Summary
How are diverse regulatory strategies integrated to impose appropriately patterned gene expression that underlie in vivo phenotypes? Here, we reveal how coordinated miRNA regulation and neural-specific alternative polyadenylation (APA) of a single locus controls complex behaviors. Our entry was the unexpected observation that deletion of Bithorax-Complex (BX-C) miRNAs converts virgin female flies into a subjective post-mated behavioral state, normally induced by seminal proteins following copulation. Strikingly, this behavioral switch is directly attributable to misregulation of homothorax (hth). We localize specific CNS abdominal neurons where de-repressed Hth compromises virgin behavior in BX-C miRNA mutants. Moreover, we use genome engineering to demonstrate that precise mutation of hth 3’UTR sites for BX-C miRNAs, or deletion of its neural 3’ UTR extension containing most of these sites, both induce post-mated behaviors in virgins. Thus, facilitation of miRNA-mediated repression by neural APA is required for virgin females to execute behaviors appropriate to their internal state.
eTOC Blurb
Garaulet et al show that two post-transcriptional regulatory pathways - miRNAs and alternative polyadenylation - intersect in the central nervous system to control a behavioral switch. Regulation of homothorax by the mir-iab-4/8 locus, preferentially via the neural 3’ UTR extension, is required for females to interpret the virgin behavioral state.
Introduction
Whether to sleep when tired, drink when thirsty, or eat when hungry, reproductive and survival success depends on the capacity for dynamic coordination of internal states with appropriate behaviors. A classic example involves the profound behavioral transformations that females of many species undergo following mating and pregnancy. Mature virgins typically exhibit receptivity to male courtship and eventually allow copulation. However, after mating, they tend to remain refractory to further copulation attempts and adapt their habits to produce, raise and protect their progeny (Clyne and Miesenbock, 2009). This behavioral transition is found across diverse phyla, from nest building or increased aggression in mice (Broida and Svare, 1982; Ogawa and Makino, 1984), to a collective behavioral remodeling in insects (Avila et al., 2011). Overall, as a result of mating or pregnancy, diverse behaviors of an inseminated female adapt to match the novel needs of the reproductive state.
Alterations of female behaviors induced by mating have been extensively studied in female fruitflies, where multiple reproductive conducts such as egg-laying and male acceptance (Connolly and Cook, 1973; Manning, 1962, 1967), but also appetite (Carvalho et al., 2006), lifespan (Fowler and Partridge, 1989), food and salt preference (Ribeiro and Dickson, 2010; Walker et al., 2015), immune system function (Peng et al., 2005b), and sleep (Isaac et al., 2010) are modulated by sexual interaction. Collectively, these behaviors comprise the post-mating switch, also referred to as the post-mating response (PMR) (Avila et al., 2011). The capacity of a male fly to manipulate female behavior resides within seminal fluids synthesized by the male accessory gland (Gillott, 2003). While many seminal fluid proteins are known (Avila et al., 2011), the 36 amino acid Sex peptide (SP) is sufficient to drive most female post-mating responses (Aigaki et al., 1991; Chen et al., 1988). SP associates with sperm and is transferred to the female genital tract during copulation (Peng et al., 2005a). It is then stored in specific female reproductive organs and released gradually, thereby conditioning female behavior for 8–10 days after copulation (Manning, 1962, 1967; Peng et al., 2005a). Considering the average fruitfly lifespan is 30–40 days, it is remarkable that an exogenous factor, transferred in a single act, can drive female behavior for ¼ of her life.
SP exerts its function through a G-coupled transmembrane receptor, SPR (Yapici et al., 2008). Although SPR is expressed throughout the abdominal ganglion of the ventral nerve cord (VNC, equivalent of the vertebrate spinal cord) and in some regions of the brain (Yapici et al., 2008), its expression in a restricted set of 6–8 pickpocket+ sensory neurons (SP sensory neurons, SPSN) adjacent to the uterus appears necessary and sufficient to drive most effects of the switch (Feng et al., 2014; Hasemeyer et al., 2009; Yang et al., 2009). While some of these cells send their processes directly to the subesophageal ganglion in the female brain, the majority of SPSN terminations target the abdominal ganglion of the VNC (Feng et al., 2014; Hasemeyer et al., 2009; Yang et al., 2009), where they contact their synaptic partners. Some of these are abdominal interneurons that express myoinhibitory peptide (Mip) (Jang et al., 2017), which input into a restricted population of ascending neurons (SAG) that project to the posterior region of the brain (Feng et al., 2014; Soller et al., 2006). SPSN-Mip-SAG define a minimal ascending axis through which information flows from the SP stored in the genital organs to the female brain. However, beyond this simple, linear afferent circuit, larger populations of less characterized neurons are also involved either in the interpretation of mating information or the execution of mated responses (Haussmann et al., 2013). Most of these neurons express fruitless, doublesex and/or pickpocket genes, and configure an intricate neuronal network underlying female sexual behavior (Bussell et al., 2014; Feng et al., 2014; Haussmann et al., 2013; Monastirioti, 2003; Rezaval et al., 2012).
Nonetheless, despite intense investigations of the behavioral, neuronal, and molecular nature of the PMR, the genetic determinants of the virgin state remain largely unknown. Here, we report that developmental repression of an individual transcription factor by microRNAs (miRNAs) is critical to establish virgin behavior. This regulatory axis was revealed by deletion of the bidirectionally transcribed Bithorax Complex (BX-C) miRNA locus, which constitutively induce multiple post-mated behaviors in virgins. The BX-C hairpin locus expresses distinct miRNAs (miR-iab-4/miR-iab-8) in adjacent domains of the ventral nerve cord, which contains specific neurons that mediate the post-mating switch. Deletion of the BX-C miRNAs strongly derepresses homothorax (hth), which is directly targeted by both BX-C miRNAs to suppress post-mated behavior in virgins. Importantly, targeted mutation of the array of BX-C miRNA binding sites in the hth 3’ UTR similarly abrogates virgin behavior. Finally, we integrate this with the regime of neural alternative polyadenylation, since specific deletion of the hth neural 3’ UTR extension that bears most of these miRNA binding sites also blocks the virgin state.
Overall, we utilize miRNAs as an entry point to reveal critical post-transcriptional biology that interfaces with alternative polyadenylation control. The failure of these regulatory events has profound consequences on the ability of female flies to integrate sexual internal state with appropriate behaviors.
Results
BX-C miRNAs suppress the post-mating switch in virgins
Egg-laying is a defining characteristic of mated behavior in female flies. Although old virgins are able to lay some unfertilized eggs, oviposition is largely suppressed in young (0–3 day old) virgins (Figure 1A and Supplementary Figure 1B, C). Instead, mating and SP reception induces females to lay eggs (Manning, 1962, 1967). Surprisingly, young virgin females deleted for the bidirectionally transcribed BX-C miRNA locus encoding miR-iab-4 and miR-iab-8 (mir[Δ]) exhibit inappropriate, robust, egg-laying activity (Figure 1A). Since many behavioral phenotypes are influenced by unknown genetic background factors, we used CRISPR/Cas9 to generate a new BX-C miRNA deletion (mir[C11], Supplementary Figure 2), and extensively backcrossed both alleles into a common wildtype reference strain (Canton-S). We consistently observed activation of egg-laying in all BX-C miRNA deletion allele combinations in comparison to multiple wildtype strains (Figure 1A and Supplementary Figure 1B,C).
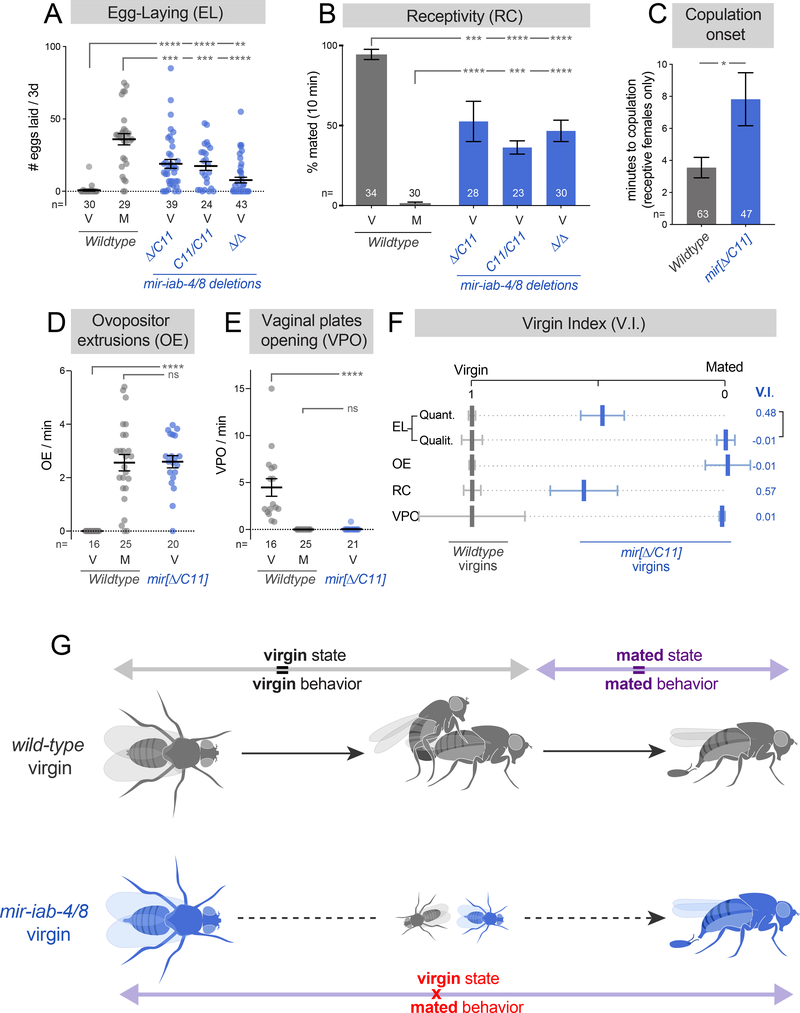
Figure 1. BX-C miRNAs prevent post-mating behaviors in virgins.
