Abstract
RNA-Seq transcriptome data for the wild type and phoP mutant strains of Pectobacterium versatile is described. P. versatile is a recently introduced name for a species of plant pathogenic bacteria that unites a group of strains previously embedded within the Pectobacterium carotovorum clade [1,2]. Little detail is available about how this pathogen adapts to changing environmental conditions, including those within its host plant. The PhoP/PhoQ two-component system is an important sensor responding to several stimuli and is present in most species of enteric bacteria. It usually controls large regulons, which vary greatly even between closely related species [3]. This dataset enables the discovery of the genes under direct or indirect transcriptional control by PhoP in P. versatile and should help to understand the physiology of this plant pathogen.
Keywords: Pectobacterium versatile, RNA sequencing, Transcriptome, PhoP, PhoQ
Specifications Table
| Subject | Biochemistry, Genetics and Molecular Biology (General) |
| Specific subject area | Molecular Biology |
| Type of data | Transcriptome sequences |
| How data were acquired | Illumina HiSeq 2500 sequencing platform |
| Data format | Raw Illumina data in FastQ format |
| Parameters for data collection | Wild type and phoP mutant Pectobacterium versatile cultures grown in synthetic media to mid-log phase |
| Description of data collection | mRNA was extracted from eight independent cultures (four wild type and four mutant) and subjected to cDNA sequencing |
| Data source location | Belarusian State University, Minsk, Belarus |
| Data accessibility | Repository name: NCBI Sequence Read Archive Data identification number: PRJNA627079 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA627079 |
Value of the Data
-
•
This dataset is, to our knowledge, the first RNA-seq one for P. versatile and will be valuable for the Pectobacterium sp. research community for characterizing the highly divergent regulon controlled by the global transcription factor PhoP.
-
•
The data may be useful for researchers studying the adaptation of P. versatile to changing environment, including plant colonisation.
-
•
The data can be used to define PhoP regulon and to establish PhoP role in the control of P. versatile virulence.
-
•
This dataset can be used to study operon organisation in P. versatile.
1. Data description
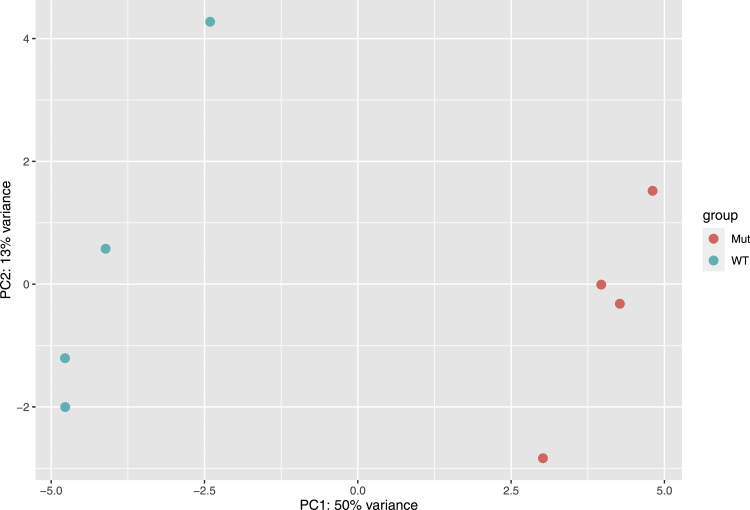
The dataset contains sequencing data obtained through the transcriptome sequencing of two P. versatile strains: JN42 and its phoP mutant derivative UK1 grown in the synthetic medium supplemented with polygalacturonic acid. Samples for transcriptome profiling were collected at the exponential growth phase. FASTQ files were deposited in NCBI Sequence Read Archive and are accessible through the BioProject PRJNA627079. Information about bacterial culture samples, statistics of sequence reads and sequence coverage data is shown in Table 1. PCA plot of RNA-seq data presented in Fig. 1 demonstrates the variance between sample groups and sample replicates according to gene expression levels. Each dot in the Fig. 1 indicates a particular sample.
Table 1.
Details of RNA-seq data submitted to the NCBI Sequence Read Archive (SRA).
| Reads |
|||||
|---|---|---|---|---|---|
| Strain | Sample ID | Biosample accession no. | SRA accession no. | Total number | Mapped to reference |
| JN42 (wild type) | wt_rep1 | SAMN14651075 | SRR11581681 | 11006472 | 99.16% |
| wt_rep2 | SAMN14651076 | SRR11581680 | 9867857 | 99.08% | |
| wt_rep3 | SAMN14651077 | SRR11581679 | 11415728 | 99.18% | |
| wt_rep4 | SAMN14651078 | SRR11581678 | 11835173 | 99.15% | |
| UK1 (phoP mutant) | phoP_rep1 | SAMN14651079 | SRR11581677 | 7841926 | 99.12% |
| phoP_rep2 | SAMN14651080 | SRR11581676 | 10640935 | 99.11% | |
| phoP_rep3 | SAMN14651081 | SRR11581675 | 10833222 | 99.12% | |
| phoP_rep4 | SAMN14651082 | SRR11581674 | 10780714 | 99.07% | |
Fig. 1.
Principal component analysis (PCA) of the general transcriptome characteristics. The first principal component (PC1) accounted for 50% and the second principal component (PC2) for 13% of the total variance in the dataset. Legend description: “WT”– samples of cultures of P. versatile strain JN42, wild type; “Mut” – samples of cultures of the UK1 strain, phoP insertional mutant of JN42.
