Abstract
Antimicrobial resistance frequently carries a fitness cost to a pathogen, measured as a reduction in growth rate compared to the sensitive wild-type, in the absence of antibiotics. Existing empirical evidence points to the following relationship between cost of resistance and virulence. If a resistant pathogen suffers a fitness cost in terms of reduced growth rate it commonly has lower virulence compared to the sensitive wild-type. If this cost is absent so is the reduction in virulence. Here we show, using experimental evolution of drug resistance in the fungal human pathogen Candida glabrata, that reduced growth rate of resistant strains need not result in reduced virulence. Phenotypically heterogeneous populations were evolved in parallel containing highly resistant sub-population small colony variants (SCVs) alongside sensitive sub-populations. Despite their low growth rate in the absence of an antifungal drug, the SCVs did not suffer a marked alteration in virulence compared with the wild-type ancestral strain, or their co-isolated sensitive strains. This contrasts with classical theory that assumes growth rate to positively correlate with virulence. Our work thus highlights the complexity of the relationship between resistance, basic life-history traits and virulence.
Keywords: growth rate, virulence, drug resistance, fungal populations
1. Introduction
The rise of antimicrobial resistance presents a major challenge to modern healthcare [1] but it is ultimately an ecological problem [2]. In natural environments, be it within or outside a host, susceptible and resistant microorganisms compete with each other [3]. While the use of antimicrobials promotes the growth of resistant strains or species [4], resistance frequently carries a fitness cost that arises when the antimicrobial is withdrawn [5–8]. The question we address here is this: how do virulence and resistance traits interact, costs of resistance in particular?
For a pathogenic microorganism, resistance costs are commonly measured as reductions in growth rate [5,7,9] which are often associated with reductions in virulence [8,10,11]. For example, resistant small colony variants (SCVs) isolated from otherwise susceptible populations [12] have lower growth rate than the susceptible isolates they were found among in the absence of antibiotics but are also less virulent [13]. By contrast, resistant strains that do not suffer reduction in growth rates, either through ‘no-cost' or compensatory mutations, have been found not to have impaired virulence [8,10,14,15]. Therefore, the existing empirical evidence suggests that the existence of virulent resistant or multi-resistant infections (as observed in [8]) is only possible when resistance costs are absent.
Here, we challenge this reasoning by providing a counter example. While theory [16,17] and within-species empirical studies [18,19] argue that growth rate positively correlates with virulence, a between-species meta-analysis suggests that growth rate and virulence might not be positively correlated [20]. Thus, we hypothesize that virulence could be maintained following the evolution of costly resistance if a reduction in within-species growth rate does not lead to a reduction in virulence.
We deployed an experimental system using the human fungal pathogen Candida glabrata, ideally placed to test this hypothesis. In particular, the opportunistic pathogen C. glabrata undergoes rapid acquisition of resistance to the main classes of clinical antifungals [21,22] and resistance can impart a growth fitness cost [11]. Crucially for testing our hypothesis, we recently observed attenuated virulence in a fast-growing C. glabrata strain [23,24] which would suggest the potential lack of a positive relationship between growth rate and virulence.
Candida glabrata is a pathogen of clear clinical importance as it is the second most commonly isolated Candida species in bloodstream infections [25] with a greater than 50% death rate [26]. It is more closely related to the well-established model yeast Saccharomyces cerevisiae than to other species of the Candida genus [27]. As such, molecular, genetic and evolutionary techniques can readily be transferred from S. cerevisiae to C. glabrata making it an increasingly popular model organism to study infection [28–31].
After 14 seasons of transfer in post-minimum inhibitory concentration echinocandin (caspofungin) treatment, we found independent evolution of two C. glabrata heterogeneous populations consisting of diverse co-isolated colony morphologies with divergent resistance and growth properties. In both populations, a SCV phenotype was associated with a low growth rate and high resistance level. Interestingly, the small colony phenotype retained virulence in a wax moth larval infection model, comparable to the wild-type ancestor and co-isolated population colonies experiencing faster growth. These results suggest that virulence could be maintained in the presence of costly resistance that results in a reduced growth rate. In addition, we found no evidence of a correlation between in vitro growth rate and virulence across sub-population colony variants. Our work therefore questions the current understanding of the relationship between growth rate and virulence.
2. Methods
(a). In vitro evolution of Candida glabrata populations on a gradient of caspofungin concentrations
The reference C. glabrata strain ATCC 2001 [32] was used as the wild-type ancestor of all evolving replicate populations and was denoted as ‘2001WT'. Triplicate populations were evolved across a gradient of eight caspofungin concentrations of clinical relevance [33] and drug-free condition in a 96-well microtiter plate (electronic supplementary material, figure S1a) in 10 mg ml−1 glucose synthetic complete (SC) medium (0.67% w/v yeast nitrogen base without amino acids and 0.079% w/v synthetic complete supplement mixture (Formedium)). Caspofungin concentrations in SC were prepared from stock solution (5 mg ml−1). Populations were serially transferred (1 : 30 dilution) every 24 h to fresh media and drug conditions for 14 days. The 96-well plate was incubated at 30°C over 24 h with shaking. Optical density (OD) was read at 650 nm wavelength in a microtiter plate reader. The whole experiment was repeated on three separate 14-day periods (experiments A, B and C).
OD data was blank-corrected and percentage relative growth at 24 h, for each drug-treated population, was presented relative to the mean OD of the no drug-treated populations [34] (electronic supplementary material, figure S2). We used the lme4 package [35] with R version 3.4.3 [36] to conduct a linear mixed effects analysis of the fixed effects of day of the evolutionary experiment and caspofungin concentration on relative growth of C. glabrata. ‘Population' was included as a nested random effect within ‘experiment' as each population was repeatedly measured across days of the evolutionary experiment. We obtained p-values from likelihood ratio tests.
(b). Growth profiling of caspofungin-evolved endpoint colonies
All nine C. glabrata populations (experiments A-C) that were evolved at 0.78, 1.37 and 2.40 µg ml−1 of caspofungin were revived from day 14 (electronic supplementary material, figure S1b). We identified two distinct colony size variants hereby named SCV and regular colony variant (RCV) in a single population from each of experiments A and B but not C, and only at 0.78 µg ml−1, shown in the electronic supplementary material, figure S3. Colony variants from experiment A were growth profiled in SC 1% w/v glucose media across replicate wells of a 96-well plate (n = 4 wells × 3 separate day repeats = 12). OD was measured over 24 h. Data from experiment A are plotted in figure 1a,b. Growth was measured in four replicate wells (n = 4) of a 96-well plate for each of the SCV and RCV from experiment B with data plotted in the electronic supplementary material, figure S4a and b.
