mediator of SNAIL-induced invasion in CRC cell lines.
Repression of miR-34a After CSF1R Activation Is Mediated by STAT3
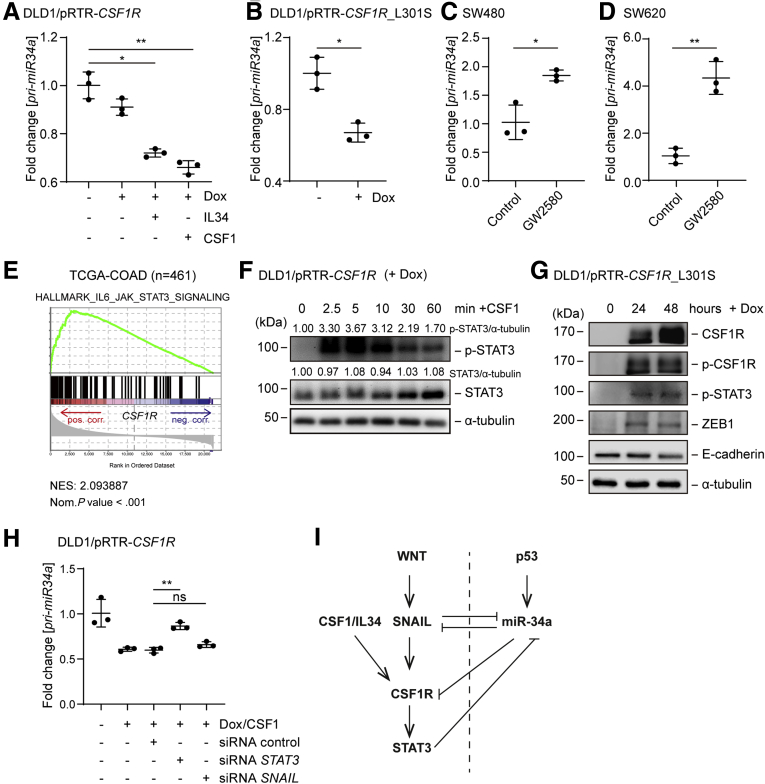
We have previously found that miR-34a often forms double-negative feedback loops with its targets.20, 21, 22, 23, 24 Also here, we observed a downregulation of pri-miR-34a expression after activation of its target CSF1R either by CSF1 or IL34 (Figure 7A). Also after ectopic expression of CSF1R_L301S, a constitutively active form of CSF1R, which was described previously,37 the expression of pri-miR-34a was downregulated in DLD1 CRC cells (Figure 7B). Furthermore, inhibition of CSF1R by the small molecule inhibitor GW258038 resulted in an upregulation of pri-miR-34a in SW480 and SW620 cells (Figure 7C and D). Because GSEAs showed a positive correlation between CSF1R expression and the IL6_JAK_STAT3 pathway hallmark gene signature (Figures 3D and 7E), we asked whether STAT3 activation may mediate the repression of miR-34a after CSF1R activation. Indeed, treatment of DLD1 cells ectopically expressing CSF1R with CSF1 resulted in increased phosphorylation of STAT3 at residue S727, which indicates STAT3 activation (Figure 7F). Also, the ectopic expression of the constitutively active CSF1R_L301S allele resulted in STAT3 phosphorylation (Figure 7G). Of note, RNAi-mediated downregulation of STAT3 significantly reversed the suppression of pri-miR-34a observed after CSF1R activation, whereas silencing of SNAIL only led to a minor de-repression (Figure 7H). Therefore, the downregulation of miR-34a by CSF1R is, at least in part, mediated by STAT3 activation. This effect is presumably mediated via a conserved STAT3-binding site in the miR-34a promoter, which we have characterized previously.22 Taken together, miR-34a, CSF1R and STAT3 therefore form a double-negative feed-back loop. In combination with the coherent feed-forward loop described above these regulatory circuitries may allow cells to integrate antagonistic mitogenic (CSF1, WNT) and antiproliferative (p53) signals (see model in Figure 7I).
Figure 7.
CSF1R activation represses miR-34a via STAT3. (A) qPCR analysis of pri-miR-34a in DLD1/pRTR-CSF1R treated with Dox for 96 hours. The last 72 hours also treated with CSF1 or IL34. (B) qPCR analysis of pri-miR-34a in DLD1/pRTR-CSF1R_L301S after addition of Dox for 48 hours. qPCR analysis of indicated mRNAs in (C) SW480 and (D) SW620 cells after treatment with GW2580 (1 nM) for 72 hours. (E) Genes were preranked by expression correlation coefficient (Pearson r) with CSF1R in descending order from left (positive correlation) to right (negative correlation) based on RNA expression data obtained from TCGA-COAD and analyzed by GSEA. (F) Western blot analysis of STAT3 phosphorylation at residue S727 and STAT3 expression after addition of Dox for 24 hours and subsequent exposure to CSF1 for indicated periods in DLD1/pRTR-CSF1R cells. (G) Western blot analysis of DLD1/pRTR-CSF1R_L301S cells after addition of Dox for indicated periods. (H) qPCR analysis of pri-miR-34a expression. DLD1/pRTR-CSF1R cells were transfected with indicated siRNAs. After 6 hours they were treated with Dox and CSF1 for 48 hours. The STAT3- and SNAIL-specific siRNAs used here have been validated previously.22 (I) Model of the regulations characterized in Figures 2 and 3. The dashed line separates p53 on (right) and p53 off states (left). In panels A, B, C, D, and H, mean values ± SD are provided. ∗P < .05 and ∗∗P < .01.
CSF1R Activation Induces EMT, Migration, and Invasion
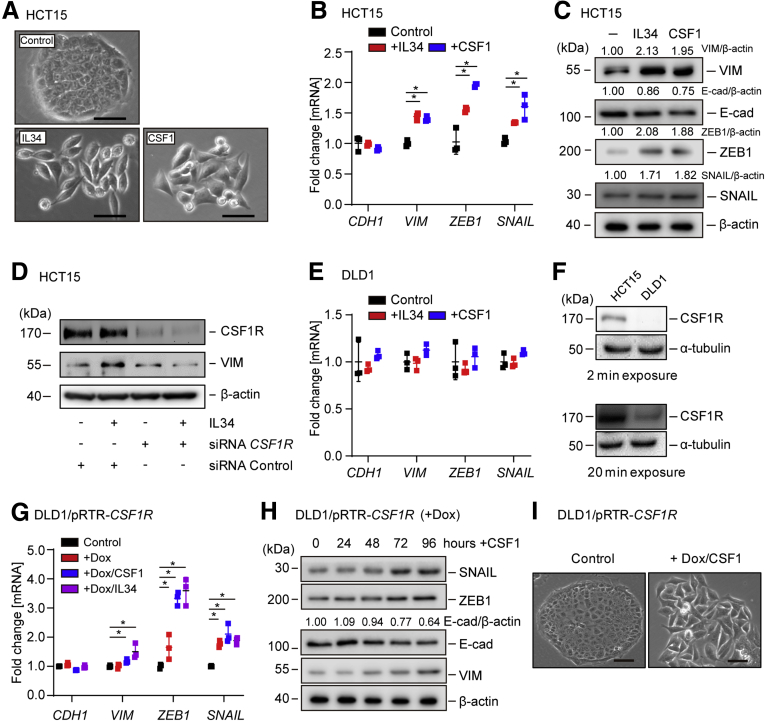
Next, we asked whether CSF1R activation is sufficient to induce EMT. Therefore, we treated the epithelial-like CRC cell line HCT15 with CSF1 or IL-34 for 72 hours. Indeed, CSF1 and IL34 induced the transition from an epithelial morphology with dense islands of cobblestone-shaped cells to a mesenchymal morphology with spindle-shaped cells forming protrusions and displaying a scattered growth pattern (Figure 8A). In addition, mesenchymal markers, such as VIM, SNAIL, and ZEB1, were induced on mRNA and protein levels, while CDH1 protein expression decreased (Figure 8B and C). Silencing of CSF1R expression by siRNAs prevented the induction of VIM by IL34 (Figure 8D), excluding the possibility that IL34-induced EMT in HCT15 cells is mediated by protein-tyrosine phosphatase ζ, which represents an alternative IL34 receptor.39 However, treatment of DLD1 CRC cells with CSF1 or IL34 did not significantly affect the expression of epithelial or mesenchymal markers (Figure 8E). This nonresponsiveness is presumably due to the relatively low expression of CSF1R protein in DLD1 cells when compared with HCT15 cells (Figure 8F). Indeed, ectopic CSF1R expression restored the responsiveness of DLD1 cells to CSF1, as CSF1 induced the hallmarks of EMT in these cells (Figure 8G–I). Taken together, these results demonstrate that CSF1R activation induces EMT in CRC cells.
Figure 8.
Activation of CSF1R induces EMT in CRC cells. (A) Representative phase-contrast pictures of HCT15 cells after treatment with IL34 or CSF1 for 72 hours. Scale bar = 25 μm. (B) qPCR analysis of the indicated EMT markers after treatment of HCT15 cells with IL34 or CSF1 for 48 hours. (C) Western blot analysis of indicated EMT markers after treatment of HCT15 cells with IL34 or CSF1 for 72 hours. (D) Western blot analysis of HCT15 transfected with siRNA CSF1R or siRNA Control oligonucleotide for 24 hours and/or subsequently treated with IL34 for 72 hours. (E) qPCR analysis of DLD1 cells after treatment with CSF1 or IL34 for 48 hours. (F) Western blot analysis of CSF1R expression in DLD1 and HCT15 cells. (G) qPCR analysis of DLD1/pRTR-CSF1R cells that were treated with Dox for 24 hours and then exposed to CSF1 or IL34 for another 48 hours. (H) Western blot analysis of DLD1/pRTR-CSF1R cells treated with Dox for 24 hours and subsequently exposed to CSF1 for the indicated periods. (I) Representative phase-contrast pictures of DLD1/pRTR-CSF1R cells after treatment with Dox for 24 hours, and then exposed to CSF1 for 72 hours. Scale bar = 25 μm. In panels B and G, mean values ± SD are provided. ∗P < .05 and ∗∗P < .01.