(A-E) Deletion of mir-iab-4/8 suppresses multiple virgin behaviors (B,E) and induces mated behaviors (A,D) in virgin females. (A) Quantification of egg-laying by individual females that are either virgin (V) or mated (M). (B) Analysis of sexual receptivity, assessed by the fraction of individual females that are copulating at 10 minutes across multiple genotypes. (C) Timing of copulation onset, analyzed in the subset of females that are receptive to mating. (D) Quantification of ovipositor extrusions by individual females. Virgins are 1 day old. (E) Quantification of vaginal plates openings by individual females. (F) A metric summarizing all of the behaviors studied (virgin index), showing that deletion of mir-iab-4/8 shifts virgin behavior to mated values. Egg-laying is presented in both quantitative (Quant.) and qualitative (Qualit., yes/no) measures. (G) Schematic representation of internal state and behavior in wildtype (gray) and BX-C miRNA mutant females (blue). While behavior is constantly coupled to mating state in wildtype females, deletion of mir-iab-4/8 in virgins suppresses virgin behaviors and triggers post-mating responses before copulation. Statistical significance was evaluated using Fisher’s exact test for receptivity (B), and Mann-Whitney non parametric test for all other behaviors. ns=not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Error bars= SEM. Wildtype flies are Canton-S strain. See also Supplementary Figures 1–3 and Supplementary Movies 1,2.
Of note, mated mir[Δ] females were previously documented to have defective egg-laying, which was previously speculated to be due to egg blockage (Bender, 2008; Gummalla et al., 2012) and/or reduced oviduct innervation (Garaulet et al., 2014). However, the robust oviposition by young virgins indicates functional neuromuscular control at the genital tract, and instead suggests a subjective shift to the mated state. Therefore, we sought evidence for alterations in other switch-influenced behaviors in miRNA mutant female virgins.
We first assayed female receptivity, which is maximal in mature virgins and allows the male to copulate shortly after courtship initiation. Receptivity is strongly suppressed following mating, as females remain refractory to male copulatory attempts for days (Connolly and Cook, 1973) (Figure 1B and Supplementary Figure 1A). mir-iab-4/8 null virgins show compromised receptivity across all mutant combinations tested, evaluating both mating success and time elapsed until copulation in the subset of females that eventually mate (Figure 1B,C, Supplementary Figure 1A, Supplementary Figure 3 and Supplementary Movies 1 and 2).
Male acceptance or rejection is achieved by distinct female genital responses, which change according to the female mating state. Mature virgins typically open their vaginal plates sporadically, while mated females extrude their ovipositor to reject male copulatory attempts (Aranha and Vasconcelos, 2018; Bussell et al., 2014; Connolly and Cook, 1973). For both of these behaviors, mir[Δ/C11] virgins were indistinguishable from mated wildtype flies (Figure 1D, E and Supplementary Figure 1D, E). Given the reliability of these different phenotypic readouts, we focused subsequent analyses of the miRNA null state using trans-heterozygous deletions (mir[Δ/C11]).
Taken together, the overall performance of BX-C miRNA knockout virgins is shifted to a canonical post-mated response (Figure 1F,G), indicating that miR-iab-4/8 are crucial for the subjective interpretation of internal state by virgin females. Importantly, this is not only revealed by the failure to perform characteristic virgin behaviors, but also by the selective activation of behavioral programs that are solely executed by mated flies (Figure 1F,G). Moreover, while virgin behaviors are commonly presumed to be a default state that must be converted to the mated state, the loss of BX-C-miRNAs reveal that virgin behavior is genetically specified.
BX-C miRNAs are expressed in VNC subpopulations that control the post-mating switch
Behavior on either side of the switch is determined by an ascending neural circuit that underlies SP sensing. Although the precise architecture of the circuit has not been fully mapped, most of its neuronal substrates reside in the abdominal ganglion (AbG) of the ventral nerve cord (VNC), and can be identified by expression of fruitless (fru), pickpocket (ppk) or doublesex (dsx) genes (Feng et al., 2014; Hasemeyer et al., 2009; Jang et al., 2017; Rezaval et al., 2012; Yang et al., 2009). While these are broadly expressed markers, some subsets of more restricted neuronal populations labeled by select VT-Gal4 lines (hereafter VT-switch neurons) have also shown to be necessary and sufficient to fully induce post-mating behaviors (Feng et al., 2014). We studied how BX-C miRNAs are deployed with respect to these neural subpopulations. These miRNAs are expressed in adjacent domains within the developing abdominal ganglion of the larval VNC, with miR-iab-4 deployed anteriorly to miR-iab-8 (Bender, 2008; Garaulet et al., 2014; Garaulet and Lai, 2015; Tyler et al., 2008). In the VNC of adult females, we used tub-GFP-miR-iab-4 and miR-iab-8 sensors to register the domains of miRNA activity with respect to Abd-A protein (Figure 2A, B).
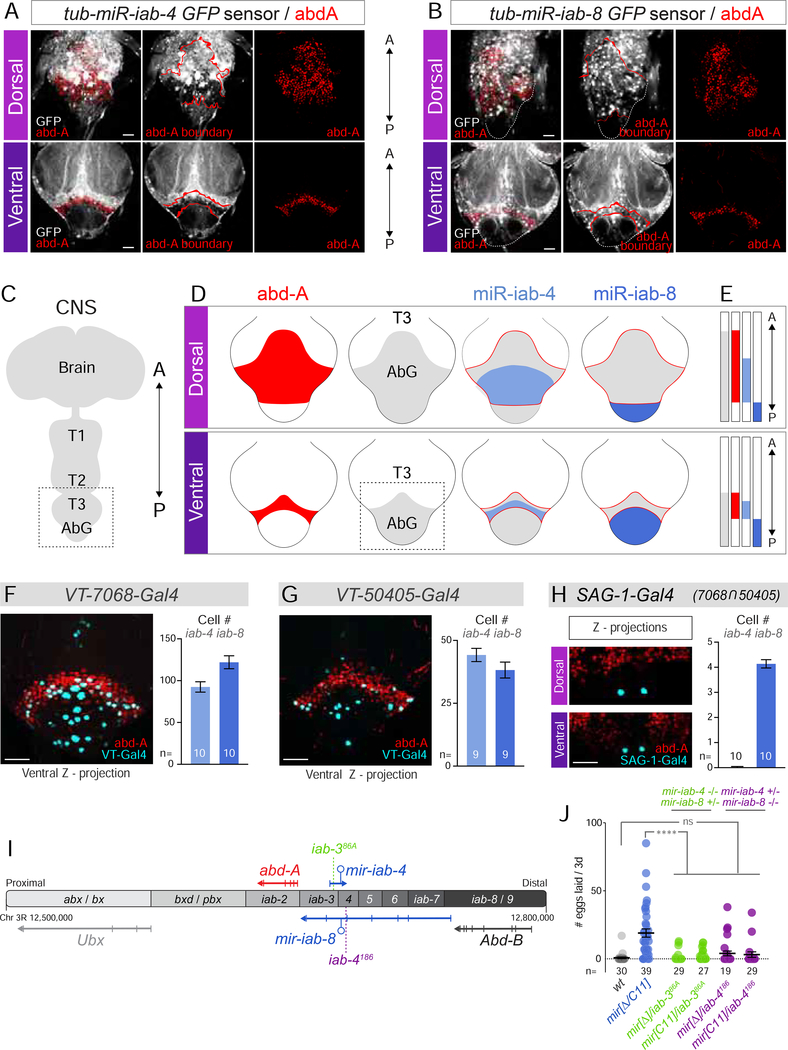
Figure 2. BX-C miRNA functional domains in the VNC.
Shown are immunostainings of the Drosophila adult VNC, focused on the abdominal ganglion (AbG, box in C). (A,B) tub-GFP activity sensors to reveal domains of active miR-iab-4 (A) and miR-iab-8 (B), as inferred from reduced GFP accumulation. Co-staining with Abdominal-A (red) reveals collinear, abdominal-exclusive activity of both miRNAs along the A/P axis. (C) Schematic of the Drosophila CNS. Boxed region corresponds to T3-AbG shown in A, B and D. (D, E) Abdominal patterning of the VNC. Although the size of each domain differs in the dorsoventral axis, the relative A/P positioning of each domain is maintained. Boxed region in (D) corresponds to the ventral side of AbG shown in F-H. (F-H) Expression pattern and quantification of VT-switch lines. Co-expression with Abd-A reveals an enrichment of Gal4+ cells in the AbG, within the domains of expression of mir-iab-4 and mir-iab-8. (I) Diagram of the Bithorax-Complex. abx=anterobithorax, bx=bithorax, bxd= bithoraxoid, pbx=postbithorax, iab=infra-abdominal, Ubx=Ultrabithorax, abd-A=abdominal-A, Abd-B=Abdominal-B. Note that the mir-iab-4 and mir-iab-8 hairpins are encoded by the same genomic DNA, but are transcribed from the top and bottom strands of the BX-C, respectively. (J) Egg-laying performance of individual mir-iab-4 (iab386A/mir[Δ], green) or mir-iab-8 mutants (iab-4186/mir[Δ], purple). A=anterior, P=posterior, CNS=central nervous system, AbG=abdominal ganglion, T= thoracic segments. Error bars= SEM. Scale bar= 25μm. mir-iab-4/8−/− mutants=mir[Δ/C11] transheterozygotes. See also Supplementary Figure 4.
In these analyses, it is relevant to consider the three-dimensional structure of the VNC, as the domains of these individual markers are substantially shifted between the dorsal and ventral surfaces. Still, the relative positions of Abd-A and the miRNAs are preserved between the dorsal and ventral regions of the adult VNC. Abd-A is present anterior to, and within the domain of active miR-iab-4, while active miR-iab-8 is exclusively posterior to Abd-A (Figure 2A–E). These relative relationships also apply in the embryo and larva (Bender, 2008; Garaulet et al., 2014; Gummalla et al., 2012; Tyler et al., 2008). Thus, we can use Abd-A (1) as a proxy for the transition of the two miRNA domains, and (2) to demarcate the abdominal ganglion.
We used co-labeling to define the domains of the VT-switch lines with respect to the BX-C miRNAs (Figure 2F–H). The most restricted driver, the split-Gal4 combination SAG-1, which derives from the regulatory intersection of the VT-7068 and VT-50405 lines, labels only four neurons that reside strictly within the miR-iab-8 domain (Figure 2H). However, the remainder of VT-switch lines are active in both miR-iab-4 and miR-iab-8 functional domains. We quantified the number of neurons labeled by each VT-switch Gal4 line in the respective miRNA domains in Figure 2F–H and Supplementary Figure 5E. These data suggest that both miRNA loci might be involved in maintaining virgin state.
