Summary
Glioblastoma (GBM) may arise from astrocytes through a multistep process involving a progressive accumulation of mutations. We explored whether GBM-derived extracellular vesicles (EVs) may facilitate neoplastic transformation and malignant growth of astrocytes. We utilized conditioned media (CM) of cultured glioma cells, its sequential filtration, diverse cell-based assays, RNA sequencing, and metabolic assays to compare the effects of EV-containing and EV-depleted CM. GBM EVs facilitated the neoplastic growth of pre-transformed astrocytes but not normal human or mouse astrocytes. They induced proliferation, self-renewal, and colony formation of pre-transformed astrocytes and enhanced astrocytoma growth in a mouse allograft model. GBM EVs appear to reprogram astrocyte metabolism by inducing a shift in gene expression that may be partly associated with EV-mediated transfer of full-length mRNAs encoding ribosomal proteins, oxidative phosphorylation, and glycolytic factors. Our study suggests an EV/extracellular RNA (exRNA)-mediated mechanism that contributes to astrocyte transformation via metabolic reprograming and implicates horizontal mRNA transfer.
Subject Areas: Biological Sciences, Cell Biology, Cancer
Graphical Abstract
Highlights
-
•
Extracellular vesicles (EVs) shed by glioma cells are taken up by astrocytes
-
•
Glioma EVs facilitate astrocyte transformation and tumor growth
-
•
EVs reprogram glycolysis and oxidative phosphorylation of transformed astrocytes
-
•
mRNAs coding ribosomal proteins and other factors are dispersed via EVs
Biological Sciences; Cell Biology; Cancer
Introduction
Extracellular vesicles (EVs) released by various types of cells attract considerable interest owing to their involvement in cell-to-cell communication via “horizontal” RNA and protein transfer and has potential as biomarkers (Thery et al., 2018). Two principal categories of extracellular packaging complexes, plasma membrane-derived microvesicles (MVs, with a diameter of 0.20–0.80 μm, that constitute a major part of large EVs), and endosome-origin exosomes (Exos, 0.02–0.20 μm, that represent the majority of small EVs), mediate this transfer along with extracellular non-vesicular ribonucleoprotein complexes (exRNPs) (Vickers et al., 2011; Valadi et al., 2007; Russell et al., 2019; Graner, 2019). The RNA-encapsulating EVs have been originally isolated from patient-derived glioma cells (Valadi et al., 2007; Skog et al., 2008), and since that time, glioma served as a fruitful model for the investigation of EV release, cargo, functions in intercellular communications, and biomarker potential. It has been recognized that extracellular RNA (exRNA) carried by EVs plays important roles in intercellular communication, opening up a new fundamental field of extracellular RNA biology for the extensive ongoing investigation.
High-grade gliomas, the most common class of primary malignant brain tumors in adults, are notorious in their interactions with resident cells of the brain microenvironment. Among gliomas, evidenced by microvascular proliferation and pseudopalisading necrosis, glioblastoma (GBM) is the most malignant type of all, resulting in dismal prognosis for patients. The median survival of patients with GBM remains 15–17 months despite the highly aggressive protocols of standard care (Wen et al., 2020). As the most critical feature contributing to this lack of therapeutic benefits, malignant gliomas nearly invariably recur in the resection margin and progress into higher-grade malignancy (Hochberg and Pruitt, 1980). It is thought that incomplete resection, as well as the presence of therapy-resistant glioma initiating stem cells, are the root cause of this recurrence; however, the broader mechanism may include the supportive pro-tumorigenic environment established by the tumor before resection. A related critical question that concerns the origin of glioma cells is also incompletely investigated, with astrocytes, neural stem cells, and oligodendrocyte precursor cells emerging as the major candidates (Cahill and Turcan, 2018). Astrocytes, the native cells of the brain that are phenotypically most similar to the bulk of glioma, present potentially the amplest and most accessible source of glioma-initiating cells. A variety of genes, such as P53, TERT, and RAS that are often mutated or activated in glioma can transform astrocytes to neoplastic cells in vitro and in vivo. These data led us to question whether the communication between GBM and surrounding reactive astrocytes can promote the process of transformation and thereby feed the tumor with newly recruited neoplastic cells.
