Abstract
The heart possesses a complex three-dimensional (3D) laminar myofiber organization; however, because engineering physiologically relevant 3D tissues remains a technical challenge, the effects of cardiomyocyte alignment on excitation-contraction coupling, shortening and force development have not been systematically studied. Cellular shape and orientations in 3D can be controlled by engineering scaffold microstructures and encapsulating cells near these geometric cues. Here, we show that a novel method of cell encapsulation in 3D methacrylated gelatin (GelMA) scaffolds patterned via Microscale Continuous Optical Printing (μCOP) can rapidly micropattern neonatal mouse ventricular cardiomyocytes (NMVCMs) in photocrosslinkable hydrogels. Encapsulated cardiomyocytes preferentially align with the engineered microarchitecture and can display morphology and myofibril alignment phenotypic of myocardium in vivo. Utilizing the μCOP system, an asymmetric, multi-material, cantilever-based scaffold was directly printed, so that the force produced by the microtissue was transmitted onto a single deformable pillar. Aligned 3D encapsulated NMVCM scaffolds produced nearly 2 times the force compared to aligned 2D seeded samples. To further highlight the flexibility of μCOP, NMVCMs were encapsulated in several patterns to compare the effects of varying degrees of alignment on tissue displacement and synchronicity. Well aligned myofiber cultured patterns generated 4–10 times the contractile force of less anisotropically patterned constructs. Finally, normalized fluo-4 fluorescence of NMVCM-encapsulated structures showed characteristic calcium transient waveforms that increased in magnitude and rate of decline during treatment with 100 nM isoproterenol. This novel instrumented 3D cardiac microtissue serves as a physiologically relevant in vitro model system with great potential for use in cardiac disease modeling and drug screening.
Keywords: 3D bioprinting, hydrogel, organ-on-a-chip, cardiomyocytes, tissue engineering
Introduction:
The pumping action of the heart arises from the contraction of anisotropic layers of cardiomyocytes arranged in distinct orientations[1]. Previous tissue engineering approaches have attempted to recapitulate these native microstructures in vitro by seeding cardiomyocytes onto nano- to micro-scale features such as grooves, patterned biochemical cues, or aligned nanofibers formed via traditional microfabrication techniques[2–5]. Owing to the mechanical cues produced by topographical patterning, these approaches have substantial control over the shape and alignment of the cardiomyocytes. However, surface patterns or grooves can only induce cellular alignment in two dimensions (2D) limiting their relevance to in vivo physiology.
To address this limitation, several groups have reported methods for producing 3D cardiac tissues for mechanical[6–10] and electrical interrogation[11,12]. However, most of these methods rely on seeding cells into prefabricated scaffolds. This can yield inconsistencies in cell distribution due to cell aggregation, particularly within wells and channels, thereby confounding results. Furthermore, cell morphology and responses including cell-cell communication and cellsubstrate interactions, can be starkly different for cells encapsulated into a 3D microarchitecture compared with 2D[13–16]. Thus, it is important to have a high degree of control over 3D microarchitecture and myocyte distribution. 3D printing is able to address these issues, and has become an emerging tool within the field of tissue engineering, allowing biomimetic scaffolds that better recapitulate native tissue to be designed[17–19]. Unlike traditional extrusion-based 3D printing that requires 1D raster-scanning of a 3D object, Microscale Continuous Optical Printing (μCOP) projects a 2D image into a volume of prepolymer solution, pairing stage movement with changes in digital masks, providing greater z-resolution and greatly increasing the speed of printing[20–22]. The user-defined patterns are loaded onto the digital micro-mirror device (DMD) chip, designating mirrors as “on” or “off”. Mirrors that are “on” reflect UV light, which is subsequently focused by the projection optics onto the prepolymer solution, immediately initiating polymerization to produce a 3D structure replicating the designated pattern. Using appropriate bioinks, cells can be directly patterned into a 3D hydrogel, allowing for control over the alignment and concentration of cells in the printed tissue construct.
In this study, we present a novel method of using μCOP to produce a scalable 3D in vitro model mimicking the microarchitecture and function of ventricular myocardium. Utilizing μCOP, we designed an asymmetric, customizable force measuring system with direct patterning of encapsulated cardiomyocytes. Neonatal mouse ventricular cardiomyocytes (NMVCMs) progressively aligned with the engineered microarchitecture and formed a synchronously contracting tissue with force output determined via the deflection of an integrated, 3D-printed force gauge. Since diseases such as hypertrophic cardiomyopathy are associated with varying degrees of myofiber disarray, we investigated the role of myocyte alignment in these structures by using the 3D printing system to fabricate slab, grid, dispersed, and randomized fiber patterns, and measuring the resulting contractile force development. Direct 3D printing of cells into an anisotropic pattern improves sarcomere alignment and force production of the cardiac tissue, compared to samples seeded on matched geometry. Complex patterning of cells disrupts the ability of cardiomyocytes to align, contract along the main axis of the scaffold, thus reducing the contractile forces of the tissues. We demonstrated that the 3D microtissues could be maintained long-term, produce large forces, and respond to inotropic agents at low concentrations. This highly customizable 3D cardiac tissue is a potential tool for future disease modeling and drug discovery.
Materials and Methods
Pre-polymer Synthesis
Photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was synthesized according to the previously published methods [23]. Methacrylated gelatin (GelMA) and hyaluronic acid glycidyl methacrylate (HAGM) were also synthesized according to previously published procedures [24–26].
Prepolymer Solution Preparation
Solutions were measured as wt/vol percentages, unless specified. A 4% LAP stock solution was dissolved in Dulbecco’s Phosphate Buffered Saline (DPBS) and filtered using a 0.22 μm filter. Lyophilized GelMA and HAGM foams were reconstituted with DPBS to produce stock solutions of 20% and 4%, respectively. Both solutions were warmed and sterile filtered using a 0.22 μm filter. Stock solutions were diluted with DPBS to make 15%, 10%, 7.5%, and 5% GelMA solutions with 0.2% LAP and 2% HAGM/2% (v/v) poly(ethylene glycol) (PEGDA) with 0.2% LAP.
Neonatal Cardiomyocyte Isolation and Culture
Neonatal mouse cardiomyocytes (NMVCMs) were isolated from the hearts of neonates of CD-1 wild-type mice (Charles River Labs). In brief, hearts were surgically removed from 1-day-old pups and digested in Hank’s Balanced Salt Solution (Gibco) with 0.046% Trypsin (Affymetrix) at 4 °C overnight. Blood cells were removed from hearts by type II Collagenase (Worthington) after shaking at 37 °C for two minutes. A heterogeneous cell population containing cardiomyocytes and fibroblasts was isolated after further digestion using type II Collagenase (Worthington) at 37 °C for seven minutes. Fibroblasts were removed by pre-plating for 1.5 hours on 75 cm2 plastic tissue culture flasks (Corning) in a humidified incubator at 37 °C with 5% CO2. Isolated cardiomyocytes were resuspended in dark medium formulated by 75% Dulbecco’s Modified Eagle Medium (DMEM) and 25% M199 medium containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 10% horse serum (HyClone), 5% fetal bovine serum (Gibco), and 1% 100x Penicillin/Streptomycin/L-Glutamine solution. Media was replaced daily and on day 3, onward, a 10 μM mM solution of arabinosylcytosine (Ara-C), an antiproliferative drug to prevent noncardiomyocyte overgrowth.
3D-Printed Force Gauges and Cell Encapsulation
Utilizing the μCOP system, an array of micron-scale features was built in various biopolymers using UV polymerization in a layer-by-layer fashion as previously described[22]. The main components of the fabrication system are a UV light source (Omnicure 2000), a digital light processing (DLP) chip (Discovery 4000, Texas Instrument), and computer controlled x-y-z stages (Newport 426/433 series). A 365 nm bandpass filter with a source output of 6 W/cm2 was utilized. User-defined bitmap patterns were transferred to the DLP chip and the modulated images were projected onto a photocurable pre-polymer solution through a UV-grade optical lens (Edmunds Optics). Areas illuminated by UV light crosslinked immediately, whereas dark regions remained uncrosslinked, forming the scaffold in a specific polymerization plane designated by the mask. These patterns were irradiated for 45 seconds at a projected UV intensity of 11 mW/ cm2.