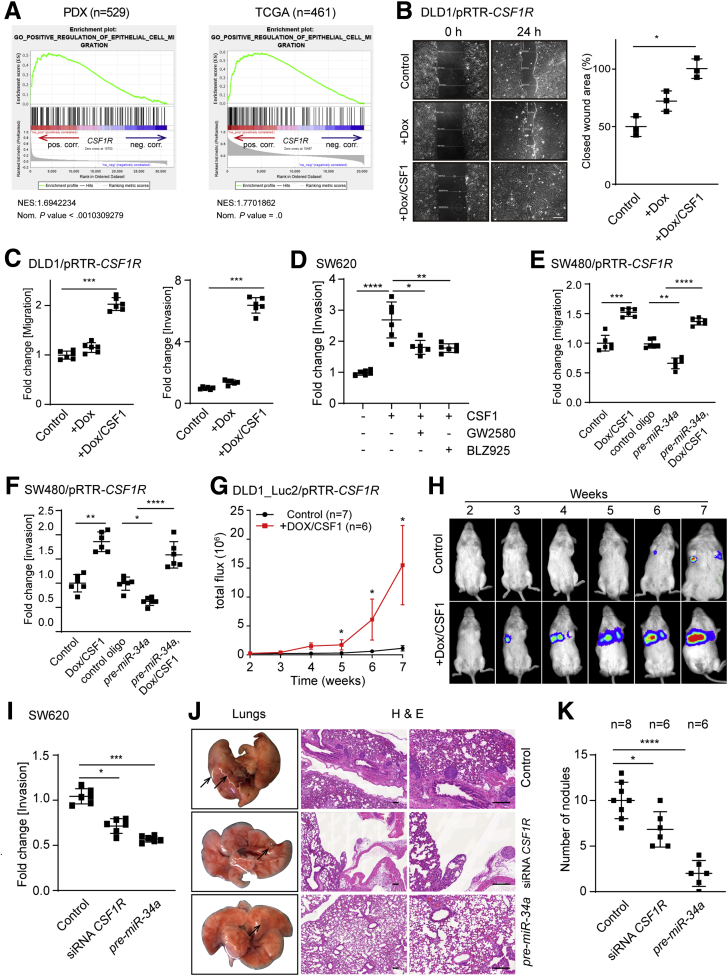
Expression of CSF1R was associated with epithelial cell migration by GSEA (Figure 9A). Therefore, we asked whether activation of CSF1R enhances cell migration, invasion, and eventually metastases formation, as these processes are functional consequences of an EMT. Indeed, activation of CSF1R accelerated the closure of a scratch in CSF1R-expressing DLD1 cells (Figure 9B). In addition, migration and invasion were enhanced after CSF1R activation as determined in a Boyden chamber assay, whereas treatment with the CSF1R inhibitors GW2580 or BLZ925 resulted in a significant decrease of cellular invasion in the mesenchymal-like cell line SW620 (Figure 9C and D). Furthermore, ectopic expression of a miR-34a–resistant CSF1R complementary DNA (cDNA) in the mesenchymal-like cell line SW480 prevented the repression of migration and invasion by pre-miR-34a (Figure 9E and F). Therefore, the repression of CSF1R by miR-34a is presumably required for inhibition of migration and invasion by miR-34a.
Figure 9.
Activation of CSF1R in CRC cells induces migration, invasion, and lung metastases. (A) Genes were preranked by expression correlation coefficient (Pearson r) with CSF1R in descending order from left (positive correlation) to right (negative correlation) based on RNA expression data and association of indicated gene signature with CSF1R expression was subsequently analyzed by GSEA. (B) Scratch assay of DLD1-pRTR-CSF1R cells treated with Dox or Dox/CSF1. Scale bar = 200 μm. (C) Boyden chamber assays of cellular migration (left) or invasion (invasion). (D) SW620 cells were pretreated with inhibitors as indicated, and subsequently treated with Dox and CSF1. After incubation with CSF1 or 48 hours, cells were subjected to a Boyden chamber assay. Cells were transfected with or without pre-miR-34a oligo 1 day before the addition of Dox and CSF1, and then subjected to a (E) migration or (F) invasion assay. (G) DLD1-Luc2/pRTR-CSF1R cells treated with or without Dox and CSF1 were injected into the tail vein of NOD/SCID mice. At the indicated time points, bioluminescence signals were recorded. Bioluminescence signals are presented as “total flux.” (H) Representative examples of bioluminescence imaging at the indicated time points after tail vein injection of DLD1-Luc2/pRTR-CSF1R cells. (I) SW620 cells were transfected with the indicated oligonucleotides for 48 hours and then subjected to an invasion assay in Boyden chambers for another 36 hours. (J) SW620 cells were transfected with the indicated oligonucleotides for 48 hours and subsequently injected into the tail vein of NOD/SCID mice. Left: lungs were resected 8 weeks after injection. Arrows indicate metastatic tumor nodules. Right: representative examples of the hematoxylin and eosin staining of resected lungs are shown. Scale bar = 200 μm. (K) Quantification of metastatic tumor nodules in the lung per mouse 8 weeks after tail-vein injection. In panels B, C, D, E, F, G, I, and K, mean values ± SD are provided. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Next, DLD1 cells harboring a luciferase marker gene and an inducible CSF1R allele were injected into mice to assess the effect of CSF1R activation on lung metastases formation. Indeed, only cells with activated CSF1R formed lung metastases in mice as evidenced by a significant increase in luciferase signal by week 5, which further increased until week 7 (Figure 9G and H). In SW620 CRC cells downregulation of CSF1R expression by transfection with CSF1R-specific siRNAs or pre-miR-34a inhibited invasion as determined in a Boyden chamber assay (Figure 9I). When SW620 cells treated similarly were injected into the tail veins of mice, a reduced number of metastatic tumor nodules were detected in the lungs 8 weeks later (Figure 9J and K). The stronger inhibitory effect of pre-miR-34a oligonucleotides, as compared with CSF1R-specific siRNAs, can be explained by the inhibition of other miR-34a targets, which promote the formation of metastases, such as the IL6R.22 Taken together, these results show that CSF1R activation is sufficient and necessary for invasion and metastases formation of CRC cells.
CSF1R Mediates Resistance to 5-FU in CRC Cells
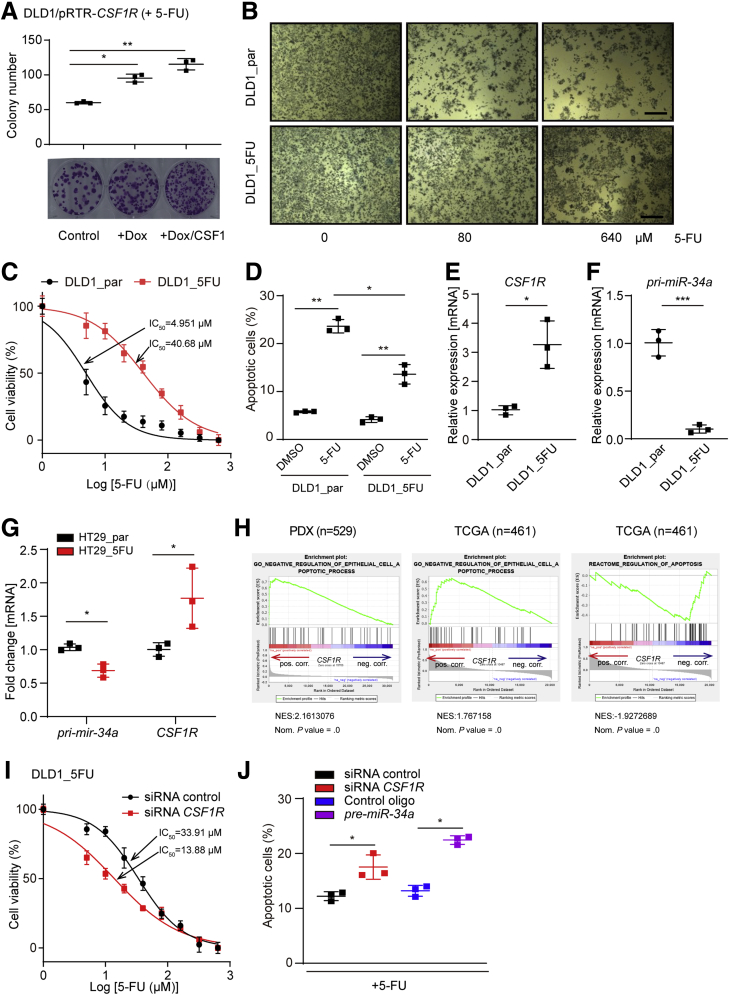
Because CSF1R activation induced EMT, which has been linked to chemoresistance,40 we determined whether CSF1R activity or expression influences the sensitivity of CRC cells to the chemo-therapeutic agent 5-FU, which is commonly used in CRC therapy. DLD1 cells ectopically expressing CSF1R were treated with CSF1 for 24 hours and subsequently exposed to 5-FU for 3 days. Cells expressing ectopic CSF1R formed more colonies and were therefore less sensitive to 5-FU when compared with control cells (Figure 10A). The addition of CSF1 further increased the number of colonies formed by cells ectopically expressing CSF1R. Next, we established a 5-FU-resistant cell pool (DLD1_5FU) by exposing DLD1 cells to increasing concentrations of 5-FU over a period of 5 months. The tolerance of DLD1_5FU cells to 5-FU was significantly higher than that of parental DLD1 cells (DLD1_par) (Figure 10B and C). The IC50 value of 5-FU for DLD1_5FU cells was increased 8-fold when compared with the parental cells (40.68 μM vs 4.951 μM; P < .01). Accordingly, DLD1_5FU cells exposed to 5-FU underwent less apoptosis than DLD1_par cells (Figure 10D). Interestingly, CSF1R expression was upregulated concomitantly with downregulation of miR-34a in DLD1_5FU cells when compared with DLD1_par cells (Figure 10E and F). Similar results were obtained with HT29 cells, that were rendered resistant to 5-FU as described previously for DLD1 cells (Figure 10G). Furthermore, GSEA showed that increased CSF1R is negatively associated with apoptosis related gene expression (Figure 10H). In addition, downregulation of CSF1R in DLD1_FU cells by specific siRNA pools resulted in decreased cell viability after 5-FU treatment (Figure 10I) and was accompanied by an increase in apoptosis (Figure 10J). Interestingly, ectopic expression of pre-miR-34a further enhanced apoptosis, indicating that miR-34a may target additional suppressors of apoptosis besides CSF1R in this context. Taken together, downregulation of miR-34a and elevated expression of CSF1R is selected for during treatment with 5-FU and confers resistance of CRC cells to 5-FU.