Several studies have suggested that glioma-derived EVs are taken up by various cells of the brain microenvironment, including astrocytes, microglia, and microvascular cells, where they appear to modulate cellular phenotypes toward the more tumor-promoting cells and may thereby support the tumor progression or recurrence (Gao et al., 2020; Oushy et al., 2018; Bian et al., 2019; Hallal et al., 2019; Broekman et al., 2018; Skog et al., 2008; Nakano et al., 2015; Taheri et al., 2018). For example, GBM-derived EVs taken up by tumor-associated microglia promote the immunosuppressive properties of microglia, partly via RNA-mediated mechanisms (Abels et al., 2019, Van Der Vos et al., 2016). GBM-derived EVs also contribute to the tumor-associated angiogenesis by reprogramming brain endothelial cells toward malignant vasculature, a hallmark of the GBM (Lucero et al., 2020).
Here, we investigated the effects of GBM-derived EVs on normal and pre-transformed astrocytes and demonstrated the EV-mediated facilitation of transformation in vitro and in vivo. For this aim, we developed a stringent protocol based on the growth of astrocytes in the complete GBM-conditioned media (CM) or, identical, except EV-depleted CM. This protocol enabled the assessment of the specific function of EV without the need of concentration, quantification, or technically challenging normalization based on vesicles derived from other sources. Monolayer glioma cells and patient-derived low-passage tumorigenic glioma spheroid cultures that represent the most therapy-resistant stem-like cell population have been utilized as donor cells. As recipient cells, we employed human and mouse low-passage astrocytes, including cultures of pre-transformed cells with oncogenic viruses and cells derived from transgenic animals. It is established that, despite genetic and expression heterogeneity of GBM, three major signaling pathways are commonly dysregulated in GBM and may drive gliomagenesis: Ras/RTKs (in 88% of GBM), P53 (87%), and RB1 (78%) (Cancer Genome Atlas Research Network, 2008). In addition, the promoter of the telomerase reverse transcriptase (TERT) is most frequently mutated in GBM (in 80%–90% cases), leading to telomerase reactivation (Killela et al., 2013; Cancer Genome Atlas Research Network, 2008). Accordingly, we established activated Ras, TERT, and SV40 large T expressing lentiviral vectors, able to transform normal human astrocytes (NHA) via activation of Ras, telomerase, or simultaneous inactivation of p53 and pRb, correspondingly. The conditions were chosen such that, although most cells were transduced by each lentiviral vector, only a small fraction were transformed, allowing for screening for cooperating factors. The transformed cells had morphological and growth features of high-grade glioma, including expanded life span, growth in soft agar and, most importantly, were tumorigenic in mice (Rich et al., 2001). We also utilized mouse transgenic astrocytes bearing characteristic mutation in EGFR (VIII), deletions in CDKN2A (INK4a/ARF) and PTEN genes that generate a fully penetrant, rapid-onset high-grade malignant glioma with prominent pathological and molecular characterization of GBM (Zhu et al., 2009). Using NHA, as well as such pre-transformed astrocytes as the recipient cells, we demonstrate that GBM EVs promote their proliferation and self-renewal ability and reprogram astrocyte metabolism toward a glycolytic tumor state. Although these effects were evident in pre-transformed astrocytes, EVs failed to induce malignant conversion of normal or untransformed astrocytes.
GBM exRNA comprises heterogeneous RNA species (Gyuris et al., 2019; Wei et al., 2017). Although these transcripts are largely non-protein-coding and fragmented, some full-length mRNAs are also found encapsulated into EVs (Wei et al., 2017). Here, we analyze the repertoire of EV-associated mRNAs and demonstrate that they are enriched in mRNAs encoding ribosomal proteins and factors involved in oxidative phosphorylation and glycolysis. We propose that direct transfer of these mRNAs from glioma to reactive astrocytes may contribute to their metabolic reprogramming and promote neoplastic transformation within the brain microenvironment.