To test this notion, we separated the function of the bidirectional miRNA locus by placing a deletion allele in trans to chromosomal rearrangements that disrupt the primary transcripts for either top or bottom strand loci (Figure 2I). This approach was used to provide evidence that certain BX-C miRNA deletion phenotypes rely primarily on mir-iab-4 (larval self-righting) (Picao-Osorio et al., 2015) or mir-iab-8 (sterility) (Bender, 2008), respectively. We focused on egg-laying to readout the post-mating switch in representative breakpoint hemizygote backgrounds. While this behavior was sporadically activated in null conditions of either miRNA, neither was significantly different from control (Figure 2J). Thus, it appears that both miR-iab-4 and miR-iab-8, and by extension, neurons resident in both of their expression domains, contribute to the virgin behavioral defect in miRNA deletion females.
Loss of BX-C miRNAs affects the activity of post-mating switch neurons
Several studies demonstrate that virgin behavior is determined by a constitutive activation of abdominal networks of neurons identified by the fru, ppk, dsx or VT-switch markers. In particular, hyperactivation of these neurons renders virgin behaviors in mated flies (Feng et al., 2014; Hasemeyer et al., 2009; Jang et al., 2017; Rezaval et al., 2012; Yang et al., 2009). As loss of miR-iab-4/8 induces post mating responses, we hypothesized that enforced neural activity might revert these conducts back to virgin state in mir-iab-4/8 knockouts.
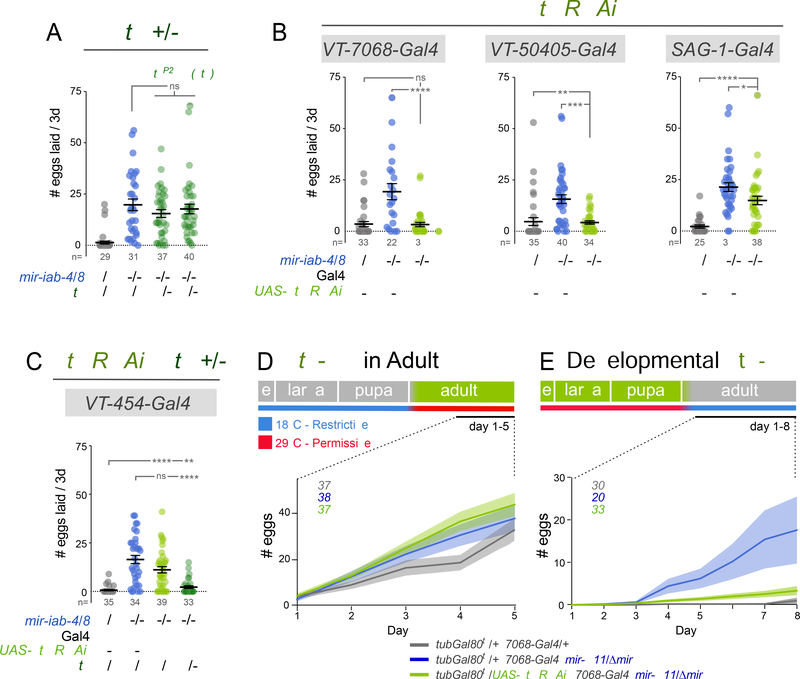
Following previous studies (Feng et al., 2014), we used the bacterial sodium channel transgene UAS-NachBac to enhance the activity of VT-switch neurons. We recapitulate prior behavior observations with the switch-Gal4 lines (Figure 3A) and show that our miRNA mutants are not rescued by inclusion of the UAS-NaChBac alone, ruling out leaky transgene effects (Figure 3B). Strikingly, activation of either VT-7068 or VT-50405 neurons substantially suppressed oviposition by miRNA deletion virgins, with VT-7068 providing near complete rescue (Figure 3C).
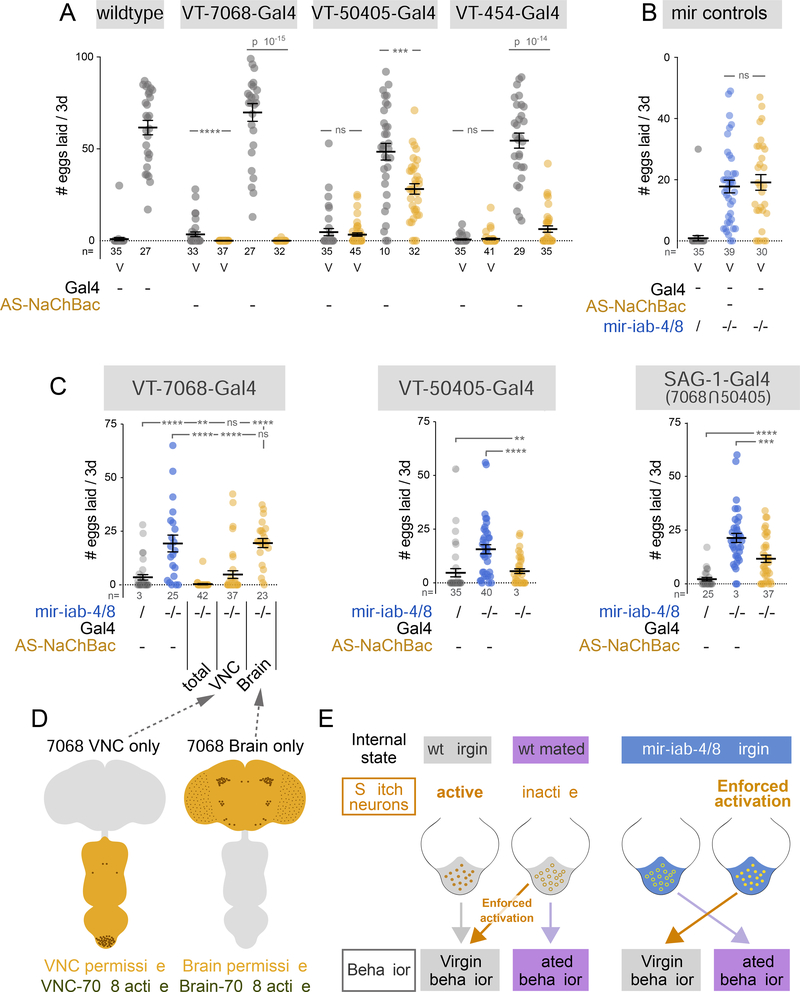
Figure 3. Enforced activation of abdominal neurons restores virgin behavior in mir-iab4/8 mutants.
(A) Hyperactivation of VT-switch neurons in wildtype mated females reverts egg-laying to virgin values. (B) UAS-NaChBac controls in mir-iab-4/8 null background. (C) Activation of multiple VT-switch neurons in mir-iab-4/8 mutant virgins suppressed their egg-laying phenotype. Activation of VNC but not Brain VT-7068 neurons alone accounts for the behavioral rescue. (D) Illustration of the VNC VS brain specific NaChBac activation of VT-7068 neurons achieved in the following genotypes: OTD-flp, tub>stop>Gal80 /+; 7068-Gal4, mir-C11/ UAS-NaChBac, Δmir and OTD-flp, >tub-Gal80> /+; 7068-Gal4, mir-C11/UAS-NaChBac, Δmir. (E) Schematic representation of the effect of neuronal activity and activity manipulations in wildtype and mir-iab-4/8 virgins. Mutant virgins behave, and respond to neuronal manipulations, in the same way as mated females. Shaded region corresponds to AbG in wildtype (grey) and mir-iab-4/8 knockouts (blue). Mann-Whitney non parametric test, ns=not significant, ** p<0.01, *** p<0.001, **** p<0.0001. Error bars= SEM. Scale bar= 25μm. mir-iab-4/8−/− mutants=mir[Δ/C11] transheterozygotes.
Despite their prominent expression in the VNC, where the BX-C miRNAs are selectively deployed, some of these Gal4 lines have certain expression in the brain (Feng et al., 2014). We sought to distinguish the contributions of VNC or brain neurons to the phenotypes of mir-iab-4/8 null females. To do so, we exploited an Otd-nls:FLPo transgene that is expressed in the central brain (Asahina et al., 2014). By using this in combination with FRT-cassettes that excise tub-Gal80 or activate Gal4, we were able to restrict NaChBac activation to either the brain or the VNC of miRNA mutant animals (Figure 3D). For these tests, we employed the VT-7068 driver, which showed the most robust rescues (Figure 3C). While activation of VT-7068-brain neurons did not affect virgin egg-laying by mir-iab-4/8 mutants, specific activation of VT-7068-VNC neurons was sufficient to rescue this mutant defect (Figure 3C). Moreover, hyperactivation of only the four VNC ascending neurons labeled by SAG-1-Gal4, which has no reported expression in the brain (Feng et al., 2014), partially reduced egg-laying in mutants (Figure 3C), although not to the extent seen with VT-7068.
Together, these experiments suggest that mir-iab-4/8 loss compromises the output of the ascending post-mating circuit of virgin females in the VNC, partially evoking the effects of SP signaling after mating (Figure 3E).
Virgin behaviors are sensitive to hth levels in switch neurons
miRNAs are generally considered to exert their function by repressing broad target networks, although most regulatory interactions are mild (Bartel, 2018). At the same time, the majority of miRNA knockouts in diverse species have at best subtle phenotypes in development or physiology (Lai, 2015). This has led to the concept that miRNAs collectively regulate multiple transcripts to buffer transcriptional noise, and that individual phenocritical targets may be rare, notwithstanding the fact that miRNAs were first identified on account of specific targets that drive miRNA mutant defects (Lee et al., 1993; Wightman et al., 1993).