Figure 10.
Interdependent deregulation of miR-34a and CSF1R mediates resistance to 5-FU. (A) For a colony-formation assay 500 cells were seeded per well of a 6-well plate and cultivated with or without Dox for 24 hours, then exposed to CSF1 for another 24 hours, and then treated with 5-FU for 72 hours. Subsequently, cells were fixed and stained with crystal violet. Quantification of colony formation (upper panel) and representative examples of crystal violet staining (lower panel). (B) The indicated cell pools were treated with 5-FU for 48 hours and subsequently subjected to an MTT assay. Micrographs show cell pools with formation of MTT formazan, which is directly proportional to the number of living cells Scale bars = 200 μm. (C) IC50 determination of DLD1_par and DLD1_5FU cells in response to 5-FU. Cells were treated with the indicated concentrations of 5-FU for 48 hours and then subjected to an MTT assay. (D) Detection of apoptotic cells by Annexin V-FITC and PI staining after treatment with 5-FU for 36 hours. qPCR analysis of (E) CSF1R and (F) pri-miR34a expression in DLD1_par and DLD1_5FU cells. (G) Detection of pri-miR-34a and CSF1R expression in HT29_par and HT29_5FU cells. (H) Genes were preranked by expression correlation coefficient (Pearson r) with CSF1R in descending order from left (positive correlation) to right (negative correlation) based on RNA expression data, and association of the indicated gene signatures with CSF1R expression was analyzed by GSEA. (I) DLD1_5FU cells were transfected with control or CSF1R-specific siRNAs for 24 hours and subsequently treated with increasing concentrations of 5-FU for 48 hours. Then the IC50 was determined by an MTT assay. (J) DLD1_5FU cells were transfected with indicated oligonucleotides, and subsequently treated with 5-FU for 36 hours, and apoptotic cells were detected by Annexin V-FITC and PI staining. In panels A, D, E, F, G, and J, mean values ± SD are provided. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
CSF1R Mediates EMT, Migration, and Invasion of 5-FU in CRC Cells
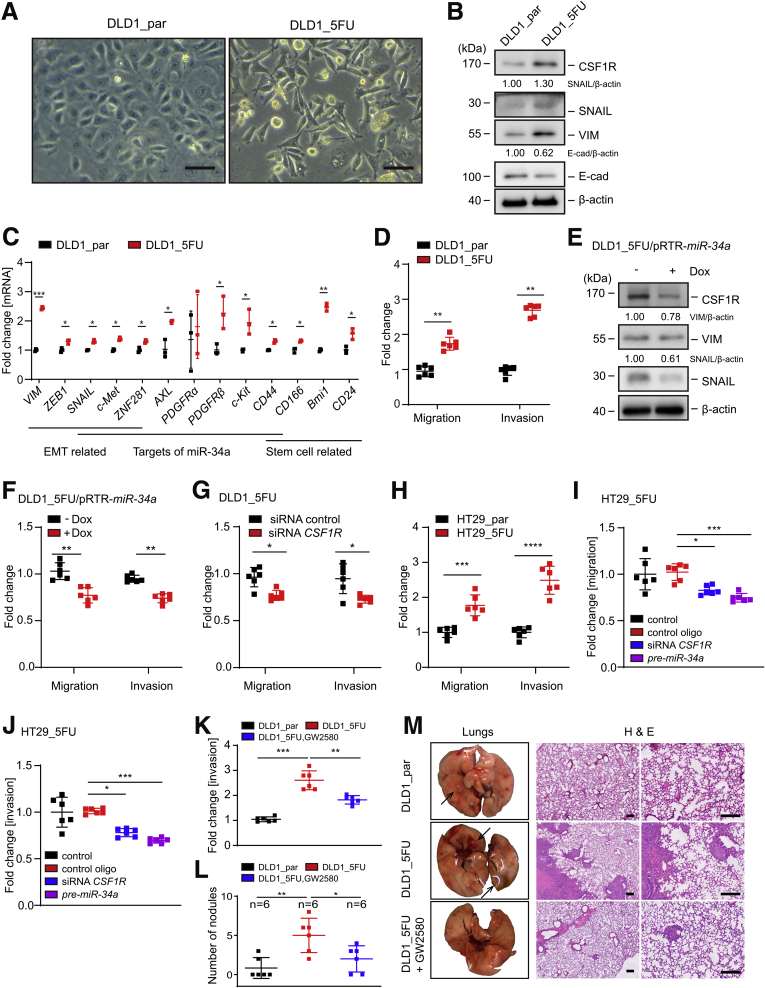
Unlike parental DLD1 cells, DLD1_5FU cells displayed a mesenchymal-like morphology (Figure 11A). Additionally, VIM, SNAIL, and ZEB1 were upregulated at the mRNA and protein levels in DLD1_5FU cells when compared with the parental DLD1 cells (Figure 11B and C). On the contrary, E-cadherin protein expression was decreased in DLD1_5FU cells. In addition to CSF1R other target mRNAs of miR-34a, such as AXL, PDGFR, c-Met, c-Kit, ZNF281, and CD44 were upregulated in DLD1_5FU cells. Consistent with the increased stemness known to be associated with EMT,41 the stemness markers CD44, CD166, BMI1, and CD24 were upregulated in DLD1_5FU cells. In line with a passage through an EMT, migration and invasion were significantly elevated in DLD1_5FU cells when compared with DLD1_par cells (Figure 11D). Furthermore, ectopic expression of pri-miR-34a reduced VIM and SNAIL expression, and significantly inhibited migration and invasion in DLD1_5FU cells (Figure 11E and F). Notably, downregulation of CSF1R expression by specific siRNAs inhibited migration and invasion in DLD1_5FU cells to a similar extent as ectopic pri-miR-34a expression (Figure 11G). Also, HT29_5FU cells displayed increased migration and invasion, which was repressed by CSF1R-specific siRNA or pre-miR-34a (Figure 11H–J). Therefore, the enhancement of migration and invasion in 5-FU-resistant CRC cells is mediated, at least in part, by downregulation of miR-34a expression and the resulting upregulation of CSF1R expression.
Figure 11.
miR-34a/CSFR1 deregulation is necessary for mesenchymal characteristics and functional properties of 5-FU-resistant CRC cells. (A) Representative phase-contrast pictures of DLD1_par and DLD1_5FU cells. Scale bars represent 25 μm. (B) Western blot analysis of indicated proteins. (C) qPCR analysis of indicated mRNAs. (D) Analysis of relative invasion and migration using Boyden chamber assays. (E) Western blot analysis of indicated proteins in DLD1_5FU/pRTR-miR-34a cells after treatment with or without Dox for 72 hours. (F) Relative invasion and migration of DLD1_5FU/pRTR-miR-34a cells after treatment with or without Dox for 72 hours. (G) DLD1_5FU cells were transfected with control or CSF1R-specific siRNAs for 24 hours and then subjected to migration and invasion assays for another 36 hours. Subsequently, cells were fixed and stained with crystal violet. (H) Relative invasion and migration of HT29_par and HT29_5FU. (I, J) DLD1_5FU cells were transfected with siRNA or pre-miR-34a oligo for 24 hours and then subjected to migration and invasion assays for another 36 hours. (K) Cells were treated with or without inhibitor GW2580 for 48 hours and then subjected to an invasion assay in Boyden chambers containing Matrigel for another 36 hours. (L) Quantification of metastatic tumor nodules in the lung per mouse 8 weeks after tail-vein injection. Cells were treated as indicated for 48 hours and subsequently injected into the tail vein of NOD/SCID mice. (M) Left: representative lungs resected 8 weeks after injection are shown. Arrows indicate metastatic tumor nodules. Right: representative examples of the hematoxylin and eosin staining of the resected lungs are shown. Scale bar = 200 μm. In panels C, D, and F–L, mean values ± SD are provided. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Treatment of DLD1_5FU cells with GW2580, a specific CSF1R inhibitor, suppressed invasion to a large extent as evidenced by a Boyden chamber assay (Figure 11K). Eight weeks after injection of DLD1_5FU cells into the tail vein of NOD/SCID mice their lungs displayed an increased number of metastases when compared with mice injected with DLD1_par cells (Figure 11L and M). Pretreatment of DLD1_5FU cells with the CSF1R-inhibitor GW2580 before injection suppressed metastasis formation in NOD/SCID mice. Taken together, these results show that elevated CSF1R expression promotes metastases formation of chemoresistant CRC cells.