Initially, a solution of 2% HAGM, 2% (v/v) PEGDA, 0.2% LAP was injected between a methacrylated coverslip held by two 500 μm polydimethylsiloxane (PDMS) spacers. The solution was exposed to the first mask corresponding to a 2mm × 3mm × 250 μm base layer (Figure 1a–b, i). The remaining solution was washed in DPBS and replaced with 15% GelMA, 0.2% LAP solution. The solution was then exposed for 18 seconds by the pillar pattern (Figure 1a–b, ii), straddling the HAGM/PEGDA slab. The 15% GelMA solution was washed with warm DPBS and a 10% GelMA, 0.2% LAP solution was mixed 1:1 with a 50 million cells/ml suspension in DM (Dark Media) to achieve a final concentration of 5% GelMA, 0.1% LAP, and 25 million cells/ml which was subsequently injected between the pillars. The cell solution was then exposed by the final line pattern (Figure 3a) for 45 seconds. The slab was made with a simple single exposure of an 1×1 × 2 mm area between the two pillars. The grid pattern used the same parallel line pattern with the addition of perpendicular lines of equal width and spacing. The dispersion pattern was designed to pass each line through the midpoint, while the random pattern connected each line randomly using a randomizing script. Two additional lines were drawn on the first and last line to ensure the volume of cells would be similar to the line pattern. The average angle of the patterned lines is 4° from the major axis.
Figure 1. μCOP printing of a 3D cardiac tissue.
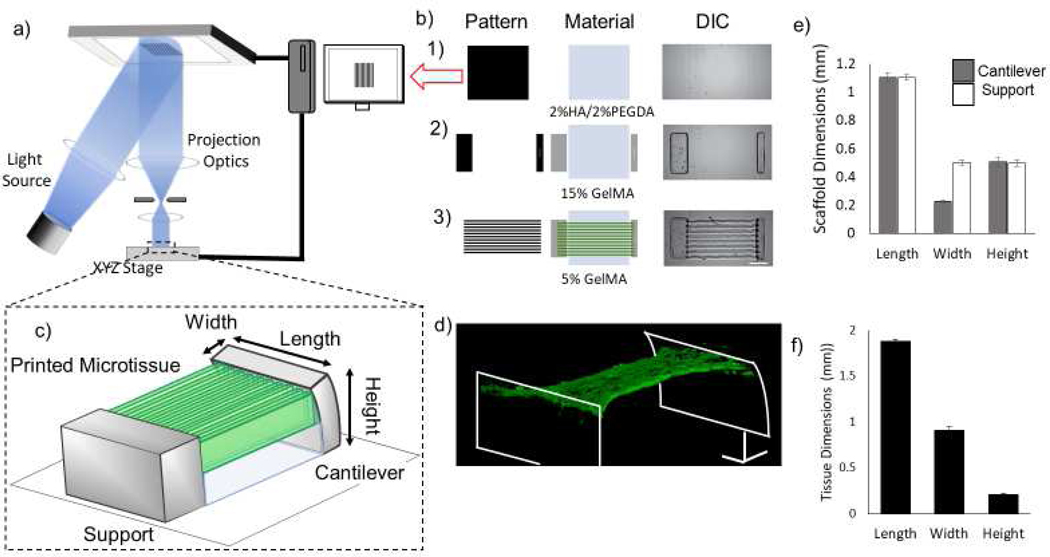
a) Schematic of the μCOP printing system. b) Mask-set and print order of: 1) 2%HAGM/2% PEGDA, 2) 15% GelMA, 3) 5% GelMA/NMVCM with DIC images (scale bar 250 μm). c) 3D schematic of full 3D tissue-measuring scaffold. d) Confocal 3D reconstruction of NMVCM cardiac tissue stained for actin (red) and nuclei (blue) across pillars. e) Measured scaffold dimensions in mm (n=19) f) Initial tissue dimensions of the 3D-printed scaffolds in mm (n=6)
Figure 3. Mechanical testing of NMVCM tissue.
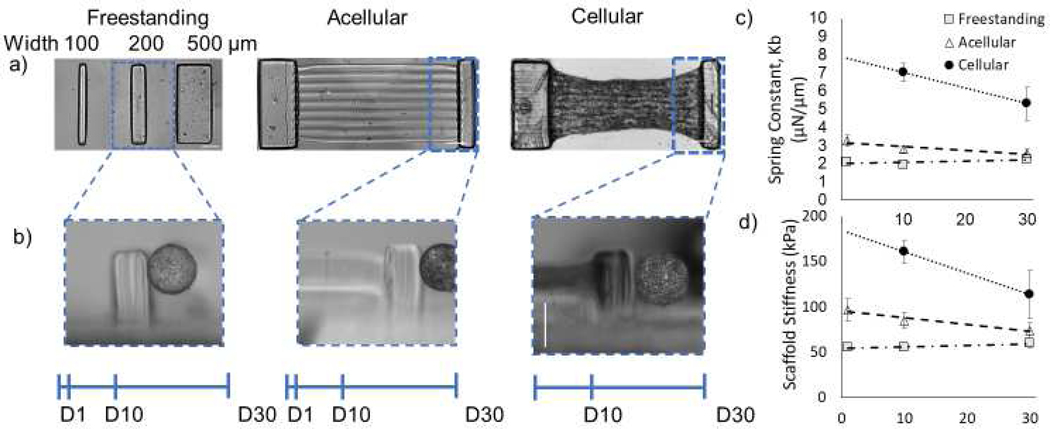
a) Mechanical testing of free standing cantilevers, acellular scaffolds and cellular scaffolds, and b) representative images of the Microsquisher pressing on each scaffold (scale bar is 500 μm). c) The spring constants of 200 μm pillars for free standing, acellular, and cellular scaffolds over 30 days. d) The calculated stiffness of each 200 μm pillar scaffolds over 30 days.
Samples were incubated in dark media at 37°C and 5% CO2 for two days, with media replaced on day 1. On day 3, samples were incubated in dark media with 10 μM Ara-C, replacing media every other day.
Calcium Imaging
To observe calcium transients, samples were first washed three times for 5 minutes at room temperature with 1.4 mM CaCl2 in Tyrode’s solution. Fluo-4 was loaded into cells using Tyrode’s with 0.1% pluronic-127 and incubated for 30 minutes at 37°C. Samples were washed three times for 5 minutes at room temperature and left in a solution of Tyrode’s with 500 nm ascorbic acid to prevent phototoxicity. These samples were then imaged with a Leica SP5 confocal microscope at 40x magnification utilizing the resonant scanner, at 128 × 128 pixel imaging area. They were stimulated with a 488 nm laser and observed at 84 frames a second. A Grass stimulator was used to stimulate the tissues. The average fluorescence was extracted from each frame into a 1D fluorescence trace. Microtissues were washed three times in Tyrode’s solution with 1.4mM CaCl2. Fluo-4 (Life Technologies) was suspended in DMSO and added into Tyrode’s solution with 0.1% (v/v) Pluronic 127 to a final working concentration of 5 μM. Constructs were incubated at 37°C and 5% CO2 for 30 minutes. Samples were washed three times for 5 minutes in Tyrode’s solution with 1.4mM CaCl2 and 500 nM L-ascorbic acid. Constructs were imaged using a Leica SP5 confocal microscope using a 40x water-immersion lens, acquiring a 128×128 image at 84 frames/second. The tissues were paced by platinum wires 1 cm apart from 0.5 to 4 Hz using a Grass pulse generator (70 V, 10 ms delay).
