Abstract
Next Generation Sequencing (NGS) using capture or amplicons strategies allows the detection of a large number of mutations increasing the rate of positive diagnosis for the patients. However, most of the detected mutations are Single Nucleotide Variants (SNVs) or small indels. Structural Variants (SVs) are often underdiagnosed in inherited genetic diseases, probably because few user-friendly tools are available for biologists or geneticists to identify them easily. We present here the diagnosis of two brothers presenting a demyelinating motor-sensitive neuropathy: a presumed homozygous c.5744_5745delAT in exon 10 of SACS gene was initially detected, while actually these patients were heterozygous for this mutation and harbored a large deletion of SACS exon 10 in the other allele. This hidden mutation has been detected thanks to the user-friendly CovCopCan software. We recommend to systematically use such a software to screen NGS data in order to detect SVs, such as Copy Number Variations, to improve diagnosis of the patients.
Keywords: Structural Variants, Diagnosis, Charcot-Marie-Tooth, NGS, CovCopCan
1. Introduction
Inherited genetic diseases are due to germline mutations. Thanks to Next Generation Sequencing (NGS), an increasing number of these mutations are detected every day improving patients’ diagnosis. Therefore, molecular diagnosis may influence patients’ care through the choice of adapted treatments, for instance in neurological diseases and particularly in genetic epilepsies [1]. However, to date, the majority of the reported mutations are Single Nucleotide Variants (SNVs) and Structural Variants (SVs) have rarely been described, probably due to the analytic methods used to analyze NGS data, comparing patients’ sequences to a reference one.
In Charcot-Marie-Tooth disease (CMT), the most common hereditary neuropathy characterized by damages of both motor and sensory peripheral nerves, the most frequent mutation involved in this disease is the PMP22 duplication, explaining around 15% of CMT patients. It has been identified by Southern-Blot in 1992 [2]. Since this date, more than other 90 genes have been identified to be involved in this disease and in associated peripheral neuropathies [3]. Most of the detected mutations are SNVs or small indels [4], [5], and Structural Variants (SVs) have rarely been described [6].
NGS techniques allow partial or total sequencing of a patient’s genome. Sequenced libraries can be prepared by capture or by amplicons. Both methods are efficient for the detection of single-nucleotide variants or short indels, however only a few tools are available for the detection of large deletions or duplications, especially with amplicon sequencing data, such as Cov’Cop and CovCopCan [7], [8]. Molecular diagnosis being an essential step of patient care, we believe it is crucial to improve the detection of SVs to increase the rate of positive diagnosis by using several bioinformatics approaches to analyze NGS data.
By presenting the diagnosis of a specific patient harboring a peripheral neuropathy, we show here how it is important to look for all kind of variants to provide an accurate diagnosis to the patients. Indeed one mutation could hide another one, such as Structural Variant.
2. Material and methods
2.1. Patients
Our current study focused on a family with two cases of peripheral neuropathy: Patients A (propositus) and B (affected brother), both with learning disabilities. Patient A is a 19 year-old man, presenting peripheral neuropathy (EMG: abolished sensitive potential and altered motor velocities, with elongated distal latencies and altered F-waves – Median nerve conduction velocity: 38 m/s). He also has pes cavus, progressive cerebellar ataxia and abolished Achilles reflex. His brother, Patient B, is a 9 year-old boy, presenting peripheral neuropathy, exhibiting progressive walking difficulties, fine motor skill disabilities, balance disorder with intermittent falls and Achilles reflex decrease. We accessed to the DNA of the four members of this family: Patients A (propositus) and B (affected brother), and patients C and D, the unaffected father and mother respectively.
2.2. DNA extraction
Blood samples were collected in EDTA tubes after providing informed consent. The protocol was in accordance with the French ethics legislation and the Declaration of Helsinki. Genomic DNA was extracted by standard methods (Illustra DNA Extraction kit BACC3, GEHC).
