Abstract
Background
Hypersensitivity pneumonitis is an immune-mediated disease triggered by exposure to organic particles in susceptible individuals. It has been reported that a subgroup of patients with hypersensitivity pneumonitis develops autoantibodies with or without clinical manifestations of autoimmune disease. However, the mechanisms involved in this process and the effect of the autoantibodies on clinical course in hypersensitivity pneumonitis is unknown. We evaluated the association between human leukocyte antigen (HLA) class II alleles and hypersensitivity pneumonitis patients with and without autoantibodies.
Methods
170 hypersensitivity pneumonitis patients were included. We analysed the presence of antinuclear antibodies, rheumatoid factor, anti-SSA/Ro, anti-SSB/La and anti-CCP at the time of diagnosis. In addition, in a subset of patients we evaluated anti-Scl-70, anti-neutrophil cytoplasmic antibody, and anti-DNA. HLA typing was performed using PCR sequence-specific primers in a high-resolution modality, including HLA-DRB1 and HLA-DQB1 loci. Statistical analysis was performed employing Epi-Info v7 and SPSS v20.
Results
60 hypersensitivity pneumonitis patients showed sera autoantibodies (HPAbs+), and 110 hypersensitivity pneumonitis patients did not (HPAbs−). The frequency of the allele HLA-DRB1*03:01 was remarkably increased in the HPAbs+ group (10.8% versus 0.45%; OR 30.14, 95% CI 3.83–237.1; p=1.65×10-4 after Bonferroni's correction). Likewise, we found that the haplotype DRB1*03:01-DQB1*02:01, which is part of the 8.1 ancestral haplotype, a major genetic determinant of autoimmune diseases, confers significant risk to develop autoantibodies (OR 19.23, 95% CI 2.37–155.9; p=0.0088 after Bonferroni's correction). In addition, the HLA-DRB1*03:01 allele was associated with higher mortality in patients with hypersensitivity pneumonitis (adjusted OR 5.9, 95% CI 1.05–33.05; p=0.043).
Conclusions
A subset of hypersensitivity pneumonitis patients presents circulating autoantibodies and higher mortality that are associated with some alleles of 8.1 ancestral haplotype.
Short abstract
Alleles from 8.1 ancestral haplotype (#HLA-DRB1 and DQB1 loci) are associated with #autoantibodies production in #hypersensitivity #pneumonitis in a cohort of Mexican mestizo patients https://bit.ly/3bprPeB
Introduction
Hypersensitivity pneumonitis is an interstitial lung disease (ILD) caused by an exaggerated immune response to the inhalation of a wide variety of organic particles in genetically susceptible individuals [1–5]. The prognosis of acute hypersensitivity pneumonitis is usually favourable; however, chronic fibrotic hypersensitivity pneumonitis often progresses, with the subsequent destruction of the lung architecture [4, 6].
The heterogeneous nature of the immune response and the clinical differences through the disease suggest that multiple molecular pathways are involved in the development and progression of the disease.
Previous studies have described the participation of some alleles from the major histocompatibility complex in hypersensitivity pneumonitis susceptibility. Particularly, human leukocyte antigen (HLA) class II alleles were identified in patients with pigeon breeder's disease and summer-type hypersensitivity pneumonitis [7–9], suggesting that genetic factors located inside the HLA region contribute to hypersensitivity pneumonitis development. Class II -DRB1 and -DQB1 alleles involved in the 8.1 ancestral haplotype seem to determine the development of specific autoantibodies, both organ-specific and non-organ-specific [10].
In 2016 a cohort of patients with chronic hypersensitivity pneumonitis was examined by Adegunsoye et al. [11] to determine the prevalence of autoimmune characteristics. They found that a significant minority of patients with hypersensitivity pneumonitis (15%) displayed autoimmune features and that the presence of autoimmunity was an independent predictor of the increased mortality.
Throughout patient evaluation with hypersensitivity pneumonitis, the presence of circulating autoantibodies is often detected without fulfilling the criteria for a connective tissue disease. To date, it is not known whether this subgroup of hypersensitivity pneumonitis patients has a different genetic susceptibility associated with HLA compared to the autoantibody-negative group.
In this context, this study aimed to examine the allele frequencies of class II HLA alleles to reveal putative differences between hypersensitivity pneumonitis with and without autoantibodies (HPAbs+ and HPAbs−, respectively) and whether some polymorphisms are associated with mortality rate.
Material and methods
Study population
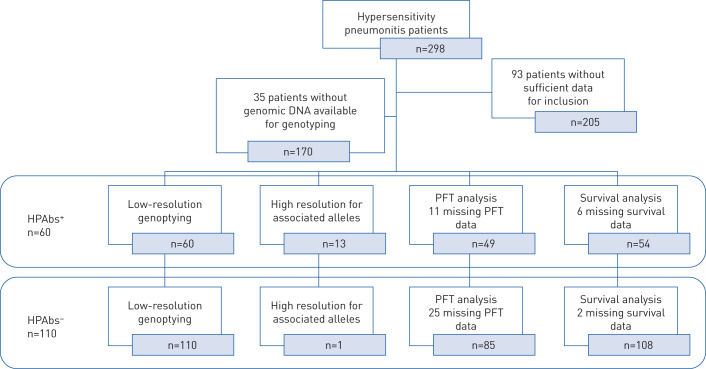
A retrospective review of adult patients (age ≥18 years) referred to the ILD clinic at the National Institute of Respiratory Diseases (Mexico City, Mexico) was conducted, and samples with enough DNA quality from 170 patients with a confirmed diagnosis of hypersensitivity pneumonitis during the period 2003–2018 were included (figure 1). Since the year 2000, DNA samples for genetic studies are obtained from patients with ILD, with clear explanation, and signed informed consent is duly collected.
FIGURE 1.
Inclusion of hypersensitivity pneumonitis with and without autoantibodies (HPAbs+ and HPAbs−, respectively), showing exclusion rates for the different analyses. PFT: pulmonary function test.