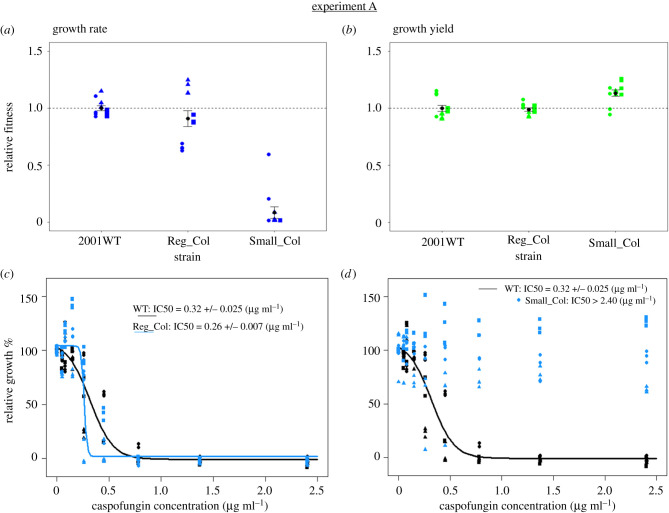
Figure 1.
Sub-population colony diversity in growth fitness and drug susceptibility in experiment A. Data are shown for the isolated RCV and SCV from the single population in which colony diversity was identified. Reg_Col (RCV), regular-sized colony variant; Small_Col (SCV), small colony variant; wild-type ancestral strain, 2001WT. In (a) and (b), growth values are calculated relative to average values of 2001WT, where a value of 1.0 signifies no change relative to ancestor. n = 12 measurements per colony variant (measurement in four individual wells of a microtiter plate, repeated separately on three days). Different symbol shapes (separated by horizontal noise on the x-axis) represent measurements from separate days. Black points and error bars show overall means and standard errors. Average relative growth rates (±s.e.): Reg_Col: 0.91 ± 0.07; Small_Col: 0.09 ± 0.05. Average relative growth yields (±s.e.): Reg_Col: 0.99 ± 0.01; Small_Col: 1.13 ± 0.03. Dose response plots (c) and (d) show final OD of drug-treated populations as a percentage of average growth of the no-drug-treated populations. The same dataset for the ancestral strain (2001WT) is plotted in (c) and (d). n = 9 measurements per colony variant for each drug concentration (measurement in three individual wells of a microtiter plate, repeated separately on three days). Different symbol shapes represent measurements from separate days. Lines represent best-fit of the logistic dose response model. Model-predicted IC50 values ± s.e. of the estimated value are shown for each dose response. (Online version in colour.)
Data was imported into MATLAB [37] to approximate intrinsic growth rate via logistic growth model fitting as described in [24], with exclusion of the lag phase parameter L. Relative growth rate and final growth yield were obtained by dividing through by the mean values of the 2001WT strain. We fitted a linear mixed effects model to relative growth rate and yield data from experiment A and experiment B using the lme4 package [35] with R version 3.4.3 [36]. We included colony variant type as a fixed factor and day of measurement as a random factor. A likelihood ratio test was used to test significance of the fixed factor.
(c). Dose response profiling
We measured the caspofungin dose response profile over 24 h for the co-isolated small and regular-sized colony variants from experiments A and B and the wild-type ancestral strain (2001WT), to calculate IC50 (drug concentration causing 50% growth inhibition). Dose responses were set-up in 96-well microtiter plates, using the same methods as described in the first season of the in vitro evolution assay (electronic supplementary material, figure S1a). For colony variants that were resistant to concentrations up to 2.40 µg ml−1, a twofold dilution series from 64 µg ml−1 was used. Microtiter plates were incubated at 30°C over 24 h with orbital shaking and OD measurement. Dose responses were repeated three times independently for experiment A (n = 3 wells × 3 repeats = 9 per caspofungin concentration) shown in figure 1c,d. Dose responses were run once for experiment B (n = 3 per caspofungin concentration) shown in the electronic supplementary material, figure S4c and d. The best-fit 4-parameter logistic dose response was plotted using R-package ‘drc' [38], as described in [34] and used to estimate IC50.
(d). Small colony phenotypes
(i). Serial passaging
We tested stability of the randomly selected SCV isolated from the single evolved population exhibiting colony diversity from each of experiments A and B. A single overnight culture of each SCV was adjusted to 6.49 × 105 cells ml−1, diluted twofold and serially passaged (1 : 30 dilution) across triplicate populations in 10 mg ml−1 glucose SC medium over 14 days. (electronic supplementary material, figure S1d). The plate was incubated at 30°C with shaking at 180 rpm and OD was measured during each 24 h season. Growth rate and yield data from the final growth cycle are plotted in figure 2 alongside 2001WT, with two-sample t-tests used to detect growth differences between 2001WT and passaged populations of each SCV. Caspofungin dose response was measured for a single-clone cultured from each preserved endpoint (day 14) population (electronic supplementary material, figure S1d), across triplicate wells for each caspofungin concentration. Endpoint passage population colony morphologies and dose response are presented in the electronic supplementary material, figure S5 for a single population (representative of three) of the passaged SCV from each of experiments A and B.
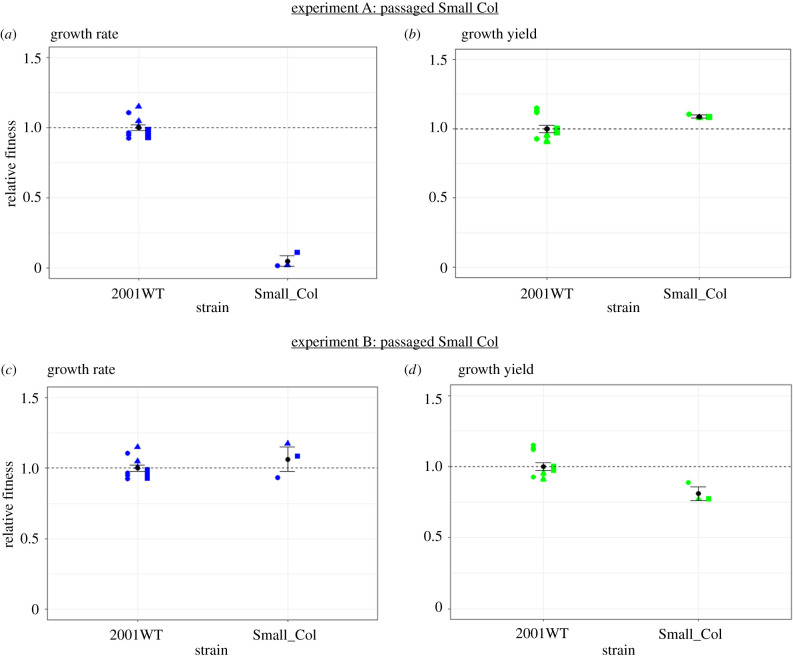
Figure 2.
