Most high-grade meningiomas show a highly perturbed copy number profile and they are enriched for TERT promoter mutations [6, 7, 11, 12]. In addition, homozygous focal deletions of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene, located at 9p21, have been observed at high frequency in anaplastic meningiomas [1, 3, 4, 9, 13]. An association of chromosome 9p21 deletion with malignant progression of meningiomas and poor prognosis, specifically in anaplastic meningiomas, has been demonstrated in 2002 by Perry et al. [9]. Here, we sought to determine the overall prognostic role of the CDKN2A/B status in a cohort of 528 meningioma patients with clinical follow-up data, covering all WHO grades and various subtypes. We thereby intended to assess the predictive power of the CDKN2A/B status, both independently and in the context of WHO grading, TERT promoter mutation status, and DNA methylation-based classification.
Tumor tissue and clinical follow-up data from 528 patients were obtained from the archives of multiple international collaborating centers and collected at the Department of Neuropathology, University Hospital Heidelberg (Heidelberg, Germany). Analysis of tissue and clinical data was performed in accordance with local ethical regulations. The clinical and pathologic characteristics of the study patients are summarized in Supplementary Table 1, online resource. DNA methylation profiling and copy number analysis of the tumors were performed using the Infinium MethylationEPIC (850k) BeadChip (Illumina, San Diego, CA, USA) or Infinium HumanMethylation450 (450k) BeadChip (Illumina) array as reported [2]. TERT promoter mutation status was assessed by Sanger sequencing or panel sequencing as previously described [5, 10]. Distribution of time to progression (as determined by imaging) or recurrence (TTP) after surgery was estimated by the Kaplan–Meier method and compared between groups with the log-rank test. Multivariable Cox proportional hazards regression was used to estimate the prognostic impact after adjusting for established prognostic factors. p values less than 0.05 were considered significant.
Among the tumors of 528 patients included in this study, 26 (4.9%) showed a homozygous deletion of CDKN2A/B as determined by DNA methylation array. Seven (27%) of these tumors were histologically graded as WHO grade II and 19 (73%) as WHO grade III. Notably, all tumors were either classified as atypical (constituting 4% of all atypical meningiomas) or anaplastic meningiomas (28% of all anaplastic meningiomas). Besides the most common grading criterion, proliferative activity, a set of morphological features also qualifies for WHO grade II or III according to the WHO classification, even in absence of high mitotic count. Interestingly, none of the WHO grade II or III meningioma variants diagnosed according to the proliferation-independent histological patterns, including chordoid, clear cell and rhabdoid meningiomas, showed a homozygous deletion of CDKN2A/B (Supplementary Table 1, online resource). In relation to the different methylation classes reported to independently stratify for risk of recurrence among meningioma [12], CDKN2A/B homozygous deletion was observed only in the methylation classes “intermediate” (n = 6; 23%) or “malignant” (n = 20; 77%). None of the tumors within the methylation class benign showed a homozygous deletion of CDKN2A/B. Of the 528 patients, 350 were female (66%) and the mean age at the time of surgery was 57 years (range 6–85 years). Median follow-up after surgery was 45 months (range 1–291 months), during which 175 patients had a progression or recurrence. Homozygous deletion of CDKN2A/B was neither significantly associated with patient age or sex nor with the tumor location. A detailed description of clinico-pathological characteristics is given in Supplementary Table 2, online resource.