Mechanical Testing
Isotropic cylinders, 1 mm diameter × 2 mm tall, were polymerized utilizing the μCOP system and were allowed to swell at 37°C for 1, 10 and 31 days. Compression testing was performed using a Cellscale Microsquisher. Samples were incubated in a DPBS bath held at 37°C. Experiment parameters include a rate of applied force to 100 μN over 1 minute. Force was calculated using a beam equation [27]:
| (1) |
where D is the displacement, F is the force, L is the length of beam, E is the modulus. Icylinder is given by:
| (2) |
where r is the radius of the beam.
Compression modulus was determined within the linear region of the stress-strain curve across 5–15% GelMA and 2% HAGM/2% PEGDA.
Immunofluorescence Staining and Imaging for the Assessment of NMVCM morphology
Scaffolds with encapsulated NMVCMs were fixed in 4% (v/v) paraformaldehyde in DPBS for 15 minutes at room temperature and subsequently blocked and permeabilized by 2% bovine serum albumin solution with 0.1% Triton X-100 for 1 hour at room temperature. Samples were incubated with primary antibodies against alpha-actinin overnight at 4°C. Secondary antibody incubation and nuclear counterstain were incubated subsequently at room temperature for 1 hour in dark. Confocal immunofluorescence images were acquired with 40X, 0.8 NA water-immersion objective lens attached to a Leica SP5 microscope to compare the cell morphology, and nuclear and sarcomere alignment in NMVCMs. Directionality of sarcomeres and nuclei were analyzed using ImageJ Fourier components [28]. Assessment of proliferation between Ara-C treated and controls was determined by counting cell nuclei of positively stained NMVCMs and non-NMVCMs.
Force Measurements
To determine the force produced by the 3D-printed microtissues, three levels of printing were measured using the Cellscale Microsquisher: the 15% GelMA freestanding pillars, the tricomponent scaffold without cells (acellular), and the tri-component scaffold with encapsulated NMVCMs. The freestanding and acellular scaffolds were incubated at 37°C 5% CO2 in DPBS with 1% antibiotic 1% antimycotic. Samples were measured on day 1, 10, and 30, and placed on their sides and submerged in a DPBS bath at room temperature. Samples were pressed using a tungsten beam, measuring the point of applied force from the base. Samples were displaced 20 μm and the force measured using Equations 1 and 2 to calculate the sample modulus utilizing[27]:
| (3) |
where ω is the displacement, F is the force, γ is the geometric modulus, I is the 2nd moment of inertia (cuboid), L is the height, and x is the height of force application.
| (4) |
where h = length and b = width.
Lastly, NMVCM encapsulated samples were submerged in a 37°C bath of Tyrode’s solution without calcium and 40 mM 2,3 butedione monoxime (BDM) to fully relax the 3D-printed microtissues. Samples were pressed using a tungsten beam, measuring the point of applied force from the base. To determine the effect of cells within the scaffold, samples were displaced 20 μm and the force measured as previously described. Forces of the aligned tissue was calculated from measured displacements of the videos using Equations 3 and 4, using the modulus of the relaxed cellular scaffolds.
Results
Direct 3D-printing of cardiac micro-tissue using micro-continuous optical printing.
Initially, to investigate the compatibility of NMVCMS with both our material and μCOP system (Figure 1a), NMVCMs were successfully encapsulated in 5% methacrylated gelatin (GelMA) hydrogels in both a 3 × 3 mm slab pattern and a parallel line pattern (line widths of 50 μm, spacing of 50 μm) on glass coverslips with cell concentrations ranging from 5 million to 40 million cells/mL. At low concentrations (5 and 10 million cells/mL) samples did not require increased exposure time to polymerize. However, at 20, 30, and 40 million cells/mL, exposure times were increased by 50% to ensure an intact and robust pattern. To determine the effect of GelMA concentration on NMVCM viability, slab samples were printed at 10 million cells/mL. Cardiomyocyte viability was evaluated on day 3, resulting in 60 ± 3% and 61 ± 5% for 5% and 7.5% GelMA, respectively, and 42 ± 5% for 15% GelMA (supplementary Figure 1a and 1b). Individual cell beats were observed as early as day 1 after printing, with NMVCMs beginning to hypertrophy by day 3. Samples were observed beating synchronously by day 3. Video capture of cardiomyocytes that were patterned in an isotropic slab and parallel lines showed synchronicity of spontaneous beating of patterned cells (supplementary video 1) compared to slab cells (supplementary video 2).
When compared to NMVCMs seeded on glass, cardiomyocytes encapsulated in the 5% GelMA slab exhibited striated α-actinin fibers, whereas the control cells did not display phenotypic expression of α-actinin unless they were seeded on top of other cells (supplementary Figure 1c and 1d). This highlights the compatibility of 5% GelMA with NMVCMs. Moreover, NMVCMs encapsulated in 5% GelMA also stained positive for α-actinin, for both slab and parallel lines, across the entire printed scaffold (supplementary Figure 1e and 1f). When analyzing the sarcomeres of the encapsulated cardiomyocytes, NMVCMs encapsulated in the slab showed a more uniform distribution of sarcomere angles, with no preference in direction, when compared with NMVCMs patterned in parallel lines (supplementary Figure 1g and 1h). This emphasizes the ability to align NMVCMs in patterned gels using the μCOP system, as NMVCMs encapsulated in the individual line pattern aligned to the pattern, and their Z-disks preferentially aligned perpendicular to that axis (supplementary Figure 1i).
To further optimize the material selection for printing, NMVCMs were printed using μCOP in parallel line patterns (line widths of 50 μm, spacing of 50 μm) in varying GelMA concentrations of 5, 7.5, and 15% (supplementary Figure 2a). Sarcomeres of cardiomyocytes encapsulated within 5% and 7.5% GelMA lines were measured, with 15% GelMA negated, as αactinin expression was minimal across 15% GelMA lines. For NMVCMs encapsulated in 5% GelMA, the average sarcomere length was 1.83 ± 0.05 μm, whereas the average sarcomere length of NMVCMS encapsulated in 7.5% GelMA was 1.72 ± 0.04 μm. (supplementary Figure 2b).
After successfully completing these initial encapsulation studies, we created a more complex structure designed to measure the force exerted by the NMVCM tissue. Using the μCOP system, we printed a hydrogel consisting of a 1) 2% HAGM/ 2% PEGDA base layer, 2) 15% GelMA cantilevers, and 3) a parallel line pattern comprised of 5% GelMA (Figure 1b). A schematic of the printed scaffold is shown in Figure 1c and a 3D reconstruction of α-actinin stained NMVCMs encapsulated between the scaffold can be seen in Figure 1d. In the final tissue scaffold, the cantilever has dimensions of 1.1 × 0.23 × 0.51 mm (Figure 1e, n=19). The elevated and patterned cardiac tissue has initial dimensions of 1.9 × 0.91 × 0.21 mm (Figure 1f, Length(L) × Width (W) × Height(H), n=6).
Encapsulation of cardiomyocytes across 3D scaffold improves alignment.
By encapsulating NMVCMs in a linear pattern, the NMVCMs have geometric cues for forming rod shaped morphology that are aligned as in mature myocardium. Confocal images stained for α-actinin were taken for samples of the isotropic slab (left) on glass, line pattern on glass (middle), and lines (right) printed across the 3D-printed pillars at day 10 (Figure 2a). The Fourier components of each the actinin filaments of each scaffold were analyzed (n=4 for each print) to compare the alignment between conditions. The slab pattern unsurprisingly had an even distribution across angles. The lines printed on glass showed an increase in frequency of fibers between 85 and 105 degrees (one-tailed t-test, p<0.05). However, samples printed on the 3D pillars showed a significant change in directionality of sarcomeres of all directions except 0–5, 60–70, 110–115, and 175–180° (two-tailed t-test, p<0.05) (Figure 2b). To further characterize the microtissue, samples were cultured out to 30 days and the degree of compaction at the midpoint of the microtissue scaffold was measured at day 1, 10, and 30 (Figure 2c). Originally, samples were 873±49 μm wide on day 1. At day 10, they contracted to 528 ± 154 μm and eventually by day 30 they contracted to 290 ± 88μm (Figure 2d, n=14). Additionally, sarcomere lengths across day 10 line patterns on glass, day 10 scaffold, and scaffold cultured for 30 days were measured (Figure 2e). The average sarcomere lengths for day 10 line patterns printed on glass was 2.2 ± 0.02 μm and for day 10 scaffold was also 2.19 ± 0.05 μm. For day 30 samples, the average sarcomere length was significantly different to both day 10 samples at 2.34 ± 0.03 μm (Figure 2f, n=4, p<0.05).
