Abstract
Müllerian mimicry strongly exemplifies the power of natural selection. However, the exact measure of such adaptive phenotypic convergence and the possible causes of its imperfection often remain unidentified. Here, we first quantify wing colour pattern differences in the forewing region of 14 co-mimetic colour pattern morphs of the butterfly species Heliconius erato and Heliconius melpomene and measure the extent to which mimicking colour pattern morphs are not perfectly identical. Next, using gene-editing CRISPR/Cas9 KO experiments of the gene WntA, which has been mapped to colour pattern diversity in these butterflies, we explore the exact areas of the wings in which WntA affects colour pattern formation differently in H. erato and H. melpomene. We find that, while the relative size of the forewing pattern is generally nearly identical between co-mimics, the CRISPR/Cas9 KO results highlight divergent boundaries in the wing that prevent the co-mimics from achieving perfect mimicry. We suggest that this mismatch may be explained by divergence in the gene regulatory network that defines wing colour patterning in both species, thus constraining morphological evolution even between closely related species.
Keywords: mimicry, WntA, developmental constraints, convergence, butterflies
1. Introduction
Adaptation is the product of natural selection on the genetic and phenotypic diversity within a population [1]. In this regard, key developmental steps that limit or bias trait variation can pose so-called constraints on the directionality of evolution [2–4]. Consequently, when populations evolve independently, lineages can accumulate changes that lead evolution along an irreversible trajectory [5]. Understanding the relative contribution (or constraint) of genetics and development to adaptation would therefore allow us to better comprehend the directionality and predictability of evolution [6].
To date, the extent to which evolutionary changes are consequential for future adaptation has been most elegantly studied using artificial selection experiments [1]. In these experiments, multiple generations can be relatively easily traced while being exposed to contrasting selection pressures. The consequences of their adaptations can then be investigated when these selection pressures are reversed. In malaria, for example, a single mutation of large effect in the K13 protein can confer drug resistance [7] but simultaneously also favours the evolution of additional epistatic mutations [8]. As a consequence of these changes, the evolved phenotype cannot be simply reversed to its ancestral state when withdrawing anti-malarial medication and thus has important consequences for resistance management strategies [8]. In butterflies, artificial selection on eyespots has been used to test for the existence of developmental constraints in wing colour patterns. Because these wing pattern elements belong to the same homologous series and share developmental pathways, covariances between them are expected in their size and shape. Nevertheless, in the butterfly Bicyclus anynana, artificial selection has suggested that there is great potential for independent change in size and shape of different eyespots [9]. This observation has been used to argue that natural selection plays a dominant role over developmental constraints in the observed eyespot diversity among Bicyclus species [9]. Similarly, in Drosophila, artificial selection experiments have shown that changes to wing shape can be rapidly induced from standing genetic variation present in the populations [10]. However, these induced phenotypes are lost when selection is suspended, presumably due to pleiotropic links to other traits that result in negative fitness consequences [11].
Not all study systems are amenable to perform artificial selection experiments. Alternatives to such controlled experiments are cases of convergent evolution that provide natural opportunities to investigate the selective, genetic, and developmental routes to adaptation [1]. For example, mimicry and the resulting phenotypic convergent evolution between distinct butterfly species provides a comparative framework to investigate the genes underlying the evolution and diversity of a wing colour pattern [12–14]. Recent studies have shown, for example, that the genes WntA, cortex, and optix are repeatedly used to control variation in wing colour patterns across Nymphalid butterflies [14–17]. Nevertheless, even in cases of Müllerian mimicry between species within the Heliconius genus, in which both partners have used the same genes to converge on an aposematic warning signal, some degree of imperfection in resemblance may exist. However, the precise extent to which Heliconius mimetic butterflies need to perfectly resemble the same phenotype to maximize the fitness value is not well understood. What may underlie these differences in resemblance are (i) conflicting or relaxed selection pressures and/or (ii) genetic and developmental constraints. Conflicting selective pressures can include variation in the mimicry community [18] and conflict between the outcomes of natural and sexual selection [19]. Relaxed selection pressures may result from coarse discrimination by predators [20,21]. On the other hand, genetic and developmental constraints can result from divergence in the genetic background or in the assembled gene regulatory network that affects the functioning and phenotypic effect of these genes.
Heliconius erato and Heliconius melpomene provide a textbook case of Müllerian mimicry. Although there is no evidence for gene flow between H. erato and H. melpomene, which split around 12–14 Mya [22], their resemblance in wing colour patterns is remarkable. This phenotypic convergence has evolved through strong selection pressures that benefit a common warning pattern that birds have learned to associate with unpalatability [23,24]. The discriminatory visual properties of birds appear to be quite precise, resulting in strong selection pressures for fine scale adjustments of the shape and size of colour patterns among local mimetic butterfly communities (for an overview of selection coefficients see [25]) [26,27].
Recently, a series of functional experiments have tested the role of the WntA gene in different Heliconius butterfly species and populations [12]. WntA is a member of the Wnt family of signalling ligands and a key molecular tool for butterfly wing colour pattern development. While the gene coding sequence is highly conserved across Lepidoptera, its cis-regulatory diversity underlies wing pattern shape variation between and within butterflies species [28,29]. Recent CRISPR/Cas9 KO experiments have shown its role in defining the ultimate colour fate of individual wing scale cells and highlighted incredible variability in the position and wing territory affected by WntA in H. erato and H. melpomene [12]. This result provides a visual representation of the effect of a divergent genetic background on a gene's function. In H. erato and H. melpomene, CRISPR/Cas9 WntA mutant phenotypes evidenced a more restricted area of modified black scales in the forewing of H. melpomene compared to H. erato [12,17]. This result demonstrates that although the two co-mimetic butterflies display identical forewing colourations, the genetic architecture and gene regulatory networks underlying their resemblance may be more complex than previously thought. These differences likely arise from divergence in the regulation of WntA and/or divergence in epistatic interactions with other genes, together defined as the gene regulatory network [12].