Final relative growth fitness following passaging without caspofungin of the single-isolated SCV from experiment A and experiment B. Plots (a) and (b) show data from the SCV isolated from experiment A; plots (c) and (d) represent the SCV isolated from experiment B. The wild-type ancestral strain (2001WT) data are as presented in figure 1a,b (n = 12) as this strain was not passaged. n = 3 per SCV (different shape symbol per replicate), representing a single endpoint measurement from each of three passaged populations seeded from a single culture of the SCV. Growth values are calculated relative to the average values of 2001WT, where a value of 1.0 signifies no change relative to the ancestor. Experiment A: average relative growth rate of SCV (±s.e.) = 0.05 ± 0.04; average relative growth yield of SCV (±s.e.) = 1.09 ± 0.01. Experiment B: average relative growth rate of SCV (±s.e.) = 1.06 ± 0.09; average relative growth yield of SCV (±s.e.) = 0.81 ± 0.05. Black points and error bars represent mean and standard error. (Online version in colour.)
(ii). Characterization of genomic targets
Genomic DNA was extracted from 2001WT and the single-isolated SCV and RCV variants from each of experiments A and B by mechanical cell lysis as described in [39]. Briefly, cells were grown overnight, centrifuged and mixed with Smash and Grab Solution (1% SDS, 2% Triton-X, 100 mM NaCl, 10 mM Tris pH 8.0), phenol-chloroform and acid-washed glass beads. The HS1 and HS2 regions of the FKS1 and FKS2 genes were amplified by polymerase chain reaction and Sanger-sequenced using primers previously described [40,41]. Amplification of genes CDC6, DOT6, MRPL11, SUI2 was performed using primers described for C. glabrata [11].
(iii). Competitive fitness assay between small colony variant and regular colony variant (experiment A)
To test competitive fitness of the stable SCV isolated from experiment A against its co-isolated RCV, the two colony variants were competed across a set of mixed strain ratios in SC 1% glucose medium supplemented with 0.78 µg ml−1 caspofungin in wells of a 96-well plate (electronic supplementary material, figure S6). Each strain was adjusted to 6.49 × 106 cells ml−1 prior to mixing and subsequent twofold dilution in SC media. Each mixed strain ratio was added to triplicate wells. The plate was incubated at 30°C with shaking at 180 rpm. Competition mixes were plated on SC 1% glucose agar plates at the start (n = 3) and end (n = 3 wells × 3 plates = 9) of 24 h growth, on which the colony morphotypes could clearly be distinguished by size. Relative fitness of the SCV was calculated as the ratio of the Malthusian growth parameters of the SCV and RCV [42]. A least-squares linear regression of relative fitness against initial SCV frequency was plotted and statistically analysed in Excel [43]. Two-tailed one-sample t-tests in R version 3.4.3 [36] tested significant deviations of relative fitness values from one.
(e). Galleria mellonella survival assays
The 2001WT strain, regular colony, unpassaged and passaged small colony variants were tested for virulence in Galleria mellonella wax moth larvae. Candida glabrata strains were grown in SC (2% glucose) media for 24 h at 30°C, cells were washed twice in phosphate buffered saline (PBS), counted on a haemocytometer and adjusted to 2.5 × 108 cells ml−1 in PBS with 20 µg ml−1 ampicillin. Groups of 20 wax moth larvae (UK Waxworms Ltd, Sheffield), selected based on their weight (0.25–0.35 g) and lack of visible melanization, were injected with 10 µl spore suspension (2.5 × 106 colony forming unit (CFU) larva−1) using a 50 µl Hamilton syringe into their last left pro-leg. Larvae were maintained at 37°C in the dark and monitored for survival. Three independent replicates of survival assays were performed on separate days (n = 20 × 3 = 60 larvae in total per C. glabrata strain). A control group of 10 larvae injected with PBS and ampicillin were run alongside each independent replicate, and no deaths were seen. Mean survival time and log-rank comparisons of survival curves were calculated using OASIS 2 [44] for each of the three replicates. As routinely done with G. mellonella survival assays [45,46], data from one of the replicates is presented in the main manuscript (figure 3) while the data from the other two are presented in the electronic supplementary material, figures S7 and S8.
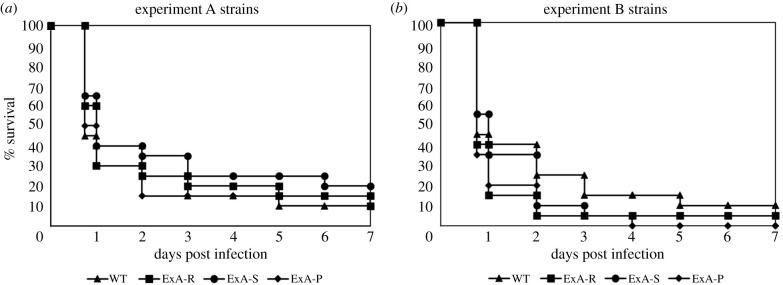
Figure 3.
Virulence of C. glabrata wild-type ancestral, small and regular colony size variants in G. mellonella larvae. Survival of groups of 20 G. mellonella wax moth larvae injected with 2.5 × 106 CFU larva−1 strain−1 over 7 days incubation at 37°C. These data represent one independent replicate of the survival analysis; the second and third replicates are presented in the electronic supplementary material, figures S7 and S8. (a) WT, 2001WT ancestral strain; ExA-R, experiment A RCV; ExA-S, experiment A SCV; ExA-P, experiment A passaged SCV. (b) ExB-S, experiment B SCV; ExB-R, experiment B RCV; ExB-P, experiment B passaged SCV (revertant). No significant differences in survival in log-rank tests were found across experiment A (p ≥ 0.3481) or experiment B strains (p ≥ 0.08).