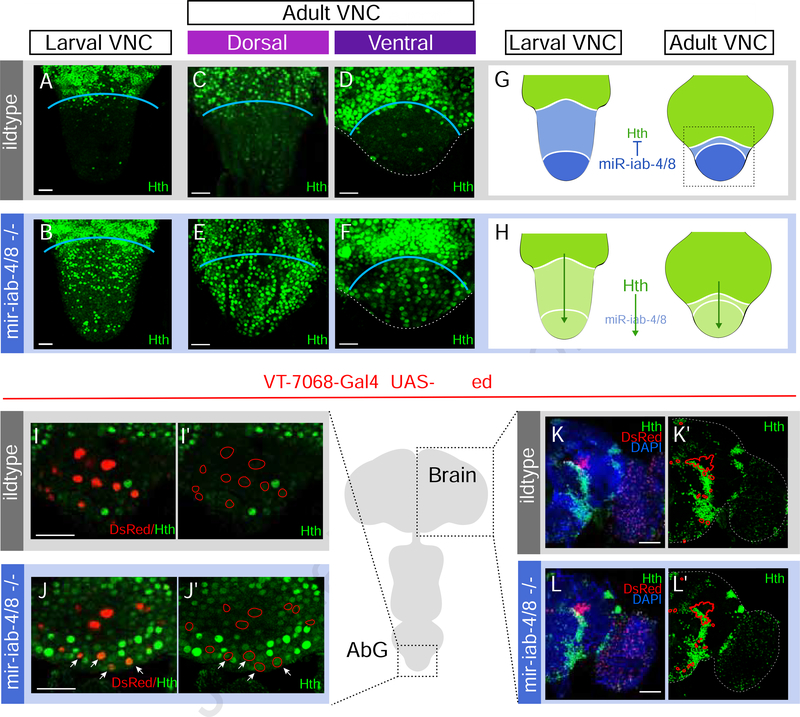
In previous studies, we identified a network of Hox genes and their TALE-class cofactors as direct in vivo targets that are misexpressed in mir-iab-4/8 mutants, and exhibit dominant phenotypic interactions with the miRNA deletion (Garaulet et al., 2014; Garaulet and Lai, 2015). We were particularly interested in the post-transcriptional regulation of homothorax (hth), which exhibits the highest number of miRNA binding sites for all the BX-C miRNAs, most of them encoded in a 2.3kb 3’ UTR extension found only in hth neural transcripts (Garaulet et al., 2014). Hth protein accumulates broadly across the thoracic neurons of the larval VNC, but is largely excluded from abdominal segments where it overlaps BX-C miRNAs. The BX-C miRNA mutants derepress Hth in abdominal neurons (Figure 4A,B and Supplementary Figure 4A) (Garaulet et al., 2014). Since we documented behavioral defects in adults, we also analyzed hth patterning in the CNS of adult females. By co-labeling Hth with the tub-GFP-miR-iab-4 sensor and Abd-A in wildtype, we can register that Hth is normally largely excluded from the abdominal ganglion region that encompasses active miR-iab-4 (Figure 4C, D, G and Supplementary Figure 4C). By extension, Hth is also mostly excluded from the miR-iab-8 domain that is located more posteriorly (Figure 4C, D, G and Supplementary Figure 4C). As in larval VNC (Figure 4B and Supplementary Figure 4A), we observed broadly ectopic Hth that extends into miR-iab-4/8 domains in mir-iab-4/8 mutants, throughout the dorsal and ventral regions of the abdominal ganglion both in adult female (Figure 4E, F, H), and male VNCs (Supplementary Figure 5A). Importantly, although there is heterogeneous distribution of nuclei with high levels of ectopic Hth (Figure 4 B,E,F and Supplementary Figure 5B), we consistently observe a lower level of derepressed Hth throughout the entire abdominal ganglion (Figure 2F–H, Figure 4J, J’ and Supplementary Figure 5C). This region encompasses populations of neurons that are functionally relevant to the post-mating switch (Feng et al., 2014), including neurons defined by the VT-7068, VT-50405 and SAG-1 drivers (Figure 4I–J and Supplementary Figure 5C,D,E). By contrast, we found no evidence for misexpression of Hth in mutant female brains, where miR-iab-4/8 are not expressed (Figure 4K–L’).
Figure 4. Hth patterning in wildtype and mir-iab-4/8 mutant CNS.
(A-F) Confocal images of female AbGs (boxed region in G). Hth (green) is mostly excluded from the abdominal segments in wildtype females in larval (A) and adult VNC (dorsal in C, ventral in D). Nevertheless, it is widely accumulated mir-iab-4/8 mutants (B, E, F). (G,H) Schematic representation of abdominal patterning of hth by BX-C miRNAs in wildtype (G) and mir-iab-4/8 mutants (H). hth is not expressed in VT-switch neurons in wildtype VNCs (I-I’) but detectable in mir[Δ/C11] mutants (arrows, J-J’). Hth is expressed in discrete clusters in the female brain, with restricted overlap with VT-7068 neurons (K,K’). Unlike abdominal neurons, no hth misexpression is observed in the brain of mir-iab-4/8 nulls (L,L’). Scale bar= 25μm (A-F, I-J’) and 60pm (K-L’). See also Supplementary Figure 4 and 5.
We used genetic strategies to evaluate if misexpression of Hth in those specific populations of neurons is responsible for the behavioral shift in mir-iab-4/8 null virgins. By monitoring egg-laying, we did not observe dominant rescue of the mutant phenotype by heterozygosity of hth[P2] null allele, or by a deficiency that uncovers hth (Figure 5A). However, we observed substantial rescue of miRNA mutant virgin egg-laying upon hth knockdown in several sets of VT-switch lines, particularly with VT-7068 and VT-50405 (Figure 5B). Suppression of hth in the intersection of these two Gal4s (SAG-1 neurons) also rescued partially, indicating involvement of these specific cells (Figure 5B). However, the weaker effect suggests that derepression of Hth in non-overlapping sets of VT-7068 and VT-50405 neurons also contributes to the miRNA phenotype. Amongst VT-switch lines tested, the strongest Hth accumulation was found in some of the VT-454 neurons (Supplementary Figure 5D). Interestingly, while VT-454>hth[RNAi] alone did not suppress egg-laying, inclusion of a hth mutant allele combined to yield nearly complete rescue (Figure 5C). This genetic threshold effect, not seen with hth heterozygosity alone, further highlights the sensitivity of this behavioral defect to Hth levels.
Figure 5. Developmental derepression of Homothorax in VT-switch neurons compromises virgin behavior.
(A-C) hth dosage manipulations. (A) Heterozygosity of hth is not sufficient to modulate increased egg-laying in miRNA mutant animals. (B) Conversely, hth knockdown in specific sets of neurons in mir-iab-4/8 mutants is sufficient to completely (VT-7068 neurons) or partially (VT-50405 and SAG-1 neurons) revert egg-laying to wildtype virgin values. (C) With VT-454, neural hth knockdown and heterozygosity act synergistically to rescue egg-laying. (D,E) Developmental vs. adult requirements for hth regulation by BX-C miRNAs in egg-laying. Adult specific knockdown of hth in VT-7068 neurons does not revert elevated egg-laying observed in mir-iab-4/8 mutants from days 1–5 after eclosion, despite the higher baseline levels observed for wildtype flies in this temperature plan (D). Conversely, developmental knockdown almost completely rescued mutant egg-laying values from days 1–8 to wildtype values (E). Mann-Whitney non parametric test, ns=not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Error bars= SEM. Shaded area (D,F)= SEM. Wildtype flies are Canton-S strain, mir-iab-4/8 −/− mutants=mir[Δ/C11] transheterozygotes. See also Supplementary Figure 6.
Although 7068-Gal4 drives specific expression in the brain (Figure 4K–L), VNC specific rescue of hth knockdown using this line was sufficient to substantially revert egg-laying in mir-iab-4/8 females (Supplementary Figure 6), highlighting the relevance of hth regulation of specific sets of abdominal neurons where miR-iab-4/8 are expressed.
Since PMRs were triggered by Hth misexpression in mir-iab-4/8 mutants, we wondered if behavior changes in mated females might involve acute changes in Hth levels in relevant abdominal neurons following copulation. However, immunostaining of the VNCs of wildtype females did not reveal dynamic elevation of Hth in abdominal segments after mating (Supplementary Figure 5F). This suggests that upregulation of Hth interferes with the proper function of mutant switch neurons, but is not involved in the normal response to copulation.
Hth upregulation in the VNC is observed throughout development and adult life. To discern the temporal requirements of miR-iab-4/8 regulation of hth, we restricted hth knockdown to either developmental stages or adult life, by inclusion of tubGal80ts transgene, and placing the flies at restrictive (18°C) or permissive temperature (29°C) before or after eclosion. (Figure 5D,E). Of note, raising the temperature after eclosion stimulates egg-laying in adult wildtype females (Figure 5D compare to Figures 1A, 3A and 5A). Nonetheless, mir-iab-4/8 mutant females still lay comparatively more eggs under these temperature conditions, and adult specific hth knockdown after eclosion does not revert this trend (Figure 5D). By contrast, temperature elevation during development did not affect egg-laying in wildtype virgins (Figure 5E). In this setting, developmental silencing of hth in VT-7068 neurons was sufficient to revert the elevated oviposition rates observed in mir-iab-4/8 nulls (Figure 5E).
Overall, while Hth is broadly misexpressed throughout the miR-iab-4/8 domains in mutants, its post-transcriptional repression in restricted sets of abdominal neurons during development mediates the capacity of virgins to integrate internal state and behavior.
Direct evidence that BX-C miRNA binding sites in the hth 3’ UTR control virgin behavior
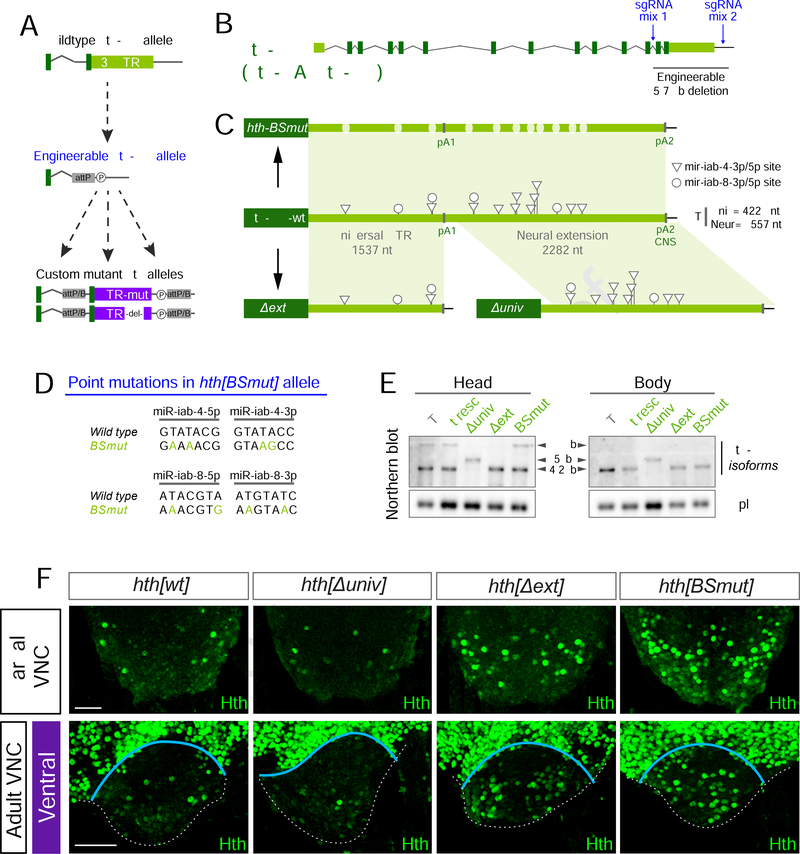
Genetic interactions can support compelling linkages amongst pathway components, but do not alone prove direct regulation. Thus, there is growing impetus to utilize genome engineering to specifically interrogate specific miRNA-target interactions in vivo (Ecsedi et al., 2015). While this is more straightforward for loci with one or two sites of interest, the full-length homeodomain-encoding isoform hth-HD bears 15 conserved sites for miR-iab-4/8 (two of which are predicted as dual sites). In order to disrupt all of these sites at the endogenous locus, we implemented a multi-step strategy that permits flexible engineering (Figure 6A, Supplementary Figure 7A). We first used CRISPR/Cas9 to replace the last two exons and 3’ UTR with an attP site (Figure 6B, Supplementary Figure 7A). The resulting hth-HD[ΔattP] allele was homozygous lethal, in accordance with known deletions of the distal end of hth isoforms (Kurant et al., 1998; Pai et al., 1998). This engineerable allele is amenable to generation of diverse downstream variants using OC31-mediated insertion (Baena-Lopez et al., 2013) (Figure 5C).