2.3. Sequencing
NGS strategy was performed on patient A using a 93-genes-custom panel designed for CMT and associated neuropathies diagnosis (Supplementary Table 1). The amplified library was prepared with Ion P1 HiQ Template OT2 200 kit (Ampliseq Custom, Life technologies), sequenced on Proton sequencer (Life technologies), and mapped to the human reference sequence hg19/GHCh37. Mutations of interest were verified by Sanger sequencing using forward and reverse primer pairs.
2.4. Bioinformatics analyses
Variants detected by targeted NGS were annotated using Ion reporter software. They were evaluated with Alamut Mutation Interpretation Software (Interactive Biosoftware, Rouen, France). Databases such as ExAC Genome browser (http://exac.broadinstitute.org), dbSNP135 (National Center for Biotechnology Information [NCBI], Bethesda, Maryland, USA, http://www.ncbi.nlm.nih.gov/projects/SNP/), ClinVar (www.ncbi.nlm.nih.gov/clinvar) and HGMD professional (www.hgmd.cf.ac.uk) were also screened. Cov’Cop and CovCopCan, interactive powerful software, were used to detect Copy Number Variations (CNV) [7], [8]. Briefly, using the read depth value of each amplicon, these software simultaneously analyze all the patients of the run. The algorithm is based on the concept that in common cases, both alleles have to be similarly amplified within each amplicon. Several normalization steps, guided by carefully chosen references amplicons, permit to compute a score for each amplicon. Theoretical score of 1 is the normal case while low (<0.5) or high (>1.5) values respectively reveal deletions or duplications. Cov’Cop and CovCopCan were used with the default settings, with all options active and we defined a minimum threshold of three successive amplicons on the same chromosome to highlight a CNV. For the tested patient, mean read depth value of the 93 tested genes was 1624 X and the mean value for the 78 amplicons covering SACS was 660 X (minimum: 64 X and maximum: 2603 X). The coverage of SACS was 100%.
2.5. Quantitative real-time Q-PCR
Primers were designed in exon 9 and 10 of SACS gene and in exon 1 of Albumin gene, chosen as reference gene. Rotor-Gene SYBR Green PCR Kit (400) (©QIAGEN) was used following the standard protocol. Reactions were performed on the Corbett Rotor-Gene 6000 Machine (© QIAGEN). The Ct values of each Real-Time reaction were normalized, using Albumin as endogenous control gene, and then compared to the normalized Ct values of three control samples.
3. Results
3.1. Detection of presumed homozygous mutation in SACS gene
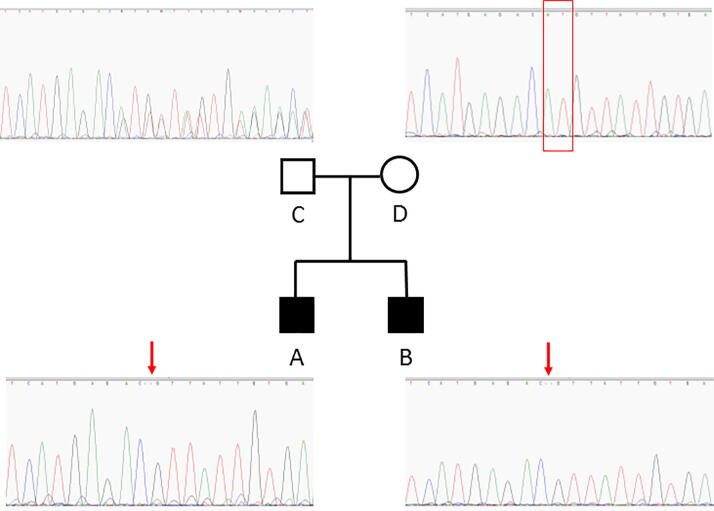
Targeted NGS of Patient A DNA, revealed the presence of mutation c.5744_5745delAT in exon 10 of SACS gene. This mutation results in a frameshift, leading to truncated protein p.His1915Argfs*19. This mutation is very rare (1/125568 in ExAc) and is predicted as pathogenic in ClinVar. The normal allele has not been sequenced suggesting the presence of mutation c.5744_5745delAT at homozygous state, confirmed by Sanger sequencing (Fig. 1-A). No other rare Single Nucleotide Variant or short InDels has been detected in this patient. Mutations in SACS have already been reported to be responsible of spastic ataxia of Charlevoix-Saguenay, an early-onset neurodegenerative disease. The transmission follows an autosomal recessive manner and two mutations are expected in a patient.