All patients had at least three prior generations born in Mexico (parents and grandparents) and were considered as Mexican mestizo. We have previously demonstrated that this criterion is a good proxy of Mexican ancestry evaluated by ancestry-informative markers [12].
Hypersensitivity pneumonitis was diagnosed according to previously described criteria [13, 14], as follows. 1) Symptoms and pulmonary function tests alterations compatible with ILD; 2) high-resolution computed tomography (HRCT) showing poorly defined nodules, ground-glass attenuation and air trapping in expiration (80% of the HPAbs+ and 54% of the HPAbs− also showed alterations compatible with fibrosis, such as irregular linear opacities and traction bronchiectasis and occasionally cystic lesions); 3) bronchoalveolar lavage lymphocytosis >30%; and 4) biopsy (performed in 30% of the HPAbs+ and 28% of the HPAbs−) compatible with hypersensitivity pneumonitis. A multidisciplinary team confirmed the diagnosis.
Clinical data were obtained from the medical records. The variables collected included demographical data, comorbidities, tobacco smoking history, environmental antigen exposure assessment, clinical laboratory studies, pulmonary function tests (including forced vital capacity, diffusing capacity of the lung for carbon monoxide (DLCO) and 6-min walk test) and mortality from all causes at 1-year follow-up.
We considered hypersensitivity pneumonitis with positive serology (HPAbs+) as positivity of at least one of the following antibodies: antinuclear antibodies (ANAs) with a specific pattern of connective tissue disease of any kind (cytoplasmic, nucleolar, centromere); ANA with homogeneous pattern, fine or coarse mottle; rheumatoid factor three or more times the upper normal limit (20 IU·mL−1); anti-cyclic citrullinated peptide ≥20 RU·mL−1 and/or at least one antibody in the autoimmunity profiles for Sjögren's syndrome (anti-Ro (SSA), anti-La (SSB)). These autoantibodies were analysed in all the patients. Additionally, autoantibodies associated with systemic sclerosis (anti-Scl-70), vasculitis (anti-neutrophil cytoplasmic antibody (ANCA)) and anti-DNA were examined in 88% of the HPAbs+ and 55% of the HPAbs−. No patient met classification criteria for connective tissue diseases according to the American College of Rheumatology/European League Against Rheumatism [15–17].
The patients were classified according to the presence of autoantibodies present in serum (HPAbs+ versus HPAbs−) (table 1).
TABLE 1.
Clinical and demographic characteristics in hypersensitivity pneumonitis subgroups
| Subjects | HPAbs+ | Subjects | HPAbs− | p-value | |
| Age years | 60 | 53±9 | 110 | 53±12 | 0.59 |
| Female | 60 | 52 (86) | 110 | 89 (81) | 0.39 |
| Duration of symptoms before diagnosis months | 60 | 25 (4–20) | 110 | 27 (2–120) | 0.30 |
| Former smokers | 60 | 14 (23) | 110 | 26 (24) | 0.96 |
| Exposure to birds | 60 | 47 (78) | 110 | 91 (83) | 0.53 |
| Diabetes mellitus | 60 | 11 (18) | 110 | 16 (15) | 0.51 |
| Systemic hypertension | 60 | 9 (15) | 110 | 22 (20) | 0.53 |
| Dead | 54 | 12 (20) | 108 | 9 (8) | 0.02 |
| Antibodies against avian antigens | 60 | 59 (94) | 110 | 57 (53) | <0.0001 |
| Unidentified antigen | 60 | 4 (6) | 110 | 50 (47) | <0.0001 |
| HRCT chronic pattern | 60 | 48 (80) | 110 | 59 (54) | <0.001 |
| C-reactive protein mg·dL−1 | 60 | 0.9 (0.02–7.1) | 110 | 0.7 (0.01–3.9) | 0.95 |
| BAL# lymphocytosis | 49 | 49±19 | 87 | 46±25 | 0.17 |
| FVC % pred | 60 | 57.8±18 | 106 | 53.8±20 | 0.20 |
| DLCO % pred | 49 | 46±22 | 85 | 51±21 | 0.24 |
| 6MWT m | 42 | 333±168 | 71 | 333±116 | 0.21 |
| PO2¶ mmHg | 42 | 51±8 | 71 | 50±13 | 0.90 |
| PAP§ mmHg | 47 | 39±14 | 69 | 38±14 | 0.40 |
Data are presented as n, mean±sd, n (%) or median (range), unless otherwise stated. HPAbs+/HPAbs−: hypersensitivity pneumonitis with and without autoantibodies, respectively; HRCT: high-resolution computed tomography; BAL: bronchoalveolar lavage; FVC: forced vital capacity; DLCO: diffusing capacity of the lung for carbon monoxide; 6MWT: 6-min walk test; PO2: oxygen tension; PAP: pulmonary artery pressure. #: performed in 81% of the HPAbs+ patients and 79% of the HPAbs− patients; ¶: measured in 42 HPAbs+ patients and 71 HPAbs− patients; §: measured by echocardiography in 47 HPAbs+ patients and 69 HPAbs− patients.
Ethics statement
This study was approved by the institutional committee for science and ethics of the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (INER; Mexico City, Mexico) (approbation codes: B20-15 and C60-17).
Genomic DNA isolation
Peripheral blood (7 mL) was obtained by venipuncture and was collected in a tube with EDTA as an anticoagulant for subsequent DNA extraction. Samples were obtained at the time of diagnosis, and patients were not receiving corticosteroids or immunosuppressive drugs.
The blood samples were handled in the HLA laboratory of INER to obtain genomic DNA employing a commercial kit (BDtract Genomic DNA Isolation Kit; Maxim Biotech, San Francisco, CA, USA). The DNA was quantified by ultraviolet spectrophotometry using a NanoDrop 2000 device (Thermo Scientific, Waltham, MA, USA). Contamination with organic compounds and proteins was defined by measuring the ratio absorbance at 280 nm and 260 nm. Samples were considered of good quality when the ratio was ∼1.8.