Results
GBM-Derived EVs Facilitate Astrocyte Transformation and Tumor Growth In Vivo
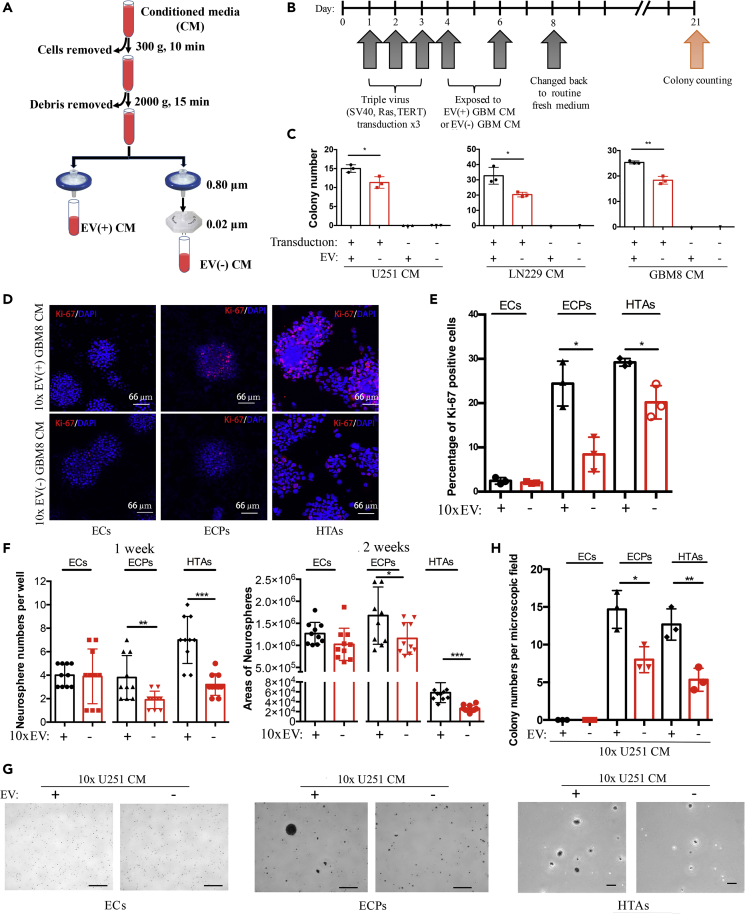
To overcome one of the major limitations of studying EV functions, e.g., co-isolation of secreted EVs with extravesicular complexes and cytokines, we used a sequential filtration (SF) protocol (Wei et al., 2017), which has the advantages of low pressure on EVs, better separation between the EVs and RNPs, and higher RNA yield and reproducibility (Figure 1A). In contrast to EV(+) CM, the EV(−) CM served as the negative control produced by the additional filtration through 0.02-μm pores that removed EVs from the flow-through. Similarly prepared EV(+) and EV(−) fractions of the fresh media allowed us to control for unspecific effects. The media conditioned over 2 days were harvested from monolayer glioma cells or spheroid cultures, and the matching pairs of EV(+) and EV(−) CM samples were collected. We first investigated the role of GBM EVs in astrocyte transformation (Figure 1B), using a combination of three oncogenic viruses (SV40, RasG12V, TERT) as the facilitator of transformation (Rich et al., 2001). Exposure of human astrocytes to glioma-derived EV(+) CM at the early stages of transformation promoted the process, as indicated by the increased number of colonies formed, and compared with the cultures exposed to EV(−) CM (Figure 1C). An additional set of colonies formed by HTAs cultured in either EV(+) or EV(−) CM exhibited the same trend (Figure S1A). No effect had EV(+) CM on normal untransformed astrocytes (Figure 1C). Also, no difference was observed between the number of colonies formed by HTA in EV(+) and EV(−) fresh medium (Figure S1B), indicating that astrocyte transformation was, indeed, induced by the GBM-derived EVs. Transmission electron microscopy confirmed the presence of EVs in EV(+) CM and the lack of thereof in EV(−) CM (Figure S1C, upper panel). Furthermore, based on the Nanoparticle Tracking Analysis (NTA) quantification of EVs in the CM (Figure S1C, lower panel), we estimated that exposure to 109 EVs in 100 μL was sufficient to enhance the transformation of recipient astrocytes plated at 100 cells/well in 96-well format, resulting in the formation of six neurospheres per well on average. Of note, fluorescent microscopy of HTA cultures exposed to EV(+) CM harvested from PalmGFP+ GBM cells demonstrated that nearly 100% of the cells were EV positive within 2–6 h (Figure S1D), indicating the efficient vesicle uptake.
Figure 1.
GBM-derived EVs Promote Transformation of Astrocytes
(A) Schematics of the preparation of EV(+) and EV(−) conditioned medium (CM).