(f). Testing for correlations between larval survival time and growth rate or yield
The mean relative growth rates or yields for each of the strains from experiments A and B (reported in figures 1a,b and 2, and electronic supplementary material, figure S4a,b) were plotted against their virulence calculated as larval survival time from either figure 3 (first replicate survival study), electronic supplementary material, figure S7 (second replicate survival study) or electronic supplementary material, figure S8 (third replicate survival study); this generated figure 4, electronic supplementary material, figures S10a and S11a, respectively. Note, as growth trait replicate measurements were not directly paired with larval survival time measurements, their mean values were plotted with standard error bars representing the variability in estimation. Bootstrapping was performed on data in figure 4, electronic supplementary material, figures S10a and S11a both for linear and Deming regressions and also for Pearson and Spearman correlations (using MATLAB's Statistics and Machine Learning Toolbox) [37]. The resulting statistics are reported in the electronic supplementary material, figures S9a and b and electronic supplementary material, figures S10–11b and c.
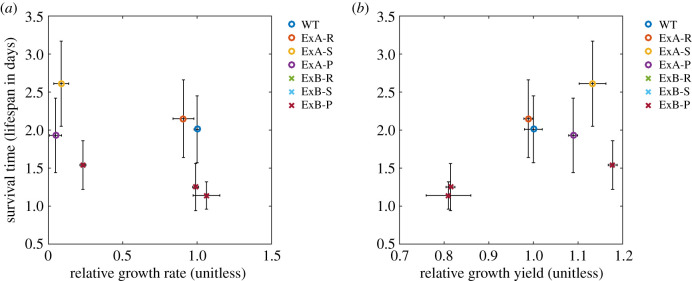
Figure 4.
Growth rate, growth yield and virulence correlations. Growth traits are plotted for all strains from experiments A and B, including RCVs and SCVs before and after passaging. Data is combined from figures 1a,b, 2 and 3 and electronic supplementary material, figure S4a and b. Plotted points represent mean values ± s.e. Growth rate and yield are plotted relative to the wild-type ancestral strain (2001WT). Strains are labelled as follows: experiment A strains: ExA-R (RCV); ExA-S (SCV); ExA-P (passaged ‘stable' small colony). Experiment B strains: ExB-R (RCV); ExB-S (SCV); ExB-P (passaged ‘unstable' small colony). Bootstrapping was performed for both linear and Deming regressions, in addition to both Pearson and Spearman but we found no evidence of correlations between virulence and either relative growth rate (electronic supplementary material, figure S9a) or relative growth yield (electronic supplementary material, figure S9b). (Online version in colour.)
3. Results
(a). Rapid evolution of sub-population phenotypic diversity with antifungal caspofungin treatment
We measured growth adaptations of populations of the reference C. glabrata strain ATCC 2001 (denoted 2001WT), serially transferred over 14 days and evolving across a gradient of eight antifungal caspofungin concentrations (see Methods). Growth across C. glabrata populations, relative to no-drug-treated controls, significantly increased over time (effect of day: likelihood ratio test: p = 4.404 × 10−12) (electronic supplementary material, figure S2). Growth was also significantly influenced by caspofungin concentration (likelihood ratio test: p < 2.2 × 10−16). The greatest relative population growth increase from 3.0 ± 2.5 (s.e.) % on day 1, to 88.8 ± 7.0 (s.e.) % on day 14 occurred at 0.78 µg ml−1 of caspofungin.
We revived triplicate endpoint (day 14) populations that were evolved at the three highest caspofungin concentrations (0.78, 1.37 and 2.40 µg ml−1), to phenotype colony morphologies (electronic supplementary material, figure S1b). We identified colony size variation in a single population from experiment A and a single population from experiment B, which had both been evolved during treatment in 0.78 µg ml−1 caspofungin. Two size variants were observed in each of these populations: a SCV and a RCV (electronic supplementary material, figure S3). We did not observe the SCV phenotype in any populations from experiment C. The small colony morphology and lower growth rate of the isolated SCV resembled the respiratory-deficient petite phenotype previously observed for C. glabrata [11]; however, our SCV strains were able to grow on a non-fermentable carbon source, indicating respiratory function (electronic supplementary material, figure S3).
We measured growth rates and final population densities (yield) of the SCV and RCV strains, relative to strain 2001WT in the absence of caspofungin. Relative growth rate significantly varied with colony type (experiment A (figure 1a): p < 2.2 × 10−16; experiment B (electronic supplementary material, figure S4a): p = 7.926 × 10−16). No significant differences were found between 2001WT and RCV, but growth rate of SCV was significantly lower than both 2001WT and RCV.
Relative growth yield significantly varied with colony type (experiment A (figure 1b): p = 2.195 × 10−5; experiment B (electronic supplementary material, figure S4b): p = 3 × 10−8). Growth yield of the SCV was significantly greater than both 2001WT and the RCV. In experiment B only, yield of the RCV was significantly lower than 2001WT.
We next measured the caspofungin dose response of the variants from experiments A and B that were growth profiled. Caspofungin susceptibility profiles of the two colony variants were highly divergent. From experiment A, the RCV (Reg_Col) had an IC50 of 0.26 ± 0.007 µg ml−1 that was slightly lower (marginally significantly) than the 2001WT strain IC50 (0.32 ± 0.0025 µg ml−1; p = 0.0454) (figure 1c). The co-isolated SCV (Small_Col), however, was not susceptible to caspofungin concentrations up to 2.4 µg ml−1 (figure 1d) and had an IC50 of 2.61 ± 0.136 µg ml−1, approximately 10-fold greater than the IC50 of the co-isolated RCV and 8.2-fold greater than the 2001WT strain.
In experiment B, the RCV (Reg_Col) had an IC50 of 0.83 ± 0.096 µg ml−1, significantly greater than the 2001WT strain (p = 0.0011; electronic supplementary material, figure S4c). The co-isolated SCV (Small_Col) was not sensitive to caspofungin concentrations up to 2.4 µg ml−1 (electronic supplementary material, figure S4d) and had an IC50 of 4.71 ± 0.236 µg ml−1, 5.7-fold greater than the regular co-isolated colony variant and 14.7-fold greater than the 2001WT strain.