Figure 2. Characterization of sarcomere alignment, maturity, and tissue patterning.
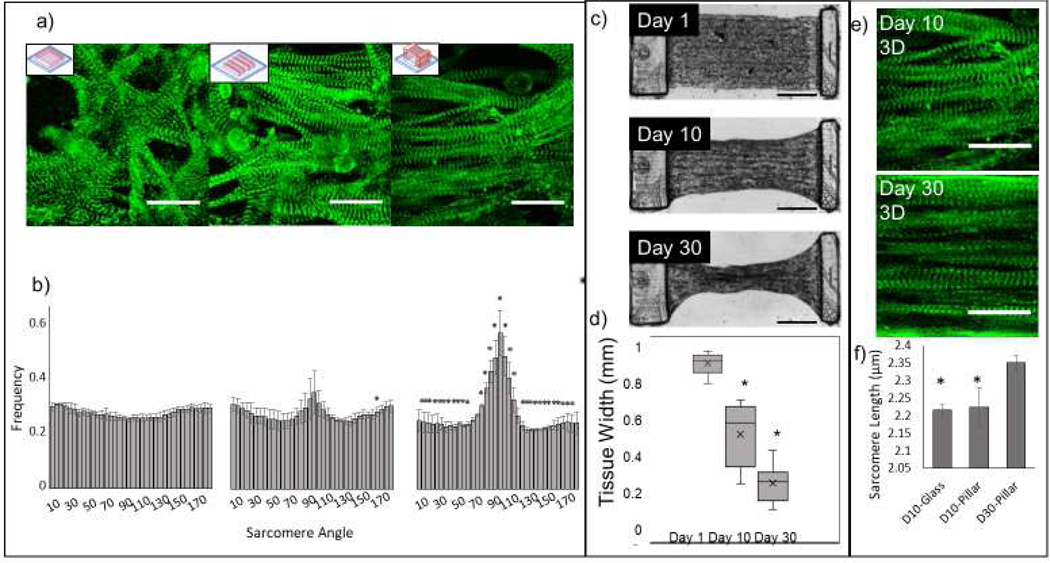
a) Representative images of α-actinin on day 10 of NMVCMs patterned in a slab on glass, lines on glass, and lines between the pillars (left to right, scale bar is 25 μm), and b) their respective sarcomere angle distributions. Significance (one-tailed p<0.05 for day 10 lines and two-tailed p<0.05 for day 10 on scaffold) is denoted with asterisks when compared to the slab control. c) DIC images of full tissue scaffolds and the compaction the tissue experiences over 30 days (scale bar is 100 μm). d) The tissue widths at the midpoint at day 1, 10 and 30 (SEM, paired two-tailed t-test, p<0.001, n=14). e) α-actinin stain of day 10 and day 30 samples (scale bar is 25 μm) and f) the average sarcomere lengths of NMVCMs aligned on glass at day 10, on the pillar at day 10 and day 30 (SEM, n=3, p<0.05).
μCOP allows for rapid tailoring of scaffolds for force measurement.
The multimaterial scaffold was designed to have two pillars; a cantilever (minor) and a support (major), where the force produced from the tissue would only deflect the cantilever. 500, 200, and 100 μm wide pillars (Figure 3a) were bent using a 0.3046, 0.2048, and 0.1024 mm diameter tungsten wire, respectively, pressed 375 μm from the base, midway through where the tissue would be printed (Figure 3b). The spring constant of each freestanding pillar is 0.084 ± 0.005, 2.10 ± 0.15, 13.8 ± 1.7 μN/μm (n=13, 20, 8), respectively, after 24 hours incubation at 37°C. Scaffolds were further incubated at 37°C for 10 and 30 days, with measured forces of 0.17 ± 0.03, 1.93 ± 0.13, 11.7 ± 0.7 μN/μm (n=8, 12, 12) on day 10, and 0.15 ± 0.07, 2.26 ± 0.16, 10.9 ± 0.6 μN/μm (n=9, 10, 14) on day 30. After empirically optimizing dimensions for the 3D cardiac tissue to display sufficient deflection, the 200 μm pillar was selected for all future scaffolds. To more accurately replicate the full system, 2% HAGM/2% PEGDA slab, both major and minor 15% GelMA pillars, and 5% GelMA lines were printed. Similarly, complete acellular scaffolds were tested using a mechanical testing instrument - “Microsquisher” under conditions stated previously. The addition of the HAGM/PEGDA base and 5% GelMA line did very little to change the spring constant of the minor pillar. The spring constant Kscaffold was 3.3 ± 0.3, 2.8 ± 0.2 and 2.6 ± 0.2 μN/μm (n=6, 10, 9) on day 1, 10, and 30 for 200 μm-width cantilevers. Finally, full scaffolds with cells encapsulated in 5% GelMA lines were measured on day 10 and day 30, having been treated in a bath of Tyrode’s solution without calcium but with glucose and 2,3 BDM to relax the cells. Pillars were pressed at the center plane of the tissue. The addition of cells nearly doubled the modulus of the scaffolds from 85 ± 9 kPa (acellular) to 148 ± 17 kPa (cellular) on day 10 and 74 ± 8 kPa (acellular) to 114 ± 26 kPa (cellular) on day 30 (Figure 3d). The calculated tissue modulus at 10 day to be 63 ± 17 kPa at day 10 and 40 ± 26 kPa at day 30. The cellular construct moduli can be used to calculate the forces generated by the aligned cardiac tissues.
Displacement and calculated forces of seeded vs encapsulated 3D micro-tissues
To evaluate the functionality of the scaffold, NMVCMs were successfully encapsulated in parallel lines, with cells seeded atop 5% GelMA lines as a control. Representative bright field images of seeded and encapsulated samples cultured for 10 days are seen in Figure 4a. Confocal microscopy z-projections were taken across the seeded scaffolds and stained for α-actinin (green, Figure 4a). The seeded scaffolds had a sparser distribution of cells expressing α-actinin compared to the encapsulated scaffold. Moreover, inlays reveal a higher compaction of cells for encapsulated samples compared to seeded samples. To evaluate force generation, samples were stimulated using platinum electrodes and a Grass pulse generator (70 V, 10 ms delay) at 1, 2, and 4 Hz. Time lapse images of the microtissues were taken at 5x and 10x magnification using a Leica inverted microscope at 20 frames per second (FPS). Using a custom MATLAB script, pillar displacement was determined by monitoring the change in position of a peak across a grayscale profile of the pillar between diastolic and systolic state over time (Figure 4b). Representative traces of both seeded and encapsulated samples are shown in Figure 4c. The maximum displacement from the relaxed position of seeded samples was calculated to be 12.0 ± 1.3 μm, 11.3 ± 1.2 μm, and 11.2 ± 1.6 μm at 1, 2, and 4 Hz stimulation, respectively (Supplementary video 3). The maximum displacement of the encapsulated samples was 2 times as great as the seeded samples, at 24.9 ± 2.2 μm, 25.0 ± 2.2 μm, and 24.4 ± 2.2 μm at 1, 2, and 4 Hz, respectively (Figure 4d, supplementary video 4, n=5, p<0.005). Using the dimensions of the scaffolds 1.1×0.23×0.51 mm mentioned previously and the modulus of a cellular construct at 148 kPa, the calculated forces of the seeded samples were 75±8 μN, 71±8 μN, and 70±10 μN at 1, 2, and 4 Hz, respectively, whereas encapsulated constructs produced 156±14 μN, 157±14 μN, and 153±14 μN at 1, 2, and 4 Hz, respectively. Thus, encapsulating NMVCMs in patterned 5% GelMA lines compared to seeding on equivalent structures increased the force by as much as two times (p<0.002 for 1, 2, and 4Hz).