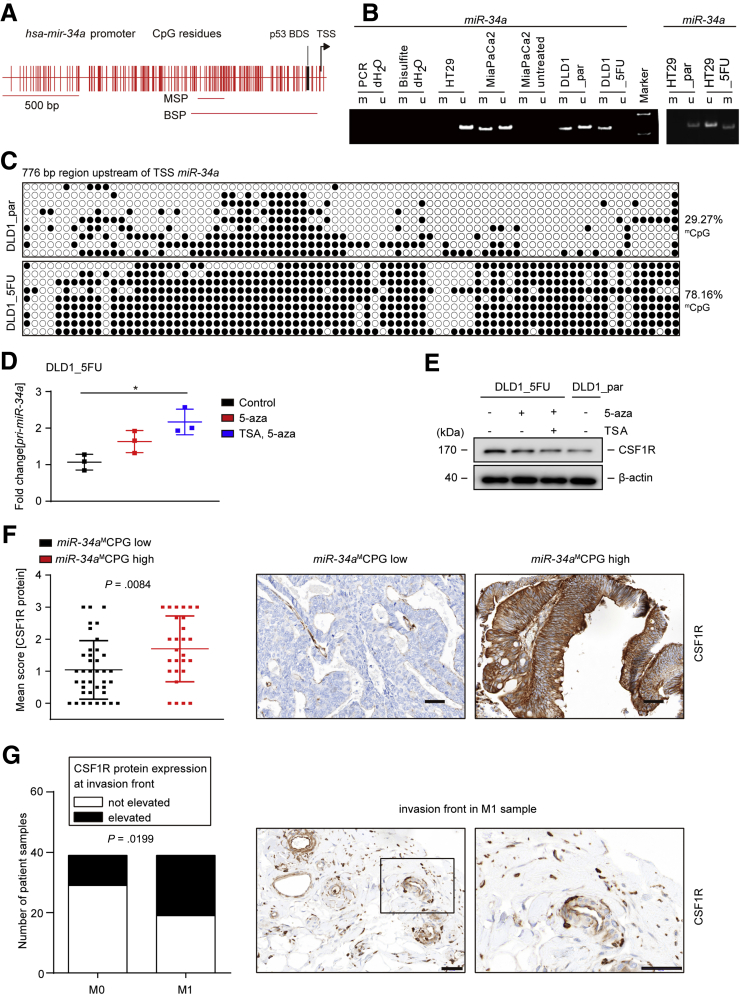
Epigenetic Silencing of miR-34a Contributes to CSF1R Upregulation, 5-FU Resistance, and CRC Progression
We have previously shown that methylation of the CpG island upstream of the miR-34a transcriptional start site results in silencing of miR-34a expression.25 Therefore, we analyzed whether the downregulation of miR-34a expression observed in DLD1_5FU cells is due to methylation of the miR-34a promoter. Whereas DLD1_par cells harbored both methylated and nonmethylated miR-34a alleles as detected by methylation-specific polymerase chain reaction (MSP), DLD1_5FU cells only displayed methylated miR-34a promoter alleles (Figure 12A and B). In HT29_par cells only nonmethylated miR-34a was detected, whereas HT29_5FU cells also showed methylated besides nonmethylated miR-34a alleles. As reported previously, MiaPaCa2 pancreatic cancer cells displayed methylated and unmethylated miR-34a alleles.25 In addition, we performed a bisulfite sequencing analysis of the miR-34a promoter region as described before.25 Overall methylation of the miR-34a promoter was significantly higher in DLD1_5FU than in DLD1_par cells (P < .0001 Figure 12C). Furthermore, DLD1_5FU cells were treated with 5-aza-2’deoxycytidine (5-aza) or trichostatin A (TSA), which are inhibitors of DNA methyltransferases and histone deacetylases, respectively, in order to reactivate the expression of miR-34a silenced by CpG methylation. Pri-miR-34a was re-expressed after treatment of DLD1_5FU cells with 5-aza and further increased by the combined treatment with 5-aza and TSA (Figure 12D). On the contrary, CSF1R expression was downregulated after treatment with 5-aza or the combination of 5-aza and TSA (Figure 12E). Therefore, hypermethylation of the miR-34a promoter decreased the expression of miR-34a and thereby presumably caused the upregulation of CSF1R expression in 5-FU resistant cells.
Figure 12.
Elevated CSF1R expression is associated with miR-34a promoter methylation and the invasion front of metastatic primary CRCs. (A) Genomic region 2.0 kbp upstream of the transcriptional start site (TSS) (position indicated by arrow) within the human miR-34a gene. Vertical bars represent CpG dinucleotides. The position of the p53 binding site (BDS) is indicated. The horizontal bars indicate PCR amplicons used for MSP and bisulfite-sequencing PCR (BSP), respectively. (B) Representative results of MSP analysis. (C) Bisulfite sequencing analysis of the miR-34a promoter in DLD1_par and DLD1_5FU cells. 9 subcloned amplification products were sequenced for each cell lines. Each horizontal line represents 1 individual clone, and each circle 1 single CpG dinucleotide. Open circles represent nonmethylated and black circles methylated CpGs. (D) qPCR analysis of pri-miR-34a in DLD1_5FU cells after treatment with 5-aza for 72 hours or alternatively with 5-aza for 72 hours combined with TSA for the last 24 hours. (E) Western blot analysis of cell lysates isolated from DLD1_5FU cells after treatment with 5-aza for 72 hours or alternatively with 5-aza for 72 hours combined with TSA for the last 24 hours. (F) Left: quantification of CSF1R protein expression in human CRC samples of 78 patients. The methylation status of miR-34a in these samples had been determined previously.65 Right: representative immunohistochemical detections of CSF1R protein in miR-34amCPG low and high tumors, respectively. Scale bar = 50 μm. (G) Evaluation of CSF1R protein expression at the invasion front in M0 and M1 CRCs (left chart) and examples of representative immunohistochemical detections (right panel). The presence of an invasion front was confirmed by DH, a certified pathologist. Results were analyzed using the chi-square test. Scale bar = 50 μm. In panels D, F, and G, mean values ± SD are provided. ∗P < .05. bisulfite dH2O, no DNA input in bisulfite reaction; HT29, negative control; M, methylation-specific PCR product; MiaPaCa2, positive control; PCR dH2O, no DNA in PCR; U, unmethylated allele spec. PCR-product; untreated, no bisulfite added;
Next, we determined whether the inverse correlation between miR-34a CpG-methylation and CSF1R expression is also present in primary CRCs. Therefore, the expression of CSF1R protein was analyzed by immunohistochemistry in 90 CRC samples, for which the methylation status of miR-34a had been determined previously.27 Notably, in CRCs with high miR-34a CPG methylation the expression of CSF1R protein was significantly higher than in CRCs with decreased miR-34a CpG methylation (Figure 12F). Furthermore, CSF1R expression was elevated at the infiltrative tumor edge of primary CRCs that were accompanied by liver metastases (M1 tumors) when compared with primary CRCs without liver metastases (M0) (Figure 12G). Therefore, the inverse correlation between miR-34a CpG methylation and CSF1R expression was also found in primary CRCs. Furthermore, increased expression of CSF1R at the invasion front of primary CRCs was associated with distant metastasis.
Discussion
Our results suggest that the reciprocal regulation between miR-34a and CSF1R controls EMT and chemosensitivity. The deregulation of this feedback loop during CRC progression may contribute to metastasis and chemoresistance (see also the graphical abstract). Because CSF1R is expressed at elevated levels in several types of tumors,42, 43, 44 the regulations identified here may also be relevant to other entities. Not only CSF1R, but also its ligands CSF1 and IL34 are expressed at elevated levels in CRCs.17,45,46 In the human colon, expression of CSF1 is significantly higher than that of IL34, suggesting that CSF1 is the main ligand for activation of CSF1R in CRC.47 Here, analysis of TCGA datasets and 2 additional cohorts of CRC patients showed that elevated mRNA levels of CSF1R, CSF1, and IL34 are associated with poor survival of CRC patients. The analysis of PDXs and single-cell sequencing data revealed tumor cell intrinsic expression of CSF1R. We determined that miR-34a directly targets CSF1R mRNA and thereby mediates the repression of CSF1R by p53. This is in line with a previous study that showed that a miR-34a mimic downregulates csf1r mRNA expression in rats.48 However, the authors did not provide evidence for a direct regulation nor did they study the miR-34a/CSF1R connection further. Because CSF1R represents a direct target of miR-34a, the elevated expression of CSF1R in CRCs may result from the epigenetic silencing of miR-34a, which frequently occurs in CRC.24, 25, 26, 27 Interestingly, ectopic expression of p53 not only repressed CSF1R via miR-34a, but also its ligand CSF1. The latter effect may be due to the induction of the microRNAs miR-148b and miR-1207 by p53, because both microRNAs are directly induced by p53 and target CSF1 mRNA.46,49 Interestingly, a recent study showed that p53 deletion results in secretion of CSF1 in a pancreatic tumor model and was suggested to influence stromal cells, such as tumor-associated macrophages.50 Our results suggest that the increased CSF1 secretion resulting from p53 inactivation or mutation may cooperate with increased CSF1R expression in a tumor cell autonomous manner.
Here, we show that CSF1R is directly and indirectly induced by SNAIL in a coherent feed-forward loop, which involves the downregulation of its repressor miR-34a by SNAIL (see also scheme in Figure 7I). The regulatory circuit characterized here also involves STAT3, which is activated by CSF1R and itself represses miR-34a. We have previously reported, that miR-34a is repressed directly by STAT3, which contributes to IL6-induced EMT and invasion in CRCs and colitis-associated colon cancer.22 Besides mediating SNAIL-induced invasion, activation of CSF1R by CSF1 or IL34 induced EMT in CRC cell lines, which was associated with increased migration, invasion and lung metastases formation in a xenograft mouse model. The induction of EMT by CSF1R presumably establishes a mesenchymal state in primary CRCs which allows invasion, intravasation, and extravasation during metastatic spread. Interestingly, CSF2/GM-CSF has recently been shown to induce EMT in colon cancer cells and may thereby contribute to CRC progression as well.51
5-FU–based chemotherapy represents the most common chemotherapeutic regime for CRC patients with metastatic tumors.52 However, long-term use of 5-FU usually results in drug resistance, which is a major cause of therapeutic failure.53 Here, we describe the establishment of 5-FU-resistant CRC cell lines that display increased mesenchymal characteristics when compared with the parental cell lines. We found that downregulation of miR-34a and increased expression of CSF1R critically contribute to 5-FU resistance. Additional targets of miR-34a, such as AXL, PDGFR, c-Met, c-Kit, ZNF281, and CD44, were also upregulated in chemoresistant DLD1 cells, suggesting that the acquisition of chemoresistance may involve several additional factors and signaling pathways. Re-expression of miR-34a or silencing of CSF1R in DLD1_5FU and HT29_5FU cells restored the sensitivity to 5-FU, indicating the importance of the dysregulation of miR-34a and CSF1R in 5-FU resistance. Therefore, inhibiting CSF1R in combination with restoring miR-34a function may have therapeutic potential for the treatment of CRC. We have previously characterized the RTK c-Kit as a miR-34a target and found that its downregulation sensitizes CRC cells to 5-FU.54 In addition, other RTKs, such as AXL and PDGFR, have been characterized as miR-34a targets.55, 56, 57 Therefore, the repression of RTKs may represent an important mechanism of tumor suppression by miR-34a.