(b). Divergent phenotypic stability of independently evolved small colony variants, in the absence of drug
We investigated stability of the resistant SCV, individually isolated from the single population from each of experiments A and B. For this, we serially passaged three replicate populations seeded from an individual culture of each SCV (from experiments A and B), over 14 days in the absence of caspofungin. For experiment A, the small colony morphology and resistance level showed no reversion via appearance of regular-sized colonies after 14 days. The ‘stable' small colony phenotype from experiment A maintained a significantly lower relative growth rate than the 2001WT strain on the 14th day of transfer in the absence of caspofungin (t13 = 21.952, p = 1.169 × 10−11) (figure 2a). However, after passage relative growth yield did not significantly differ from the wild-type (t13 = 1.7375, p = 0.1059) (figure 2b). The level of resistance displayed by the SCV was maintained, with no growth inhibition between 0–2.4 µg ml−1 of caspofungin (electronic supplementary material, figure S5: experiment A). The IC50 values for a single colony of each passaged population of the SCV from experiment A (experimental set-up as shown in the electronic supplementary material, figure S1d) were as follows: 2.40 ± 0.146 µg ml−1 (population 1), 2.33 ± 0.678 µg ml−1 (population 2) and 2.70 ± 0.480 µg ml−1 (population 3)). This indicated an underlying stable resistance mechanism and a lack of compensatory fitness improvement.
By contrast, the resistant small colony phenotype from experiment B was homogeneously reversed in all three passaged replicate populations after 14 days. The ‘unstable’ small colony phenotype from experiment B had lost its lower relative growth rate by the 14th transfer day and was not significantly different from 2001WT (t13 = 1.2265, p = 0.2418) (figure 2c). This small colony variant after passage showed a significant 1.2-fold lower relative growth yield than the wild-type (t13 = 3.5315, p = 0.003685) (figure 2d). The resistance level of the passaged populations decreased but sensitivity levels (IC50 values) were not completely restored to wild-type levels: in two populations, the IC50 values were higher than 2001WT (0.49 ± 0.023 µg ml−1; p = 0.000336 (electronic supplementary material, figure S5: experiment B, population 1) and 0.44 ± 0.019 µg ml−1; p = 0.00463 (population 2)) but were comparable in the third population (0.34 ± 0.055 µg ml−1; p = 0.797 (population 3)). Despite independent evolution of the small colony phenotype in separate populations during caspofungin treatment, the findings indicate that the SCV isolated from experiment B evolved a different but transient mechanism of resistance compared with the SCV from experiment A.
We tested whether resistance seen in the small colony phenotype from experiments A and B correlated with genetic differences in hotspot regions of the FKS genes, which are common targets associated with caspofungin resistance [11,40]. Sequenced regions were identical in all strains with no nucleotide changes detected. Furthermore, we identified no nucleotide differences between strains in an additional set of genetic targets (CDC6, DOT6, MRPL11, SUI2) putatively associated with development of caspofungin resistance [11].
We then tested competitive fitness of the isolated SCV against the RCV from the evolved population in experiment A, over five different initial frequencies of the SCV in the presence of caspofungin (0.78 µg ml−1). Relative fitness of the SCV was significantly greater than one for all initial frequencies, apart from the highest starting SCV frequency (0.95) when there was no significant difference (electronic supplementary material, figure S6). Relative fitness of the SCV was significantly negative frequency-dependent (least-squares linear regression: slope = −4.8990, t42 = −7.554, p = 2.37 × 10−9). These results lead to a longer term prediction that a low-frequency of an RCV could coexist with a high frequency of an SCV; however, the underlying mechanisms of this dynamic were not explored here.
(c). Independently-evolved drug-resistant small colony variants are not attenuated in virulence
We tested virulence of 2001WT, SCVs (experiments A and B) and co-isolated RCVs in the wax moth model G. mellonella. The ‘stable' small colony variant from experiment A was virulent in G. mellonella both before and after passaging without caspofungin (figure 3a). Mean larval survival times were 2.61 ± 0.56 days and 1.93 ± 0.49 days, respectively, and we found no significant differences from 2001WT (2.01 ± 0.44 days; log-rank test p-values = 0.3481 (ExA-S); 0.9834 (ExA-P)) or the co-isolated RCV (2.15 ± 0.51 days; p = 0.438 (ExA-S); 0.9483 (ExA-P)). For the ‘unstable' SCV from experiment B we found no significant difference in G. mellonella mean survival times when comparing states before (1.54 ± 0.32 days) and after (1.14 ± 0.18 days) loss of the phenotype (p = 0.2393) (figure 3b). No significant differences in mean larval survival time occurred between 2001WT and either ExB-S (p = 0.5338) or ExB-P (p = 0.08), nor between ExB-R (1.25 ± 0.31 days) and either ExB-S (p = 0.2825) or ExB-P (p = 0.8389). These findings were consistent with two additional independent replicate survival studies conducted on separate days (electronic supplementary material, figures S7 and S8).
Bootstrapping analysis tested for, but found no evidence of, correlations between larval survival time (measure of virulence) and either relative growth rates or yields in three replicate survival studies (figure 4, electronic supplementary material, figures S9–S11). Further, G. mellonella infection studies involving larger sample sizes of drug-evolved clones would be needed to establish evidence for an absence of correlation.
4. Discussion
Our work demonstrates that drug resistance cost in the form of reduced growth rate, does not necessarily lead to a marked reduction in virulence. This challenges the current understanding of the resistance–virulence relationship [8]. Using a clinical isolate of a deadly human pathogen C. glabrata [25,26], we conducted in vitro evolutionary studies in the presence of an antifungal drug and repeatedly evolved a resistant SCV (figure 1 and electronic supplementary material, figure S4). Despite having a significantly reduced growth rate compared to the susceptible wild-type, the SCVs did not suffer a marked alteration in virulence when tested in G. mellonella, a well-established model host for detecting virulence differences in C. glabrata strains [23]. The isolated SCVs represented a sub-population of highly resistant individuals, among susceptible sub-populations within a drug-treated population—a hallmark of heteroresistance [12]. Increasingly observed among fungal [10,47] and bacterial [12] pathogens, heteroresistance presents challenges for detection and antibiotic susceptibility testing [12]. This can lead to persistent and recurrent infections [48–50] and missed detection of resistant sub-populations [51].