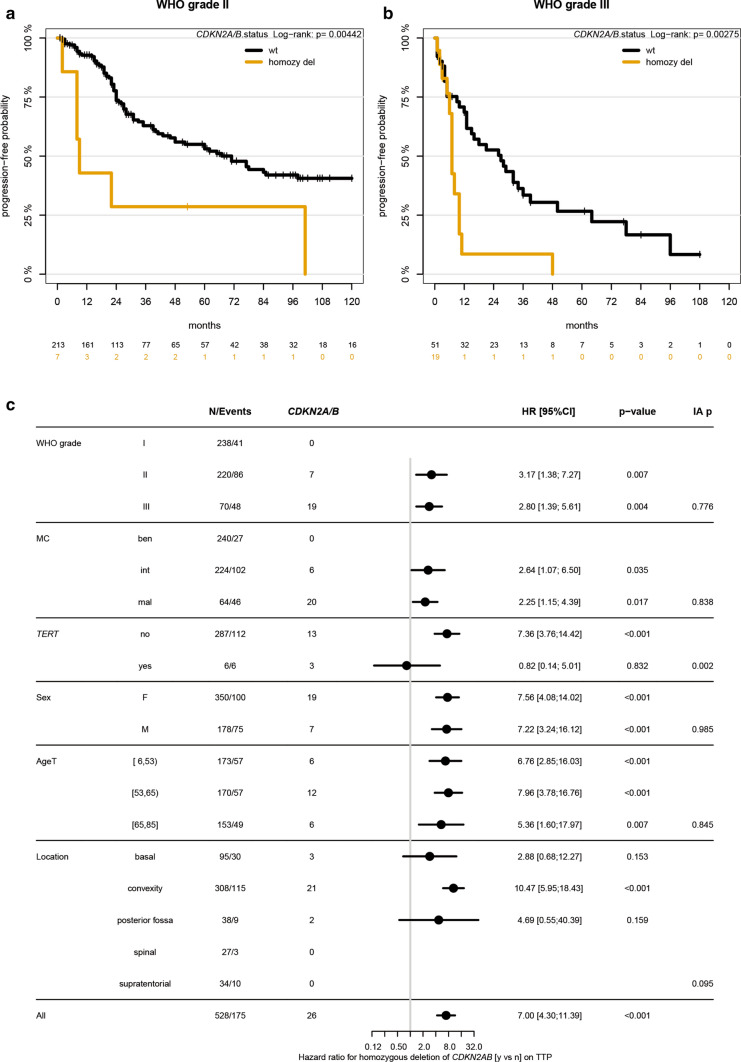
Patients whose tumors carried CDKN2A/B homozygous deletions had a significantly worse outcome and more rapid progression from the time of surgery (p < 0.001; median TTP 8 vs. 101 months, Supplementary fig. 1, online resource). Importantly, this held true even within WHO grades (WHO grade II/III: p = 0.004/0.003; Fig. 1a, b). A significant difference was also observed within DNA methylation-based subtypes (intermediate/malignant: p = 0.03/0.02, correlation of these and other parameters depicted in Fig. 1c). CDKN2A/B status remained an independent prognostic factor in Cox regression when adjusting for WHO grade, DNA methylation-based classification, tumor location, age and sex (Supplementary Table 3, online resource).
Fig. 1.
Time to progression or recurrence (TTP) of the 528 patients in the cohort stratified for CDKN2A/B homozygous deletion vs. WHO grade (a, b). Forest plot of univariable (unadjusted) hazard ratios for TTP for CDKN2A/B homozygous deletions stratified by WHO grade, methylation classes, TERT promoter mutation status, sex, age and location (c). AgeT gives age tertiles. IA p value gives p value for test on interaction, i.e., different prognostic effect of CDKN2A/B homozygous deletion in subgroups. MC methylation class, ben benign, int intermediate, mal malignant, F female, M male, TTP time to progression/recurrence
As TERT promoter mutations are associated with early recurrence [8, 11], we evaluated the effects of the TERT status on the TTP in a subset of patients (n = 293, 16 with CDKN2A/B homozygously deleted tumors) with available sequencing data, both individually and combined with the CDKN2A/B status. Tumors of 6/293 patients carried a TERT promoter mutation, three of them showed co-occurrence of a homozygous deletion of CDKN2A/B (p = 0.002). The outcome of patients with TERT promoter-mutant tumors was similarly unfavorable as that of patients with CDKN2A/B homozygously deleted tumors (median TTP for TERT, CDKN2A/B homozygous deletion, and other (i.e. none of both alterations): 11.5, 8, and 147 months, respectively; TERT status illustrated in Supplementary Fig. 2, online resource).
Our data demonstrates an independent adverse effect of CDKN2A/B homozygous deletion on the time to progression of patients with meningiomas. Thus, CDKN2A/B status can provide a useful biomarker for the identification of meningioma patients with a high risk of early recurrence. Consistent with previous studies, homozygous deletion of CDKN2A/B was found mainly in meningiomas graded as WHO grade II or III [1, 9, 13] which underlines a potential role in the malignant transformation of meningiomas. However, further studies will be needed for identification of the decisive steps in meningioma progression which will additionally assist in prioritizing the most relevant targets for novel therapy approaches. So far, CDKN2A/B homozygous deletion itself may be further clinically investigated as target for inhibitors of the CDK4/6 axis, e.g. ribociclib or palbociclib.