HLA class II molecular typing
Molecular typing for DRB loci and DQB1 locus was done using a PCR sequence-specific primers (PCR-SSP) technique using Micro SSP™ HLA DNA Typing Trays (One Lambda, Canoga Park, CA, USA). Unlike other PCR-based methods, the SSP methodology employed discriminates between the different alleles during the PCR process. The amplified DNA fragments are separated by agarose gel electrophoresis and visualised by staining and exposure to ultraviolet light. The interpretation of PCR-SSP results is based on the presence or absence of a specific amplified DNA fragment. Since amplification during the PCR reaction may be adversely affected by various factors such as pipetting errors, poor DNA quality and presence of inhibitors, an internal control primer pair was included in every PCR reaction. The control primer pair amplifies a conserved region of the human β-globin gene, which is present in all human DNA samples and is used to verify the integrity of the PCR reaction.
Initially, HLA-DRB typing was performed by low-resolution modality, which includes DRB1, DRB3, DRB4 and DRB5 alleles in 24 independent reactions. The DRB loci allele group covers DRB1*01, *03, *04, *07, *08, *11, *12, *13, *14, *15 and *16 as well as DRB3*02, DRB3*03, DRB4*01 and DRB5*01 specificities (DR51, DR52 and DR53 serological equivalents). High resolution for HLA-DRB1*03 was performed with an additional panel of 16 primer pairs to establish allele discrimination. For HLA-DQB1 typing, eight primer pairs were used, while the HLA-DQB1 high-resolution kit used to achieve DQB1 amplification, which includes 55 alleles for DQ6, DQ3, DQ4, DQ5 and DQ2.
All amplifications were performed using the Kapa Taq HotStart PCR kit (Kapa Biosystems, Cape Town, South Africa). PCR-SSP products were electrophoresed in 3% agarose gels stained using MIDORI Green Advanced DNA Stain (Nippon Genetics Europe, Dueren, Germany). Interpretation of typing results was made using the assistance of the HLA Fusion Software versus 4.3 (One Lambda).
Statistical analysis
The statistics programme SPSS (version 21; SPSS, Chicago, IL, USA) was used to describe the study population and determine the median, minimum, and maximum values for each variable and compared using a Mann–Whitney U-test. Continuous variables were reported as mean±sd and analysed using a t-test. Categorical variables were reported as counts and percentages and contrasted using Fisher's exact test.
Allele and haplotype frequencies of HLA were determined by direct counting. The observed and expected HLA class II alleles (each locus) tested for Hardy–Weinberg equilibrium using a conventional Fisher's exact test. The associations were evaluated by Fisher's exact two-tailed test, with a statistical significance value of p<0.05. Bonferroni correction was performed considering the 20 alleles identified in both loci and the 22 haplotypes found in the whole population. Odds ratios and 95% confidence intervals were calculated using 2×2 contingency tables comparing both groups by alleles and haplotypes reported using the Epi Info v4.0.1 [18]. Finally, the haplotypes were constructed employing Arlequin v3.1 software [19] using the maximum likelihood method, with an iterative expectation maximisation algorithm.
To evaluate the strength of association between mortality and the risk allele, we first estimated a crude odds ratio (cOR) using univariate regression analysis; the p-value was estimated with the Wald test, and then we estimated an adjusted odds ratio (aOR) with a multivariate logistic regression analysis that includes only variables with p<0.05, due to the small sample size. No imputation procedures were performed to handle missing data.
Results
Out of 298 patients registered in the hypersensitivity pneumonitis cohort, 93 did not have enough inclusion data for the study, including lack of autoantibodies results (before 2006, ILD patients did not undergo the measurement of autoantibodies routinely), or because our multidisciplinary team did not confirm the diagnosis of hypersensitivity pneumonitis. The other 35 were excluded due to not having enough genomic DNA to perform genotyping. 170 patients with hypersensitivity pneumonitis were included, 60 with circulating autoantibodies (HPAbs+) and 110 without (HPAbs−) (figure 1). Most HPAbs+ patients (89%) were positive for antinuclear antibodies (>1:320), and occasionally they were also positive for another of the autoantibodies examined (see methods section). 12% (seven patients) were negative for ANA, but showed increased levels of other autoantibodies (e.g. rheumatoid factor n=2, ANCA n=2, anti-Ro n=2, anti-La n=1). HPAbs− patients were negative for all examined autoantibodies.
Demographic characteristics
There were no differences between HPAbs+ and HPAbs− regarding demographic characteristics, including age, sex and smoking (table 1). Likewise, the comparison of pulmonary function tests between both groups (performed in 49 of 60 HPAbs+ patients and 85 of 110 HPAbs patients) showed no differences (table 1). However, HPAbs+ patients were more likely to have chronic/fibrotic changes on HRCT than HPAbs−. In contrast, the percentage of patients in whom we were unable to identify the antigen was significantly higher in the HPAbs− group (table 1).
During the 1-year follow-up, six HPAbs+ patients and two HPAbs− patients were lost. 12% of the entire cohort died, with a significantly higher proportion in the HPAbs+ group (20% versus 8%, p=0.02).
HLA class II alleles and haplotypes
No statistically significant differences were found in the population stratification based on the region of origin between cases and controls. The HLA allele pairs of each locus from both groups satisfied Hardy–Weinberg equilibrium (p>0.05), accounting for common alleles producing common genotypes, i.e. both heterozygous (e.g. DQB1*02:01, DQB1*05:01) and homozygous (e.g. DQB1*03:02) pairs.
The distributions of DRB1 and DQB1 alleles are listed in tables 2 and 3. 10 HLA-DRB1 alleles were identified in the HPAbs+ group, seven of them with an allele frequency >5%, and the most common were DRB1*08 (allele frequency 20.00) and DRB1*04 (allele frequency 17.50). A marked increase in the frequency of HLA-DRB1*03 was identified in the HPAbs+ group (10.8% versus 0.45%; OR 30.14, 95% CI 3.83–237.1; p=1.65×10-4 after Bonferroni's correction for all alleles (20 identified in both loci). All DRB1*03 subjects were DRB1*03:01 in the high-resolution subtyping analysis. The alleles in the DRB3 and DRB4 genes were not found in HPAbs+ patients nor in HPAbs− patients, while DRB5 alleles were identified in few patients without statistical differences.