At the first glance, the small colony morphology and lower growth rate of our isolated SCVs resembles the well-known C. glabrata petite phenotype [11,52,53]. Candida glabrata is known to rapidly evolve resistance to caspofungin [11], a front-line therapeutic for Candida infections, also used in our study. During patient treatment with caspofungin for recurring bloodstream candidemia, resistant isolates including a small colony phenotype were recovered [11]. These small colony isolates were found to be respiratory-deficient having lost mitochondrial function and were termed petite mutants. The petite phenotype was first observed in the yeast S. cerevisiae [54] and more recently identified in C. glabrata azole-resistant isolates [52,53]. However, unlike previously identified caspofungin-resistant petite mutants [11] our SCVs do not suffer a reduction in virulence (figure 3) and can grow on media containing glycerol as a non-fermentative carbon source, indicating respiratory function (electronic supplementary material, figure S3). This indicates potential presence of a different resistance mechanism that co-occurs with virulence, not previously observed for the cidal (cell-killing) echinocandin drug class [11]. In line with our findings, a prior study that isolated a C. glabrata azole-resistant petite mutant during clinical infection identified that it had enhanced virulence but reduced in vitro fitness in the absence of the drug, compared to the sensitive co-isolated type [55]. Thus we argue that detection of reduced in vitro growth rate of a sub-population clone [11] is not a reliable indicator of virulence.
Our study also indicates that small colony resistance in response to the front-line antifungals can be both reversible and non-reversible (electronic supplementary material, figure S5) when the drug is removed. So far, studies on the small colony phenotype have shown reversibility of growth rate and either reversibility or non-reversibility of resistance [56]. For example, petite C. glabrata mutants result from stable genetic alteration of mitochondrial DNA and show upregulation of drug efflux transporters when evolved with azoles [52,53,57]. Stability over passaging in the absence of drug, for the first study to isolate C. glabrata petite mutants during echinocandin treatment, was not investigated [11]. By contrast, heteroresistance has consistently proven reversible for both clinical and environmental isolates of Cryptococcus neoformans exposed to most commonly used antifungals [58]. Other studies have shown that resistance achieved through induction is reversible, while resistance achieved through selection is non-reversible in a range of Candida species [59]. Here, we show that different resistance mechanisms could be acting in parallel giving rise to either reversibility or non-reversibility. Recent studies of bacterial heteroresistance describe stable heteroresistance to be associated with minimal fitness costs, whereas unstable heteroresistance has been associated with larger fitness costs leading to compensatory changes that often reverse resistance [60]. By contrast, we find low growth rates for both our reversible and non-reversible SCVs. Moreover, we show that the resistant small colony phenotype can be stable with passaging in the absence of antibiotics, without compensatory mechanisms that restore growth rate as was previously described as a necessity for stability [56]. This highlights the need for future studies to investigate repeatability of heteroresistance development and stability across multiple parallel populations. This will lead to better predictability of evolutionary pathways to resistance development in pathogen populations [61].
Classical theory assumes that growth rate positively correlates with pathogen fitness and virulence [16,62–64] owing to debilitation of the host [65]. Despite some empirical support of this classical assumption [19,66], recent meta-analysis across 61 human pathogens found that growth rate was negatively correlated with virulence [20]. Moreover, comparisons of C. glabrata clinical strains found that a fast grower was less virulent than slower growers [23] with fast growth also being constrained by a low growth yield [24]. Here, we found no evidence of correlation between virulence and either growth rate or growth yield.
Our study further highlights the complex nature of the relationship between pathogen growth rate and virulence. In particular, existing empirical studies [20,23,66,67] including ours conducted here, compare virulence between pathogenic strains with undefined genetic differences. As a result, observed virulence variations cannot be mechanistically linked to differences in growth traits. In general, this could be owing to confounding effects such as differences in host responses to genetically diverse pathogens [68] or undefined interactions between pathogen strains [69].
Here, we argue that in order to understand the effect of resistance costs on the evolution of virulence we need to develop an in-depth, mechanistic understanding of the relationship between growth rate and virulence.
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank Richard Lindsay for helpful discussions.
Data accessibility
Additional description of methods, results and the supplementary figures are provided in the electronic supplementary material file ‘Duxbury_Methods_Figures1-11EMS.pdf.’ The datasets supporting this article have been uploaded as part of the supplementary material in file ‘Duxbury_RawDataEMS.xlsx’.
Authors' contributions
S.J.N.D. and I.G. designed the study. S.J.N.D. and S.B. performed the experiments. S.J.N.D. and R.E.B completed data presentation, analysis and statistics. S.J.N.D., R.E.B, S.B. and I.G. wrote the manuscript.
Competing interests
We declare that we have no competing interests
Funding
This work was supported by a BBSRC PhD studentship to S.J.N.D. I.G. was funded by an ERC Consolidator grant no. (MathModExp 647292) and R.E.B. was funded by an EPSRC Healthcare Technology Impact Fellowship (grant no. EP/N033671/1).