In the current study, we aimed to explore the contribution of genetic and developmental constraints to the phenotypic mismatch between co-mimetic pairs of Heliconius butterflies. We therefore first quantitatively measured and compared differences in the mid-forewing band (MFB) pattern of 14 co-mimicking populations of the butterfly species H. erato and H. melpomene from Central and South America. We then interpreted these differences in light of the recent Heliconius WntA CRISPR/Cas9 KO mutants [12]. We were specifically interested to test how species-specific divergence might impact the developmental function of WntA and limit adaptive convergence. More precisely, we argue that overlap of wild-type differences with differences in pattern boundaries as defined by WntA KOs in H. erato and H. melpomene would suggest the possible existence of genetic and developmental constraints for natural selection to achieve perfect mimicry. By combining the most recent methodological advances in functional experiments with quantitative measures, our study offers an alternative approach to artificial selection experiments to test the relative constraint of genetics and development to adaptation.
2. Materials
(a). Colour pattern analysis
We obtained 8–14 images of each of 14 mimicking colour pattern morphs (electronic supplementary material, tables S1 and S2). Images were obtained through the authors' collections, collaborations, and collections made publicly available by Cuthill et al. [30] and Jiggins et al. [31]. Individual genders were determined based on sexual dimorphism in the androconial region [32]. To align images, we used a total of 11 landmarks at vein intersections on one forewing of each sample (electronic supplementary material, figure S1). Methods and results describing additional analysis to control for interspecific changes in wing shape and sex differences are described in Supplementary materials S1 and electronic supplementary material, table S3. While WntA is involved in black scale development in both the fore- and hindwing in Heliconius [12,17], differences in the distribution of WntA have mainly been found to correlate with the position of black colour in the central part of the forewing among Heliconius colour pattern morphs, here called mid-forewing band or MFB [28]. With the interest of studying variation in the MFB, we extracted and focused on the area of the forewing in which black is absent and in which WntA is thus likely not expressed. MFB patterns were extracted and aligned using the R package patternize [33]. Depending on the MFB phenotype, we specified red, green, and blue (RGB) values for red, yellow, and/or white with a colour threshold (colOffset) chosen to fully extract the pattern. For H. e. notabilis, H. m. plesseni, and H. m. cythera, we extracted and combined both red and white to represent the MFB shape. Background noise or damaged regions in the wing that were co-extracted with the colour patterns were masked using the setMask function. Next, a thin plate spline (tps) transformation was obtained from transforming landmarks to a common reference sample. This common reference sample included the landmarks of an arbitrarily chosen sample and was used in all colour pattern analysis. The tps transformation was then used to align and compare the extracted MFB shape, size, and position.
Differences in the MFB patterns were first compared by subtracting the H. erato and H. melpomene pattern frequencies of each population, obtained with the sumRaster function in patternize (i.e. absolute MFB difference) and compared between co-mimics using a one-sample t-test. Next, the relative size of the pattern was calculated as the proportion of the total wing area in which the pattern is observed, using the patArea function (i.e. relative MFB difference) and compared between co-mimics using a two-sample t-test. Principal component analysis (PCA) was performed on the binary representation of the aligned colour pattern rasters of each sample [33]. The PCA visualizes the main variations in colour pattern boundaries among samples and groups and provides predictions of colour pattern changes along the PC axis, with positive values presenting a higher predicted presence of the MFB pattern and negative values presenting the absence of the pattern. Parts of the colour patterns that are present in all samples have a predicted value of zero, as these pixels do not contribute variance in the PCA. We tested the effect of population, sex, and species on shape variables using multivariate analysis of variance (MANOVA) and linear discriminant analysis (LDA) using only the values of samples along significant PC axes (see Supplementary Materials S1 for details).
(b). WntA CRISPR KO analysis
Five mutant butterflies (10 wings) for each of the Panamanian geographical colour pattern morphs Heliconius erato demophoon and Heliconius melpomene rosina for which a frame shift mutation was generated at the gene WntA using CRISPR/Cas9 were obtained from Concha et al. [12]. All these mutants showed symmetric changes in wing patterns on both the left and right forewings and were thus likely full KO mutants [12]. Both left and right forewings were landmarked and the mutant pattern was extracted and aligned using the R package patternize [33]. Red was extracted from the H. e. demophoon mutants. As the H. m. rosina mutants often showed a yellow spot appearing in the proximal part of the MFB, both red and yellow were extracted for H. m. rosina. The mutant patterns were superimposed on the wild-type wing pattern comparisons by aligning to the common reference sample and using the contour function of the R package raster [34].
3. Results
(a). Divergence and convergence in mid-forewing band pattern
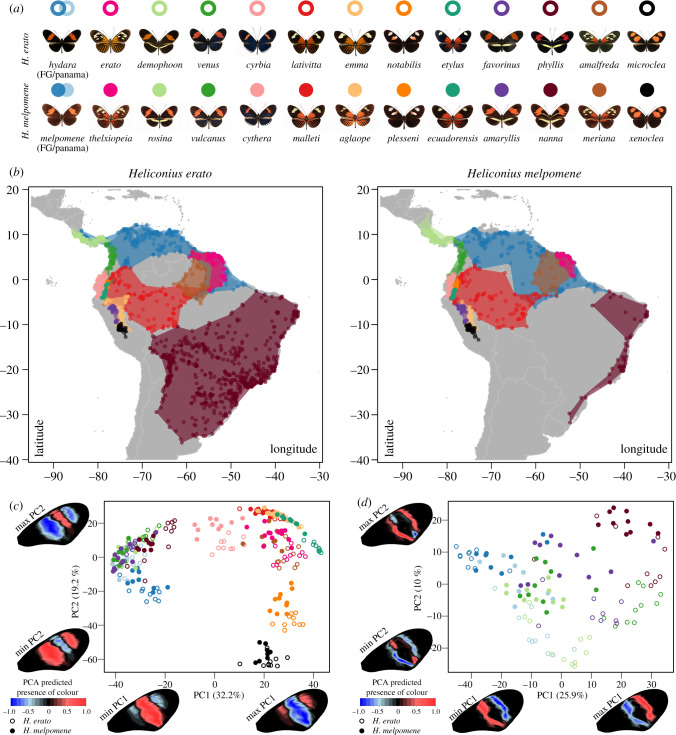
Geographical colour pattern morphs of H. erato and H. melpomene cover a wide spectrum of MFB patterns, with unique or partially overlapping pattern elements among them (figure 1a,b). In the PCA of the MFB, the first main axis of variance was dominated by the absence or presence of a broad red MFB, also typically called a ‘Postman’ phenotype (PC1, 32% of variation; figure 1c). The second axis of variation in the MFB shape was dominated by the presence of either a narrow median band, as observed in H. e. cyrbia/H. m. cythera, H. e. lativitta/H. m. malleti, and H. e. emma/H. m. aglaope, or two spots, as observed in the H. e. notabilis/H. m. plesseni and H. e. microclea/H. m. xenoclea populations (PC2, 19% of variation; figure 1c).