TABLE 2.
Frequencies of human leukocyte antigen (HLA)-DRB1 alleles in patients with hypersensitivity pneumonitis with and without autoantibodies (HPAbs+ and HPAbs−, respectively)
| Hypersensitivity pneumonitis | HPAbs+ | HPAbs− | p-value | OR (95% CI) | ||||
| Patients | AF % | Patients | AF % | Patients | AF % | |||
| Patients | 170 | 60 | 110 | |||||
| *01 | 17 | 5.000 | 5 | 4.167 | 12 | 5.455 | 0.79 | 0.74 (0.24–2.21) |
| *03:01 | 14 | 4.118 | 13 | 10.833 | 1 | 0.455 | 1.65×10-4# | 30.14 (3.83–237.1) |
| *04 | 70 | 20.588 | 21 | 17.500 | 49 | 22.273 | 0.25 | 0.67 (0.34–1.28) |
| *07 | 22 | 6.471 | 9 | 7.500 | 13 | 5.909 | 0.63 | 1.31 (0.52–3.28) |
| *08 | 71 | 20.882 | 24 | 20.000 | 47 | 21.364 | 0.74 | 0.89 (0.47–1.69) |
| *09 | 4 | 1.176 | ND | 4 | 1.818 | |||
| *10 | 5 | 1.471 | ND | 5 | 2.273 | |||
| *11 | 24 | 7.059 | 6 | 5.000 | 18 | 8.182 | 0.35 | 0.56 (0.21–1.51) |
| *12 | 2 | 0.588 | ND | 2 | 0.909 | |||
| *13 | 23 | 6.765 | 10 | 8.333 | 13 | 5.909 | 0.48 | 1.49 (0.61–3.64) |
| *14 | 59 | 17.353 | 18 | 15.000 | 41 | 18.636 | 0.4 | 0.72 (0.36–1.41) |
| *15 | 14 | 4.118 | 8 | 6.667 | 6 | 2.727 | 0.08 | 2.66 (0.87–8.08) |
| *16 | 15 | 4.412 | 6 | 5.000 | 9 | 4.091 | 0.77 | 1.24 (0.42–3.68) |
Data are presented as n, unless otherwise stated. AF: allele frequency; ND: not detected. #: p-value after Bonferroni's correction for the total alleles (n=20) identified in both loci. The comparison was made between HPAbs+ versus HPAbs− groups.
TABLE 3.
Frequencies of human leukocyte antigen (HLA)-DQB1 alleles in patients with hypersensitivity pneumonitis with and without autoantibodies (HPAbs+ and HPAbs−, respectively)
| Hypersensitivity pneumonitis | HPAbs+ | HPAbs− | p-value | OR (95% CI) | ||||
| Patients | AF % | Patients | AF % | Patients | AF % | |||
| Patients | 170 | 60 | 110 | |||||
| 02:01 | 32 | 9.412 | 18 | 15.000 | 14 | 6.364 | 0.14# | 2.93 (1.33–6.45) |
| 03:01 | 115 | 33.824 | 36 | 30.000 | 79 | 35.909 | 0.12 | 0.58 (0.30–1.14) |
| 03:02 | 68 | 20.000 | 19 | 15.833 | 49 | 22.273 | 0.14 | 0.57 (0.29–1.11) |
| 03:03 | 4 | 1.176 | ND | 4 | 1.818 | |||
| 04:02 | 72 | 21.176 | 25 | 20.833 | 47 | 21.364 | 1 | 0.95 (0.50–1.81) |
| 05:01 | 24 | 7.059 | 7 | 5.833 | 17 | 7.727 | 0.64 | 0.72 (0.28–1.85) |
| 06:01 | 25 | 7.353 | 15 | 12.500 | 10 | 4.545 | 0.12# | 3.33 (1.39–7.98) |
Data are presented as n, unless otherwise stated. AF: allele frequency; ND: not detected. #: p-value after Bonferroni's correction for the total alleles (n=20) identified in both loci. The comparison was made between HPAbs+ versus HPAbs− groups.
Seven alleles were recognised at the HLA-DQB1 locus, among them, DQB1*03:01, DQB1*03:02 and DQB1*04:02 were the most frequent in both groups. A higher frequency of the allele DQB1*02:01 (allele frequency 15.00) was detected in the HPAbs+ group compared with HPAbs− (15.0% versus 6.364%, p=0.007; OR 2.93, 95% CI 1.33–6.45); however, this association was lost after Bonferroni's correction (p=0.14). The frequency of the HLA-DQB1*06:01 allele was also more than two-fold higher in the HPAbs+ group (12.5% versus 4.5%, p=0.006; OR 3.33, 95% CI 1.39–7.98), but Bonferroni's correction revealed that this difference was not significant either (p=0.12).
In addition, we compared the haplotype frequency of the DRB1-DQB1 loci. As shown in table 4, 22 haplotypes were identified in the whole hypersensitivity pneumonitis group; interestingly, in the HPAbs+ group (n=60) there are 19 haplotypes, while in the HPAbs− group (n=110) only 14 haplotypes were determined (19 out of 22 versus 14 out of 22, p=0.08).
TABLE 4.