(B) Experimental design and timeline of the colony formation assay. Primary human astrocytes (pHAs) were pre-transformed with three oncogenic viruses (SV40, RasG12V, TERT) and then treated with glioma-derived EV(+) or EV(−) CM, resulting in the growth of single-cell-derived colonies.
(C) Quantification of the colonies demonstrates that treatment with EV(+) CM from U251 (P = 0.025), LN229 (P = 0.020), and GBM8 glioma cells (P = 0.002) promotes colony formation in oncogenes-induced cultures but not in cultures of naive pHAs. N = 3 wells per group.
(D) Fluorescent imaging of Ki-67-labeled (red) and DAPI-labeled (blue) EGFRvIII; CDKN2A-/-; and EGFRvIII; CDKN2A-/-; PTEN-/- mouse astrocytes (ECs and ECPs, correspondingly) and HTAs, treated with the 10x concentrated GBM8 CM, complete or EV-depleted (confocal microscopy; scale bar, 66 μm).
(E) Quantification of the Ki-67-positive cells shown in (D) demonstrates that glioma EVs enhance the proliferation of ECP and HTA (P = 0.012 and 0.016, respectively) but not EC cultures. N = 3 wells per group.
(F) Monitoring growth of 3D neurospheres (number, left panel, and size, right panel) indicates that self-renewal capacity of ECP and HTA cells (for count: P = 0.008 and 0.001, respectively; for size: P = 0.041 and 0.0002, respectively), but not EC cells, is increased by the GBM8 EV(+) CM. N = 3 wells per group. GBM8 CM most compatible with the neurosphere formation assay was utilized.
(G) Representative images of colonies grown by EC, ECP, and HTA cells in soft agar, in either 10x EV(+) or 10x EV(−) CM in 2 weeks (scale bar, 500, 500, 100 μm in ECs, ECPs, and HTAs panels, respectively). U251 CM most compatible with the soft agar assay was utilized.
(H) Quantification of colony numbers per microscopic field indicates that glioma EVs promote growth of ECP (P = 0.019) and HTA (P = 0.008) cells in soft agar. N = 3 wells per group.
∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; two-tailed t test. Data are represented as mean ± SEM. See also Figure S1.
To further investigate whether GBM EVs affect the proliferation of pre-transformed astrocytes, Ki-67 staining was quantified in cultures treated with either EV(+) or EV(−) CM. In addition to HTAs, we utilized mouse EGFRvIII; CDKN2A-/- (EC) and EGFRvIII; CDKN2A-/-; PTEN-/- (ECP) astrocytes, derived from the transgenic animals bearing characteristic GBM mutations. The rate of Ki-67-positive cells was increased by GBM8 EV(+) CM in ECP and HTA but not in EC cultures (Figures 1D and 1E). This was despite the efficient EV uptake by all three cell types (Figure S1D). We further investigated the effects of EVs on the self-renewal capacity of recipient astrocytic cells using the neurosphere formation assay. GBM-derived EV(+) CM significantly increased the number and size (Figures 1F and S1E) of spheroids formed by the ECP and HTA cells, whereas the ECs showed no difference. Lastly, GBM EV(+) CM increased the growth of ECP and HTA cells in soft agar (Figures 1G, 1H, and S1F), whereas, again, have not affected the EC cultures. Collectively, these data indicate that GBM EVs facilitate the transformation of astrocytes in the permissive oncogenic environment.