Figure 1.
Co-mimicking Heliconius erato and Heliconius melpomene colour pattern morphs, their distribution and PCA of MFB pattern. (a) Dorsal images of mimicking H. erato and H. melpomene colour pattern morphs. (b) Distribution areas of the mimicking populations as obtained from Rosser et al. [35]. (c) PCA of mid-forewing band (MFB) shape of the mimicking colour pattern morphs. (d) PCA of MFB shape of mimicking ‘Postman’ populations. Wing heatmaps indicate minimum and maximum predicted MFB patterns along each PC axis while considering the PC value of all other PC axes at zero. Positive values present a higher predicted presence of the MFB pattern (red), whereas negative values present the absence of the pattern (blue). (Online version in colour.)
As expected, co-mimicking populations of H. erato and H. melpomene are found to have more similar MFB phenotypes than different populations of the same species. However, significant differences in clustering can be observed in the PCA between the two species, with posterior probability of classification 88.6% and 92.2% for H. erato and H. melpomene, respectively (F1,280 = 70.8, p < 0.001; electronic supplementary material, table S4; figure 1c). When restricting our analysis to co-mimicking Postman populations, differences were also significant with posterior probability of classification 100% and 98.3% for H. erato and H. melpomene, respectively (F1,127 = 70.0, p < 0.001; electronic supplementary material, table S4; figure 1d). This difference between species was highly significant along PC2 (10% of variation; F1,127 = 131.1, p < 0.001) and suggests a general trend of expansion of both the proximal and distal area of the MFB in H. melpomene Postman phenotypes compared to H. erato. Significant differences between male and female MFB patterns were observed in both H. erato (F1,139 = 3.11, p = 0.002) and H. melpomene (F1,140 = 3.17, p = 0.001). However, sex differences had a low probability of posterior classification (55.8% and 58.3% for male and female H. erato and 78.0% and 62.5% for male and female H. melpomene, which is close to classification between random groups) and were only significant along PC axes that explain small amounts of variation among samples (electronic supplementary material, table S4).
Using pairwise comparisons we further explored interspecific differences between H. erato and H. melpomene co-mimics (figure 2a). First, among Postman phenotypes we observed an area in the distal posterior part of the MFB that consistently shows the absence of black scales in H. melpomene populations compared to H. erato populations (orange triangles in Postman (P) mimics in figure 2a). As demonstrated in the PCA on Postman phenotypes (figure 1d), we generally observed an expansion of the proximal area of the MFB in H. melpomene compared to H. erato in the pairwise comparisons (figure 2a). However, this trend was reversed between the Postman populations H. e. hydara and H. m. melpomene in French Guiana. In Colombia, the Postman phenotype showed an expansion of the MFB in the distal area in H. e. venus compared to H. m. vulcanus (figure 2a).