Human leukocyte antigen (HLA)-class II haplotypes (DRB1*-DQB1*) in patients with hypersensitivity pneumonitis with and without autoantibodies (HPAbs+ and HPAbs−, respectively)
| Hypersensitivity pneumonitis | HPAbs+ | HPAbs− | p-value | OR (95% CI) | ||||
| Patients | HF % | Patients | HF % | Patients | HF % | |||
| Patients | 170 | 60 | 110 | |||||
| 01-03:01 | 1 | 0.294 | 1 | 0.833 | ND | |||
| 01-05:01 | 16 | 4.706 | 4 | 3.333 | 12 | 5.455 | 0.42 | 0.58 (0.17–1.89) |
| 03:01-02:01 | 10 | 2.941 | 9 | 7.500 | 1 | 0.455 | 0.0088# | 19.23 (2.37–155.9) |
| 03:01-03:01 | 1 | 0.294 | 1 | 0.833 | ND | |||
| 03:01-06:01 | 3 | 0.882 | 3 | 2.500 | ND | |||
| 04-03:01 | 1 | 0.294 | 1 | 0.833 | ND | |||
| 04-03:02 | 68 | 20.000 | 19 | 15.833 | 49 | 22.273 | 0.14 | 0.57 (0.29–1.11) |
| 04-04:02 | 1 | 0.294 | 1 | 0.833 | ND | 0.000 | ||
| 07-02:01 | 22 | 6.471 | 9 | 7.500 | 13 | 5.909 | 0.63 | 1.31 (0.52–3.28) |
| 08-04:02 | 71 | 20.882 | 24 | 20.000 | 47 | 21.364 | 0.74 | 0.89 (0.47–1.69) |
| 09-03:03 | 4 | 1.176 | ND | 4 | 1.818 | |||
| 10-05:01 | 5 | 1.471 | ND | 5 | 2.273 | |||
| 11-03:01 | 16 | 4.706 | 2 | 1.667 | 14 | 6.364 | 0.05 | 0.23 (0.05–1.07) |
| 11-06:01 | 8 | 2.353 | 4 | 3.333 | 4 | 1.818 | 0.45 | 1.89 (0.45–7.85) |
| 12-03:01 | 2 | 0.588 | ND | 2 | 0.909 | |||
| 13-03:01 | 18 | 5.294 | 5 | 4.167 | 13 | 5.909 | 0.6 | 0.67 (0.22–2.0) |
| 13-05:01 | 3 | 0.882 | 3 | 2.500 | ND | |||
| 13-06:01 | 2 | 0.588 | 2 | 1.667 | ND | |||
| 14-03:01 | 59 | 17.353 | 18 | 15.000 | 41 | 18.636 | 0.4 | 0.72 (0.36–1.41) |
| 15-03:01 | 2 | 0.588 | 2 | 1.667 | ND | |||
| 15-06:01 | 12 | 3.529 | 6 | 5.000 | 6 | 2.727 | 0.34 | 1.92 (0.59–6.25) |
| 16-03:01 | 15 | 4.412 | 6 | 5.000 | 9 | 4.091 | 0.77 | 1.24 (0.42–3.68) |
Data are presented as n, unless otherwise stated. HF: haplotype frequency; ND: not detected. #: p-value after Bonferroni correction for the total haplotypes (n=22) identified. The comparison was made between HPAbs+ versus HPAbs− groups.
We observed a significant higher frequency of the haplotype DRB1*03:01-DQB1*02:01 in patients with HPAbs+ (7.5% versus 0.45%; OR 19.23, 95% CI 2.37–155.9; p=0.0088 after Bonferroni's correction (applying the 22 haplotypes identified in the whole population)).
Logistic regression
To evaluate the strength of the association between mortality and the risk allele, we estimated the cOR using univariate logistic regression analysis of mortality. The risk allele HLA-DRB1*03:01 was associated to mortality (cOR 4.5, 95% CI 1.19–16.9; p=0.026). To adjust for confounding, we performed three multivariate logistic regression models (table 5). In model 1, we included the four variables that showed significant association with mortality by univariate analysis: HLA-DRB1*03:01, DLCO, C-reactive protein (CRP) and HPAbs+. We found that DLCO and CRP remained significant. In model 2 when we excluded HPAbs+, the allele HLA-DRB1*03:01 was strongly associated with mortality (aOR 5.9, 95% CI 1.05–33.0; p=0.043). By contrast, when we excluded the allele HLA-DRB1*03:01, HPAbs+ lost significance, supporting the strong association with the risk allele. Importantly, HLA-DRB1*03:01 and HPAbs+ were strongly associated (OR 9.52, 95% CI 1.85–92), indicating collinearity between these two variables.
TABLE 5.
Univariate analysis adjusting the risk allele (DRB1*03:01) by diffusing capacity of the lung for carbon monoxide (DLCO) and C-reactive protein (CRP)
| Nonsurvivors | Survivors | cOR (95% CI) | p-value | Model 1 aOR (95% CI) | p-value | Model 2 aOR (95% CI) | p-value | Model 3 aOR (95% CI) | p-value | |
| Patients# | 21 | 141 | ||||||||
| Age at baseline evaluation years | 52.8±9.9 | 52.6±11.3 | 1.0 (0.96–1.04) | 0.94 | ||||||
| Male | 6 (29) | 22 (16) | 2.1 (0.75 –6.10) | 0.15 | ||||||
| DRB1*03:01 | 4 (19) | 7 (5) | 4.5 (1.19–16.90) | 0.026 | 3.9 (0.69–24.83) | 0.14 | 5.9 (1.05–33.05) | 0.043 | ||
| Current smokers | 4 (19) | 33 (24) | 0.7 (0.23–2.40) | 0.62 | ||||||
| Arterial hypertension | 3 (14) | 26 (20) | 0.6 (0.18–2.40) | 0.54 | ||||||
| Diabetes mellitus | 3 (14) | 20 (15) | 0.9 (0.25–3.40) | 0.91 | ||||||
| Baseline FVC % pred | 48 (32–66) | 53 (41–73) | 0.9 (0.96–1.01) | 0.27 | ||||||
| Baseline DLCO % pred | 26.5 (20–47) | 48.5 (37–64) | 0.9 (0.92–0.98) | 0.006 | 0.94 (0.91–0.98) | 0.008 | 0.94 (0.91–0.98) | 0.007 | 0.94 (0.91–0.98) | 0.006 |
| PO2 mmHg | 50.9±13.1 | 50.8±11.0 | 1.0 (0.95–1.04) | 0.99 | ||||||
| PCO2 mmHg | 33.7 (30.8–36.0) | 35 (32.0–39.9) | 0.9 (0.85–1.02) | 0.13 | ||||||
| PAP¶ mmHg | 42 (31–50) | 35 (25–45) | 1.0 (0.99–1.06) | 0.09 | ||||||
| 6MWT m | 333 (220–400) | 350 (245–434) | 0.9 (0.98–1.00) | 0.48 | ||||||
| CRP mg·dL−1 | 0.52 (0.37–1.16) | 0.43 (0.23–0.84) | 1.6 (1.02–2.55) | 0.037 | 2.09 (1.13–3.86) | 0.02 | 2.2 (1.2–3.9) | 0.01 | 2.03 (1.12–3.6) | 0.02 |
| BAL lymphocytosis % | 40 (33–54) | 47 (30–62) | 0.9 (0.96–1.01) | 0.36 | ||||||
| HRCT chronic pattern | 13 (62) | 91 (61) | 1.03 (0.4–2.65) | 0.94 | ||||||
| HPAbs+ | 12 (57) | 42 (30) | 3.14 (1.23–8.01) | 0.02 | 2.9 (0.83–10.19) | 0.09 | 3.08 (0.88–10.7) | 0.076 |