Figure 4. Comparison of the force and displacement of seeded and encapsulated NMVCMs tissues.
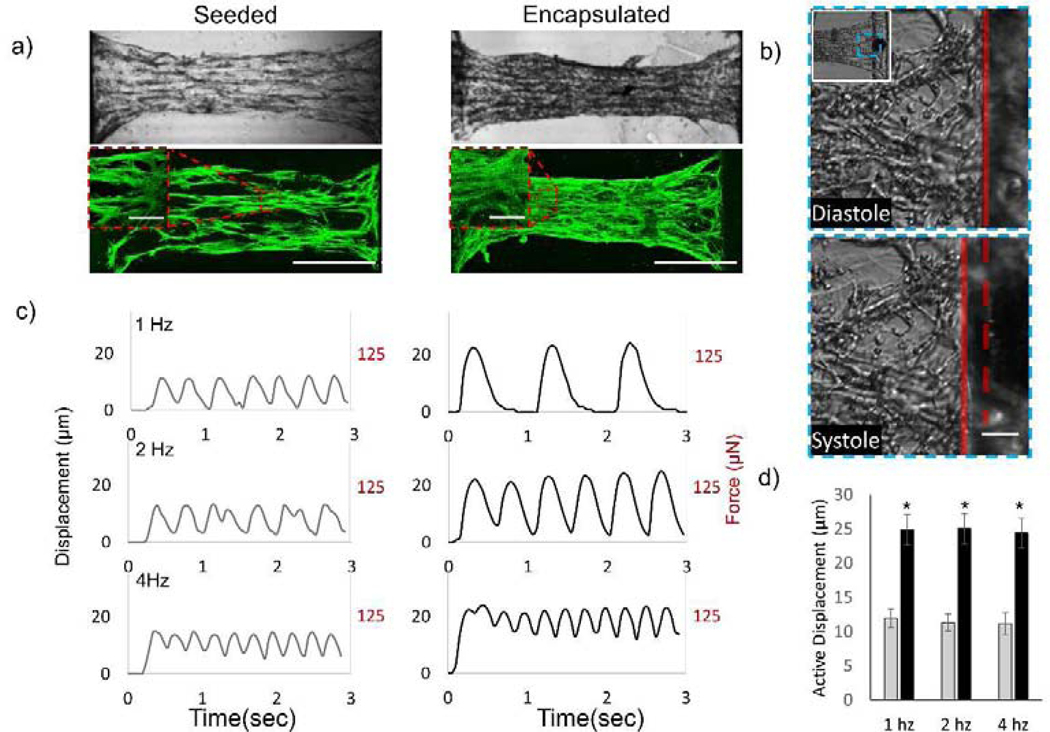
a) DIC and fluorescent images of α-actinin stained for both scaffolds with seeded NMVCMs and encapsulated NMVCMs within the patterned hydrogel at day 10 (scale bar is 500 μm, inlay is 25 μm). b) DIC image of the pillar showing its position at diastole (dotted red line) and at systole (solid red line), c) displacement and force traces of seeded (left) and encapsulated (right) samples at various stimulating frequencies, d) and a summary of max displacement of seeded (gray) and encapsulated (black) samples (n=5, SEM, p<0.005).
Encapsulating cardiomyocytes in complex geometries using μCOP.
In addition to parallel lines, we investigated how the encapsulation pattern could impact the cardiomyocytes’ ability to remodel their environment and resulting force generation. Using the μCOP system we printed four additional patterns; slab, grid, dispersion, and a randomly connected set of lines (Figure 5a). Differential interference contrast (DIC) images of each pattern type is shown in figure 5b. Videos taken at 10x magnification of each pattern are shown in supplementary videos 5–9. Confocal images taken of each pattern on day 10 were stained for αactinin at 40x (Figure 5c) and merged to show the distribution of sarcomeres. For all conditions, the cells appeared align within and along the GelMA lines. Of particular note is the slab and grid, whose patterns disrupted the alignment of cardiomyocytes along the major axis of the scaffold. Force generation of the tissues in each of these conditions was evaluated by the displacement of the cantilever. As seen in Table 1, across all conditions, the line pattern was the only one to significantly improve the displacement of the cantilever. A one-way ANOVA conducted to compare the effect of the patterns on displacement and force production showed that the effect of the line pattern on displacement and force production was significant compared to the other patterns (F(4,24)=56.5 p<0.000001). By varying the levels of patterning, we were able to tailor the level of displacement and thus force produced by each tissue.
Figure 5. Micropatterning of cardiac tissue affects displacement.
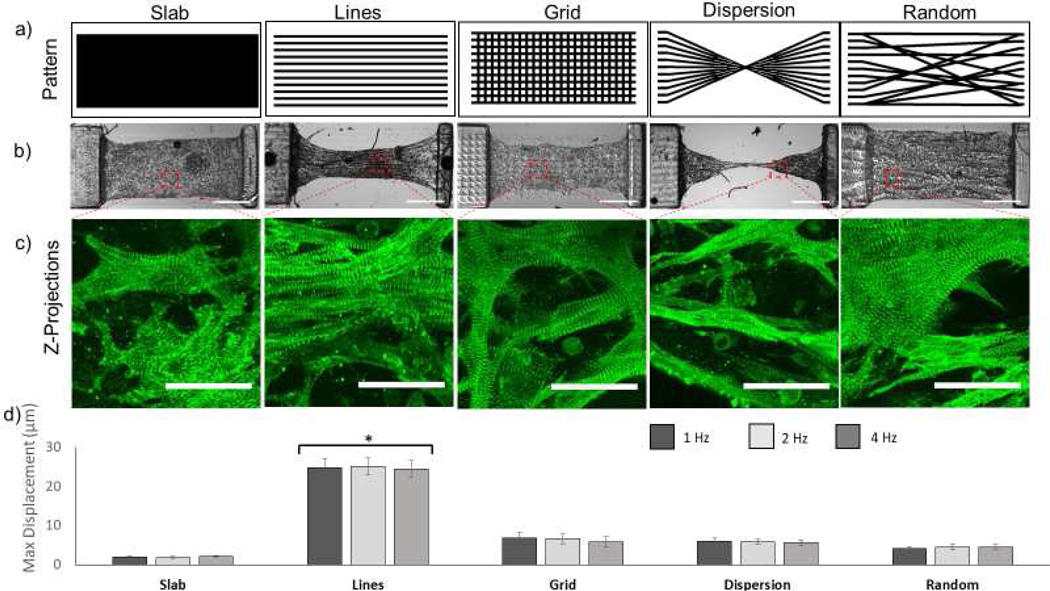
a) Masks of various complex patterns including (from left to right) slab, line, grid, dispersion, and random designs. b) DIC images showing each patterned tissue at day 10 (scale bar is 500 μm). c) Fluorescent images of NMVCMS stained for α-actinin for each scaffold type showing alignment of sarcomeres (scale bar is 25 μm). d) Summary of max displacement for various patterned tissues at 1, 2, and 4 hz (SEM, n=3).
Table 1.
Pillar Displacement in μm and force of various patterns
| Slab | Lines (encapsulated) | Grid | Dispersion | Random | |
|---|---|---|---|---|---|
| 1 Hz | 2.10 ± 0.19 μm 13 ± 1 μN | 24.9 ± 2.2 μm 156± 14 μN | 6.8 ± 1.3 μm 43 ± 8 μN | 6.1 ± 0.8 μm 38 ± 5 μN | 4.3 ± 0.8 27 ± 5 μN |
| 2 Hz | 1.91 ± 0.24 μm 12 ± 1 μN | 25.0 ± 2.2 μm 157 ± 14 μN | 6.7 ± 1.3 μm 42 ± 8 μN | 5.9 ± 0.6 μm 37 ± 4 μN | 4.6± 0.8 29 ± 5 μN |
| 4 Hz | 2.21 ± 0.16 μm 14 ± 1 μN | 24.4 ± 2.2 μm 153 ± 14 μN | 5.8 ± 0.8 μm 36 ± 8 μN | 5.6 ± 0.1 μm 35 ± 1 μN | 4.6 ± 0.6 29 ± 4 μN |
Calcium transients in cardiac microtissues.