Figure 2.
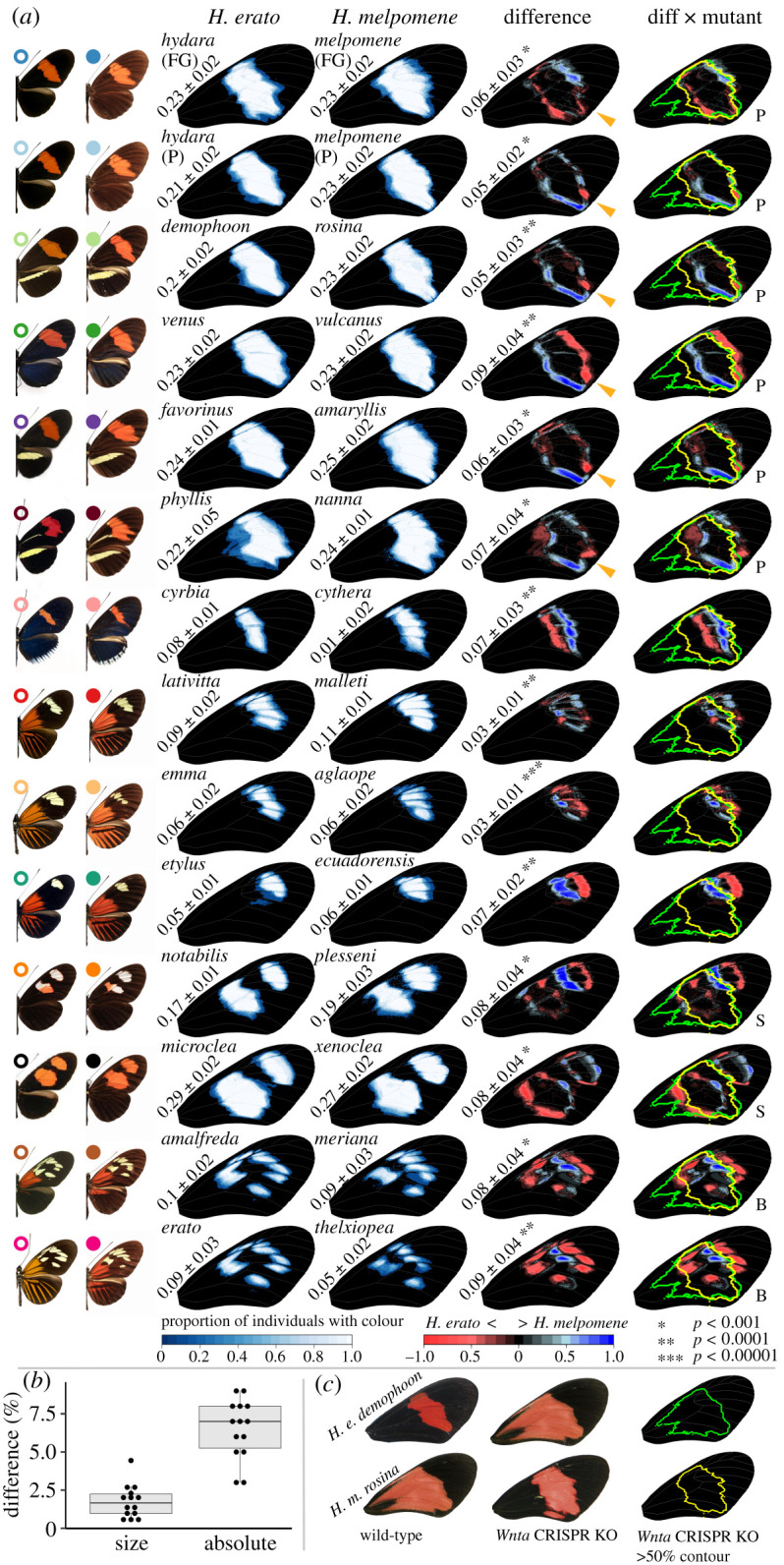
Quantification of mid-forewing band (MFB) pattern in co-mimicking Heliconius erato and Heliconius melpomene colour pattern morphs. (a) Heatmaps demonstrate the consistency of MFB within colour pattern morphs with white indicating consistent presence of MFB patterns and blue gradient indicating less consistent presence. Inter-species differences in MFB are shown on the right, with red indicating higher presence of MFB in H. erato and blue indicating higher presence of MFB in H. melpomene. Values next to wings indicate the average proportion of the wing in which the MFB is present within colour pattern morphs and in which differences are found between co-mimics. As a positional reference of the MFB pattern variation, the column on the right overlays the H. e. demophoon (green outline) and H. m. rosina (yellow outline) WntA CRISPR KO phenotype as found in at least 50% of the KO samples with the differences between co-mimics. ‘P’, ‘B’, and ‘S’ indicate phenotypes commonly referred to as the ‘Postman’, ‘Broken’, and ‘Split’ band morphs, respectively. Coloured circles next to butterfly wing images correspond to distribution areas in figure 1. (b) Comparison of the difference in relative size of the MFB (as proportion of the wing in which MFB is present) and absolute mismatch between H. erato and H. melpomene MFB. (c) Wild-type and WntA CRISPR KO phenotype of H. e. demophoon and H. m. rosina. (Online version in colour.)
As seen for the Postman populations, all other co-mimicking Heliconius colour pattern morphs showed marked differences in their MFB (figure 2a). Comparison of the similarly patterned H. e. cyrbia with H. m. cythera from West Ecuador suggests that the position of the MFB is shifted proximally in H. e. cyrbia compared to H. m. cythera. In East Ecuador, the H. m. malleti generally showed a larger MFB than the co-mimetic H. e. lativitta populations. Interestingly, differences in the comparison between the co-mimics H. e. etylus and H. m. ecuadorensis, which have a single distal spot, resembled differences in the distal spot between H. e. notabilis and H. m. plesseni. These latter populations have the so-called ‘Split’ MFB phenotype, which consists of two white/red spots in the MFB area (‘S’ in figure 2a). Finally, the so-called ‘Broken’ MFB phenotypes H. e. erato and H. e. amalfreda, which consist of multiple yellow spots in the MFB area (‘B’ in figure 2), consistently differed from the H. m. thelxiopeia and H. m. meriana populations by a proximal shift of the distal margin.
In all co-mimetic comparisons of H. erato and H. melpomene, the average absolute difference which includes the position of the MFB pattern was larger than the average difference in the relative size of the MFB pattern (i.e. proportion of the wing in which MFB is present; figure 2b). Significant differences in the size of the MFB were only observed between the co-mimics H. e. hydara and H. m. melpomene from Panama (p = 0.013), H. e. demophoon and H. m. rosina (p = 0.013), H. e. cyrbia and H. m. cythera (p = 0.009) and H. e. erato and H. m. thelxiopeia (p = 0.001).