During the establishment of 5-FU-resistant CRC cells, miR-34a expression was downregulated as a consequence of CpG methylation of its promoter. This event and the resulting upregulation of CSF1R expression critically contributed to resistance toward 5-FU. We have previously shown that the silencing of miR-34a in primary tumors is associated with metastasis in CRC patients and in combination with the elevated expression of c-Met and β-Catenin predicts a poor outcome.27 Here, CSF1R was expressed at elevated levels at the invasion front of primary CRCs. Therefore, upregulation of CSF1R expression due to miR-34a silencing may promote CRC progression and result in decreased survival of CRC patients. The results presented here suggest that targeting the miR-34a/CSF1R pathway might be a feasible approach to inhibit CRC metastasis and overcome resistance to 5-FU-based therapy. Taken together, targeting CSF1R may not only affect the tumor microenvironment and boost immune cells targeting the tumor,15 but may also directly inhibit tumor initiation and progression via the mechanisms described here.
Materials and Methods
Cell Culture and Treatments
The CRC cell lines HCT15, RKO, and DLD1 were maintained in McCoy’s 5A Medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen), SW480 and SW620 cell lines were maintained in Dulbecco’s modified Eagle medium (Invitrogen) containing 10% fetal bovine serum. p53–/– and p53+/+ RKO cell lines were kindly provided by Bert Vogelstein (Johns Hopkins University, Baltimore, MD). All cells were cultivated in presence of 100 units/mL penicillin and 0.1 mg/mL streptomycin at 20% O2, 5% CO2, and 37°C. Dox (Sigma-Aldrich, St. Louis, MO) was dissolved in water (100-μg/mL stock solution) and always used at a final concentration of 100 ng/mL. Recombinant human CSF1 (BioLegend, San Diego, CA) was dissolved in water and used at a final concentration of 50 ng/mL with daily refreshment. Pre-miRNA mimics (PM11030; Ambion, Austin, TX), miRNA antagomirs, and respective negative controls (Ambion-Applied Biosystems, Foster City, CA) were transfected using HiPerfect (Qiagen, Hilden, Germany). siRNAs (Ambion silencer siRNA: negative control [ID#4611], STAT3 [ID#6880], and Dharmacon: siRNA CSF1R [SMART pool]) were transfected at a final concentration of 20 nM using lipofectamine 2000 (Invitrogen). The sequences of pre-miR-34a and the miR-34a antagomir are listed in Table 3.
Table 3.
Oligonucleotides (pre-miR-34a, Antagomir miR-34a)
| Product | Sequence (5′-3′) | Company |
|---|---|---|
| Pre-miR-34a | GGCCAGCUGUGAGUGUUUCUUUGGCAGUGUCUUAGCUGGUUGUUGUGAGCAAUAGUAAGGAAGCAAUCAGCAAGUAUACUGCCCUAGAAGUGCUGCACGUUGUGGGGCCC | Thermo Fisher Scientific |
| Antagomir miR-34a | UUGCCAGGCAGUGUAGUUAGCUGAUUGACGAGGCAACAGUCACUAACAACACGGCCAGGUGA | Thermo Fisher Scientific |
Modified Boyden-Chamber Assay
Migration and invasion analyses were performed as described previously.22 In brief, cells were serum-starved for 24 hours. For the migration assay, 5 × 104 cells were seeded in the upper chamber (8.0-μm pore size membrane; Corning, Corning, NY) in serum-free medium. For invasion assays, chamber membranes were first coated with Matrigel (BD Biosciences, East Rutherford, NJ) at a dilution of 3.3 ng/mL in medium without serum. Then, 5 × 105 cells were seeded on the Matrigel (Corning) in the upper chamber in serum-free medium. As chemoattractant 10% fetal calf serum was placed in the lower chamber. After cells were cultured for 36 hours, nonmotile cells at the top of the filter were removed and the cells in the bottom chamber were fixed with methanol and stained with crystal violet. Relative invasion or migration was normalized to the corresponding control.
Western Blot Analysis
Sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis and Western blot analyses were performed as described previously.23 Cells were lysed in RIPA lysis buffer (50-mM Tris/HCl, pH 8.0, 250-mM NaCl, 1% NP40, 0.5% [w/v] sodium deoxycholate, 0.1% SDS, complete mini protease inhibitors [Roche, Basel, Switzerland] and PhosSTOP Phosphatase Inhibitor Cocktail Tablets [Roche]). Lysates were sonicated and centrifuged at 16.060 g for 20 min at 4°C. A total of 30- to 80 μg protein were separated on 7.5%, 10%, or 12% SDS-acrylamide gels. Gel electrophoresis and transfer to Immobilon PVDF membranes (Millipore, Burlington, MA) was carried out using standard protocols (Bio-Rad Laboratories, Hercules, CA). Primary antibodies were used in combination with horseradish peroxidase–coupled secondary antibodies. ECL (Millipore) signals were recorded with a 440-CF imaging system (Kodak, Rochester, NY). Antibodies used here are listed in Table 4.
Table 4.
List of Antibodies
| Epitope | Species | Catalog No. | Company | Use | Dilution | Source |
|---|---|---|---|---|---|---|
| Primary antibodies | ||||||
| α-tubulin | human | # T-9026 | Sigma-Aldrich | WB | 1:1000 | mouse |
| β-actin | human | # A2066 | Sigma-Aldrich | WB | 1:1000 | rabbit |
| p53 | human | # sc-126 | Santa Cruz | WB | 1:1000 | mouse |
| E-cadherin | human | # 334000 | Invitrogen | WB | 1:1000 | mouse |
| CSF1R | human | # HPA012323 | Sigma-Aldrich | WB | 1:1000 | rabbit |
| Vimentin | human | # 2707-1 | Epitomics | WB | 1:1000 | rabbit |
| SNAIL | human | # 3879S | Cell Signaling | WB | 1:500 | rabbit |
| ZEB1 | human | # sc-25388 | Santa Cruz | WB | 1:1000 | rabbit |
| STAT3pS727 | human | # 9134 | Cell Signaling | WB | 1:1000 | rabbit |
| STAT3 | human | # sc-482 | Santa Cruz | WB | 1:1000 | rabbit |
| VSV | human | # V4888 | Sigma-Aldrich | WB; ChIP | 1:1000 | rabbit |
| CSF1R | human | # sc-692 | Santa Cruz | WB | 1:500 | rabbit |
| CSF1R | human | # ab183316 | Abcam | IHC | 1:100 | rabbit |
| Secondary antibodies | |||||
|---|---|---|---|---|---|
| Name | Ordering No. | Company | Use | Dilution | Source |
| anti-mouse HRP | # W4021 | Promega | WB | 1:10.000 | goat |
| anti-rabbit HRP | # A0545 | Sigma | WB | 1:10.000 | goat |
ChIP, chromatin immunoprecipitation; IHC, immunohistochemistry; WB, Western blot analysis.
Colony Formation Assay
For low-density, colony-formation assays, 500 cells were seeded into a 6-well plate and cultivated for 24 hours in the presence or absence of Dox or CSF1 for 24 hours, and subsequently treated with or without 5-FU for 72 hours. Cells were washed once with Hank’s Balanced Salt Solution, new medium was added and cells were allowed to recover for 2 days before fixation and crystal violet staining.
“Wound Healing” Assay
Mitomycin C (10 ng/mL) was added 2 hours before generating a scratch using a Culture-Insert (80241; IBIDI, Martinsried, Germany). Cells were allowed to close the “wound” for the indicated periods and images were captured on an Axiovert Observer Z.1 microscope connected to an AxioCam MRm camera using the Axiovision software (Axiovs 40 Version 4.8.0.0, Zeiss, Oberkochen, Germany) at the respective time points.
Detection of Apoptosis
Apoptosis rates were determined by flow cytometry after staining with Annexin V-FITC (apoptotic cell marker) and PI (necrotic cell marker) according to the Annexin V-FITC/PI staining kit (556570; BD Pharmingen, San Diego, CA). In brief, treated and control cells were harvested by addition of trypsin (without EDTA) and washed twice with Hank’s Balanced Salt Solution. Then cells were resuspended in 1× binding buffer (0.01-M HEPES/NaOH [pH 7.4], 0.14-M NaCl, 2.5-mM CaCl2) at a concentration of 1 × 106 cells/mL. A total of 100 μL of the solution (1 × 105 cells) was incubated with 5 μL of FITC Annexin V and 5 μL propidium iodide. Cells were gently agitated and incubated for 15 minutes at room temperature in the dark. Then 400 μL of the one binding buffer was added to each tube and the samples were analyzed within 1 hour by flow cytometry (CFlow6; Accuri, Ann Arbor, MI).
MTT Assay
Cell viability was measured with a modified MTT assay.58 In brief, CRC cells were seeded in 96-well plates and treated with different doses of 5-FU for 48 hours, and MTT was added at concentration of 0.5 μg/μL 4 hours before addition of formazan solvents (10% SDS in 0.01-M HCL). Following overnight incubation in the dark, plates were agitated and the absorbance was measured at 570 nm.