To evaluate excitation-contraction coupling in the cardiac scaffolds, we measured calcium transients of the parallel constructs under various conditions. Normalized fluo-4 fluorescence (ΔF/Fo) of NMVCM-encapsulated structures showed characteristic calcium transient waveforms (Figure 6a) that increased in magnitude and rate of decline during treatment with 100 nM isoproterenol (ISO) for 5 minutes at room temperature. After ISO, the appearance of ectopic beats at 0.5 Hz were observed for day 10 and day 30 samples. Moreover, one of five samples experienced an increase in the stimulation frequency it could catch from 4 to 5 Hz while under ISO treatment. Beyond 5 Hz, all day 10 samples could not capture stimulation frequencies above 5 Hz. Due to their increased maturity, all 30 samples experienced an increase in stimulation frequencies that could be captured (7 to 8 Hz, 4 to 6 Hz, and 8 to 9 Hz, respectively). For day 10 samples, total peak fluorescence only increased significantly for samples stimulated at 0.5 Hz by 55±29%, however the change in fluorescence at higher frequency, i.e. 4 Hz also increased after ISO treatment by 46±22% (p<0.05, n=5). The maximum depolarization rate increased by 53±25% and 42±19% for 0.5 and 1 Hz (p<0.05, n=5) and the maximum rate of repolarization increased by 93±43%, 74±28%, and 56±33% for 0.5, 1, and 2 Hz respectively (Figure 6 b, p<0.05, n=5). For day 30 samples, the max fluorescence significantly increased by 30±11, 24±4, and 37 ± 9% for 1 Hz, 2 Hz, and 3 Hz, along with rate of depolarization for 2 and 3 Hz (Figure 6b, p<0.01, n=3).
Figure 6. Calcium staining of NMVCM tissues.
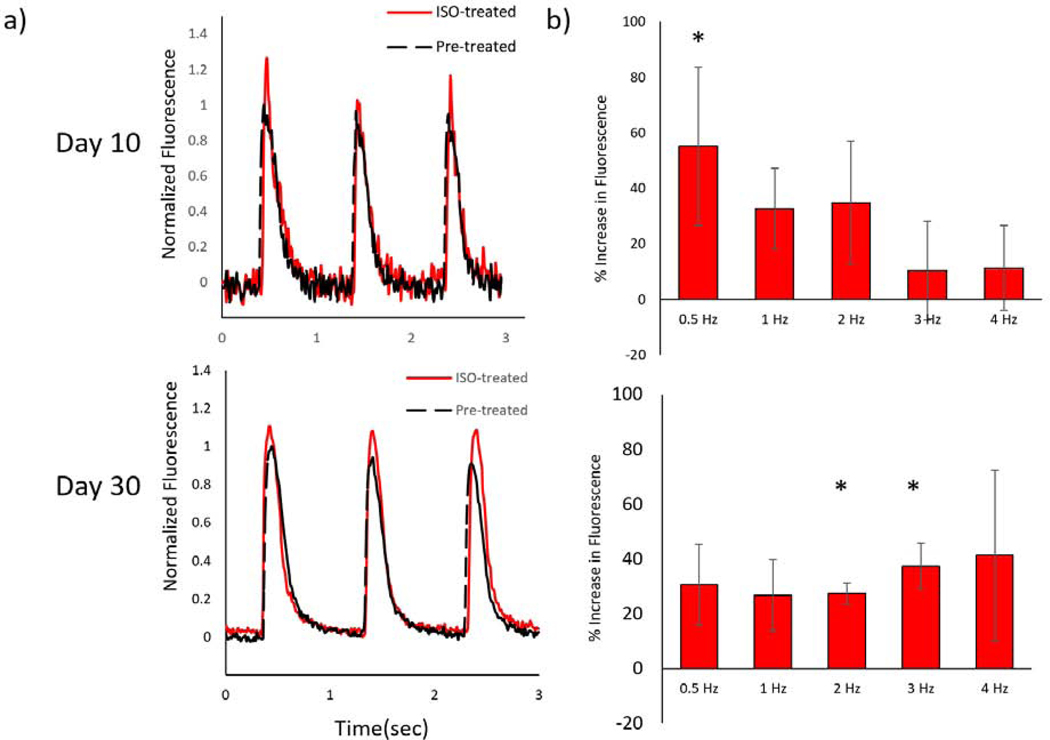
a) Representative normalized fluorescence traces ΔF/Fo traces of day 10 (top) and day 30 (bottom), as well as before (black) and after (red) 100 nM ISO treatment. b) Percent increase in ΔF/Fo (asterisks denote significance ) for day 10 (SEM, n=5 p<0.05) and day 30 sample (SEM, n=3 p<0.05)
Discussion
Engineering a physiological in vitro cardiac tissue model has been the subject of intense research [9,29–32], but there is still a need to produce 3D constructs with structures that resemble normal and disease myocardium in vivo. Our cardiac microtissue model was designed with pre-patterned NMVCMs and showed both improved alignment and physiological responsiveness to ionotropic stimulation. In addition, we used GelMA, a collagen-based photopolymerizable material, shown previously to be a favorable material for micropatterning tissue constructs and promoting cardiomyocyte attachment and spreading [33], and LAP, a minimally toxic photoinitiator [23]. The μCOP method allows one to rapidly iterate, without the need for a physical mask or mold, and design unique systems that could be tailored for various parameters, including but not limited to complex spatial patterning, tissue and geometries, multiple materials, and mechanical stiffnesses. Originally 2% HAGM/2% PEGDA was chosen as a negatively charged material [34] to prevent cell interaction with the bottom substrate, allowing us to wash away those cells that were not specifically printed.
By directly encapsulating cells within the pattern, cells elongated near physical cues and had improved alignment when compared with an isotropic slab. Sarcomere alignment of linepattern tissues printed on glass between ±15 degrees only increased by 3% when compared with isotropic slab on glass, whereas printed tissues on scaffolds increased by 12%. The occurrence of noise in the form of sarcomeres near the perimeter of the cell contributed to a dispersion of angles. Owing to the Frank-Starling mechanism, an increase in sarcomere length should increase force generation [35]. When comparing with mouse trabeculae and papillary muscle preparations, which have slack sarcomere lengths of <2 μm [7,36,37], sarcomere lengths in our scaffolds at day 10 (2.2 μm) and day 30 (2.35 μm) were significantly higher, consistent with a physiological diastolic load due to the effects of gel contraction and pillar deformation.
These scaffolds were extensively characterized to determine the relationship of pillar size, addition of the full scaffold, along with cells to the stiffness of the printed cantilever. Thus, the mechanical properties can be used to estimate the forces generated per 3D printed NMVCM tissue. Furthermore, utilizing the μCOP system, the cantilever could be easily tailored to the user’s needs, e.g. reducing the width or decreasing the material concentration to decrease the overall stiffness of the mechanical tester, making it more sensitive to tissues that generate less force such as human embryonic stem cell cardiomyocytes or human induced pluripotent stem cell derived cardiomyocytes [38,39]
By directly printing the tissue sample within the predefined mechanical tester, the samples lay within the same area and z-position, making it easier to measure and calculate force generation. When comparing 2D and 3D samples, a greater number of cells were used per sample to seed than to encapsulate. During seeding, many cells were seeded on the glass and well floor. Also, for some cells falling within the patterned grooves, they were still not incorporated in the tissue. Although compaction did occur, there were still patches of gel unoccupied by cells on day 10. This may suggest the necessity of a secondary seeding to ensure complete coverage of the samples. However, by day 3, samples with single seeding already required media exchange daily to ensure cell viability and continuous contraction as opposed to every 48 hrs. Thus, seeding at an even higher number of cells per well to ensure proper coverage can lead to practical challenges in maintaining the samples over time. This leaves much to be desired for a seeding methodology of this scaffold. However, when comparing the seeded scaffolds with the various scaffold designs that were discussed in Figure 5, the alignment of NMVCMs on a simple pattern of parallel lines produced greater average displacements of the cantilever when compared to slab, dispersion, and random patterns. Thus, this corroborates the concept that alignment of NMVCMs is paramount to the contractile properties of the scaffold.