(b). Mismatch between co-mimics partly coincides with developmental WntA boundaries
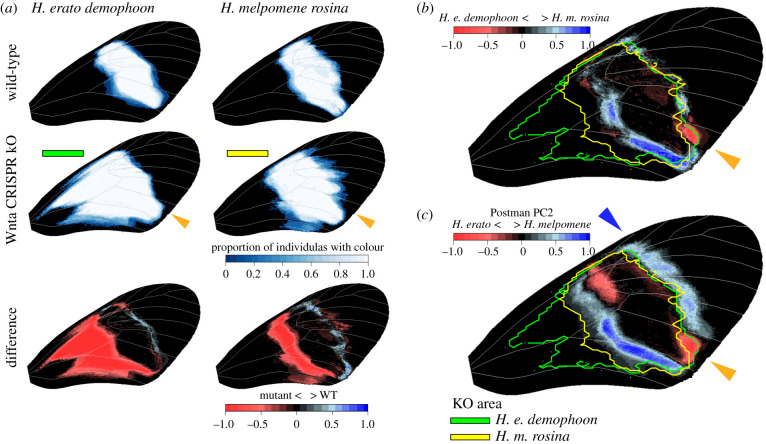
An important aspect of our study was to determine the possible link between wing pattern variation observed between wild-type butterflies and the WntA CRISPR/Cas9 KO phenotypes. We focused on the Panamanian co-mimics H. e. demophoon and H. m. rosina for which the largest WntA KO dataset is available and compared the WntA boundaries defined by these mutants with wild-type variation in the Postman phenotypes. As recently described by Concha et al. [12], WntA CRISPR/Cas9 KOs of the Postman phenotype H. e demophoon show a strong proximal expansion of the MFB pattern due to the development of red scales instead of black scales in this area of the wing (figure 3a, left; impacting 21.5 ± 11.3% of the forewing). In contrast, the WntA KOs of the co-mimic H. m. rosina showed a less pronounced proximal expansion (impacting 9.5 ± 3.9% of the forewing) and an observable distal expansion of the MFB pattern in several of the mutants (figure 3a, right; impacting 0.7 ± 0.1% of the forewing). By comparing the WntA mutant phenotypes for both species, we observed an area at the posterior of the distal margin of the MFB where WntA affects scale colouration in H. e. demophoon but not in H. m. rosina (figure 3a, orange triangles; covering 0.6 ± 0.1% of the forewing). This area overlaps with a forewing region in which wild-type individuals of H. e. demophoon and H. m. rosina show consistent difference (figure 2 and figure 3b, orange triangle). Moreover, MFB variation along the PC axis that strongly differentiated pattern variation between H. erato and H. melpomene Postman populations (figure 1d, PC2) recapitulated the distal spot difference (figure 3c, orange triangle; covering 1.0 ± 0.2% of the forewing). In line with the observation by Concha et al. [12] of a distal expansion of the MFB in H. m. rosina WntA mutants, this PC axis also demonstrated a general distal margin expansion in H. melpomene Postman populations compared to H. erato populations (figure 3c, blue triangle; covering 2.9 ± 1.0% of the forewing).
Figure 3.
Comparison of mid-forewing band (MFB) differences between co-mimicking Heliconius erato and Heliconius melpomene Postman phenotypes and WntA CRISPR/Cas9 KO phenotypes. (a) Heatmaps showing the presence of MFB in wild-type, WntA KO phenotypes and their difference for H. e. demophoon and H. m. rosina (obtained from 10 wings of five KO butterflies each [12]). (b) Comparison of WntA KO area to wild-type difference between H. e. demophoon and H. m. rosina. (c) Comparison of WntA KO area to Postman phenotype variation (figure 1). Orange and blue arrows indicate overlap in mismatch between wild-type co-mimicking populations and WntA KO patterning area. The yellow and green outlines show the WntA KO area as present in at least 50% of the KO samples. (Online version in colour.)
4. Discussion
In Heliconius butterflies, convergence between co-mimicking populations has been broadly defined as nearly identical. In accordance with these phenotypic similarities, genetic work has suggested that convergence is governed by a small and shared set of genes [13,15,17,28,36]. For example, WntA has been linked to wing colour pattern variation in the forewing of both H. erato and H. melpomene butterflies [28]. Recently, the availability and implementation of CRISPR/Cas9 functional approaches in butterfly wing colour pattern development has allowed to further investigate the role of wing patterning genes [12,17,37]. One of the most striking results that emerged from the KO phenotypes is an apparent divergent developmental architecture underlying butterfly wing colour pattern convergence, despite WntA being involved in forewing band variation in both species. Hence, while the difference in KO phenotypes reflects distinct WntA wing colour pattern domains in the two divergent co-mimetic butterflies H. e. demophoon and H. m. rosina [12], it also highlights the power of natural selection in driving convergent adaptive phenotypes despite a divergent genetic and developmental background. However, the extent to which divergence in WntA functioning may have affected the evolution of natural populations of mimetic phenotypes in Heliconius has not been investigated. By doing so, our study provides new insights into the dynamics of selection and how adaptation may be constrained to find alternative routes.
(a). Patterns of advergence in mid-forewing band
A question that arises from the observed differences in MFB patterns between co-mimics is whether it is necessary for H. erato and H. melpomene to perfectly mimic each other and whether selective forces could work against convergence [19,38]. In our comparisons we observed that even though the position of forewing pattern elements may not be perfectly identical between co-mimics, the relative amount of black versus red or yellow is generally more similar than the absolute difference. This improved match of the size of the MFB seems to result from compensatory changes in the MFB pattern and are in line with phenotypic evolution being driven by predation pressure. A similar ‘advergence’ process has previously been demonstrated for the mimicry ring including H. timareta thelxinoe, H. e. favorinus, and H. m. amaryllis in Peru [26] and the mimicry ring including H. e. phyllis, H. besckei, H. m. burchelli, and H. m. nanna in Brazil [27]. They thus indicate fine scale pattern adaptation in non-homologous regions of the wing to obtain a better match in the shape and size of the pattern even though they have a shifted position in the wings. Differences in MFB patterns between sexes were the same for both species and suggest that sexual conflicts for species recognition are likely not driving species differences in MFB patterns.
The compensatory evolution to obtain a more similar area of the MFB despite its mismatch in position may suggest imperfect discrimination in the visual range of their predators, or the relative importance of overall features of colour contrast distribution rather than the exact position of pattern elements [20,21]. A remarkable example of this are the co-mimetic Ecuadorian butterflies H. e. notabilis and H. m. plesseni which both have red and white in their proximal forewing element but have the relative positions of white versus red colour inversed. Notably, these wing colour patterns are the results of complex epistatic interactions between WntA and other genes such as the transcription factor optix, which controls white and red scale development [13,36].
We also found detectable within-species differences between populations that are generally considered identical. Among Postman populations, the greatest MFB differences were observed between several H. erato populations (e.g. between H. e. hydara from Panama and French Guiana). Notably, these H. erato Postman populations also had the most apparent differences with its H. melpomene co-mimic (i.e. H. e. hydara versus H. m. melpomene from French Guiana). As H. erato is often suggested to be the more abundant co-mimic and, thus, the model which H. melpomene mimics [21,22], the presence of a less perfect mimetic signal in these cases may reflect a ‘lag’ in the evolution of better mimicry of the H. melpomene populations. This inference potentially fits a signal of recent adaptive evolution (i.e. selective sweep signal) across the regulatory regions in the first intronic region of WntA of H. e. hydara populations from French Guiana, but not H. e. hydara populations from Panama, or H. m. melpomene populations from French Guiana [25]. What drives the divergence in MFB pattern between the H. erato Postman populations may include local changes in the composition of the mimicry and/or predator community [18,24,26].