Establishment of a 5-FU–Resistant Cell Pool
5-FU–resistant cell pools were established by exposure to stepwise increasing concentrations of 5-FU. Initially, DLD1 and HT29 cells were cultured in medium containing 0.1 μmol/L 5-FU. The drug concentration was then increased in steps of 1.25× increases from 0.1 μmol/L up to 30 μmol/L. Cells were cultured for at least 1 week at each step, with medium exchange every 3 days. The 5-FU–resistant cell pools were designated DLD1_5FU and HT29_5FU, respectively. The tolerance toward 5-FU was determined with an MTT assay.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction Analysis
Total RNA was isolated with the High Pure RNA Isolation Kit (Roche) according to the manufacturer's protocol. cDNA was generated from 1 μg of total RNA per sample using the Verso cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA). Quantitative real-time polymerase chain reaction (qPCR) was performed with the Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA) by using the LightCycler 480 (Roche). Expression was normalized using detection of GAPDH or β-actin using the ΔΔCt method.59 Results are represented as fold induction of the treated or transfected condition compared with the control condition. Experiments were performed in triplicates. The sequences of oligonucleotides used as qPCR primers are listed in Table 5.
Table 5.
Oligonucleotides Used for qPCR
| mRNA | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| β-actin | TGACATTAAGGAGAAGCTGTGCTAC | GAGTTGAAGGTAGTTTCGTGGATG |
| CSF1R | CCTCGCTTCCAAGAATTGCA | CCCAATCTTGGCCACATGA |
| CSF1 | GCAAGAACTGCAACAACAGC | ATCAGGCTTGGTCACCACAT |
| pri-miR-34a | CGTCACCTCTTAGGCTTGGA | CATTGGTGTCGTTGTGCT |
| CDH1 | CCCGGGACAACGTTTATTAC | GCTGGCTCAAGTCAAAGTCC |
| VIM | TACAGGAAGCTGCTGGAAGG | ACCAGAGGGAGTGAATCCAG |
| SNAIL | GCACATCCGAAGCCACAC | GGAGAAGGTCCGAGCACAC |
| ZEB1 | TCAAAAGGAAGTCAATGGACAA | GTGCAGGAGGGACCTCTTTA |
| STAT3 | GGGAAGAATCACGCCTTCTAC | ATCTGCTGCTTCTCCGTCAC |
mRNA, messenger RNA.
Methylation-Specific PCR
Genomic DNA was isolated from cell lines using the DNeasy Blood & Tissue Kits (Qiagen). 400 ng of gDNA was treated with bisulfite using the EZ DNA methylation kit (D5001 & D5002; Zymo Research, Irvine, CA). The modified DNA was eluted with a final volume of 10 μL elution buffer. A total of 3 μL were amplified by PCR. The MSP primers used for detection of CpG methylation of the miR-34a promoter are depicted in Table 6 and were previously established.25 The PCR protocol entailed 5 min at 95°C; 2 cycles of 95°C for 20 seconds, 68°C for 30 seconds, and 72°C for 30 seconds, followed by 2 cycles with 66°C annealing temperature, then 34 cycles with 65°C annealing temperature, and a final elongation step at 72°C for 10 minutes. For the methylated allele, a 122-bp fragment and for the unmethylated allele a 126-bp fragment were obtained. The PCR products were separated by electrophoresis on 8% polyacrylamide gels and then visualized by ethidium bromide staining.
Table 6.
Oligonucleotides used for MSP and BSP
| Forward (5′-3′) | Reverse (5′-3′) | |
|---|---|---|
| MSP_M | GGTTTTGGGTAGGCGCGTTTC | TCCTCATCCCCTTCACCGCCG |
| MSP_U | (Inosine)(Inosine)GGTTTTGGGTAGGTGTGTTTT | AATCCTCATCCCCTTCACCACCA |
| BSP_1 | TAGAGATAATAGGTTTTGATTCGGGATAGA | CAAAACTCCCACAAAATCTCCAAA TACCCCC |
| BSP_2 | TAGAGATAATAGGTTTTGATTTGGGATAGA | CAAAACTCCCGCAAAATCTCCAAA TACCCCC |
BSP, bisulfite-sequencing polymerase chain reaction; MSP, methylation-specific polymerase chain reaction.
Bisulfite Sequencing
A total of 5 μL of bisulfite-treated genomic DNA was used as a template to amplify fragments of a 776-bp region upstream of the miR-34a promoter encompassing the transcription start site and p53 binding site with a high CpG content.25 The bisulfite-sequencing PCR primers used here are depicted in Table 6, with PCR settings of 95°C for 5 minutes, followed by 38 cycles of 95°C for 20 seconds, 65°C for 30 seconds, and 72°C for 60 seconds, with a final elongation step at 72°C for 10 minutes. Amplification products were purified using a QIAquick Gel Extraction Kit, and then subcloned into the shuttle vector pGEM-T-Easy (Promega, Madison, WI). For each cell line, at least 9 individual clones were sequenced on both strands using SP6 and T7 sequencing primers. The sequencing reactions were analyzed on a capillary sequencer (ABI 3130; Applied Biosystems). Clones with a cytosine conversion rate of <90% were excluded. Methylation data from bisulfite sequencing were trimmed, aligned and displayed as lollipop graphs using QUMA.
Chromatin Immunoprecipitation
DLD1/pRTR-SNAIL-VSV cells were cultured as described previously. Before crosslinking, cells were treated with Dox (100 ng/mL) for 24 hours to induce ectopic expression of VSV-tagged proteins. Crosslinking was conducted with formaldehyde (Merck, Kenilworth, NJ) at 1% final concentration and terminated after 5 minutes by addition of glycine at a final concentration of 0.125 M. Cells were harvested in SDS buffer (50 mM Tris, pH 8.1, 0.5% SDS, 100 mM NaCl, 5 mM EDTA), pelleted and resuspended in immunoprecipitation buffer (2 parts of SDS buffer and 1 part Triton dilution buffer [100 mM Tris-HCl, pH 8.6, 100 mM NaCl, 5 mM EDTA, pH 8.0, 0.2% NaN3, 5.0% Triton X-100]). Chromatin was sheered by sonication (HTU SONI 130, G.Heinemann Ultraschall und Labortechnik, Schwäbisch Gmünd, Germany) to generate DNA fragments with an average size of 500 bp. Preclearing and incubation with polyclonal VSV antibody (V4888; Sigma) for 16 hours was performed as previously described.21,60 Washing and reversal of cross-linking was performed as described.61 Immunoprecipitated DNA was analyzed by qPCR and the enrichment was expressed as percentage of the input for each condition. The sequences of oligonucleotides used as quantitative chromatin immunoprecipitation primers are listed in Table 7.
Table 7.
Oligonucleotides used for qChIP
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| CSF1R | ACAACTTTCCCACCAGTCCT | GGGGTGAGTAGTTTGGTGGG |
| MiR-200c | CAGGAGGACACACCTGTGC | TCCCCTGGTGGCCTTTAC |
| AchR | CCTTCATTGGGATCACCACG | AGGAGATGAGTACCAGCAGGTTG |
qChIP, quantitative chromatin immunoprecipitation.
Generation of Cell Pools Stably Expressing Conditional Alleles
Open reading frames that were inserted into the episomal, inducible pRTR vectors67 were validated by sequencing. Stably transfected cells were generated by transfection of pRTR vectors using Fugene6 (Roche) and selected with incrementally increasing concentrations of Puromycin (0.5–6.0 μg/mL) for 10 days.20 The frequency of green fluorescent protein–positive cells was determined 48 hours after addition of Dox at a final concentration of 100 ng/mL by flow cytometry.
Dual 3′-UTR Luciferase Reporter Assays
The full-length 3′-UTRs of the human CSF1R mRNA were PCR amplified from cDNA of human diploid fibroblasts. The PCR product was cloned into the shuttle vector pGEM-T-Easy (Promega), and then transferred into the pGL3-control-MCS vector62 and verified by sequencing. For mutagenesis of the miR-34a seed-matching sequences the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, San Diego, CA) was used according to the manufacturer’s instructions and verified by sequencing. H1299 cells were seeded in 12-well plate at 3×104 cells/well for 24 hours and transfected for 72 hours with 100 ng of the indicated firefly luciferase reporter plasmid, 20 ng of Renilla reporter plasmid as a normalization control and 25 nM of miR-34a pre-miRNA oligonucleotide (PM11030; Ambion, Austin, TX), or a negative control oligonucleotide (neg. control #1; Ambion) with HiPerFect Transfection Reagent (Qiagen) for 48 hours. The analysis was performed with the Dual Luciferase Reporter assay (Promega) according to manufacturer’s instructions. Fluorescence intensities were measured with an Orion II luminometer (Berthold, Bad Wildbad, Germany) in 96-well format and analyzed with the SIMPLICITY software package (DLR, Stuttgart, Germany). The sequences of oligonucleotides used for cloning and mutagenesis of human 3′-UTR are listed in Table 8.
Table 8.
Oligonucleotides Used for Cloning and Mutagenesis
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Human CSF1R 3′UTR | CGGAATTCGGAGTTGACGACAGGGAGTACCACTC | CGCTGCAGATGTGGACAGAGACATCCCAC |
| Human CSF1R 3′UTR mutant | CCTGAGCATGGGCCATCAGTCGGAGTCAGGGGCTGGGGG | CCCCCAGCCCCTGACTCCGACTGATGGCCCATGCTCAGG |
| Human CSF1R (L301S) | GAGAGTGCCTACTCGAACTTGAGCTCT | AGAGCTCAAGTTCGAGTAGGCACTCTC |
Bioinformatic Analysis of Online Databases
TCGA gene expression data and follow-up information of COADs were downloaded from the National Cancer Institute’s Genomic Data Commons (https://gdc.cancer.gov/).28 Normalized RSEM counts were used to determine the expression of relevant mRNAs. Pearson's correlation analyses of gene expressions were performed with the Prism5 program (Graph Pad Software, San Diego, CA). Association of patient samples with the different CMS categories was obtained from the Cancer Subtyping Consortium (www.synapse.org). The CMS subtypes were described in Guinney et al.31 CMS-specific signature gene sets were obtained from Sveen et al.63 PDX RNA expression data of human CRC specimens (GSE76402), the classification of CRC intrinsic subtypes (CRIS) and the respective signature genes for each CRIS subtype were obtained from Isella et al.34 Single-cell RNA expression data of normal colonic and CRC cells (GSE81861) were obtained from Li et al35 and analyzed with the RCA R package as described (R3.6.0, R Foundation for Statistical Computing, Vienna, Austria). Expression and clinical data of GSE37892, GSE39582, and GSE48267 datasets were downloaded from National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo). GSEA was performed on preranked gene lists based on expression correlation coefficients (Pearson, London, UK) with CSF1R using the GSEA software obtained from http://software.broadinstitute.org/gsea/index.jsp.64 Hallmark gene sets were obtained from the Molecular Signatures database (MSigDB).65 Heatmaps were generated with GENE-E (Broad Institute, Cambridge, MA).