Current 2D culture methods, especially on tissue culture plastic, are not sufficient to elicit phenotypic 3D cell morphologies and connectivity or responses to external stimuli. Whereas most worker rely on the self-alignment of cardiomyocytes encapsulated in extracellular matrix around stiff cantilevers [6,8,31,32] and 3D printing studies have been limited to log pile structures [40,41], this is the first instance in which the effects of directly patterning various 3D microstructures on cellular alignment and tissue mechanics have been observed. Unsurprisingly, the aligned tissue patterned with parallel lines displaced the most, leading to over a 10-fold increase in tissue force compared to slab tissues, which lack directional contraction. Similarly, the disruption caused by the grid pattern disrupted alignment of NMVCMs and caused isotropic compression of the scaffold when beating. By disrupting the aligned NMVCMs with perpendicular lines in the grid pattern, we effectively reduced the force produced by the 3D- printed tissue over 3.5 times. However, when normalizing for the volume of printed tissue (mm displacement/mm3), assuming the same thickness, the dispersion pattern also showed an increase in displacement compared with the slab (F(4,24)=44.4, p<0.005) and random patterns (F(4,24)=44.4, p<0.05); however, the displacement was still lower than the line pattern (F(4,24)=44.4, p<0.00005). The concept of prepatterning cells in specific geometries may be of significant interest in developing disease models where current models cannot recapitulate disease phenotypes that consist of myocyte disarray i.e. hypertrophic cardiomyopathy [42]. Across all patterns, there was very little change in displacement with increased stimulation frequency suggesting a force-frequency of zero. One way to further investigate this is to use a higher magnification objective (10x to 20x). At the current acquisition, the pixel to μm ratio is nearly 1 μm/ pixel. Increased resolution and increased acquisition time may be able to discern the force-frequency relationship of the printed scaffold in the future.
For the parallel lines pattern, we calculated a force of 156 μN for the encapsulated NMVCMs. Previous studies showed generated forces of only 16 μN [7], an order of magnitude less due to cross-sectional areas 10 times smaller than our 3D-printed tissues. Though, cardiac tissue tension normalized by cross-sectional area may be a better metric. Isolated human muscle strip has been reported to produce 17 mN/mm2 [43]. The tension of our 3D-printed NMVCMs tissues were calculated to be 14.5 mN/mm2 at 1 Hz stimulation, a nearly comparable model. In comparison previous studies have produced 0.5, 0.8, and 0.1 mN/mm2 tension for rat neonatal CMs, two to three times less tension from a larger animal model [8,44,45].
Further work is required to fully characterize the calcium handling of 3D-printed scaffolds. Finding imaging areas with minimal motion artifacts was difficult and in some cases choosing areas with significant motion was unavoidable. Prior attempts to mechanically decouple samples using BDM and calcium transients data could be successfully acquired. Another point of interest that made acquisition difficult was that after treatment of ISO, the passive tension of the tissue would increase, and the imaging plane would shift. However, given these shortcomings, an ionotropic effect was measured 0.5 Hz for day 10 and for 2 and 3 Hz for day 30 samples. An increase in samples for day 30 may elicit a statistically significant increase as both 0.5 and 1 Hz stimulations are p<0.055. Furthermore, a chronotropic effect was observed when treated with ISO in the form of ectopic beats at low frequencies for both day 10 and day 30 samples, while day 30 samples showed an increase in achievable stimulation capturing up to 9 Hz (from 8 Hz). Beyond treatment with ISO, the effects of other drugs could be investigated with our model.
Conclusions
This work highlights the ability to rapidly print complex scaffolds tailored to user’s needs for cardiac tissue engineering via the μCOP system. With its high tension, the proposed aligned 3D-printed cardiac tissue is a good 3D tissue model for potential drug screening. Furthermore, the initial investigation of printing complex microarchitectures is a promising tool in studying and producing cardiac disease modeling. The long survival times in culture (>30 days) may allow for the potential to see how cardiomyopathies may manifest in early development, and how cell-cell, cell-microarchitecture, and electrical coupling affect tissue function and how certain drugs may affect tissue function long-term.
Supplementary Material
Acknowledgements
The work was supported by grants from the California Institute for Regenerative Medicine (RT3–07899), National Institutes of Health (R01EB021857, R21AR074763), and National Science Foundation (1644967, 1903933). ADM is a co-founder of and has an equity interest in Insilicomed, Inc., and he serves on the scientific advisory board. Some of his research grants, including those acknowledged here, have been identified for conflict of interest management based on the overall scope of the project and its potential benefit to Insilicomed, Inc. The author is required to disclose this relationship in publications acknowledging the grant support, however the research subject and findings reported here did not involve the company in any way and have no relationship with the business activities or scientific interests of the company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. SC is a co-founder of and has an equity interest in Allegro 3D, Inc., and he serves on the scientific advisory board. Some of his research grants, including those acknowledged here, have been identified for conflict of interest management based on the overall scope of the project and its potential benefit to Allegro 3D, Inc. The author is required to disclose this relationship in publications acknowledging the grant support, however the research subject and findings reported here did not involve the company in any way and have no relationship with the business activities or scientific interests of the company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. The other authors have no competing interests to declare.