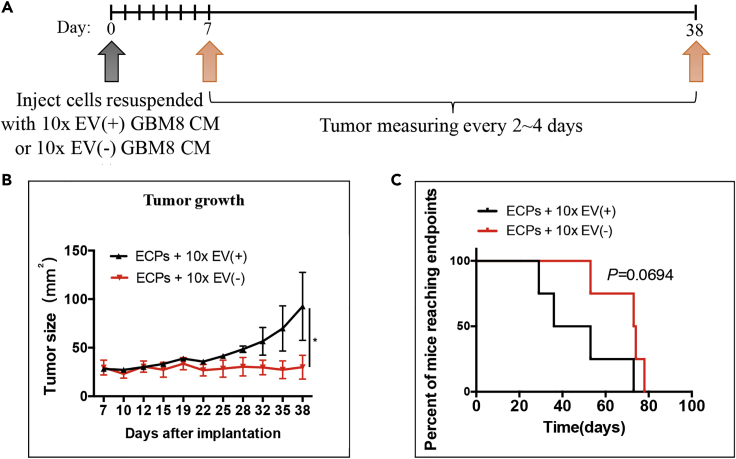
To examine the effects of GBM EVs on glioma formation and growth in vivo, we utilized mouse transgenic EC and ECP cells. It has been previously demonstrated that double-transgenic EC cells were not tumorigenic, whereas ECP allografts produce malignant glioma. EC and ECP cells resuspended in GBM8 10x EV(+) or 10x EV(−) CM were implanted into the flanks of the nude mice, and tumor growth was monitored by caliper every 2–4 days (Figure 2A). Glioma EVs stimulated the growth of the ECP allografts, as compared with the growth of ECPs treated with EV(−) CM (Figure 2B). The mice injected with EV-treated ECPs exhibited shortened survival, with a marginal significance (Figure 2C). Neither EV(+) nor EV(−) CM-treated ECs were tumorigenic in nude mice (Figure S2).
Figure 2.
GBM EVs Facilitate Tumorigenesis of Transformed Astrocytes In Vivo
(A) Experimental design and timeline of the in vivo experiment. ECP cells resuspended with either concentrated EV(+) or EV(−) CM were inoculated subcutaneously to both flanks of immuno-deficient mice, and xenograft growth monitored.
(B) The mice injected with ECPs resuspended in EV(+) CM formed tumors faster and exhibited elevated growth rate relative to animals injected with ECPs resuspended in EV(−) CM (N = 8 injections per condition, P < 0.05).
(C) Kaplan-Meier analysis of mice injected with ECPs resuspended in EV(+) or EV(−) CM were performed (N = 4 mice per condition). Endpoint: death or tumor diameter >2.0 cm.
∗P < 0.05; Kaplan-Meier test. Data are represented as mean ± SEM. See also Figure S2.
GBM EVs Reprogram Oncogenic Metabolism of Transformed Astrocytes
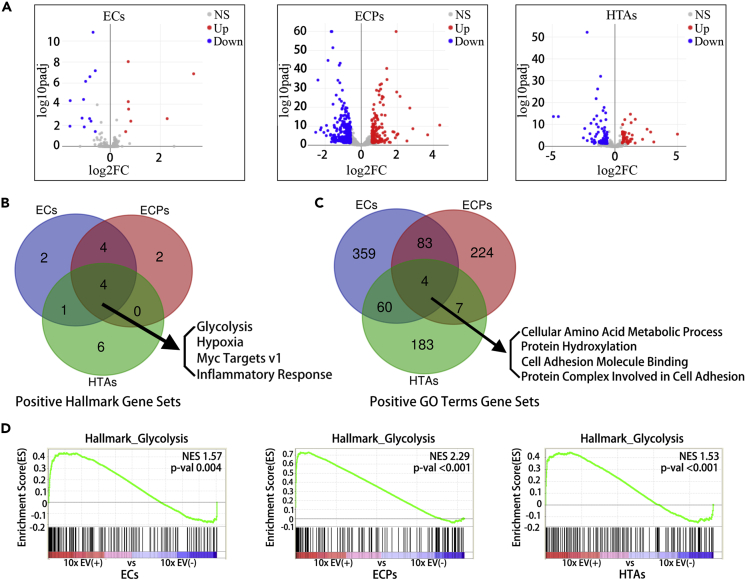
Since a short exposure to GBM EVs significantly affected the proliferation, self-renewal, and colony formation of the pre-transformed astrocytes, we examined the transcriptomes of EC, ECP, and HTA cells treated for 6 days with the 10x concentrated GBM EV(+) and EV(−) CM. As evidenced by volcano plots of differentially expressed genes, we found that EV-treated ECP and HTA cultures demonstrated global alterations of their gene expression programs (449 and 123 genes, respectively, changed >1.5-fold, adjusted P < 0.05), whereas ECs exhibited more limited changes (19 genes) (Figure 3A). The gene set enrichment analysis (GSEA) revealed that EVs-treated cultures activated several cancer-related pathways, based on the GSEA Hallmark and GO Term gene sets (Table S2). Of the commonly and significantly upregulated Hallmark gene sets (P < 0.05), many were associated with glycolysis, hypoxia, Myc targets, and inflammatory response (Figure 3B). Of note, three of these four gene sets are tightly associated with the characteristic metabolic changes in cancer. Furthermore, commonly and significantly upregulated (P < 0.05) GO Term gene sets included “cellular amino acid metabolic process” and “protein hydroxylation” (Figure 3C). The genes associated with glycolysis and implicated in Warburg effect (Liberti and Locasale, 2016) and metabolic reprogramming in brain tumors (Venneti and Thompson, 2017) were the most remarkably enriched in the group of upregulated mRNAs in all three types of astrocytes (Figure 3D).