Clinical Samples and Immunohistochemistry
CSF1R expression was evaluated using formalin-fixed, paraffin-embedded colon cancer samples of 90 patients who underwent surgical tumor resection at the Ludwig-Maximilians University Munich. Tissue microarrays were generated with 6 representative 1 mm cores of each case, for which the methylation status of miR-34a had been determined previously.27 The tissue microarray sections were deparaffinized and stained with human CSF1R antibody (ab183316; Abcam, Cambridge, UK) on a Benchmark XT Autostainer with UltraView Universal DAB and alkaline phosphatase detection kits (Ventana Medical Systems, Oro Valley, AZ). The staining intensity was scored is 0 for absent, 1 for low, 2 for intermediate, and 3 for strong signal.
Metastasis Formation in NOD/SCID Mice
Immune-compromised NOD/SCID mice were obtained from the Jackson Laboratory (Bar Harbor, ME). DLD1 cells stably expressing Luc2 were generated as described previously.66 DLD1-Luc/pRTR-CSF1R cells were generated by stable transfection of pRTR plasmids and maintained in medium with puromycin. A total of 1 × 106 cells were resuspended in 0.2-mL Hank’s Balanced Salt Solution and injected into the lateral tail vein of a 6- to 8-week-old age-matched male NOD/SCID mouse using a 25-gauge needle. For monitoring of the injected cells, anesthetized mice were injected intraperitoneally with D-luciferin (150 mg/kg) and imaged with the IVIS Illumina System (Caliper Life Sciences, Waltham, MA) 10 minutes after injection. The acquisition time was set to 2 min and imaging was preformed once a week. After 8 weeks, mice were sacrificed and the whole lungs were resected and subjected to hematoxylin and eosin staining. All studies involving mice were performed with approval by the local Animal Experimentation Committee (Regierung of Oberbayern). All experiments were conducted in accordance with relevant guidelines and regulations.
Statistics
Calculations of significant differences between 2 groups of samples were analyzed by a Student’s t test (2-tailed; unpaired or paired where indicated). For the comparison of multiple groups, a 1-way analysis of variance followed by a Tukey multiple comparisons post hoc test was performed. Log-rank test was used for the statistical analysis of the Kaplan-Meier curves. Cox proportional hazards models were applied for multiple regression analyses of survival data. Association of CSF1R expression with clinical parameters was analyzed using chi-square tests. For mRNA expression correlation analyses, a Pearson’s correlation was applied. P values ≤ 0.05 were considered as significant, with asterisks generally indicating levels (∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001). Statistics were calculated with Prism5 (Graph Pad Software ) and SPSS (version 25, IBM, Armonk, NY).
Study Approval
All experimentations involving mice were approved by the Government of Upper Bavaria, Germany (AZ-ROB-55.2-2532.Vet_02-18-57). Because the human tumor biopsies analyzed in Figure 12G and 12H underwent dual anonymization a specific approval was not deemed necessary by the ethics committee of the Medical Faculty, Ludwig-Maximilians-University Munich.
Acknowledgments
We are grateful to Bert Vogelstein for providing p53+/+ and p53-/- RKO cell lines. We also thank Ursula Götz for technical assistance.
Footnotes
Author Contributions XS: Designed and performed experiments, analyzed results, wrote the paper; MK: bioinformatics analysis and initial experiments; MR: xenograft analysis; TK and DH: designed and provided the M0/M1 cohort of CRC samples, DH: IHC analysis of human CRC samples; HH conceived and supervised the study, planned experiments and wrote the paper.
Conflicts of Interest The authors disclose no conflicts.
Funding Xiaolong Shi is a recipient of a China Scholarship Council fellowship. This work has been funded by a grant of the Wilhelm-Sander-Stiftung (2013.108.1) to Heiko Hermeking.
References
- 1.Anderson R.L., Balasas T., Callaghan J., Coombes R.C., Evans J., Hall J.A., Kinrade S., Jones D., Jones P.S., Jones R., Marshall J.F., Panico M.B., Shaw J.A., Steeg P.S., Sullivan M., Tong W., Westwell A.D., Ritchie J.W.A., Cancer Research UK, Cancer Therapeutics CRCAMWG A framework for the development of effective anti-metastatic agents. Nat Rev Clin Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Cutsem E., Oliveira J., Group E.G.W. Advanced colorectal cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):61–63. doi: 10.1093/annonc/mdp130. [DOI] [PubMed] [Google Scholar]
- 3.Punt C.J., Koopman M., Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 4.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat Rev Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 5.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heisterkamp N Fau - Groffen J., Groffen J Fau - Stephenson J.R., Stephenson J.R. Isolation of v-fms and its human cellular homolog. Virology. 1983;126:248–258. doi: 10.1016/0042-6822(83)90476-2. [DOI] [PubMed] [Google Scholar]
- 7.Roussel M.F., Sherr C.J., Barker P.E., Ruddle F.H. Molecular cloning of the c-fms locus and its assignment to human chromosome 5. J Virol. 1983;48:770–773. doi: 10.1128/jvi.48.3.770-773.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yarden Y., Ullrich A. Growth factor receptor tyrosine kinases. Annu Rev Biochem. 1988;57:443–478. doi: 10.1146/annurev.bi.57.070188.002303. [DOI] [PubMed] [Google Scholar]
- 9.Coussens L., Van Beveren C., Smith D., Chen E., Mitchell R.L., Isacke C.M., Verma I.M., Ullrich A. Structural alteration of viral homologue of receptor proto-oncogene fms at carboxyl terminus. Nature. 1986;320:277–280. doi: 10.1038/320277a0. [DOI] [PubMed] [Google Scholar]
- 10.Roussel M.F., Dull T.J., Rettenmier C.W., Ralph P., Ullrich A., Sherr C.J. Transforming potential of the c-fms proto-oncogene (CSF-1 receptor) Nature. 1987;325:549–552. doi: 10.1038/325549a0. [DOI] [PubMed] [Google Scholar]
- 11.Ullrich A., Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Szretter K.J., Vermi W., Gilfillan S., Rossini C., Cella M., Barrow A.D., Diamond M.S., Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novak U., Harpur A.G., Paradiso L., Kanagasundaram V., Jaworowski A., Wilks A.F., Hamilton J.A. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86:2948–2956. [PubMed] [Google Scholar]
- 15.Cannarile M.A., Weisser M., Jacob W., Jegg A.M., Ries C.H., Ruttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer. 2017;5:53. doi: 10.1186/s40425-017-0257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardsen E., Uglehus R.D., Johnsen S.H., Busund L.T. Macrophage-colony stimulating factor (CSF1) predicts breast cancer progression and mortality. Anticancer Res. 2015;35:865–874. [PubMed] [Google Scholar]
- 17.Mroczko B., Szmitkowski M., Okulczyk B. Hematopoietic growth factors in colorectal cancer patients. Clin Chem Lab Med. 2003;41:646–651. doi: 10.1515/CCLM.2003.098. [DOI] [PubMed] [Google Scholar]
- 18.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 19.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 20.Siemens H., Jackstadt R., Hunten S., Kaller M., Menssen A., Gotz U., Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 21.Hahn S., Jackstadt R., Siemens H., Hunten S., Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rokavec M., Oner M.G., Li H., Jackstadt R., Jiang L., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S., Slotta-Huspenina J., Bader F.G., Greten F.R., Hermeking H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Rokavec M., Jiang L., Horst D., Hermeking H. Antagonistic Effects of p53 and HIF1A on microRNA-34a Regulation of PPP1R11 and STAT3 and Hypoxia-induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells. Gastroenterology. 2017;153:505–520. doi: 10.1053/j.gastro.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Oner M.G., Rokavec M., Kaller M., Bouznad N., Horst D., Kirchner T., Hermeking H. Combined Inactivation of TP53 and MIR34A Promotes Colorectal Cancer Development and Progression in Mice Via Increasing Levels of IL6R and PAI1. Gastroenterology. 2018;155:1868–1882. doi: 10.1053/j.gastro.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 26.Vogt M., Munding J., Gruner M., Liffers S.T., Verdoodt B., Hauk J., Steinstraesser L., Tannapfel A., Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Archiv. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 27.Siemens H., Neumann J., Jackstadt R. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19:710–720. doi: 10.1158/1078-0432.CCR-12-1703. [DOI] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marisa L., de Reynies A., Duval A., Selves J., Gaub M.P., Vescovo L., Etienne-Grimaldi M.C., Schiappa R., Guenot D., Ayadi M., Kirzin S., Chazal M., Flejou J.F., Benchimol D., Berger A., Lagarde A., Pencreach E., Piard F., Elias D., Parc Y., Olschwang S., Milano G., Laurent-Puig P., Boige V. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laibe S., Lagarde A., Ferrari A., Monges G., Birnbaum D., Olschwang S., Project C.O.L. A seven-gene signature aggregates a subgroup of stage II colon cancers with stage III. Omics. 2012;16:560–565. doi: 10.1089/omi.2012.0039. [DOI] [PubMed] [Google Scholar]