References
- 1.O'Neill J. 2016. Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations. London, UK: HM Government. [Google Scholar]
- 2.Levy SB. 1997. Antibiotic resistance: an ecological imbalance. Ciba Found Symp. 207, 1–14. ( 10.1002/9780470515358.ch1) [DOI] [PubMed] [Google Scholar]
- 3.Gillespie SH. 2001. Antibiotic resistance in the absence of selective pressure. Int. J. Antimicrob. Agents 17, 171–176. ( 10.1016/S0924-8579(00)00340-X) [DOI] [PubMed] [Google Scholar]
- 4.Kolář M, Urbánek K, Látal T. 2001. Antibiotic selective pressure and development of bacterial resistance. Int. J. Antimicrob. Agents 17, 357–363. ( 10.1016/S0924-8579(01)00317-X) [DOI] [PubMed] [Google Scholar]
- 5.Andersson DI. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9, 461–465. ( 10.1016/j.mib.2006.07.002) [DOI] [PubMed] [Google Scholar]
- 6.Hall AR, Angst DC, Schiessl KT, Ackermann M. 2015. Costs of antibiotic resistance: separating trait effects and selective effects. Evol. Appl. 8, 261–272. ( 10.1111/eva.12187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat . Rev. Microbiol. 8, 260–271. ( 10.1038/nrmicro2319) [DOI] [PubMed] [Google Scholar]
- 8.Beceiro A, Tomás M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26, 185–230. ( 10.1128/CMR.00059-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen T, Sommers B, Murray M. 2003. The effect of drug resistance on the fitness of Mycobacterium tuberculosis. Lancet Infect. Dis. 3, 13–21. ( 10.1016/S1473-3099(03)00483-3) [DOI] [PubMed] [Google Scholar]
- 10.Ferreira GF, Santos DA. 2017. Heteroresistance and fungi. Mycoses 60, 562–568. ( 10.1111/myc.12639) [DOI] [PubMed] [Google Scholar]
- 11.Singh-Babak SD, et al. 2012. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 8, e1002718 ( 10.1371/journal.ppat.1002718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207. ( 10.1128/CMR.00058-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sifri CD, Baresch-Bernal A, Calderwood SB, von Eiff C. 2006. Virulence of Staphylococcus aureus small colony variants in the Caenorhabditis elegans infection model. Infect. Immun. 74, 1091–1096. ( 10.1128/IAI.74.2.1091-1096.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durante-Mangoni E, Del Franco M, Andini R, Bernardo M, Giannouli M, Zarrilli R. 2015. Emergence of colistin resistance without loss of fitness and virulence after prolonged colistin administration in a patient with extensively drug-resistant Acinetobacter baumannii. Diagn. Micr. Infec. Dis. 82, 222–226. ( 10.1016/j.diagmicrobio.2015.03.013) [DOI] [PubMed] [Google Scholar]
- 15.Hurdle JG, O'Neill AJ, Chopra I. 2004. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J. Antimicrob. Chemother. 53, 102–104. ( 10.1093/jac/dkh020) [DOI] [PubMed] [Google Scholar]
- 16.Anderson RM, May R. 1982. Coevolution of hosts and parasites. Parasitology 85, 411–426. ( 10.1017/S0031182000055360) [DOI] [PubMed] [Google Scholar]
- 17.Bull JJ. 1994. Virulence. Evolution 48, 1423–1437. ( 10.1111/j.1558-5646.1994.tb02185.x|) [DOI] [PubMed] [Google Scholar]
- 18.Paisley D, Robson GD, Denning DW. 2005. Correlation between in vitro growth rate and in vivo virulence in Aspergillus fumigatus . Med. Mycol. 43, 397–401. ( 10.1080/13693780400005866) [DOI] [PubMed] [Google Scholar]
- 19.de Roode JC, et al. 2005. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl Acad. Sci. USA 102, 7624–7628. ( 10.1073/pnas.0500078102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leggett HC, Cornwallis CK, Buckling A, West SA. 2017. Growth rate, transmission mode and virulence in human pathogens. Phil. Trans. R. Soc. B 372, 20160094 ( 10.1098/rstb.2016.0094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaller MA, Castanheira M, Lockhart SR, Jones RN. 2012. Candida glabrata: multidrug resistance and increased virulence in a major opportunistic fungal pathogen. Curr. Fung. Infect. Rep. 6, 154–164. ( 10.1007/s12281-012-0091-0) [DOI] [Google Scholar]
- 22.Alexander BD, et al. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of fks mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56, 1724–1732. ( 10.1093/cid/cit136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ames L, Duxbury S, Pawlowska B, Ho HL, Haynes K, Bates S. 2017. Galleria mellonella as a host model to study Candida glabrata virulence and antifungal efficacy. Virulence 8, 1909–1917. ( 10.1080/21505594.2017.1347744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reding-Roman C, Hewlett M, Duxbury S, Gori F, Gudelj I, Beardmore R. 2017. The unconstrained evolution of fast and efficient antibiotic-resistant bacterial genomes. Nat. Ecol. Evol. 1, 0050 ( 10.1038/s41559-016-0050) [DOI] [PubMed] [Google Scholar]
- 25.Odds FC. 1996. Epidemiological shifts in opportunistic and nosocomial Candida infections: mycological aspects. Int. J. Antimicrob. Agents 6, 141–144. ( 10.1016/0924-8579(95)00049-6) [DOI] [PubMed] [Google Scholar]
- 26.Gudlaugsson O, Gillespie S, Lee K, Berg JV, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37, 1172–1177. ( 10.1086/378745) [DOI] [PubMed] [Google Scholar]
- 27.Roetzer A, Gabaldon T, Schuller C. 2011. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 314, 1–9. ( 10.1111/j.1574-6968.2010.02102.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunke S, et al. 2015. Of mice, flies–and men? Comparing fungal infection models for large-scale screening efforts. Dis. Model Mech. 8, 473–486. ( 10.1242/dmm.019901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enkler L, Richer D, Marchand AL, Ferrandon D, Jossinet F. 2016. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-CAS9 system. Sci. Rep. 6, 35766 ( 10.1038/srep35766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galocha M, Pais P, Cavalheiro M, Pereira D, Viana R, Teixeira MC. 2019. Divergent approaches to virulence in C. albicans and C. glabrata: two sides of the same coin. Int. J. Mol. Sci. 20, 2345 ( 10.3390/ijms20092345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Usher J, Haynes K. 2019. Attenuating the emergence of anti-fungal drug resistance by harnessing synthetic lethal interactions in a model organism. PLoS Genet. 15, e1008259 ( 10.1371/journal.pgen.1008259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitada K, Yamaguchi E, Arisawa M. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165, 203–206. ( 10.1016/0378-1119(95)00552-H) [DOI] [PubMed] [Google Scholar]
- 33.Espinel-Ingroff A, et al. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. Using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob. Agents Chemother. 57, 5836–5842. ( 10.1128/AAC.01519-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roemhild R, Barbosa C, Beardmore RE, Jansen G, Schulenburg H. 2015. Temporal variation in antibiotic environments slows down resistance evolution in pathogenic Pseudomonas aeruginosa. Evol. Appl. 8, 945–955. ( 10.1111/eva.12330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 36.R Development Core Team. 2017. R: a language and environment for statistical computing: R Foundation for Statistical Computing. See https://www.R-project.org Vienna, Austria. [Google Scholar]
- 37.MATLAB and Statistics Toolbox Release 2012a and 2020a.2012, 2020 Natick, MA: The MathWorks Inc. [Google Scholar]
- 38.Ritz C, Baty F, Streibig JC, Gerhard D. 2015. Dose-response analysis using R. PLoS ONE 10, e0146021 ( 10.1371/journal.pone.0146021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Libuda D.2007. Genomic DNA extraction S. Cerevisiae and S. Pombez. See https://www.princeton.edu/genomics/botstein/protocols/yeast_DNA.pdf .