(b). Indications of genetic and developmental constraints
Some of the pattern differences we observed between co-mimetic populations were shared among several geographically distinct populations and phenotypes. Such shared differences thus likely result from a shared genetic and developmental background of specific MFB areas among these geographical populations. For example, differences in MFB between H. e. etylus and H. m. ecuadorensis strongly resembled differences in the distal part of the ‘Split’ MFB of H. e. notabilis and H. m. plesseni (‘S’ in figure 2a). Similarly, the ‘Broken’ MFB phenotypes H. e. erato and H. e. amalfreda showed differences in the same areas of the wings compared to H. m. thelxiopeia and H. m. meriana, respectively (‘B’ in figure 2a). These pattern differences were observed regardless of interspecific wing shape differences, suggesting that differences in MFB phenotype between H. erato and H. melpomene are largely independent from shape differences in their wings.
Regarding the inter-species Postman population comparisons, our analyses showed a correlation between wild-type differences and differences in the WntA CRISPR/Cas9 KO phenotypes of H. e. demophoon and H. m. rosina. This correspondence between the H. e. demophoon and H. m. rosina WntA KO phenotypes and their wild-type observed mismatch was most obvious in the posterior spot at the distal margin of the MFB, where the H. erato Postman populations and KOs had red scales and the H. melpomene Postman populations and KOs always had black scales (figure 3, orange triangles). This observation highlights the existence of a WntA KO boundary and suggests that the observed imperfections in mimicry might be to some extent imposed by divergence in the gene regulatory network involved in the development of the MFB. As discussed earlier, it is possible that conflicting selection pressures can explain imperfections in mimicry. However, our quantitative wing colour pattern analyses on wild-type individuals and WntA KO phenotypes suggests the likely role of divergent genetic backgrounds in constraining the observed imperfect mimicry. While not obvious in the pairwise comparisons of the Postman co-mimics, the PCA analysis identified that the distal anterior margin of the MFB is generally expanded in H. melpomene compared to H. erato. Finally, the difference observed in the distal anterior margin of the MFB between co-mimicking Postman populations matched the distal boundary of WntA affected wing area as identified in the H. e. demophoon WntA KOs (figure 3c) and may potentially further highlight areas of the wing that are constrained by divergence in the gene regulatory network between H. erato and H. melpomene.
(c). Candidates of divergence in the gene regulatory network
Divergence in the gene regulatory network that is involved in the development of the MFB may include a multitude of changes in upstream factors that regulate spatial and/or temporal expression of WntA [39], the cis-regulatory elements of WntA itself [29], as well as additional genes that define the fate of wing scale cells. A few of these diverged elements are likely loci or genes that have previously been implicated in wing colour pattern variation in various Heliconius species. For example, the distal part of the MFB pattern that is expanded in H. e. etylus/H. e. notabilis compared to H. m. ecuadorensis/H. m. plesseni (figure 2) matches the distal area of the wing that is described to be affected by an additional locus called Ro [29,40–42]. Next, differences in the mismatch between the ‘Split’ MFB phenotypes H. e. notabilis/H. m. plesseni and H. e. xenoclea/H. m. microclea potentially result from epistatic interactions with the optix gene, affecting the size and shape of the MFB pattern. This has been previously suggested by looking at MFB patterns in hybrid butterflies that have the same WntA alleles but absence/presence of optix expression in the MFB [33]. Further, contrasting results have been found regarding the number of loci that affect MFB shape in H. erato compared to H. melpomene. For example, in H. erato the genetic architecture of MFB pattern variation has so far been mapped to the so-called Sd, St, and Ly loci that each affect a particular part of the MFB shape [40] and these loci have been demonstrated to include cis-regulatory elements of WntA [29]. In H. melpomene, on the other hand, regulatory variation at the so-called N locus, which includes the gene cortex and a few additional candidate genes, seems to underlie MFB shape variation together with WntA [40]. Potentially, this latter locus provides a candidate that explains the absence of a WntA effect on black wing scale development in the proximal area of the wing in H. melpomene. Finally, apart from the major effect loci involved in Heliconius colour patterns, quantitative trait loci (QTL) studies of pattern variation in H. erato and H. melpomene have demonstrated the existence of minor effect loci associated with quantitative changes in wing colour pattern [42–44]. This larger set of genetic variants controlling quantitative variation is additional to the regulatory complexity that modulates the expression of the major colour pattern genes [12,29,37].
5. Conclusion
Studying the evolutionary dynamics of adaptation is complicated due to the interplay of selection, genetics, and development. Adaptive radiations and mimicry systems have long been powerful study systems to unravel the genetic basis of adaptive traits. Here, we used Heliconius butterflies to investigate the interplay of selection and genetics and provide a tentative explanation on the causes that limit perfect wing mimicry after several million years of strong natural selection towards a mutual anti-predatory signal. We propose that phenotypic patterns of adaptation and convergence between Heliconius co-mimics is biased by divergence in the gene regulatory network underlying the mimicry trait. These networks can be highly polygenic and interconnected and evolution may not have been able to perfectly rewire them in the same identical way after the accumulation of changes across their genomes. In the case of the observed differences between the WntA mutant phenotypes of H. erato and H. melpomene, these constraints may exist due to independently evolved genetic elements that interact with the WntA gene or protein. These elements may include trans factors, cis-regulatory elements, or additional genes with a complementary role to WntA and may not have the genetic architecture to easily be detached from potential developmental interactions with other genes [45,46]. While we quantified exact wing colour pattern differences between H. erato and H. melpomene co-mimics, we also observed that the warning signal represented by the relative amount of colours is more consistent. Hence, changes to the MFB can be observed that compensate the developmental bias in other areas of the wing, indicating that selection has used an alternative route to convergence. Overall, our results provide a quantitative measure of the imperfection of mimicry and propose a novel view into the interplay between selective conflicts compared to genetic and developmental constraints in the production of similar phenotypes.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We kindly thank Ananda Martins, Owen McMillan, Joe Hanly, Carolina Concha, Tim Thurman, Jennifer Cuthill, Markus Möst, Camilo Salazar, Mathieu Joron, Jake Morris, and Claire Mérot for sharing images and Chris Jiggins, Ian Warran, Patricio Salazar and Gabriela Montejo-Kovacevich for the earthcape repository (https://heliconius.ecdb.io). We thank Frederik Nijhout and Claire Mérot for thoughtful comments on the manuscript.