Data are presented as n, mean±sd, n (%) or median (range), unless otherwise stated. The crude odds ratio (cOR) of the strength of association of DLCO with mortality was estimated with the data of 118 patients. The multivariate logistic regression model was obtained with the data of 103 patients, the goodness of fit of the model is p=0.69, and the area under receiver operating characteristic curve=0.82. HLA DRB1*03 and HP Abs+ were strongly associated (OR 9.52, 95% CI 1.85–92.0), suggesting collinearity between these two variables in model 1. aOR: adjusted odds ratio; FVC: forced vital capacity; PO2: oxygen tension; PCO2: carbon dioxide tension; PAP: pulmonary artery pressure; 6MWT: 6-min walk test; BAL: bronchoalveolar lavage; HRCT: high-resolution computed tomography; HPAbs+: hypersensitivity pneumonitis with and without autoantibodies. #: survival was obtained in 162 out of the 170 patients; ¶: measured by echocardiography.
The multivariate logistic regression model was obtained with the data of 103 patients, the goodness of fit of model 2 is p=0.69, and the area under receiver operating characteristic curve=0.82 (table 5).
Discussion
Studies about hypersensitivity pneumonitis genetic susceptibility are limited, but HLA class II genes have been identified as critical factors that contribute to disease development [8, 9, 20].
In the past few years, a growing body of evidence indicates that a subgroup of patients with hypersensitivity pneumonitis develop serum or even clinical features of autoimmunity, although the underlying mechanisms are still unknown. Moreover, except for higher mortality, this “phenotype” has not been characterised.
Similarly, in recent years a subset of patients has been described, who have idiopathic interstitial pneumonia (IIP) and autoimmune features (IPAF), who do not meet the criteria for a connective tissue disease (CTD) but have at least one sign or symptom suggestive of a CTD and at least one serological test reflective of an autoimmune process [21]. These patients have been better characterised, and some studies also suggest that patients who met the IPAF criteria had a significantly worse survival than those with IIP without autoimmune features [22], although substantial variability in outcome has been reported [23]. As in hypersensitivity pneumonitis, the mechanisms and genetic susceptibility triggering the autoimmune process is unknown.
In this study, we found that approximately a third of the patients with hypersensitivity pneumonitis may present autoantibodies. This proportion is higher than reported by Adegunsoye et al. [11], to our knowledge the only study to have shown this association, although the reasons are currently unknown.
In this study, we aimed to identify putative genetic factors associated with the HLA class II system that may increase the risk of developing autoimmune features in patients with hypersensitivity pneumonitis. Our results revealed, for the first time, some of the genetic alleles conferring risk of developing autoantibodies in these patients. Thus, a significant increase in the frequency of the HLA-DRB1*03:01 allele as well as the haplotype DRB1*03:01-DQB1*02:01 was observed in the group of patients with autoantibodies compared with patients without them.
Interestingly, these alleles have been described in the formation of autoantibodies in classic and rare autoimmune disorders. For example, alleles HLA-DRB1*03:01 and DQB1*06:01 are associated with the development of autoantibodies in systemic lupus erythematosus [24, 25]. Likewise, the presence of anti-Jo-1 or anti-PM-Scl antibodies was found to be strongly associated with HLA-DRB1*03:01 and DQA1*05:01 alleles in a cohort of patients with adult and juvenile myositis [26–28]. The DQB1*02:01 allele is linked genetically to DQA1*05:01 and is classically described in coeliac disease, type 1 diabetes and other autoimmune disorders [29].
Haplotype analysis revealed a significant increase in the frequency of HLA-DRB1*03:01-DQB1*02:01, commonly observed in the European population [30] and associated with several autoimmune disorders, such as primary Sjögren's syndrome with autoantibody production [31]. This haplotype contains two main alleles that are part of 8.1 ancestral haplotype. The ancestral haplotype is an exceptionally long (more than four megabases) conserved combination of alleles: HLA-A1, Cw7, B8, TNFAB*a2b3, TNFN*S, C2*C, Bf*s, C4A*Q0, C4B*1, DRB1*03:01, DRB3*01:01, DQA1*05:01 and DQB1*02:01. It is accepted as the most critical immunological determinant conferring risk to several autoimmune diseases [10]. Moreover, even healthy carriers of this haplotype show high values of autoantibodies, blood-activated T-cells and blood immune complexes. Therefore, the 8.1 ancestral haplotype enhances immune dysfunctions and autoimmune disorders, and our results suggest that it may also contribute to the development of autoimmunity in hypersensitivity pneumonitis.