Figure 6. Engineering of homothorax 3’UTR.
(A) Flexible 3’ UTR engineering pipeline. (B) sgRNA cocktails were designed to delete the last 2 exons and the full 3’UTR of hth-HD isoform, which encodes the full-length homeodomain (HD) factor. (C) hth-HD 3’UTR alleles generated by CRISPR/ΦC31 mediated deletion/replacement. (D) Two point mutations were introduced per predicted binding site for either mature species of miR-iab-4 and miR-iab-8. (E) Northern blotting of the hth-HD engineered alleles. Deletion of the neural extension (hth[ΔextJ) eliminates the 6.6 kb isoform, only present in heads. Deletion of the universal segment (hth[Δuniv]) generates a novel isoform of 5kb, which constitutively incorporates the UTR extension both in heads and bodies (5kb). hth[wt] replacement and binding sites mutant (hth[BSmut]) do not affect the isoforms detected. kb=kilobase. (F) Hth pattern in the posterior region of the larval VNC and the AbG of hth-HD 3’ UTR alleles. hth[BSmut] and hth[Δext] fail to repress hth in abdominal segments. Scale bar= 50 μm. See also Supplementary Figure 7 and Supplementary Tables 1,2.
Here, we re-introduced either the deleted segment of the native gene (wildtype rescue, hth[wt]), or a 3’ UTR variant in which two seed bases of all conserved miR-iab-4 and miR-iab-8 binding sites were mutated (hth-[BSmut], Figure 6C–D). Thus, we obtained directly comparable control and modified hth alleles with only minimal genomic scars. Northern blotting confirmed that these replacement alleles preserved the biogenesis of proximal and distal hth isoforms of expected sizes (Figure 6E). In both cases, full viability of hth-HD[ΔattP] was restored, and no external developmental defects were observed. By contrast, immunostaining of the adult VNC revealed substantial defects specifically in the hth[BSmut] replacement allele. In particular, Hth was derepressed in the mutant abdominal ganglion compared to the wildtype 3’ UTR replacement, both during larval stages (Supplementary Figure 7B) and in adult females (Figure 6F and Supplementary Figure 7C). The adult de-repression of Hth was more overt in the ventral VNC relative to dorsal VNC (Figure 6F and Supplementary Figure 7C). Overall, these experiments yield stringent evidence that specific endogenous miRNA sites are essential to exclude Hth from the abdominal ganglion (Figure 4G, H).
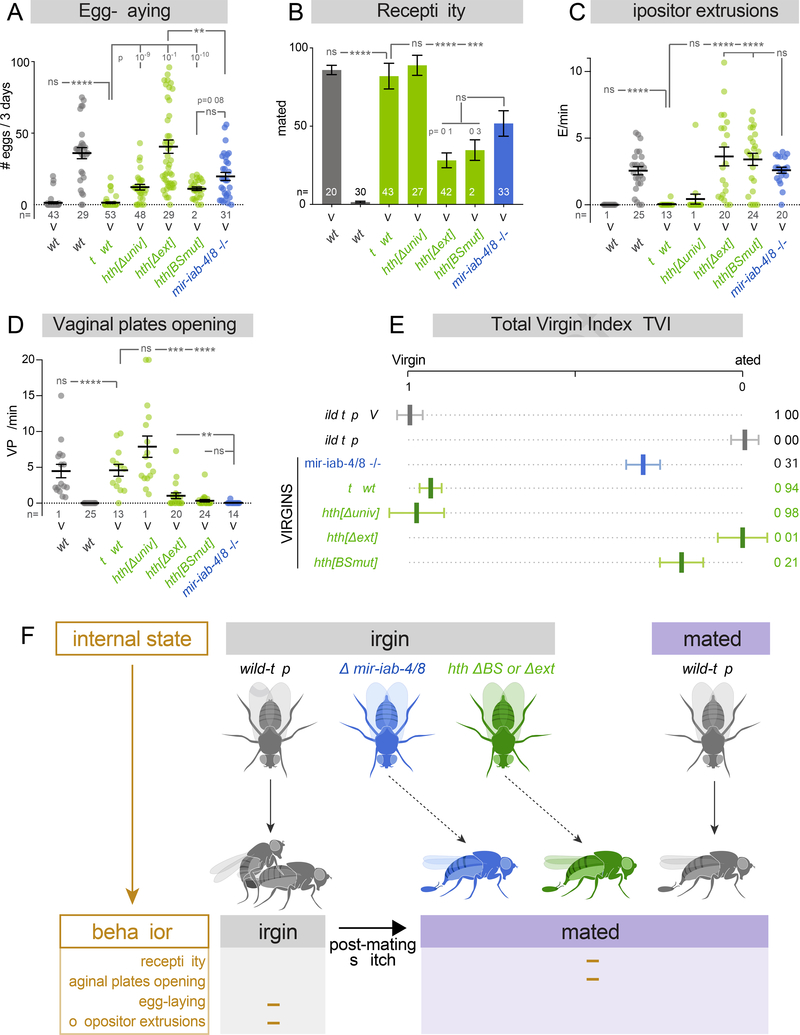
Since the spatial derepression of Hth was less in hth[BSmut] (Figure 6F and Supplementary Figure 7C) than with deletion of the miRNAs per se (Figure 4E–F), it was reasonable to wonder if this is sufficient to drive behavioral changes. Strikingly, we found that hth[BSmut] females nearly phenocopy the switch to the mated state observed in miRNA nulls. Compared to hth[wt], which exhibits normal virgin behaviors, hth[BSmut] virgins lay substantial numbers of eggs, exhibit reduced receptivity to courtship, fail to open their vaginal plates, but perform ovipositor extrusions instead (Figure 7A–D). Thus, in spite of other evident misregulation of Hth that occurs in the miRNA deletion, perhaps reflecting indirect effects involving other Hox gene targets (Garaulet et al., 2014), failure of its direct repression by miR-iab-4/8 can account for the uncoupling of internal state and behavioral output in virgin females with respect to multiple PMR readouts (Figure 6E).
Figure 7. Functional study of hth-HD 3’UTR elements.
(A-D) Virgin and mated behaviors assayed in wildtype, mir-iab-4/8 and hth-HD 3’ UTR alleles. (A) Egg-laying, (B) receptivity, (C) ovipositor extrusions, and (D) vaginal plates openings. Deletion hth-HD 3’ UTR neural extension or mutation of miR-iab-4/8 binding sites inhibit virgin behaviors and induce mated behaviors in virgins. (E) Total virgin index, integrating the values of all four behaviors analyzed in A-D. (F) Summary illustrating how neural specific, post-transcriptional regulation of homothorax is required for appropriate female behavior. Multiple behaviors in virgin females are altered after mating. The mated state is induced in virgin females deleted for mir-iab-4/8, by specific mutation of their binding sites in endogenous hth, or by deletion of the hth neural 3’ UTR extension. Thus, the integration of both miRNA control and neural APA is required to execute behaviors that are appropriate to the internal state of virgin female flies. (B) Fisher’s exact test. (A, C, D) Kruskal-Wallis t test with Dunn’s correction. ns=not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Error bars= SEM. Wildtype flies are Canton-S strain, mir-iab-4/8 −/− mutants= mir[Δ/C11] transheterozygotes.
Integration of neural APA with miRNA regulation adjusts virgin behavior to internal state
Neural 3’ UTR extensions induced by tissue-specific polyadenylation adds substantial regulatory real estate to hundreds of transcript isoforms (Sanfilippo et al., 2017). However, there are few in vivo examples of such extensions controlling phenotypically overt processes (An et al., 2008; Andreassi et al., 2010; Zhang et al., 2019). There are conserved miR-iab-4 and miR-iab-8 sites in both the universal and extended 3’ UTR segments of hth (Garaulet et al., 2014), although more total sites reside in the neural extension (Figure 6C). We prepared two additional hth alleles, in which we deleted either the universal or extended regions of the hth-HD 3’ UTR (Figure 6C). We confirmed that these alleles yielded expected transcript sizes, that is, deletion of the universal portion (Δuniv) resulted in a single common isoform bearing the extended 3’ UTR in head and body RNA, whereas deletion of the extension region (Δext) yielded a single short isoform in both RNA samples (Figure 6E).
Notably, these alleles had disparate effects on Hth protein expression and animal behavior. In particular, hth[Δext] caused far more abdominal neurons to accumulate Hth than did the hth[Δuniv] allele (Figure 6F and Supplementary Figure 7 B,C). Because the universal hth 3’ UTR segment contains multiple conserved miR-iab-4/8 sites, it is conceivable that loss of this regulatory region impacts VNC development. We found that virgin hth[Δuniv] mutants exhibited elevated egg-laying, but otherwise had normal sexual receptivity and genital behaviors. Thus, virgin behaviors seemed mostly intact in hth[Δuniv] mutants. By contrasT, hth[Δext] virgins displayed full PMRs with respect to all behavioral parameters, namely egg-laying, sexual receptivity, ovipositor extrusions, and frequency of vaginal plate openings (Figure 7A–D).
We integrated their performances in multiple assays into a single value, total virgin index (TVI). We assigned equal contributions to the different PMR readouts, which we acknowledge is arbitrary, but provides a simple metric to aid in the comparison of overall behaviors across mutants. This comparison emphasizes that the specific hth engineered mutants cause stronger induction of mated responses in virgins than does deletion of the miRNA locus itself (Figure 7F). In fact, hth[Δext] virgins are nearly indistinguishable from wildtype mated females. Since deletion of the miRNA locus may alter other direct and potentially indirect targets (Garaulet et al., 2014), we may not expect a complete correspondence of phenotypes, although these are qualitatively similar. On the other hand, as all the hth mutants are controlled with respect to a precise replacement of the wildtype 3’ UTR, these data highlight the strength of precision genetics to assay in vivo requirements at an individual phenocritical locus. Overall, these data demonstrate that miRNA regulation and neural APA are integrated at the hth locus to enable the virgin behavioral state in Drosophila females.