- 31.Guinney J., Dienstmann R., Wang X., de Reynies A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., Bot B.M., Morris J.S., Simon I.M., Gerster S., Fessler E., De Sousa E.M.F., Missiaglia E., Ramay H., Barras D., Homicsko K., Maru D., Manyam G.C., Broom B., Boige V., Perez-Villamil B., Laderas T., Salazar R., Gray J.W., Hanahan D., Tabernero J., Bernards R., Friend S.H., Laurent-Puig P., Medema J.P., Sadanandam A., Wessels L., Delorenzi M., Kopetz S., Vermeulen L., Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isella C., Terrasi A., Bellomo S.E., Petti C., Galatola G., Muratore A., Mellano A., Senetta R., Cassenti A., Sonetto C., Inghirami G., Trusolino L., Fekete Z., De Ridder M., Cassoni P., Storme G., Bertotti A., Medico E. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 33.Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M., Sevillano M., Palomo-Ponce S., Tauriello D.V., Byrom D., Cortina C., Morral C., Barcelo C., Tosi S., Riera A., Attolini C.S., Rossell D., Sancho E., Batlle E. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 34.Isella C., Brundu F., Bellomo S.E., Galimi F., Zanella E., Porporato R., Petti C., Fiori A., Orzan F., Senetta R., Boccaccio C., Ficarra E., Marchionni L., Trusolino L., Medico E., Bertotti A. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun. 2017;8:15107. doi: 10.1038/ncomms15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Courtois E.T., Sengupta D., Tan Y., Chen K.H., Goh J.J.L., Kong S.L., Chua C., Hon L.K., Tan W.S., Wong M., Choi P.J., Wee L.J.K., Hillmer A.M., Tan I.B., Robson P., Prabhakar S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 36.Li E., Ji P., Ouyang N. Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol. 2014;45:587–594. doi: 10.3892/ijo.2014.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roussel M.F., Downing J.R., Rettenmier C.W., Sherr C.J. A point mutation in the extracellular domain of the human CSF-1 receptor (c-fms proto-oncogene product) activates its transforming potential. Cell. 1988;55:979–988. doi: 10.1016/0092-8674(88)90243-7. [DOI] [PubMed] [Google Scholar]
- 38.Bencheikh L., Diop M.K., Riviere J., Imanci A., Pierron G., Souquere S., Naimo A., Morabito M., Dussiot M., De Leeuw F., Lobry C., Solary E., Droin N. Dynamic gene regulation by nuclear colony-stimulating factor 1 receptor in human monocytes and macrophages. Nat Commun. 2019;10:1935. doi: 10.1038/s41467-019-09970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandi S., Cioce M., Yeung Y.G., Nieves E., Tesfa L., Lin H., Hsu A.W., Halenbeck R., Cheng H.Y., Gokhan S., Mehler M.F., Stanley E.R. Receptor-type protein-tyrosine phosphatase zeta is a functional receptor for interleukin-34. J Biol Chem. 2013;288:21972–21986. doi: 10.1074/jbc.M112.442731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sale M.J., Balmanno K., Saxena J., Ozono E., Wojdyla K., McIntyre R.E., Gilley R., Woroniuk A., Howarth K.D., Hughes G., Dry J.R., Arends M.J., Caro P., Oxley D., Ashton S., Adams D.J., Saez-Rodriguez J., Smith P.D., Cook S.J. MEK1/2 inhibitor withdrawal reverses acquired resistance driven by BRAF(V600E) amplification whereas KRAS(G13D) amplification promotes EMT-chemoresistance. Nat Commun. 2019;10:2030. doi: 10.1038/s41467-019-09438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cioce M., Canino C., Goparaju C., Yang H., Carbone M., Pass H.I. Autocrine CSF-1R signaling drives mesothelioma chemoresistance via AKT activation. Cell Death Dis. 2014;5:e1167. doi: 10.1038/cddis.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patsialou A., Wang Y., Pignatelli J., Chen X., Entenberg D., Oktay M., Condeelis J.S. Autocrine CSF1R signaling mediates switching between invasion and proliferation downstream of TGFbeta in claudin-low breast tumor cells. Oncogene. 2015;34:2721–2731. doi: 10.1038/onc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menke J., Kriegsmann J., Schimanski C.C., Schwartz M.M., Schwarting A., Kelley V.R. Autocrine CSF-1 and CSF-1 receptor coexpression promotes renal cell carcinoma growth. Cancer Res. 2012;72:187–200. doi: 10.1158/0008-5472.CAN-11-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franze E., Dinallo V., Rizzo A., Di Giovangiulio M., Bevivino G., Stolfi C., Caprioli F., Colantoni A., Ortenzi A., Grazia A.D., Sica G., Sileri P.P., Rossi P., Monteleone G. Interleukin-34 sustains pro-tumorigenic signals in colon cancer tissue. Oncotarget. 2018;9:3432–3445. doi: 10.18632/oncotarget.23289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang W., Qin Z., Fan S., Wen Q., Lu Y., Wang J., Zhang X., Wei L., He W., Ye Q., Yan Q., Li G., Ma J. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7:32421–32432. doi: 10.18632/oncotarget.8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwicker S., Martinez G.L., Bosma M., Gerling M., Clark R., Majster M., Soderman J., Almer S., Bostrom E.A. Interleukin 34: a new modulator of human and experimental inflammatory bowel disease. Clin Sci. 2015;129:281–290. doi: 10.1042/CS20150176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., Cao S., Xu P., Han W., Shan T., Pan J., Lin W., Chen X., Wang X. Changes in the Expression of miR-34a and its Target Genes Following Spinal Cord Injury In Rats. Med Sci Monit. 2016;22:3981–3993. doi: 10.12659/MSM.900893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cimino D., De Pitta C., Orso F., Zampini M., Casara S., Penna E., Quaglino E., Forni M., Damasco C., Pinatel E., Ponzone R., Romualdi C., Brisken C., De Bortoli M., Biglia N., Provero P., Lanfranchi G., Taverna D. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 50.Blagih J., Zani F., Chakravarty P., Hennequart M., Pilley S., Hobor S., Hock A.K., Walton J.B., Morton J.P., Gronroos E., Mason S., Yang M., McNeish I., Swanton C., Blyth K., Vousden K.H. Cancer-Specific Loss of p53 Leads to a Modulation of Myeloid and T Cell Responses. Cell Rep. 2020;30:481–496.e6. doi: 10.1016/j.celrep.2019.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y., Zhao Z., Chen Y., Lv Z., Ding X., Wang R., Xiao H., Hou C., Shen B., Feng J., Guo R., Li Y., Peng H., Han G., Chen G. An epithelial-to-mesenchymal transition-inducing potential of granulocyte macrophage colony-stimulating factor in colon cancer. Sci Rep. 2017;7:8265. doi: 10.1038/s41598-017-08047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong M., Zheng W., Li H., Li X., Ao L., Shen Y., Liang Q., Li J., Hong G., Yan H., Cai H., Li M., Guan Q., Guo Z. Multi-omics landscapes of colorectal cancer subtypes discriminated by an individualized prognostic signature for 5-fluorouracil-based chemotherapy. Oncogenesis. 2016;5:e242. doi: 10.1038/oncsis.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Housman G., Byler S., Heerboth S., Lapinska K., Longacre M., Snyder N., Sarkar S. Drug resistance in cancer: an overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siemens H., Jackstadt R., Kaller M., Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4:1399–1415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaller M., Liffers S.T., Oeljeklaus S., Kuhlmann K., Roh S., Hoffmann R., Warscheid B., Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.010462. M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silber J., Jacobsen A., Ozawa T., Harinath G., Pedraza A., Sander C., Holland E.C., Huse J.T. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garofalo M., Jeon Y.J., Nuovo G.J., Middleton J., Secchiero P., Joshi P., Alder H., Nazaryan N., Di Leva G., Romano G., Crawford M., Nana-Sinkam P., Croce C.M. MiR-34a/c-Dependent PDGFR-alpha/beta Downregulation Inhibits Tumorigenesis and Enhances TRAIL-Induced Apoptosis in Lung Cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Septisetyani E.P., Ningrum R.A., Romadhani Y., Wisnuwardhani P.H., Santoso A. Optimization of Sodium Dodecyl Sulphate as a Formazan Solvent and Comparison of 3-(4,-5-Dimethylthiazo-2-Yl)-2,5-Diphenyltetrazolium Bromide (Mtt) Assay with Wst-1 Assay in Mcf-7 Cells. Indonesian J Pharm. 2014;25:245. [Google Scholar]
- 59.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.Menssen A., Epanchintsev A., Lodygin D., Rezaei N., Jung P., Verdoodt B., Diebold J., Hermeking H. c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle. 2007;6:339–352. doi: 10.4161/cc.6.3.3808. [DOI] [PubMed] [Google Scholar]
- 61.Amati B., Frank S.R., Donjerkovic D., Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- 62.Welch C., Chen Y., Stallings R.L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 63.Sveen A., Bruun J., Eide P.W., Eilertsen I.A., Ramirez L., Murumagi A., Arjama M., Danielsen S.A., Kryeziu K., Elez E., Tabernero J., Guinney J., Palmer H.G., Nesbakken A., Kallioniemi O., Dienstmann R., Lothe R.A. Colorectal Cancer Consensus Molecular Subtypes Translated to Preclinical Models Uncover Potentially Targetable Cancer Cell Dependencies. Clin Cancer Res. 2018;24:794–806. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liberzon A., Birger C., Thorvaldsdottir H. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi L., Jackstadt R., Siemens H., Li H., Kirchner T., Hermeking H. p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2014;74:532–542. doi: 10.1158/0008-5472.CAN-13-2203. [DOI] [PubMed] [Google Scholar]
- 67.Jackstadt R., Röh S., Neumann J. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J Experimental Medicine. 2013;210:1331–1350. doi: 10.1084/jem.20120812. [DOI] [PMC free article] [PubMed] [Google Scholar]