Hypersensitivity pneumonitis is a complex disease, where the interaction of environmental and genetic factors influences the development of the disease and the phenotype, e.g. fibrotic or nonfibrotic disease [4]. For example, it has been demonstrated that the minor allele frequency of MUC5B rs35705950 is higher in patients with chronic hypersensitivity pneumonitis than in healthy controls, and more importantly, that the extent of radiographic fibrosis is associated with this common variant [32].
Our findings suggest that the presence of specific alleles and haplotypes of the HLA system may lead to the development of autoimmune features, modifying the course and outcome of hypersensitivity pneumonitis. Moreover, to carry the risk allele, HLA-DRB1*03:01 was associated with reduced survival. Interestingly, immune dysfunctions associated with the 8.1 ancestral haplotype appear to be a factor to early morbidity and mortality in older females [33]; females were markedly more frequent in our hypersensitivity pneumonitis cohort.
In addition, we found that mortality in the group of HPAbs+ was higher than in the group without autoantibodies, coinciding with a previous study in which the presence of autoimmunity was found to be associated with poor prognosis [11]. However, after adjusting by the allele HLA-DRB1*03:01, the presence of autoantibodies lost significance. Surprisingly, mortality did not show differences between chronic fibrotic and nonfibrotic hypersensitivity pneumonitis patients, probably because of the short time of follow-up and the small sample size.
Our research has some limitations, including the relatively small sample size. Besides, we excluded patients with hypersensitivity pneumonitis and confirmed connective tissue disease or a history of inflammatory disorders.
Further studies are needed to validate these findings and to determine the molecular mechanisms of the underlying autoimmune process in patients with hypersensitivity pneumonitis.
Conclusion
In patients with hypersensitivity pneumonitis, the allele HLA-DRB1*03:01 and the haplotype DRB1*03:01-DQB1*02:01, part of the 8.1 ancestral haplotype, a major genetic determinant of autoimmune diseases, are associated with the presence of autoantibodies. HLA-DRB1*03:01 also increases the risk of mortality in patients with hypersensitivity pneumonitis.
Shareable PDF
Supplementary Material
Footnotes
Conflict of interest: I. Buendía-Roldán has nothing to disclose.
Conflict of interest: L. Santiago-Ruiz has nothing to disclose.
Conflict of interest: G. Pérez-Rubio has nothing to disclose.
Conflict of interest: M. Mejía has nothing to disclose.
Conflict of interest: J. Rojas-Serrano has nothing to disclose.
Conflict of interest: E. Ambrocio-Ortiz has nothing to disclose.
Conflict of interest: G. Benítez-Valdez has nothing to disclose.
Conflict of interest: M. Selman has nothing to disclose.
Conflict of interest: R. Falfán-Valencia has nothing to disclose.
References
- 1.Ohshimo S, Bonella F, Guzman J, et al. . Hypersensitivity pneumonitis. Immunol Allergy Clin North Am 2012; 32: 537–556. doi: 10.1016/j.iac.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 2.Selman M, Pardo A, King TE. Hypersensitivity pneumonitis: insights in diagnosis and pathobiology. Am J Respir Crit Care Med 2012; 186: 314–324. doi: 10.1164/rccm.201203-0513CI [DOI] [PubMed] [Google Scholar]
- 3.Spagnolo P, Rossi G, Cavazza A, et al. . Hypersensitivity pneumonitis: a comprehensive review. J Investig Allergol Clin Immunol 2015; 25: 237–250. [PubMed] [Google Scholar]
- 4.Vasakova M, Selman M, Morell F, et al. . Hypersensitivity pneumonitis: current concepts of pathogenesis and potential targets for treatment. Am J Respir Crit Care Med 2019; 200: 301–308. doi: 10.1164/rccm.201903-0541PP [DOI] [PubMed] [Google Scholar]
- 5.Selman M, Buendía-Roldán I. Immunopathology, diagnosis, and management of hypersensitivity pneumonitis. Semin Respir Crit Care Med 2012; 33: 543–554. doi: 10.1055/s-0032-1325163 [DOI] [PubMed] [Google Scholar]
- 6.Vasakova M, Morell F, Walsh S, et al. . Hypersensitivity pneumonitis: perspectives in diagnosis and management. Am J Respir Crit Care Med 2017; 196: 680–689. doi: 10.1164/rccm.201611-2201PP [DOI] [PubMed] [Google Scholar]
- 7.Camarena A, Juárez A, Mejía M, et al. . Major histocompatibility complex and tumor necrosis factor-α polymorphisms in pigeon breeder's disease. Am J Respir Crit Care Med 2001; 163: 1528–1533. doi: 10.1164/ajrccm.163.7.2004023 [DOI] [PubMed] [Google Scholar]
- 8.Falfán-Valencia R, Camarena Á, Pineda CL, et al. . Genetic susceptibility to multicase hypersensitivity pneumonitis is associated with the TNF-238 GG genotype of the promoter region and HLA-DRB1*04 bearing HLA haplotypes. Respir Med 2014; 108: 211–217. doi: 10.1016/j.rmed.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 9.Ando M, Arima K, Yoneda R, et al. . Japanese summer-type hypersensitivity pneumonitis: geographic distribution, home environment, and clinical characteristics of 621 cases. Am Rev Respir Dis 1991; 144: 765–769. doi: 10.1164/ajrccm/144.4.765. [DOI] [PubMed] [Google Scholar]
- 10.Gambino CM, Aiello A, Accardi G, et al. . Autoimmune diseases and 8.1 ancestral haplotype: an update. HLA 2018; 92: 137–143. doi: 10.1111/tan.13305. [DOI] [PubMed] [Google Scholar]
- 11.Adegunsoye A, Oldham JM, Demchuk C, et al. . Predictors of survival in coexistent hypersensitivity pneumonitis with autoimmune features. Respir Med 2016; 114: 53–60. doi: 10.1016/j.rmed.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Rubio G, Silva-Zolezzi I, Fernández-López J, et al. . Genetic variants in IL6R and ADAM19 are associated with COPD severity in a Mexican mestizo population. COPD 2016; 13: 610–615. doi: 10.3109/15412555.2016.1161017 [DOI] [PubMed] [Google Scholar]
- 13.García de Alba C, Buendia-Roldán I, Salgado A, et al. . Fibrocytes contribute to inflammation and fibrosis in chronic hypersensitivity pneumonitis through paracrine effects. Am J Respir Crit Care Med 2015; 191: 427–436. doi: 10.1164/rccm.201407-1334OC [DOI] [PubMed] [Google Scholar]
- 14.Barrera L, Mendoza F, Zuñiga J, et al. . Functional diversity of T-cell subpopulations in subacute and chronic hypersensitivity pneumonitis. Am J Respir Crit Care Med 2008; 177: 44–55. doi: 10.1164/rccm.200701-093OC [DOI] [PubMed] [Google Scholar]
- 15.Shiboski CH, Shiboski SC, Seror R, et al. . 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjögren's syndrome: a consensus and data-driven methodology involving three international patient cohorts. Ann Rheum Dis 2017; 76: 9–16. doi: 10.1136/annrheumdis-2016-210571 [DOI] [PubMed] [Google Scholar]
- 16.Aletaha D, Neogi T, Silman AJ, et al. . 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 2010; 62: 2569–2581. doi: 10.1002/art.27584 [DOI] [PubMed] [Google Scholar]
- 17.Baron M, Pope JE, Van Den Hoogen F, et al. . Specificity of systemic sclerosis classification criteria. J Rheumatol 2015; 42: 2512. doi: 10.3899/jrheum.150694 [DOI] [PubMed] [Google Scholar]
- 18.Dean AG, Arner TG, Sunki GG, et al. 2004 www.cdc.gov/epiinfo/index.html Epi Info, a Database and Statistics Program for Public Health Professionals.