Discussion
Intersection of miRNA control and neural alternative polyadenylation
miRNAs are known to influence diverse aspects of development and adult physiology, and some miRNA mutants have been suggested to affect post-mated responses (Bender, 2008; Fricke et al., 2014; Garaulet et al., 2014; Maeda et al., 2018). Nevertheless, many miRNA knockout effects are subtle and challenging to comprehend using classical genetics and epistatic analysis. These strategies were critical to initially uncover the biological impact of miRNAs and to determine key individual miRNA targets whose regulation/dysregulation mediates in vivo phenotypes (Lai et al., 1998; Lee et al., 1993; Leviten et al., 1997; Wightman et al., 1993). Although we have learned much about the quantitative influence of miRNA targeting across the transcriptome (Agarwal et al., 2015; Kim et al., 2016), computational and genomic strategies still cannot predict which miRNAs and targets are involved in phenotypically overt regulation, and what settings these might manifest in. The finding that miRNA targeting is highly conditional on 3’ UTR utilization, via alternative polyadenylation (APA), raises further complexities into considering miRNA biology (Gruber and Zavolan, 2019). Nevertheless, it has also been reported that at least in some contexts, such as cultured fibroblasts, APA has limited impacts on gene expression and translation (Spies et al., 2013).
In this study, we dissect the biological impact of miRNAs and APA, which intersect to regulate an individual critical target in the nervous system. In particular, we use genetic engineering and neurobiological strategies to reveal a biological imperative for alternative polyadenylation that extends the hth 3’ UTR in the CNS. In general, many hundreds of genes express extended 3’ UTRs in the nervous system, which presumably increase post-transcriptional capacities of these neural isoforms (Miura et al., 2014). However, there are few in vivo reports that such neural 3’ UTR APA extensions mediate biologically substantial effects (An et al., 2008; Terenzio et al., 2018). By taking advantage of our engineerable allele platform, we were able to delete the universal and extended hth 3’ UTR in the intact fly. These mutants provide evidence that the role of neural APA is to unveil an array of BX-C miRNA sites that are crucial for hth suppression in the posterior VNC, where the miRNAs are normally expressed. Since BX-C miRNAs do not appear to be active in other settings where Hth is involved in patterning, e.g., in imaginal discs, the APA mechanism allows for heightened sensitivity of Hth to miRNAs in the CNS. Other BX-C targets of the BX-C miRNAs similarly undergo neural APA (Garaulet et al., 2014; Thomsen et al., 2010). Although phenotypic demonstration of their regulatory impacts of these extensions remain to be elucidated, the engineering platform described here is suitable. Moreover, it will be of considerable interest to determine the mechanism by which neural APA extensions of these and other genes are achieved (Hilgers et al., 2012).
Genetic control of the virgin behavioral state
We exploited the entry point of a miRNA knockout, which unexpectedly bypasses the virgin behavioral state of females. Multiple loss-of-function mutants are known (e.g., of SP and SP receptor) in which females cannot transition to normal mated behaviors following copulation. However, while certain transgenic interventions can induce mated conducts in virgins, such as inhibiting PKA activity downstream of the SP receptor in SPSNs (Yang et al., 2009), deletion of the BX-C miRNAs appears to be the first loss-of-function mutant that induces the mated state in virgins. Moreover, this effect does not appear to be the consequence of broad derepression of a large target cohort, but instead can be largely attributed to a single key target (hth).
These findings have important implications for understanding genetic impacts on the post-mating switch. While it has been frequently assumed that the virgin state is a default state, our results reveal genetic programming of virgin behavior. We use precision genetics to identify two new factors that are critical for the execution of virgin behaviors. Moreover, we reveal how the neural exclusive 3’ UTR extension of hth installs critical post-transcriptional elements, principally BX-C miRNA sites, which restrict Hth expression in the CNS and mediate appropriate adult behaviors. We observe derepression of the causal target Hth during both larval and in adult stages, but our spatio-temporally controlled experiments demonstrate that misexpression of Hth in the developing VNC is critical for the defective post-mating response. We note that the strong misexpression of Hth in adult VNC neurons of miRNA mutants likely has consequences for gene regulation, directed misexpression of this transcription factor has powerful detrimental consequences. However, it remains to be seen how its adult-specific misexpression affects behavior, since the phenotypes we analyzed here appear to have mostly developmental origins.
In any case, it is clear that both BX-C miRNAs and Hth comprise an unanticipated developmental regulatory axis that is required for adult virgin behavior. The failure of appropriate Hth regulation has two consequences for female virgin flies. First, they do not readily copulate, although we do observe that they can be inseminated. However, they also inappropriately execute reproductive activities without having mated. As these behaviors such as egg-laying are energetically demanding and reduce female fitness, it is necessary to avoid these before insemination, when they would be otherwise unsuccessful (Figure 7F). We reveal that females encode dual post-transcriptional regulatory mechanisms that are required to execute behaviors that are appropriate to the internal state of virgin flies.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric Lai (laie@mskcc.org. tel: 212-639-5578), or by Daniel L. Garaulet (lopezgad@mskcc.org).
Materials availability
Plasmid and transgenic flies generated in this study are available from the corresponding authors on request.
Data and code availability
Original behavior data for Figures 1,2,3,5,7 and Supplementary Figures 1,6 in the paper are available from the corresponding authors on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly strains and maintenance
This study used male and female flies of wildtype and genetically engineered strains of Drosophila melanogaster. Virgin and mated parameters refer to assays of female behavioral performances.
Larval and adult flies were raised on cornmeal/molasses media recipe: 83.8% water. 0.6% agar. 4.6% cornmeal. 2.3% dried yeast. 7.8% molasses solids. 0.3% propionic acid. 0.1% tegosept. 0.5% ethanol. They were kept at 25°C (unless mentioned otherwise). 55% humidity and under 12h:12h LD cycles.
Drosophila lines used in this study: mir[Δ] (Bender. 2008). mir [C11] (see Method Details). Canton-S (gift of Karla Kaun). tub-GFP-sensors for miR-iab-4 and miR-iab-8 (Garaulet et al.. 2014). VT-lines and SAG-1 split Gal4 line (Feng et al.. 2014). hth[P2] (Pai et al.. 1998). Otd-nls:FLPo (Asahina et al.. 2014). UAS-hth-RNAi (Vienna Drosophila RNAi Center), Df[hth] (Exelixis Collection). The following lines were obtained from Bloomington Drosophila Stock Center: Oregon R and Crimea strains, UAS-Red-Stinger, nos-cas9 (attP40), nos-ΦC31 (X), nos-ΦC31(X); attP2(3), TM6B-hs-Cre, >tubGal80>, tub>stop>Gal80 and tubGal80ts. All the lines used in this study have been backcrossed at least 8 generations to the Canton-S wildtype strain.
METHOD DETAILS
CRISPR-Cas9 engineered alleles
mir-iab-4/8 [C11]
mir-iab-4/8 deletion [C11] was generated using transgenic CRISPR (Kondo and Ueda, 2013). The sgRNA guide: 5’-ATACTGAAGGTATACCGGAT-3’ (chr3R:12682031-12682045) was cloned in pCFD4 under the U6.2 promoter (Port et al., 2014 and https://www.crisprflydesign.org/), and then injected in nos-ΦC31(X); attP2(3) flies (BDRC 25710). Transformant flies were identified by vermillion+ eyes and then crossed to nos-cas9 flies (BDRC #78781) to mutagenize mir-iab-4/8 locus (Kondo and Ueda, 2013). Individual offspring were balanced and analyzed by PCR (Fwd primer; 5’-AACGCTTGTAAAATCGGTCG-3’, Rev primer; 5’-AATTGCCGCTTGTTGAAGTT-3’) and sequencing to detect mutant events. We isolated a 24nt deletion uncovering the distal arm of the mir-iab-4/8 hairpin. The potential structures of this deletion, as well as the mir[Δ] allele (Bender, 2008), were predicted using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi).
hth-HD[ΔattP] allele
The hth-HD[ΔattP] allele is a 5.7kb deletion uncovering exons 13, 14 and the full 3’ UTR of hth-HD isoform (deletion coordinates chr3R: 6332160-6337923). We designed 8 different sgRNA guides targeting hth intron 12 and intergenic DNA downstream of the most distal pA site identified for hth-HD 3’ UTR (see guide sequences and chromosomal coordinates of targeted sequences in Supplementary Table 1). sgRNAs were cloned in 4 different pCFD4 plasmids (Port et al., 2014). A pHD-attP-DsRed donor plasmid (Gratz et al., 2014) was also cloned using two homology arms of 1081bp (chr3R: 6337924-6339004) and 1326 bp (chr3R: 6330834-6332159), adjacent to the outermost designed DNA double-strand breaks (Gratz et al., 2014, https://flycrispr.org/). The primers used to clone homology arms are in Supplementary Table 1. A CRISPR mix containing the pCFD4 plasmids at 75 ng/μL each and the pHD-attP-DsRed donor at 100 ng/μL was injected in nos-Cas9 expressing flies (BDRC #78781). Mutant events were identified by fluorescent red eyes and verified by PCR and sequencing (see primers in Supplementary Table 1).
hth-HD reintegration alleles
Reintegration of custom hth-HD 3’ UTR sequences was performed using pRIV-attB vectors (Baena-Lopez et al., 2013). An intermediate pRIV-attB-white vector (pRIV-w-CR/DS) was prepared carrying the intronic/exonic sequences (chr3R: 6337099-6337923) and the intergenic region downstream of hth-HD 3’UTR (chr3R: 6332160-6333279) deleted in the hth-HD allele, separated by a double SapI site (GAAGAGC-ccgctcgagcgg-GCTCTTC). This plasmid contains all the sequences that remain invariable in all of our engineered hth-HD 3’ UTR alleles, and allows flexible, scar-less integration of specific 3’ UTR variants. Wildtype, universal and extension 3’UTR fragments for hth[wt], hth[Δext] and hth[Δuniv] alleles, respectively, were obtained by PCR (see primer sequences and chromosomal locations in Supplementary Table 1), and cloned in the double SapI site of pRIV-w-CR/DS. The hth[BSmut] 3’ UTR was designed incorporating 2 point mutations per putative binding site for any of the 4 miRNA species emanating from the mir-iab-4/8 locus, as shown in Figure 6D and Supplementary Table 1. The resulting hth[BSmut] 3’ UTR sequence was synthesized as a g-block - Gene Fragment by Integrated Gene Technologies (https://www.idtdna.com/pages), and cloned in pRIV-w-CR/DS as described above. The reintegration plasmids were then injected in nos-OC31(X); hth-HD[ΔattP] flies. Red eyed (white+) transformants were recovered and sequenced using the same PCR primers used for hth-HD[ΔattP] verification (See Supplementary Table 1). pRIV-attB-white backbone was excised from the new alleles by balancing the 3rd chromosome over TM6B-hs-Cre (BDRC #1501).