2. Experimental design, materials and methods
2.1. Bacterial strains and growth conditions
P. versatile strains JN42 (wild type) and UK1 (phoP insertional mutant of JN42) were used and grown in minimal medium composed of K2HPO4 (10,5 g/l), KH2PO4 (4,5 g/l), (NH4)2SO4 (1 g/l), sodium citrate (0,6 g/l), 0.5 mM MgSO4, 10 µM CaCl2, 0.2% glycerol and 0.5% Sodium polypectate (Sigma). For RNA isolation, four separate cultures of each strain were grown at 28 °C with aeration (180 rpm) to mid-log phase (OD600 = 0.4).
2.2. RNA isolation, cDNA library preparation and sequencing
Bacterial cells in mid-log phase cultures were fixed by adding phenol/ethanol (1/20 v/v) solution to 20% and kept on ice for 30 min. The fixed cells were harvested (8000 g, 5 min, 4 °C) and resuspended in 1 mL of ExtractRNA Reagent (Evrogen, Russia) and the subsequent procedures were performed according to the manufacturer's instructions. Residual DNA was eliminated by treatment of RNA samples with DNAse I (Thermo Fischer, USA). Total RNA was processed using Ribo-Zero rRNA Removal Kit (Gram-Negative Bacteria) (Illumina, USA) and NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA) according to manufacturer's instructions. The quality and quantity of the cDNA libraries during processing before sequencing were monitored using the Agilent 2100 Bioanalyser (Agilent Technologies, USA) and CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, USA). Sequencing was conducted by a HiSeq 2500 Sequencing System (Illumina) at Joint KFU-Riken Laboratory, Kazan Federal University (Kazan, Russia).
2.3. Sequence QC and filtering
84,222,027 reads were obtained in total with a length of 57 nucleotides (Table 1). FastQC software (Version 0.11.5) [4] was used to assess the quality of the raw Fastq files and clean reads. Raw reads were filtered using BBDuk (v. 37.23, http://jgi.doe.gov/data-and-tools/bb-tools/) to remove Illumina adapters, NEB indexes and to quality-trim right end to Q20 (ktrim 1⁄4 r k 1⁄4 23 mink 1⁄4 11 hdist 1⁄4 1 tpe tbo minlen 1⁄4 25 qtrim 1⁄4 r trimq 1⁄4 20). Thereafter, the rRNA reads were eliminated by using SortMeRNA v2.1 program [5]. DESeq2 [6] was used to assess variance between sample groups and sample replicates using principal component analysis (PCA). PCA plot shown in the Fig. 1 demonstrates the overall quality of our sample collection, library preparation, and sequencing.
2.4. Reads alignment to the reference genome
The reads were mapped onto the genome sequence of P. versatile strain 3-2 (GenBank accession CP024842) which is the wild type parent of the laboratory strain JN42. BWA version 0.7.16a [7] was used to build the index of the reference genome and align the reads to the reference genome with default aligner parameters. SAM files of alignments created by BWA were converted to sorted BAM files with SAMtools v. 1.10 [8] using samtools sort command. Reads mapping statistics are presented in Table 1.
Author's contribution and ethics statement
Natalia Gogoleva: Investigation, Methodology. Uljana Kravchenko: Investigation, Software. Yevgeny Nikolaichik: Conceptualization, Data curation, Writing - Original draft preparation, Review & Editing. Yuri Gogolev: Conceptualization, Supervision, Writing - Original draft preparation, Review & Editing, Funding acquisition. All ethical requirements for such studies were observed in the preparation of the publication. The work was not related to the use of human objects and did not include experiments with animals.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
This work was supported by grants from Russian Foundation for Basic Research (project no. 18-54-00021) and Belarusian Republican Foundation for Basic Research. The RNA-Seq analysis was supported by the Russian Science Foundation (project no. 17-14-01363). The cDNA-library preparation was worked out at the financial support of the Ministry of Science and Higher Education of the Russian Federation (grant no. 075-15-2019-1881). DNA sequencing was performed within the frameworks of the government assignment for FRC Kazan Scientific Center of RAS. The study was carried out by using the equipment of the CSF-SAC FRC KSC RAS.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106123.
Appendix. Supplementary materials
References
- 1.Shirshikov F.V., Korzhenkov A.A., Miroshnikov K.K., Kabanova A.P., Barannik A.P., Ignatov A.N., Miroshnikov K.A. Draft genome sequences of new genomospecies “Candidatus Pectobacterium maceratum” strains, which cause soft rot in plants. Genome Announc. 2018;6:e00260. doi: 10.1128/genomeA.00260-18. -18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Portier P., Pédron J., Taghouti G., Fischer-Le Saux M., Caullireau E., Bertrand C., Laurent A., Chawki K., Oulgazi S., Moumni M., Andrivon D., Dutrieux C., Faure D., Hélias V., Barny M.-A. Elevation of Pectobacterium carotovorum subsp. odoriferum to species level as Pectobacterium odoriferum sp. nov., proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., emended description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 2019;69:3207–3216. doi: 10.1099/ijsem.0.003611. [DOI] [PubMed] [Google Scholar]
- 3.Perez J.C., Shin D., Zwir I., Latifi T., Hadley T.J., Groisman E.A. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 2009:5. doi: 10.1371/journal.pgen.1000428.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews S. Babraham Institute; Cambridge, United Kingdom: 2011. Fast QC: a Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed 30 April 2020) [Google Scholar]
- 5.Kopylova E., Noé L., Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 6.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(550):1e21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM, ArXiv:1303.3997 [q-Bio]. (2013). http://arxiv.org/abs/1303.3997(accessed July 20, 2018).
- 8.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.