Notably, CDKN2A/B homozygous deletion allowed to further discriminate patients with unfavorable outcome within WHO grade II and III cases. This may suggest CDKN2A/B deletion as independent criterion for identification of highly aggressive (i.e. WHO grade III) meningiomas. However, a major limitation in the value of testing for CDKN2A/B homozygous deletion as well as TERT promoter mutation, both relevant to identify high-risk cases, is the low frequency of cases harboring such alterations. Furthermore, there seem to be a very small number of cases showing concordant alterations of CDKN2A/B and TERT which necessitates evaluation of both markers for a reliable risk prediction.
Interestingly, CDKN2A/B homozygous deletion further stratified for cases with highest risk of recurrence even within the methylation classes intermediate and malignant (Supplementary Fig. 3, online resource). This indicates that risk prediction based on methylation classes can be further refined when incorporating CDKN2A/B status, which will typically be available concurrently with generating methylation array data. Assessment of CDKN2A/B alone, however, cannot achieve the same prediction accuracy as methylation, since cases without homozygous deletion can still fall in any of the epigenetic classes.
In conclusion, our study demonstrates that homozygous deletion of CDKN2A/B is highly prognostic in meningiomas and may be a useful, independent molecular biomarker for grading of these tumors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Open Access funding provided by Projekt DEAL. For excellent technical support we sincerely thank the Microarray Unit of the German Cancer Research Center (DKFZ) Genomics and Proteomics Core Facility, as well as L. Dörner and A. Habel (Department of Neuropathology, Institute of Pathology, University Hospital Heidelberg, Heidelberg, Germany). This study was supported by the Hertie Network of Excellence in Clinical Neuroscience, the German Cancer Aid (70112956), and the Else Kröner-Fresenius Stiftung (EKFS 2015_A60). P. Sievers is a fellow of the Hertie Academy of Excellence in Clinical Neuroscience. F. Sahm is a fellow of the Else Kröner Excellence Program of the Else Kröner-Fresenius Stiftung (EKFS 2017_EKES.24).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Philipp Sievers and Thomas Hielscher shared authorship.
References
- 1.Bostrom J, Meyer-Puttlitz B, Wolter M, Blaschke B, Weber RG, Lichter P, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159:661–669. doi: 10.1016/S0002-9440(10)61737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goutagny S, Yang HW, Zucman-Rossi J, Chan J, Dreyfuss JM, Park PJ, et al. Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res. 2010;16:4155–4164. doi: 10.1158/1078-0432.CCR-10-0891. [DOI] [PubMed] [Google Scholar]
- 4.Guyot A, Duchesne M, Robert S, Lia AS, Derouault P, Scaon E, et al. Analysis of CDKN2A gene alterations in recurrent and non-recurrent meningioma. J Neurooncol. 2019;145:449–459. doi: 10.1007/s11060-019-03333-6. [DOI] [PubMed] [Google Scholar]
- 5.Koelsche C, Sahm F, Capper D, Reuss D, Sturm D, Jones DT, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126:907–915. doi: 10.1007/s00401-013-1195-5. [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Maillo A, Orfao A, Sayagues JM, Diaz P, Gomez-Moreta JA, Caballero M, et al. New classification scheme for the prognostic stratification of meningioma on the basis of chromosome 14 abnormalities, patient age, and tumor histopathology. J Clin Oncol. 2003;21:3285–3295. doi: 10.1200/JCO.2003.07.156. [DOI] [PubMed] [Google Scholar]
- 8.Mirian C, Duun-Henriksen AK, Juratli T, Sahm F, Spiegl-Kreinecker S, Peyre M, et al. Poor prognosis associated with TERT gene alterations in meningioma is independent of the WHO classification: an individual patient data meta-analysis. J Neurol Neurosurg Psychiatry. 2020;91:378–387. doi: 10.1136/jnnp-2019-322257. [DOI] [PubMed] [Google Scholar]
- 9.Perry A, Banerjee R, Lohse CM, Kleinschmidt-DeMasters BK, Scheithauer BW. A role for chromosome 9p21 deletions in the malignant progression of meningiomas and the prognosis of anaplastic meningiomas. Brain Pathol. 2002;12:183–190. doi: 10.1111/j.1750-3639.2002.tb00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahm F, Schrimpf D, Jones DT, Meyer J, Kratz A, Reuss D, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131:903–910. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
- 11.Sahm F, Schrimpf D, Olar A, Koelsche C, Reuss D, Bissel J, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016 doi: 10.1093/jnci/djv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 13.Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G, et al. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA. 1997;94:14719–14724. doi: 10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.