Figure 3.
GBM-Derived EVs Alter the Global Transcriptome of Astrocytes
(A) Volcano plots depict the alterations in RNA expression between the cells treated with 10x EV(+) and 10x EV(−) CM, with the log2-transformed fold change (FC) as x axis and the log10-transformed adjusted p value (padj) as y axis. In EC, ECP, and HTA cells, 19, 449, and 123 genes showed significant transcriptional changes upon EV treatment, respectively. Red and blue dots represent genes upregulated and downregulated with the cutoff of FC > 1.5 and adjusted P < 0.05 (DEseq2 analysis), respectively.
(B and C) Venn diagrams depict the number of common significantly upregulated gene sets, based on the Gene set enrichment analysis (GSEA) Hallmark (B) and GO Terms (C), in EC, ECP, and HTA cultures treated with 10x EV(+) CM, relative to those treated with 10x EV(−) CM.
(D) GSEA Hallmark Glycolysis gene set is enriched in EC, ECP, and HTA cultures treated with 10x EV(+) CM relative to those treated with 10x EV(−) CM. NES, normalized enrichment score.
GBM EVs Accelerate Oxidative Phosphorylation and Glycolysis in Transformed Astrocytes
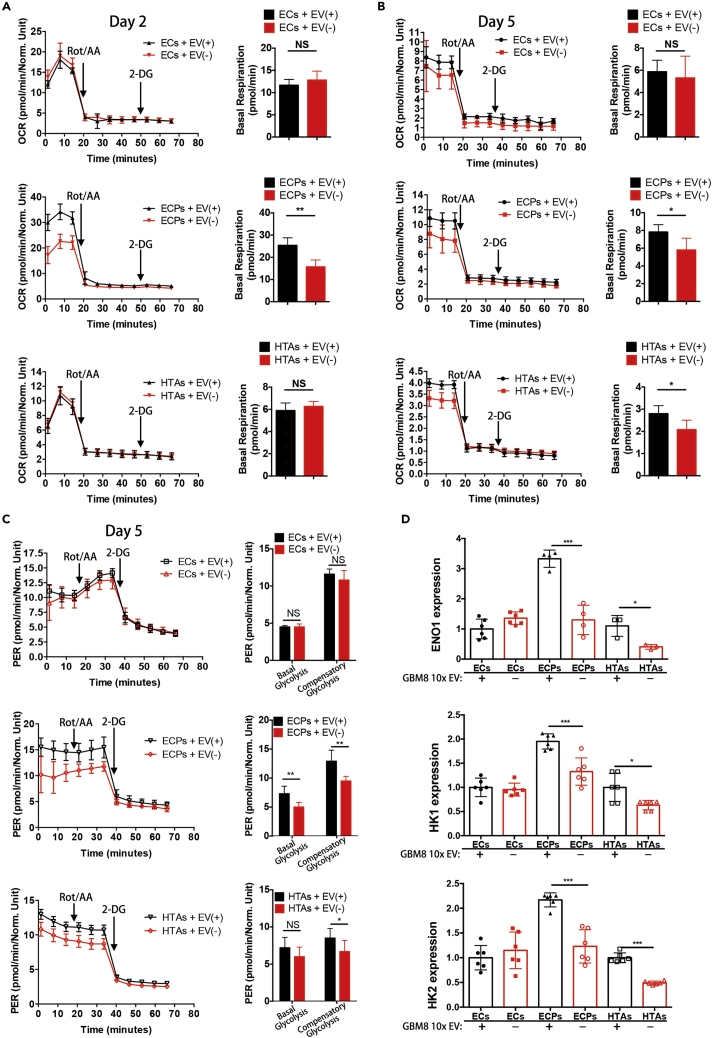
To directly investigate the effects of GBM EVs on the metabolism of the pre-transformed mouse and human astrocytes, we evaluated mitochondrial respiration and glycolysis by measuring the basal mitochondrial respiration and proton efflux rate (PER), respectively. Basal mitochondrial respiration was calculated by subtracting non-mitochondrial respiration reveled by Rot/AA administration from the baseline cellular oxygen consumption rate (OCR). These parameters have been measured in the recipient EC, ECP, and HTA cultures treated with GBM-derived EV(+) and EV(−) CM for either 2 or 5 days. EV exposure significantly increased basal mitochondrial respiration in the ECP cells (Figures 4A and 4B, middle panel), with nearly 2-fold induction observed at day 2 (Figure 4A, middle panel), whereas no significant change was observed in the EC cultures (Figures 4A and 4B, upper panel). EV exposure also increased the basal respiration in HTAs, albeit the response was more delayed and less pronounced than in ECPs (Figures 4A and 4B, lower panel). Furthermore, ECP and HTA cultures exposed to GBM EVs over 5 days demonstrated activated glycolysis (Figure 4C, middle and lower panels). In addition to basal glycolysis, the compensatory glycolysis (the rate of glycolysis in the cells following the addition of Rot/AA reflecting maximum glycolysis capacity) was also affected in these cells by GBM EV(+) CM. In contrast, no difference was observed in EC cultures (Figure 4C, upper panel). Consistent with the activation of glycolysis, we also observed that mRNA levels of the key glycolytic enzymes ENO1, HK1, and HK2 (Chen et al., 2018) were markedly upregulated in ECP and HTA cultures by EVs, whereas no increase was detected in the recipient EC cells (Figure 4D). These data indicate that GBM EVs promote both mitochondrial respiration and glycolysis in the pre-transformed astrocytes.