Data accessibility
All code to build the figures, landmarks, and images are available through GitHub repository (https://github.com/patternize-projects/Heliconius_forewing_band).
Competing interests
We declare we have no competing interests.
Funding
This work was funded by NSF EPSCoR RII Track-2 FEC (grant no. OIA 1736026) to R.P. and B.A.C. and NSF IOS 1656389 to R.P. S.M.V.B. was supported by a Puerto Rico Science, Technology & Research Trust catalyzer award (#2020-00142) and the NSF OIA 1736026. P.A.A.R. was supported by the Interdisciplinary & Quantitative Biology Summer Research Experience for Undergraduates (IQ BIO REU) Program funded by the National Science Foundation (grant no. DBI 1852259). H.C.G. was supported by The Puerto Rico Space Grant Consortium by NASA (grant no. NNX15AI11H).
References
- 1.Blount ZD, Lenski RE, Losos JB. 2018. Contingency and determinism in evolution: replaying life's tape. Science 362, eaam5979 ( 10.1126/science.aam5979) [DOI] [PubMed] [Google Scholar]
- 2.Maynard SJ, Burian R, Kauffman S, Alberch P, Campbell J, Goodwin B, Lande R, Raup D, Wolpert L. 1985. Developmental constraints and evolution: a perspective from the Mountain Lake Conference on Development and Evolution. Q. Rev. Biol. 60, 265–287. ( 10.1086/414425) [DOI] [Google Scholar]
- 3.Schluter D. 1996. Adaptive radiation along genetic lines of least resistance. Evolution 50, 1766 ( 10.1111/j.1558-5646.1996.tb03563.x) [DOI] [PubMed] [Google Scholar]
- 4.Brakefield PM. 2006. Evo-devo and constraints on selection. Trends Ecol. Evol. 21, 362–368. ( 10.1016/j.tree.2006.05.001) [DOI] [PubMed] [Google Scholar]
- 5.Bridgham JT, Ortlund EA, Thornton JW. 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461, 515–519. ( 10.1038/nature08249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould S. 1989. Wonderful life. New York, NY: Norton. [Google Scholar]
- 7.Menard D, Dondorp A. 2017. Antimalarial drug resistance: a threat to malaria elimination. Cold Spring Harb. Perspect. Med. 7, 1–24. ( 10.1101/cshperspect.a025619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogbunugafor CB, Hartl D. 2016. A pivot mutation impedes reverse evolution across an adaptive landscape for drug resistance in Plasmodium vivax. Malar. J. 15, 1–10. ( 10.1186/s12936-016-1090-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beldade P, Koops K, Brakefield PM. 2002. Developmental constraints versus flexibility in morphological evolution. Nature 416, 844–847. ( 10.1038/416844a) [DOI] [PubMed] [Google Scholar]
- 10.Weber K. 1992. How small are the smallest selectable domains of form? Genetics 353, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolstad GH, Cassara JA, Márquez E, Hansen TF, Van Der Linde K, Houle D, Pélabon C. 2015. Complex constraints on allometry revealed by artificial selection on the wing of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 112, 13 284–13 289. ( 10.1073/pnas.1505357112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Concha C, et al. 2019. Interplay between developmental flexibility and determinism in the evolution of mimetic Heliconius wing patterns. Curr. Biol. 29, 3996–4009. ( 10.1016/j.cub.2019.10.010) [DOI] [PubMed] [Google Scholar]
- 13.Reed RD, et al. 2011. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141. ( 10.1126/science.1208227) [DOI] [PubMed] [Google Scholar]
- 14.VanKuren N, Massardo D, Nallu S, Kronforst MR. 2019. Butterfly mimicry polymorphisms highlight phylogenetic limits of gene reuse in the evolution of diverse adaptations. Mol. Biol. Evol. 36, 2842–2853. ( 10.1093/molbev/msz194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadeau N, et al. 2016. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534, 106–110. ( 10.1038/nature17961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallant JR, et al. 2014. Ancient homology underlies adaptive mimetic diversity across butterflies. Nat. Commun. 5, 1–10. ( 10.1038/ncomms5817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazo-Vargas A, Concha C, Livraghi L, Massardo D, Wallbank RWR, Zhang L. 2017. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc. Natl Acad. Sci. USA 114, 10 701–10 706. ( 10.1073/pnas.1708149114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joron M, Mallet JLB. 1998. Diversity in mimicry: paradox or paradigm? Trends Ecol. Evol. 13, 461–466. ( 10.1016/S0169-5347(98)01483-9) [DOI] [PubMed] [Google Scholar]
- 19.Estrada C, Jiggins CD. 2008. Interspecific sexual attraction because of convergence in warning colouration: is there a conflict between natural and sexual selection in mimetic species? J. Evol. Biol. 21, 749–760. ( 10.1111/j.1420-9101.2008.01517.x) [DOI] [PubMed] [Google Scholar]
- 20.Ihalainen E, Rowland HM, Speed MP, Ruxton GD, Mappes J. 2012. Prey community structure affects how predators select for Müllerian mimicry. Proc. R. Soc. B 279, 2099–2105. ( 10.1098/rspb.2011.2360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penney HD, Hassall C, Skevington JH, Abbott KR, Sherratt TN. 2012. A comparative analysis of the evolution of imperfect mimicry. Nature 483, 461–464. ( 10.1038/nature10961) [DOI] [PubMed] [Google Scholar]
- 22.Kozak KM, Wahlberg N, Neild AFE, Dasmahapatra KK, Mallet J, Jiggins CD. 2015. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst. Biol. 64, 505–524. ( 10.1093/sysbio/syv007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chouteau M, Arias M, Joron M. 2016. Warning signals are under positive frequency-dependent selection in nature. Proc. Natl Acad. Sci. USA 113, 2164–2169. ( 10.1073/pnas.1519216113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallet J, Joron M. 1999. Evolution of diversity in warning color and mimicry: polymorphisms, shifting balance and speciation. Annu. Rev. Ecol. Syst. 30, 201–233. ( 10.1146/annurev.ecolsys.30.1.201) [DOI] [Google Scholar]