- 40.Thompson GR, Wiederhold NP, Vallor AC, Villareal NC, Lewis JS, Patterson TF. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 52, 3783–3785. ( 10.1128/AAC.00473-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimbeck AJ, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Chiller T, Lockhart SR. 2010. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from us population-based surveillance. Antimicrob. Agents Chemother. 54, 5042–5047. ( 10.1128/AAC.00836-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341. ( 10.1086/285289) [DOI] [Google Scholar]
- 43.Zaiontz C.2015. Real statistics using Excel. See www.real-statistics.com .
- 44.Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SV, Kim S. 2016. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56 147–56 152. ( 10.18632/oncotarget.11269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flanagan PR, Fletcher J, Boyle H, Sulea R, Moran GP, Sullivan DJ. 2018. Expansion of the TLO gene family enhances the virulence of Candida species. PLoS ONE 13, e0200852 ( 10.1371/journal.pone.0200852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh YC, Wang HY, Lan CY. 2020. Candida albicans Aro1 affects cell wall integrity, biofilm formation and virulence. J. Microbiol. Immunol. Infect. 53, 115–124. (doi:1016/j.jmii.2018.04.002) [DOI] [PubMed] [Google Scholar]
- 47.Ben-Ami R, Zimmerman O, Finn T, Amit S, Novikov A, Wertheimer N, Lurie-Weinberger M, Berman J. 2016. Heteroresistance to fluconazole is a continuously distributed phenotype among Candida glabrata clinical strains associated with in vivo persistence. MBio 7, e00655-16 ( 10.1128/mBio.00655-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proctor RA, van Langevelde P, Kristjansson M, Maslow JN, Arbeit RD. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20, 95–102. ( 10.1093/clinids/20.1.95) [DOI] [PubMed] [Google Scholar]
- 49.Singh R, Ray P, Das A, Sharma M. 2009. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J. Med. Microbiol. 58, 1067–1073. ( 10.1099/jmm.0.009720-0) [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg A, et al. 2018. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 9, 2470 ( 10.1038/s41467-018-04926-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183, 1674–1679. ( 10.1164/rccm.201009-1430OC) [DOI] [PubMed] [Google Scholar]
- 52.Bouchara J-P, Zouhair R, Le Boudouil S, Renier G, Filmon R, Chabasse D, Ballet J-N, Defontaine A. 2000. In vivo selection of an azole-resistant petite mutant of Candida glabrata. J. Med. Microbiol. 49, 977–984. ( 10.1099/0022-1317-49-11-977) [DOI] [PubMed] [Google Scholar]
- 53.Brun S, Dalle F, Saulnier P, Renier G, Bonnin A, Chabasse D, Bouchara J-P. 2005. Biological consequences of petite mutations in Candida glabrata. J. Antimicrob. Chemother. 56, 307–314. ( 10.1093/jac/dki200) [DOI] [PubMed] [Google Scholar]
- 54.Bernardi G. 1979. The petite mutation in yeast. Trends Biochem. Sci. 4, 197–201. ( 10.1016/0968-0004(79)90079-3) [DOI] [Google Scholar]
- 55.Ferrari SSM, De Bernardis F, Torelli R, Posteraro B, Vandeputte P, Sanglard D. 2011. Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 55, 1852–1860. ( 10.1128/AAC.01271-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao S, Huseby DL, Brandis G, Hughes D. 2017. Alternative evolutionary pathways for drug-resistant small colony variant mutants in Staphylococcus aureus. MBio 8, e00358-17 ( 10.1128/mBio.00358-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanglard D, Ischer F, Bille J. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45, 1174–1183. ( 10.1128/AAC.45.4.1174-1183.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6, e1000848 ( 10.1371/journal.ppat.1000848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Claudino ALR, Peixoto Junior RF, Melhem MS, Szeszs MW, Lyon JP, Chavasco JK, Franco MC. 2009. Mutants with heteroresistance to amphotericin b and fluconazole in Candida. Braz. J. Microbiol. 40, 943–951. ( 10.1590/S1517-83822009000400028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson DI, Nicoloff H, Hjort K. 2019. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 17, 479–496. ( 10.1038/s41579-019-0218-1) [DOI] [PubMed] [Google Scholar]
- 61.van Dijk T, Hwang S, Krug J, de Visser JAGM, Zwart MP. 2017. Mutation supply and the repeatability of selection for antibiotic resistance. Phys. Biol. 14, 055005 ( 10.1088/1478-3975/aa7f36) [DOI] [PubMed] [Google Scholar]
- 62.Nowak MA, May RM. 1994. Superinfection and the evolution of parasite virulence. Proc. R. Soc. B 255, 81–89. ( 10.1098/rspb.1994.0012) [DOI] [PubMed] [Google Scholar]
- 63.Levin BR, Bull JJ. 1994. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 2, 76–81. ( 10.1016/0966-842X(94)90538-X) [DOI] [PubMed] [Google Scholar]
- 64.Frank SA. 1996. Models of parasite virulence. Q. Rev. Biol. 71, 37–78. ( 10.1086/419267) [DOI] [PubMed] [Google Scholar]
- 65.Perlman RL. 2009. Life histories of pathogen populations. Int. J. Infect. Dis. 13, 121–124. ( 10.1016/j.ijid.2008.07.003) [DOI] [PubMed] [Google Scholar]
- 66.Ben-Ami F, Mouton L, Ebert D. 2008. The effects of multiple infections on the expression and evolution of virulence in a Daphnia-endoparasite system. Evolution 62, 1700–1711. ( 10.1111/j.1558-5646.2008.00391.x) [DOI] [PubMed] [Google Scholar]
- 67.Buckling A, Brockhurst MA. 2008. Kin selection and the evolution of virulence. Heredity 100, 484–488. ( 10.1038/sj.hdy.6801093) [DOI] [PubMed] [Google Scholar]
- 68.Taylor LH, Mackinnon MJ, Read AF. 1998. Virulence of mixed-clone and single-clone infections of the rodent malaria Plasmodium chabaudi. Evolution 52, 583–591. ( 10.1111/j.1558-5646.1998.tb01656.x) [DOI] [PubMed] [Google Scholar]
- 69.Davies CM, Fairbrother E, Webster JP. 2002. Mixed strain schistosome infections of snails and the evolution of parasite virulence. Parasitology 124, 31–38. ( 10.1017/S0031182001008873) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional description of methods, results and the supplementary figures are provided in the electronic supplementary material file ‘Duxbury_Methods_Figures1-11EMS.pdf.’ The datasets supporting this article have been uploaded as part of the supplementary material in file ‘Duxbury_RawDataEMS.xlsx’.