Fig. 1.
Family tree associated with Sanger sequencing of SACS exon 10.
3.2. Problematic familial segregation
Sanger Sequencing surrounding the mutation was then performed on the affected brother B and on father C and mother D, both asymptomatic. As expected, only the c.5744_5745delAT mutation was detected in patient B (Fig. 1-B), heterozygous mutation was detected in the father (Fig. 1-C), but no mutation was detected in the mother (Fig. 1-D), showing that only the normal allele could be sequenced in this case and suggesting the potential presence of a deletion of this area, that had to be defined more precisely.
3.3. Detection of Structural Variant using CovCopCan
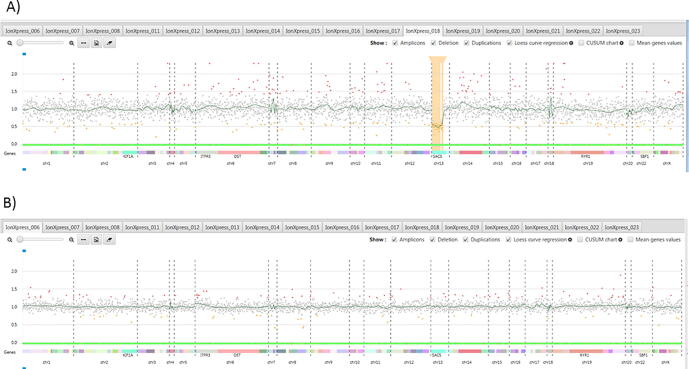
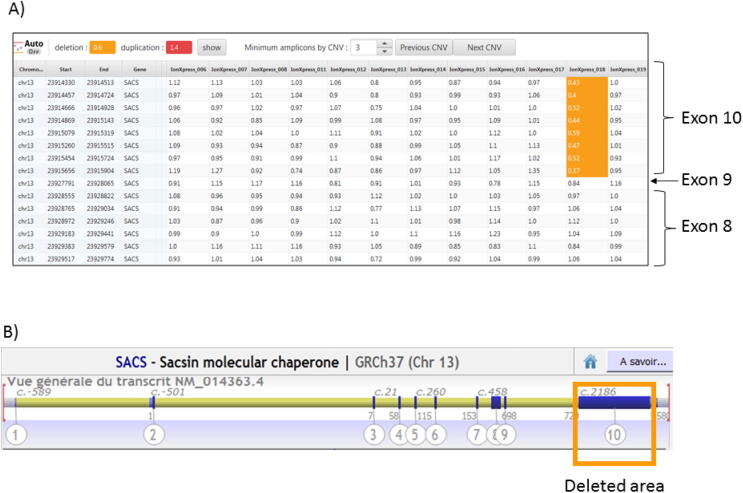
We then used the new CovCopCan software, a user-friendly tool, based on the read-depth analysis of NGS data, that allows the rapid and easily detection of Structural Variants (SVs) in inherited diseases but also in Cancer [8]. Using CovCopCan, we identified easily the presence of a heterozygous deletion in SACS gene (Fig. 2A) in comparison to a control (Fig. 2B). We could also define, thanks to CovCopCan, the boundaries of the deletion. Indeed, we could see that amplicons on exon 9 were not deleted (value around 1 in CovCopCan) and that amplicons on exon 10 were deleted (value around 0.5) (Fig. 3A and 3B). This new SV has never been described (SV-GnomAD) and it could lead to the production of a truncated protein.
Fig. 2.