- 25.Moest M, et al. 2020. Selective sweeps on novel and introgressed variation shape mimicry loci in a butterfly adaptive radiation. PLoS Biol. 18, e3000597 ( 10.1371/journal.pbio.3000597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mérot C, Le Poul Y, Théry M, Joron M. 2016. Refining mimicry: phenotypic variation tracks the local optimum. J. Anim. Ecol. 85, 1056–1069. ( 10.1111/1365-2656.12521) [DOI] [PubMed] [Google Scholar]
- 27.Rossato DO, Boligon D, Fornel R, Kronforst MR, Gonçalves GL, Moreira GRP. 2018. Subtle variation in size and shape of the whole forewing and the red band among co-mimics revealed by geometric morphometric analysis in Heliconius butterflies. Ecol. Evol. 8, 3280–3295. ( 10.1002/ece3.3916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin A, et al. 2012. Diversification of complex butter flywing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl Acad. Sci. USA 109, 12 632–12 637. ( 10.1073/pnas.1204800109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Belleghem SM, et al. 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1, 52 ( 10.1038/s41559-016-0052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyal CJF, Guttenberg N, Ledger S, Crowther R, Huertas B. 2019. Deep learning on butterfly phenotypes tests evolution's oldest mathematical model. Sci. Adv. 5, eaaw4967 ( 10.1126/sciadv.aaw4967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiggins C, Warren I, Salazar P, Montejo-Kovacevich G. 2019. Heliconiine butterfly collection records from University of Cambridge. Dep. Zool. Cambridge. See GBIF.org.
- 32.Darragh K, et al. 2017. Male sex pheromone components in Heliconius butterflies released by the androconia affect female choice. PeerJ 2017, 1–23. ( 10.7287/peerj.3953v0.1/reviews/3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Belleghem SM, Papa R, Ortiz-zuazaga H, Hendrickx F, Jiggins CD, Mcmillan WO, Counterman BA. 2018. patternize: an R package for quantifying colour pattern variation. Methods Ecol. Evol. 9, 390–398. ( 10.1111/2041-210X.12853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hijmans RJ. 2016. raster: Geographic data analysis and modeling. R package version 2.5-8 See http://cran.r-project.org/package=raster.
- 35.Rosser N, Dasmahapatra KK, Mallet J. 2014. Stable Heliconius butterfly hybrid zones are correlated with a local rainfall peak at the edge of the Amazon basin. Evolution 68, 3470–3484. ( 10.1111/evo.12539) [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Mazo-Vargas A, Reed RD. 2017. Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proc. Natl Acad. Sci. USA 114, 10 707–10 712. ( 10.1073/pnas.1709058114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis JJ, Geltman RC, Pollak PC, Rondem KE, Belleghem SM. 2019. Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc. Natl Acad. Sci. USA 116, 24 174–24 183. ( 10.1073/pnas.1907068116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherratt TN, Peet-Paré CA. 2017. The perfection of mimicry: an information approach. Phil. Trans. R. Soc. B 372, 20160340 ( 10.1098/rstb.2016.0340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanly JJ, Wallbank RWR, McMillan WO, Jiggins CD. 2019. Conservation and flexibility in the gene regulatory landscape of heliconiine butterfly wings. Evodevo 10, 1–14. ( 10.1186/s13227-019-0127-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC. 1985. Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Phil. Trans. R. Soc. Lond. B 308, 433–610. ( 10.1098/rstb.1985.0066) [DOI] [Google Scholar]
- 41.Nadeau NJ, et al. 2014. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 24, 1316–1333. ( 10.1101/gr.169292.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris J, Navarro N, Rastas P, Rawlins LD, Sammy J, Mallet J, Dasmahapatra KK. 2019. The genetic architecture of adaptation: convergence and pleiotropy in Heliconius wing pattern evolution. Heredity 123, 138–152. ( 10.1038/s41437-018-0180-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papa R, Kapan DD, Counterman BA, Maldonado K, Lindstrom DP, Reed RD, Nijhout HF, Hrbek T, McMillan WO. 2013. Multi-allelic major effect genes interact with minor effect QTLs to control adaptive color pattern variation in Heliconius erato. PLoS ONE 8, e57033 ( 10.1371/journal.pone.0057033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baxter SW, Johnston SE, Jiggins CD. 2009. Butterfly speciation and the distribution of gene effect sizes fixed during adaptation. Heredity 102, 57–65. ( 10.1038/hdy.2008.109) [DOI] [PubMed] [Google Scholar]
- 45.Jiggins CD, Wallbank RWR, Hanly JJ. 2017. Waiting in the wings: what can we learn about gene co-option from the diversification of butterfly wing patterns? Phil. Trans. R. Soc. B 372, 20150485 ( 10.1098/rstb.2015.0485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis JJ, Belleghem SMV. 2020. Mechanisms of change: unraveling the roles of modularity and pleiotropy in diversification. Preprints 2020020068.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code to build the figures, landmarks, and images are available through GitHub repository (https://github.com/patternize-projects/Heliconius_forewing_band).