Figure 4.
GBM EVs Reprogram Recipient Astrocytes to Activate Oxidative Phosphorylation and Glycolysis
(A and B) Alterations in basal mitochondrial respiration, monitored as oxygen consumption rate (OCR), in EC, ECP, and HTA cells treated with 10x EV(+) CM and those treated with EV(−) CM for 2 (A) and 5 days (B). Quantification of the differences in basal mitochondrial respiration is shown in the right panel (N = 5 wells per condition).
(C) Alterations in the proton efflux rate (PER) in EC, ECP, and HTA cells treated with 10x EV(+) CM and those treated with EV(−) CM for 5 days. Quantification of basal and compensatory glycolysis is shown in the right panel (N = 5 wells per condition).
(D) Analysis of ENO1, HK1, HK2 mRNA expression in EC, ECP, and HTA cells upon their treatment with either concentrated EV(+) or EV(−) CM for 5 days. qRT-PCR analysis normalized to the expression of actin mRNA (N = 3 wells per condition).
∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; two-tailed t test. Data are represented as mean ± SEM.
GBM Disseminates Abundant Oncogenes via EVs
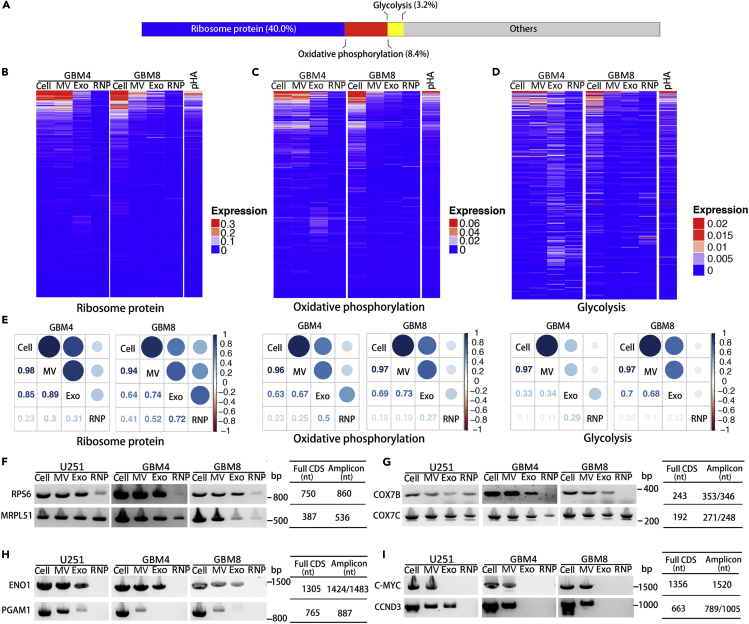
To further explore the molecular mechanisms underlying the phenotype switch triggered in the pre-transformed astrocytes by GBM EVs, we examined the RNA content of the GBM EVs, by re-analyzing our previously reported RNA sequencing-based datasets of large 200–800 nm and small EVs <200 nm (called in the text below MVs and exosomes, correspondingly), and RNPs derived from four heterogeneous types of GSC cultures (Wei et al., 2017). Since the majority of exRNA reads in exRNA libraries represent fragmented rRNA (Wei et al., 2017; Jenjaroenpun et al., 2013; Turchinovich et al., 2019), we employed rRNA depletion to increase the sequencing depth of other exRNA classes. We selected 200 top abundant mRNAs for each MV and Exosome dataset and identified among them the top 92 mRNAs common in all four types of MVs and 37 mRNAs common in exosomes (Figures S3A and S3B). A combination of these two lists provided a list of 97 mRNAs highly and commonly enriched in the GSC EVs. Notably, many of these abundant EV mRNAs corresponded to relatively short transcripts, consistent with the previously reported analysis (Wei et al., 2017). A