- 19.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 2007; 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 20.Camarena Á, Aquino-Galvez A, Falfán-Valencia R, et al. . PSMB8 (LMP7) but not PSMB9 (LMP2) gene polymorphisms are associated to pigeon breeder's hypersensitivity pneumonitis. Respir Med 2010; 104: 889–894. doi: 10.1016/j.rmed.2010.01.014 [DOI] [PubMed] [Google Scholar]
- 21.Fischer A, Antoniou KM, Brown KK, et al. . An official European Respiratory Society/American Thoracic Society research statement: interstitial pneumonia with autoimmune features. Eur Respir J 2015; 46: 976–987. doi: 10.1183/13993003.00150-2015 [DOI] [PubMed] [Google Scholar]
- 22.Dai J, Wang L, Yan X, et al. . Clinical features, risk factors, and outcomes of patients with interstitial pneumonia with autoimmune features: a population-based study. Clin Rheumatol 2018; 37: 2125–2132. doi: 10.1007/s10067-018-4111-5 [DOI] [PubMed] [Google Scholar]
- 23.Lee CT, Oldham JM. Interstitial pneumonia with autoimmune features: overview of proposed criteria and recent cohort characterization. Clin Pulm Med 2017; 24: 191–196. doi: 10.1097/CPM.0000000000000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Motwee S, Jawdat D, Jehani GS, et al. . Association of HLA-DRB1*15 and HLADQB1*06 with SLE in Saudis. Ann Saudi Med 2013; 33: 229–234. doi: 10.5144/0256-4947.2013.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris DL, Fernando MMA, Taylor KE, et al. . MHC associations with clinical and autoantibody manifestations in European SLE. Genes Immun 2014; 15: 210–217. doi: 10.1038/gene.2014.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chinoy H, Payne D, Poulton K V, et al. . HLA-DPB1 associations differ between DRB1*03 positive anti-Jo-1 and anti-PM-Scl antibody positive idiopathic inflammatory myopathy. Rheumatology 2009; 48: 1213–1217. doi: 10.1093/rheumatology/kep248 [DOI] [PubMed] [Google Scholar]
- 27.Chinoy H, Adimulam S, Marriage F, et al. . Interaction of HLA-DRB1*03 and smoking for the development of anti-Jo-1 antibodies in adult idiopathic inflammatory myopathies: a European-wide case study. Ann Rheum Dis 2012; 71: 961–965. doi: 10.1136/annrheumdis-2011-200182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chinoy H, Salway F, Fertig N, et al. . In adult onset myositis, the presence of interstitial lung disease and myositis specific/associated antibodies are governed by HLA class II haplotype, rather than by myositis subtype. Arthritis Res Ther 2006; 8: R13. doi: 10.1186/ar1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muro M, Mondejar-López P, Moya-Quiles MR, et al. . HLA-DRB1 and HLA-DQB1 genes on susceptibility to and protection from allergic bronchopulmonary aspergillosis in patients with cystic fibrosis. Microbiol Immunol 2013; 57: 193–197. doi: 10.1111/1348-0421.12020 [DOI] [PubMed] [Google Scholar]
- 30.Onengut-Gumuscu S, Chen W-M, Robertson CC, et al. . Type 1 diabetes risk in African-ancestry participants and utility of an ancestry-specific genetic risk score. Diabetes Care 2019; 42: 406–415. doi: 10.2337/dc18-1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anaya J-M, Mantilla RD, Correa PA. Immunogenetics of primary Sjögren's syndrome in Colombians. Semin Arthritis Rheum 2005; 34: 735–743. doi: 10.1016/J.SEMARTHRIT.2004.11.008 [DOI] [PubMed] [Google Scholar]
- 32.Ley B, Newton CA, Arnould I, et al. . The MUC5B promoter polymorphism and telomere length in patients with chronic hypersensitivity pneumonitis: an observational cohort-control study. Lancet Respir Med 2017; 5: 639–647. doi: 10.1016/S2213-2600(17)30216-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caruso C, Candore G, Romano GC, et al. . Immunogenetics of longevity. Is major histocompatibility complex polymorphism relevant to the control of human longevity? A review of literature data. Mech Ageing Dev 2001; 122: 445–462. doi: 10.1016/s0047-6374(00)00255-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01380-2019.Shareable (331.2KB, pdf)