Footnotes
Data Availability
The raw/processed data required to reproduce these findings can be shared by the authors upon request.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
☐The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Sengupta PP, Korinek J, Belohlavek M, Narula J, Vannan MA, Jahangir A, Khandheria BK, Left Ventricular Structure and Function, J. Am. Coll. Cardiol. 48 (2006) 1988–2001. [DOI] [PubMed] [Google Scholar]
- [2].Kim D-H, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh K-Y, Tung L, Levchenko A, Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs, Proc. Natl. Acad. Sci. 107 (2010) 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zong X, Bien H, Chung C-Y, Yin L, Fang D, Hsiao BS, Chu B, Entcheva E, Electrospun fine-textured scaffolds for heart tissue constructs, Biomaterials. 26 (2005) 5330–5338. [DOI] [PubMed] [Google Scholar]
- [4].Alford PW, Feinberg AW, Sheehy SP, Parker KK, Biohybrid thin films for measuring contractility in engineered cardiovascular muscle, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ma Z, Koo S, Finnegan MA, Loskill P, Huebsch N, Marks NC, Conklin BR, Grigoropoulos CP, Healy KE, Three-dimensional filamentous human diseased cardiac tissue model, Biomaterials. 35 (2014) 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Turnbull IC, Karakikes I, Serrao GW, Backeris P, Lee JJ, Xie C, Senyei G, Gordon RE, Li RA, Akar FG, Hajjar RJ, Hulot JS, Costa KD, Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium, FASEB J. 28 (2014) 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van Spreeuwel ACC, Bax NAM, Bastiaens AJ, Foolen J, Loerakker S, Borochin M, van der Schaft DWJ, Chen CS, Baaijens FPT, Bouten CVC, The influence of matrix (an)isotropy on cardiomyocyte contraction in engineered cardiac microtissues, Integr. Biol. 6 (2014) 422–429. [DOI] [PubMed] [Google Scholar]
- [8].Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS, A Microfabricated Platform to Measure and Manipulate the Mechanics of Engineered Cardiac Microtissues, Tissue Eng. Part A. 18 (2012) 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Agarwal A, Farouz Y, Nesmith AP, Deravi LF, McCain ML, Parker KK, Micropatterning Alginate Substrates for In Vitro Cardiovascular Muscle on a Chip, Adv. Funct. Mater. 23 (2013) 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Grosberg A, Alford PW, McCain ML, Parker KK, Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip, Lab Chip. 11 (2011) 4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang D, Shadrin IY, Lam J, Xian H-Q, Snodgrass HR, Bursac N, Biomaterials, Biomaterials. 34 (2013) 5813–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim SB, Nikkhah M, Khabiry M, Azize M, Kong J, Wan KT, Palacios T, Dokmeci MR, Bae H, Tang XS, Khademhosseini A, Carbon-nanotube-embedded hydrogel sheets for engineering cardiac constructs and bioactuators, ACS Nano. 7 (2013) 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fraley SI, Feng Y, Krishnamurthy R, Kim D-H, Celedon A, Longmore GD, Wirtz D, A distinctive role for focal adhesion proteins in three-dimensional cell motility, Nat. Cell Biol. 12 (2010) 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cukierman E, Pankov R, Yamada KM, Cell interactions with three-dimensional matrices, Curr. Opin. Cell Biol. 14 (2002) 633–639. [DOI] [PubMed] [Google Scholar]
- [15].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc. Natl. Acad. Sci. 113 (2016) 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Soares CP, Midlej V, de Oliveira MEW, Benchimol M, Costa ML, Mermelstein C, 2D and 3D-Organized Cardiac Cells Shows Differences in Cellular Morphology, Adhesion Junctions, Presence of Myofibrils and Protein Expression, PLoS One. 7 (2012) e38147–e38147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].V Murphy S, Atala A, 3D bioprinting of tissues and organs., Nat. Biotechnol. 32 (2014) 773–785. [DOI] [PubMed] [Google Scholar]
- [18].Atala A, Kasper FK, Mikos AG, Engineering Complex Tissues, Sci. Transl. Med. 4 (2012) 160rv12−−160rv12. [DOI] [PubMed] [Google Scholar]
- [19].Wang Z, Lee SJ, Cheng H-J, Yoo JJ, Atala A, 3D bioprinted functional and contractile cardiac tissue constructs, Acta Biomater. 70 (2018) 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang AP, Qu X, Soman P, Hribar KC, Rapid fabrication of complex 3D extracellular microenvironments by dynamic optical projection stereolithography, Advanced. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Soman P, Chung PH, Zhang AP, Chen S, Digital microfabrication of user-defined 3D microstructures in cell-laden hydrogels, Biotechnol. Bioeng. 110 (2013) 3038–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu J, Hwang HH, Wang P, Whang G, Chen S, Direct 3D-printing of cell-laden constructs in microfluidic architectures, Lab Chip. 16 (2016) 1430–1438. [DOI] [PubMed] [Google Scholar]
- [23].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS, Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility, Biomaterials. 30 (2009) 6702–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gauvin R, Chen YC, Lee JW, Soman P, Zorlutuna P, Nichol JW, Bae H, Chen S, Khademhosseini A, Microfabrication of complex porous tissue engineering scaffolds using 3D projection stereolithography, Biomaterials. 33 (2012) 3824–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Cell-laden microengineered gelatin methacrylate hydrogels, Biomaterials. 31 (2010) 5536–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suri S, Han L-H, Zhang W, Singh A, Chen S, Schmidt CE, Solid freeform fabrication of designer scaffolds of hyaluronic acid for nerve tissue engineering, Biomed. Microdevices. 13 (2011) 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Carrera E, Giunta G, Petrolo M, Beam Structures, John Wiley & Sons, Ltd, Chichester, 2011. [Google Scholar]
- [28].Liu ZQ, Scale space approach to directional analysis of images., Appl. Opt. 30 (1991) 1369–1373. [DOI] [PubMed] [Google Scholar]
- [29].Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS, Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues, Proc. Natl. Acad. Sci. 106 (2009) 10097–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ribeiro AJS, Ang Y-S, Fu J-D, Rivas RN, Mohamed TMA, Higgs GC, Srivastava D, Pruitt BL, Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness., Proc. Natl. Acad. Sci. U. S. A. 112 (2015) 12705–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M, Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes, Nat. Methods. 10 (2013) 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang D, Shadrin IY, Lam J, Xian H-QQ, Snodgrass HR, Bursac N, Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes, Biomaterials. 34 (2013) 5813–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, Akhyari P, Khademhosseini A, Directed 3D cell alignment and elongation in microengineered hydrogels, Biomaterials. 31 (2010) 6941–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Maharjan AS, Pilling D, Gomer RH, High and low molecular weight hyaluronic acid differentially regulate human fibrocyte differentiation., PLoS One. 6 (2011) e26078–e26078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].ter Keurs HE, Rijnsburger WH, van Heuningen R, Nagelsmit MJ, Tension development and sarcomere length in rat cardiac trabeculae. Evidence of length-dependent activation., Circ. Res. 46 (1980) 703–714. [DOI] [PubMed] [Google Scholar]
- [36].Feinberg AW, Ripplinger CM, Van Der Meer P, Sheehy SP, Domian I, Chien KR, Parker KK, Functional differences in engineered myocardium from embryonic stem cell-derived versus neonatal cardiomyocytes, Stem Cell Reports. 1 (2013) 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang D, Shadrin IY, Lam J, Xian H-Q, Snodgrass HR, Bursac N, Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes., Biomaterials. 34 (2013) 5813–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kensah G, Lara AR, Dahlmann J, Zweigerdt R, Schwanke K, Hegermann J, Skvorc D, Gawol A, Azizian A, Wagner S, Maier LS, Krause A, Dräger G, Ochs M, Haverich A, Gruh I, Martin U, Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro, Eur. Heart J. 34 (2013) 1134–1146. [DOI] [PubMed] [Google Scholar]
- [39].Morimoto Y, Mori S, Sakai F, Takeuchi S, Human induced pluripotent stem cellderived fiber-shaped cardiac tissue on a chip, Lab Chip. 16 (2016) 2295–2301. [DOI] [PubMed] [Google Scholar]
- [40].Pati F, Jang J, Ha D-H, Kim SW, Rhie J-W, Shim J-H, Kim D-H, Cho D-W, Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink, Nat. Commun. 5 (1AD) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gao L, Kupfer ME, Jung JP, Yang L, Zhang P, Da Sie Y, Tran Q, Ajeti V, Freeman BT, Fast VG, Campagnola PJ, Ogle BM, Zhang J, Myocardial tissue engineering with cells derived from human-induced pluripotent stem cells and a native-like, high-resolution, 3-dimensionally printed scaffold, Circ. Res. 120 (2017) 1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tsybouleva N, Zhang L, Chen S, Patel R, Lutucuta S, Nemoto S, DeFreitas G, Entman M, Carabello BA, Roberts R, Marian AJ, Aldosterone, through novel signaling proteins, is a fundamental molecular bridge between the genetic defect and the cardiac phenotype of hypertrophic cardiomyopathy., Circulation. 109 (2004) 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Christian H, Thorsten R, J. GD, C. AR, Werner N, Burkert P, Katja P, Jens L, Sandra W, Gerd H, Herbert P, Hanjörg J, Daniel B, Existence of the Frank-Starling Mechanism in the Failing Human Heart, Circulation. 94 (1996) 683–689. [DOI] [PubMed] [Google Scholar]
- [44].Pok S, Benavides OM, Hallal P, Jacot JG, Use of Myocardial Matrix in a Chitosan-Based Full-Thickness Heart Patch, Tissue Eng. Part A. 20 (2014) 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hirt MN, Boeddinghaus J, Mitchell A, Schaaf S, Börnchen C, Müller C, Schulz H, Hubner N, Stenzig J, Stoehr A, Neuber C, Eder A, Luther PK, Hansen A, Eschenhagen T, Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation, J. Mol. Cell. Cardiol. 74 (2014) 151–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