Visualization of CovCopCan results. Each dot corresponds to an amplicon. The amplicons are distributed on the x-axis according to their genomic position. The y-axis corresponds to the normalized values. Grey dots indicate a “normal” value, whereas red or orange dots indicate duplicated and deleted amplicons, respectively. A) Patient A analysis in which one can see a partial deletion of SACS gene, highlighted in orange; B) A control sample analysis in which no deletion or duplication can be detected. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Detection of SACS Exon 10 deletion – A) Overview of CovCopCan table results, values around 0.5 (highlighted in yellow) show a deletion of this area in patient A (X-press 18), but not in the other patients tested in the same NGS Run; B) Alamut Overview of SACS gene and of the deleted area. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Confirmation of Structural Variant and correct segregation
We confirmed the presence of exon 10 deletion in Patient A by Real-Time qPCR. We investigated the other individuals of the family: as expected, the affected brother and the mother presented the exon 10 deletion, but not the father. Normal value has been obtained for exon 9 in all the family members confirming that the deletion does not overlap the first exons of SACS gene.
4. Discussion
We showed here the importance of complete bioinformatics analysis of NGS data to perform an accurate diagnosis. In this family, the presumed homozygous mutation c.5744_5745delAT has been detected in SACS gene by classical variants detection software such as Ion Reporter in Patient A. This conclusion was actually not accurate. Indeed, Patient A presents in fact one allele with c.5744_5745delAT mutation and one allele with a Structural Variation: a large deletion of SACS exon 10. This SV has been detected thanks to CovCopCan Software, a new user-friendly tool allowing the detection of Copy Number Variations (CNVs) in NGS data generated from amplicons sequencing.
Few user-friendly tools are easily usable for biologists and geneticists to detect SVs, such as CNVs, in amplicons sequencing data. In addition to CovCopCan, ExomeDepth [9], IonCopy [10], DeviCNV [11], Cov’Cop [7], are also available and have to be tested by the users to define their preferred tools to detect these SVs. We believe tools detecting SVs, such as CNVs, have to be used systematically on NGS data analysis in addition to the classical variants detection tools highlighting only Single Nucleotide Variants and small indels.
However, it is important to notice that these tools using the read-depth to detect CNVs, will not be able to detect easily SVs such as inversion or translocation. To date, we do not know the percent of CNVs responsible for inherited diseases in comparison to others SVs, such as inversion or translocation, because all of them were poorly detected until now. In addition, we do not know the percent of pathogenic CNVs in comparison to SNVs or some indels.
Nevertheless, we estimated in our cohort of 695 CMT patients analysed by NGS using an amplicon targeted sequencing panel of 93 genes of Charcot-Marie-Tooth disease and associated neuropathies, that CNVs were present in 107 patients (15.4% of the patients), showing a large amount of CNVs in our cohort. Twenty-eight were small deletions (3–6 amplicons), 10 were large deletions, 49 were small duplications and 20 were large duplications. We now investigate the pathogenicity of these new SVs and we presented one of them in this article: a large deletion of SACS exon 10, which appeared to be pathogenic.
Currently, in the diagnosis of peripheral neuropathy, we reach between around 40% of positive diagnosis using only classical variants detection software highlighting SNVs or some indels. By using, in addition, tools such as CovCopCan, allowing the detection of SVs, such as CNVs, we hope to increase the rate of positive diagnosis for our patients. Indeed, to date, a patient with a homozygous SV was not diagnosed positively using the classical tools, this was also the case for male patients harboring, for example, a deletion on X chromosome gene, while these kind of SVs could be pathogenic mutations. Thanks to CovCopCan, or equivalent tools, all these patients will be diagnosed positively. Of course, CovCopCan can detect CNVs on all the inherited diseases. This tool works on data generated from Ion Designer (Life Technologies, CA, USA) as well as that from Illumina DesignStudio (Illumina Inc., San Diego, CA, USA). The user-friendly interface associated with our 2D visualization facilitates data exploration.
5. Conclusion
Structural Variants are probably underdiagnosed and should be more looked for to improve inherited diseases diagnosis. It is crucial for physicians to be aware that a potential homozygous variation can hide a Structural Variant. In addition, if no pathogenic SNV was found by NGS sequencing, SVs should be systematically investigated. Detection of such variants would then help to better understand the physiopathology involved in inherited diseases, in order to develop, in fine, therapeutic approaches.