Injections were performed at Bestgene, Inc. (https://www.thebestgene.com/) or Rainbow Transgenic Flies, Inc. (https://www.rainbowgene.com/). All new alleles were isogenized to the Canton-S strain prior to behavior analysis.
Northern Blotting
Body and head samples were prepared using liquid nitrogen to freeze, followed by vortexing and separation of heads from bodies using a sieve. Total RNAs were extracted from either heads or bodies of all genotypes using TRIzol (Invitrogen). Poly(A)+ RNAs were enriched from total RNAs using Oligo d(T)25 magnetic beads (New England Biolabs), according to the manufacturer’ s instructions. Northern blot analysis was performed essentially as previously described (Smibert et al., 2012) using 1.5 μg of poly(A)+ RNA per lane. Millennium RNA Marker (Invitrogen) was used as RNA size standard for each blot. Northern blot probes to detect different isoforms of homothorax were prepared by Amersham Megaprime DNA Labeling System (GE Healthcare Life Sciences), according to manufacturer’s instructions. Oligonucleotide sequences used to generate northern probes are listed in Supplementary Table 2.
Behavioral assays
Virgin males and females were collected after eclosion and were kept isolated in vials at 25°C, 55% humidity and 12h:12h LD cycles until the time of the behavior test. Virgin behaviors were assayed 3 days after eclosion unless specifically mentioned otherwise. For the analysis of mated females, they were mated a first time at day 3, separated from male after copulation, and kept isolated for 24h until the performance of the mated responses.
Receptivity, ovipositor extrusions and vaginal plates openings were analyzed from video recordings of custom made, 18x multiplex mating arenas (chamber size: 10mm diameter). Single males and females were placed in a half of each arena and let accommodate for 5 min before the assay. Egg-laying was calculated as the number of eggs laid in the first 3 days after eclosion. For receptivity; courtship initiation, mating start, and mating end were manually annotated. Receptivity was calculated as the proportion of animals mating at each time during the course of an hour (Supplementary Figure 1), or cumulative proportion of animals mated at 10 min (rest of the figures). Replicates of at least 8 individuals were analyzed. Ovipositor extrusions and vaginal plates openings were examined during the first 4 minutes after courtship copulation or until mating. Counts of either behavior were normalized to time. For ovipositor extrusions of mir-iab-4/8 mutants we used 1 day old virgins, since quantitative levels of extrusions were slightly higher at this age (Supplementary Figure 1). Nonetheless, nearly all flies displayed some levels of extrusions at any age from day 1 to 3. All tests were performed at ZT 7–11 and at least at four different occasions. Wildtype and heterozygous control females are always Canton-S strain unless otherwise specified. All males are Canton-S.
Temperature shifts were induced immediately after eclosion. Both at restrictive (18°C) and permissive (29°C) temperature, 55% humidity and 12h:12h LD cycles were maintained.
Virgin Indexes (VI) were calculated by interpolating the behavioral value obtained for each mutant fly within the rank defined from the average wildtype virgin value (1) and the average wildtype mated value (0). The plots show the average and SEM of all the individual values per behavior (Virgin Index), or the average of all behaviors (Total Virgin Index).
Immunohistochemistry, imaging, and image quantification analysis
Larval and adult CNS were dissected in cold 1x PBS and fixed for 1h in 4% paraformaldehyde + 0.1% Triton. Primary and secondary antibodies were incubated for >36h at 4°C in wash buffer (PBS 1% BSA) and mounted in Vectashield (Vector Labs). Antibodies used were rabbit and guinea pig anti-Hth (1:500) (Salvany et al., 2009), mouse anti-Abd-A (1:500), and Alexa-488, −555, −647 conjugated goat and/or donkey antibodies from Thermo Fisher Scientific.
Imaging was performed in a Leica TCS SP5 confocal microscope. Each VNC was typically scanned in 55 planes (Z step ~2nm). When image quantification or comparison was performed (Figures 2,4,6), all different genotypes used were dissected at the same time, fixed and incubated together in the same well. To identify the genotype of each VNC while mounting, different parts of the head were left attached or removed from the VNC during dissection. Then, the same number of VNCs from different genotypes were arranged in a known fashion per slide, to avoid differences in the quantification due to the mounting process. Laser power and offset were maintained identically for all the samples being compared. Gain was slightly adjusted to an internal control in each case. All adult VNC images are 0–24hr old virgin females.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was evaluated using Fisher’s exact test for receptivity (Figures 1,7 and Supplementary Figure 1), copulation onset (Figure 1), qualitative analyses of oviposition and ovipositor extrusions (Supplementary Figure 1), and number of neurons (Figure 2 and Supplementary Figure 5), and Mann-Whitney non parametric test for Egg-laying, quantitative ovipositor extrusions and vaginal plates openings (Figures 1,3,5,7 and Supplementary Figures 1,6). ns=not significant, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Error bars in Figures 1,2,3,5,7, and Supplementary Figures 1,5,6, and shaded areas in Figure 5 and Supplementary Figure 1 represent SEM. All n values are displayed on the figures.
Supplementary Material
Supplementary movie 1 (related to Figure 1). Mating assay. Wildtype virgins and wildtype males.
Supplementary movie 2 (related to Figure 1). Mating assay. mir[Δ/C11] virgins and wildtype males
KEY RESOURCES TABLE (KRT).
| SOURCE | IDENTIFIER | |
|---|---|---|
| Antibodies | ||
| rabbit anti-Hth (1:500) | (Salvany et al, 2009) | N/A |
| guinea pig anti-Hth (1:500) | (Salvany et al, 2009) | N/A |
| Mouse anti-Abd-A (1:500) | Gift of Ian Duncan | Department of Biology. Washington University in St. Louis |
| Donkey anti-rabbit Alexa fluor 488 | Thermo Fisher Scientific | A32790 |
| Goat anti-rabbit Alexa fluor 647 | Thermo Fisher Scientific | A32733 |
| Donkey anti-mouse Alexa fluor 594 | Thermo Fisher Scientific | A32744 |
| Donkey anti-mouse Alexa fluor 647 | Thermo Fisher Scientific | A32787 |
| Goat anti-ginea pig Alexa fluor 488 | Thermo Fisher Scientific | A-11073 |
| Bacterial and Virus Strains | ||
| TOP10 Chemically Competent E. coli | Custom made - Lai Lab | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Penicillin-Streptomycin | Life Technologies | 15070063 |
| T4 DNA Ligase | NEB | M0202L |
| Gel Loading Buffer II | Invitrogen | AM8547 |
| AccuPrime pfx DNA Polymerase | Thermo Fisher | 12344024 |
| Trizol reagent | Life Technologies | 15596018 |
| Oligo d(T)25 Magnetic Beads | New England Biolabs | S1419S |
| Millennium™ RNA Markers | Thermo Fisher | AM7150 |
| Amersham Megaprime DNA Labeling System | Cytiva | RPN1606 |
| Plasmids | ||
| pHD-attP-DsRed | (Gratz et al., 2014) | N/A |
| pCFD4 | (Port et al., 2014) | N/A |
| pRIV-attB vectors | (Baena-Lopez et al., 2013) | N/A |
| Experimental Models: Organisms/Strains | ||
| mir[Δ] | (Bender, 2008) | N/A |
| Canton-S | gift of Karla Kaun | Department of Neuroscience. Brown University. |
| VT-lines and SAG-1 split Gal4 line | (Feng et al., 2014) | N/A |
| hth[P2] | (Pai et al., 1998) | N/A |
| Df[hth] | Exelixis Collection | 6158 |
| Otd-nls:FLPo, | (Asahina et al., 2014) | N/A |
| tub-GFP-sensors for miR-iab-4 and miR-iab-8 | (Garaulet et al., 2014) | N/A |
| UAS-hth-RNAi | Vienna Drosophila RNAi Center | 100630 |
| Oregon R | Bloomington Drosophila Stock Center | 5 |
| Crimea | Bloomington Drosophila Stock Center | 4266 |
| UAS-Red-Stinger | Bloomington Drosophila Stock Center | 8547 |
| nos-cas9 (attP40) | Bloomington Drosophila Stock Center | 78781 |
| nos-ΦC31 (X) | Bloomington Drosophila Stock Center | 34771 |
| TM6B-hs-Cre | Bloomington Drosophila Stock Center | 1501 |
| >tubGal80> | Bloomington Drosophila Stock Center | 38880 |
| tub>stop>Gal80 | Bloomington Drosophila Stock Center | 38878 |
| tubGal80ts | Bloomington Drosophila Stock Center | 7108 |
| nos-ΦC31(X); attP2(3) | Bloomington Drosophila Stock Center | 25710 |
| Oligonucleotides | ||
| sgRNAs for hth-HD deletion, see Table S1 | This paper | N/A |
| Primers for pHD-attP-DsRed cloning, see Table S1 | This paper | N/A |
| Primers for hth-HD[ΔattP] verification, see Table S1 | This paper | N/A |
| Primers for hth-HD 3’UTR alleles, see Table S1 | This paper | N/A |
| sgRNA for mir[C11]: ATACTGAAGGTATACCGGAT | This paper | N/A |
| Fwd primer for mir-iab-4/8-HP amplification: AACGCTTGTAAAATCGGTCG | This paper | N/A |
| Rev primer for mir-iab-4/8-HP amplification: AATTGCCGCTTGTTGAAGTT | This paper | N/A |
| Primers used to generate Northern probes, see Table S2 | This paper | N/A |
Highlights.
Mutants of Bithorax-Complex miRNAs cause virgin females to adopt mated behaviors
The miRNAs act within the ventral nerve cord to suppress homothorax (hth)
Genome engineering links miRNA sites and neural APA of hth with female behavior
Acknowledgments
We thank Welcome Bender, Barry Dickson, Natalia Azpiazu, Richard Mann, Carlos Ribeiro, Carolina Rezaval, Stephen Goodwin, Young-Joon Kim, Leslie Vosshall, Shu Kondo, Karla Kaun, Luis Alberto Baena-Lopez, Mariana Wolfner, Ernesto Sánchez-Herrero, Paloma Martin, Michelle Arbeitman, Filip Port, Kate O’Connor-Giles, David J. Anderson and the Bloomington Stock Center for fly strains, plasmids, and antibodies used in this study. Ricardo Toledo-Crow helped fabricate mating chambers. We are grateful to Alex Panzarino and Silvia Rodriguez for helping with initial behavioral assays, and Jennifer Bussell and Leslie Vosshall for advice on assaying female mating behaviors. Work in E.C.L.’s group was supported by the NIH (R01-GM083300 and R01-NS083833) and MSK Core Grant P30-CA008748.
Footnotes
Declaration of Interests:
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal V, Bell GW, Nam JW, and Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigaki T, Fleischmann I, Chen PS, and Kubli E (1991). Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 7, 557–563. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. (2008). Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, and Riccio A (2010). An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nature neuroscience 13, 291–301. [DOI] [PubMed] [Google Scholar]
- Aranha MM, and Vasconcelos ML (2018). Deciphering Drosophila female innate behaviors. Curr Opin Neurobiol 52, 139–148. [DOI] [PubMed] [Google Scholar]
- Asahina K, Watanabe K, Duistermars BJ, Hoopfer E, Gonzalez CR, Eyjolfsdottir EA, Perona P, and Anderson DJ (2014). Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell 156, 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, and Wolfner MF (2011). Insect seminal fluid proteins: identification and function. Annu Rev Entomol 56, 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez LA, Alexandre C, Mitchell A, Pasakarnis L, and Vincent JP (2013). Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 140, 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W (2008). MicroRNAs in the Drosophila bithorax complex. Genes & development 22, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broida J, and Svare B (1982). Strain-typical patterns of pregnancy-induced nestbuilding in mice: maternal and experiential influences. Physiology & behavior 29, 153–157. [DOI] [PubMed] [Google Scholar]
- Bussell JJ, Yapici N, Zhang SX, Dickson BJ, and Vosshall LB (2014). Abdominal-B neurons control Drosophila virgin female receptivity. Curr Biol 24, 1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho GB, Kapahi P, Anderson DJ, and Benzer S (2006). Allocrine modulation of feeding behavior by the Sex Peptide of Drosophila. Curr Biol 16, 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, and Bohlen P (1988). A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54, 291–298. [DOI] [PubMed] [Google Scholar]
- Clyne JD, and Miesenbock G (2009). Postcoital finesse. Neuron 61,491–493. [DOI] [PubMed] [Google Scholar]
- Connolly K, and Cook R (1973). Rejection responses by female Drosophila melanogaster: Their ontogeny, causality and effects upon behaviour of courting male. Behavior 44, 142–166. [Google Scholar]
- Ecsedi M, Rausch M, and Grosshans H (2015). The let-7 microRNA Directs Vulval Development through a Single Target. Developmental cell 32, 335–344. [DOI] [PubMed] [Google Scholar]
- Feng K, Palfreyman MT, Hasemeyer M, Talsma A, and Dickson BJ (2014). Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron 83, 135–148. [DOI] [PubMed] [Google Scholar]
- Fowler K, and Partridge L (1989). A cost of mating in fruitflies. Nature 338, 760–761. [Google Scholar]
- Fricke C, Green D, Smith D, Dalmay T, and Chapman T (2014). MicroRNAs influence reproductive responses by females to male sex peptide in Drosophila melanogaster. Genetics 198, 1603–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet DL, Castellanos MC, Bejarano F, Sanfilippo P, Tyler DM, Allan DW, Sanchez-Herrero E, and Lai EC (2014). Homeotic Function of Drosophila Bithorax-Complex miRNAs Mediates Fertility by Restricting Multiple Hox Genes and TALE Cofactors in the CNS. Developmental cell 29, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet DL, and Lai EC (2015). Hox miRNA regulation within the Drosophila Bithorax complex: Patterning behavior. Mechanisms of development 138 Pt 2, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillott C (2003). Male accessory gland secretions: modulators of female reproductive physiology and behavior. Annu Rev Entomol 48, 163–184. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AJ, and Zavolan M (2019). Alternative cleavage and polyadenylation in health and disease. Nature reviews Genetics. [DOI] [PubMed] [Google Scholar]
- Gummalla M, Maeda RK, Castro Alvarez JJ, Gyurkovics H, Singari S, Edwards KA, Karch F, and Bender W (2012). abd-A Regulation by the iab-8 Noncoding RNA. PLoS genetics 8, e1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemeyer M, Yapici N, Heberlein U, and Dickson BJ (2009). Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron 61, 511–518. [DOI] [PubMed] [Google Scholar]
- Haussmann IU, Hemani Y, Wijesekera T, Dauwalder B, and Soller M (2013). Multiple pathways mediate the sex-peptide-regulated switch in female Drosophila reproductive behaviours. Proc Biol Sci 280, 20131938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Lemke SB, and Levine M (2012). ELAV mediates 3’ UTR extension in the Drosophila nervous system. Genes & development 26, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac RE, Li C, Leedale AE, and Shirras AD (2010). Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc Biol Sci 277, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YH, Chae HS, and Kim YJ (2017). Female-specific myoinhibitory peptide neurons regulate mating receptivity in Drosophila melanogaster. Nature communications 8, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Sung YM, Park J, Kim S, Kim J, Park J, Ha H, Bae JY, Kim S, and Baek D (2016). General rules for functional microRNA targeting. Nature genetics 48, 1517–1526. [DOI] [PubMed] [Google Scholar]
- Kondo S, and Ueda R (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, and Salzberg A (1998). Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 125, 1037–1048. [DOI] [PubMed] [Google Scholar]
- Lai EC (2015). Two decades of miRNA biology: lessons and challenges. RNA 21, 675–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Burks C, and Posakony JW (1998). The K box, a conserved 3’ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development 125, 4077–4088. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, and Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Leviten MW, Lai EC, and Posakony JW (1997). The Drosophila gene Bearded encodes a novel small protein and shares 3’ UTR sequence motifs with multiple Enhancer of split Complex genes. Development 124, 4039–4051. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Sitnik JL, Frei Y, Prince E, Gligorov D, Wolfner MF, and Karch F (2018). The lncRNA male-specific abdominal plays a critical role in Drosophila accessory gland development and male fertility. PLoS genetics 14, e1007519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A (1962). A Sperm factor affecting the receptivity of Drosophila melanogaster females. Nature 194, 252–253. [Google Scholar]
- Manning A (1967). The control of sexual receptivity in female Drosophila. Anim Behav 15, 239–250. [DOI] [PubMed] [Google Scholar]
- Miura P, Sanfilippo P, Shenker S, and Lai EC (2014). Alternative polyadenylation in the nervous system: to what lengths will 3’ UTR extensions take us? BioEssays 36, 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastirioti M (2003). Distinct octopamine cell population residing in the CNS abdominal ganglion controls ovulation in Drosophila melanogaster. Developmental biology 264, 38–49. [DOI] [PubMed] [Google Scholar]
- Ogawa S, and Makino J (1984). Aggressive behavior in inbred strains of mice during pregnancy. Behavioral and neural biology 40, 195–204. [DOI] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, and Sun YH (1998). The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes & development 12, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Chen S, Busser S, Liu H, Honegger T, and Kubli E (2005a). Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Curr Biol 15, 207–213. [DOI] [PubMed] [Google Scholar]
- Peng J, Zipperlen P, and Kubli E (2005b). Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr Biol 15, 1690–1694. [DOI] [PubMed] [Google Scholar]
- Picao-Osorio J, Johnston J, Landgraf M, Berni J, and Alonso CR (2015). MicroRNA-encoded behavior in Drosophila. Science 350, 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, and Bullock SL (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 111, E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaval C, Pavlou HJ, Dornan AJ, Chan YB, Kravitz EA, and Goodwin SF (2012). Neural circuitry underlying Drosophila female postmating behavioral responses. Curr Biol 22, 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C, and Dickson BJ (2010). Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr Biol 20, 1000–1005. [DOI] [PubMed] [Google Scholar]
- Salvany L, Aldaz S, Corsetti E, and Azpiazu N (2009). A new role for hth in the early pre-blastodermic divisions in drosophila. Cell cycle 8, 2748–2755. [DOI] [PubMed] [Google Scholar]
- Sanders LE, and Arbeitman MN (2008). Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Developmental biology 320, 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo P, Wen J, and Lai EC (2017). Landscape and evolution of tissue-specific alternative polyadenylation across Drosophila species. Genome biology 18, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads B, Carlson J, Brown JB, et al. (2012). Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell reports 1, 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, Haussmann IU, Hollmann M, Choffat Y, White K, Kubli E, and Schafer MA (2006). Sex-peptide-regulated female sexual behavior requires a subset of ascending ventral nerve cord neurons. Curr Biol 16, 1771–1782. [DOI] [PubMed] [Google Scholar]
- Spies N, Burge CB, and Bartel DP (2013). 3’ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome research 23, 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzio M, Koley S, Samra N, Rishal I, Zhao Q, Sahoo PK, Urisman A, Marvaldi L, Oses-Prieto JA, Forester C, et al. (2018). Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen S, Azzam G, Kaschula R, Williams LS, and Alonso CR (2010). Developmental RNA processing of 3’UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development 137, 2951–2960. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, and Lai EC (2008). Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes & development 22, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SJ, Corrales-Carvajal VM, and Ribeiro C (2015). Postmating Circuitry Modulates Salt Taste Processing to Increase Reproductive Output in Drosophila. Curr Biol 25, 2621–2630. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, and Ruvkun G (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Yang CH, Rumpf S, Xiang Y, Gordon MD, Song W, Jan LY, and Jan YN (2009). Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici N, Kim YJ, Ribeiro C, and Dickson BJ (2008). A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451,33–37. [DOI] [PubMed] [Google Scholar]
- Zhang Z, So K, Peterson R, Bauer M, Ng H, Zhang Y, Kim JH, Kidd T, and Miura P (2019). Elav-Mediated Exon Skipping and Alternative Polyadenylation of the Dscam1 Gene Are Required for Axon Outgrowth. Cell reports 27, 3808–3817 e3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary movie 1 (related to Figure 1). Mating assay. Wildtype virgins and wildtype males.
Supplementary movie 2 (related to Figure 1). Mating assay. mir[Δ/C11] virgins and wildtype males
Data Availability Statement
Original behavior data for Figures 1,2,3,5,7 and Supplementary Figures 1,6 in the paper are available from the corresponding authors on request.