6. Author statement
All authors have reviewed and edited the manuscript.
FM: participated to the bioinformatics study, performed experiments and proofread the manuscript
PAF: participated to the bioinformatics study, participated to the writing and proofread the manuscript
IP: participated to the bioinformatics study and performed experiments
SB: performed experiments
PD: participated to the bioinformatics study and proofread the manuscript
MH: reported clinical study
FF: performed a critical revision of the manuscript
FS: contributed to the interpretation of the results and proofread the manuscript
CM: planned the experiments, contributed to the interpretation of the results and proofread the manuscript.
ASL: initiated the work, designed and planned the experiments, analyzed the results and wrote most of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to thank the Sequencing GenoLim team of the Limoges CHU, especially Emilie Guerin, Valentin Tilloy and Sophie Alain.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.07.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Striano P., Vari M., Mazzocchetti C., Verrotti A., Zara F. Management of genetic epilepsies: from empirical treatment to precision medicine. Pharmacol Res. 2016;107:426–429. doi: 10.1016/j.phrs.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Timmerman V., Nelis E., Van Hul W., Nieuwenhuijsen B.W., Chen K.L., Wang S. The peripheral myelin protein gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat Genet. 1992;1(3):171–175. doi: 10.1038/ng0692-171. Erratum in: Nat Genet 1992 Sep;2(1):84. [DOI] [PubMed] [Google Scholar]
- 3.Timmerman V., Strickland A.V., Züchner S. Genetics of Charcot-Marie-Tooth (CMT) disease within the frame of the human genome project success. Genes (Basel) 2014;5(1):13–32. doi: 10.3390/genes5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerat J., Cintas P., Beauvais-Dzugan H., Magdelaine C., Sturtz F., Lia A.S. A complex homozygous mutation in ABHD12 responsible for PHARC syndrome discovered with NGS and review of the literature. J Peripher Nerv Syst. 2017;22(2):77–84. doi: 10.1111/jns.12216. [DOI] [PubMed] [Google Scholar]
- 5.Lerat J., Magdelaine C., Roux A.F., Darnaud L., Beauvais-Dzugan H., Naud S. Hearing loss in inherited peripheral neuropathies: molecular diagnosis by NGS in a French series. Mol Genet Genomic Med. 2019;7(9) doi: 10.1002/mgg3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mortreux J., Bacquet J., Boyer A., Alazard E., Bellance R., Giguet-Valard A.G. Identification of novel pathogenic copy number variations in Charcot-Marie-Tooth disease. J Hum Genet. 2020;65(3):313–323. doi: 10.1038/s10038-019-0710-5. [DOI] [PubMed] [Google Scholar]
- 7.Derouault P., Parfait B., Moulinas R., Barrot C.C., Sturtz F., Merillou S., Lia A.S. 'COV'COP' allows to detect CNVs responsible for inherited diseases among amplicons sequencing data. Bioinformatics. 2017;33(10):1586–1588. doi: 10.1093/bioinformatics/btx017. [DOI] [PubMed] [Google Scholar]
- 8.Derouault P., Chauzeix J., Rizzo D., Miressi F., Magdelaine C., Bourthoumieu S. CovCopCan: an efficient tool to detect Copy Number Variation from amplicon sequencing data in inherited diseases and cancer. PLoS Comput Biol. 2020;16(2) doi: 10.1371/journal.pcbi.1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plagnol V., Curtis J., Epstein M., Mok K.Y., Stebbings E., Grigoriadou S. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28(21):2747–2754. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budczies J., Pfarr N., Stenzinger A., Treue D., Endris V., Ismaeel F. Ioncopy: a novel method for calling copy number alterations in amplicon sequencing data including significance assessment. Oncotarget. 2016;7(11):13236–13247. doi: 10.18632/oncotarget.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y., Nam S.H., Park K.S., Kim Y., Kim J.W., Lee E. DeviCNV: detection and visualization of exon-level copy number variants in targeted next-generation sequencing data. BMC Bioinf. 2018;19(1):381. doi: 10.1186/s12859-018-2409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.