total of 38 ribosomal protein (RP) mRNAs account for 40.0% (38/95) of these abundant EV mRNAs (Figure 5A). Intersecting the list of 115 mRNAs abundant in EVs with 200 OXPHOS and 200 glycolysis mRNAs from GSEA gene sets, we found 8 (8/95, 8.4%) OXPHOS and 3 (3/95, 3.2%) glycolysis associated mRNAs among the most abundant GSC EV (MV and Exo) mRNAs (Figure 5A). The analysis of mRNAs in GSCs and the corresponding MV, Exo, and RNP fractions indicated that extracellular RP and OXPHOS mRNAs were primarily associated with the EV fractions, with MVs exhibiting the highest enrichment (Figures 5B and 5C). Similarly, the glycolysis mRNAs were also mostly EV associated as compared with the RNP fraction (Figure 5D). The correlation analysis of RP, OXPHOS, and glycolysis mRNAs performed on GSCs and the corresponding extracellular complexes (MVs, Exos, and RNPs) further confirmed the highest degree of similarity between the cellular transcriptome and MV content, suggesting that mRNAs are encapsulated mainly in the MVs (Figure 5E). Notably, the abundance of mRNAs encoding RPs in EVs, and especially in MVs, suggested that EVs might carry a transcriptome encoding the ribosomal machinery. Further TCGA analysis demonstrated the global upregulation of RP, OXPHOS, and glycolysis genes in GBM relative to the normal brain tissues (Figure S3C). These data suggest that glioma-derived vesicles may modulate the translational and metabolic status, and thus, transformation and proliferation of astrocytes, by direct transfer of RP, OXPHOS, and glycolysis mRNAs.
Figure 5.
GBM EVs Carry Protein-Coding Transcripts for Ribosomal Proteins, Oxidative Phosphorylation, and Glycolytic Factors to Recipient Cells
(A) The chart exhibiting the composition of most abundant mRNA species in GBM MVs and/or Exos, demonstrates that RP, OXPHOS, and glycolysis mRNAs totally account for more than 50% of them.
(B–D) Heatmap analysis of intercellular and extracellular RP (B), OXPHOS (C), and glycolysis (D) mRNAs indicates the abundance of exRNAs within EVs.
(E) Correlation matrix of the transcript composition of the cells, MVs, Exos, and RNPs for RP, OXPHOS, and glycolysis mRNAs, in GBM4 and GBM8 GSCs.
(F–I) Full CDS RT-PCR analysis of selected RP (F), OXPHOS (G), glycolysis (H), and oncogene (I) encoding mRNAs demonstrates the presence of the transcripts with protein-coding potential in EVs. Sizes of amplicons that contain the CDSs are indicated.
See also Figure S3.
To test whether EVs carry the full-length mRNAs with protein-coding potentials, PCR primers were designed for the selected mRNAs abundant in exRNA and amplification of the complete coding sequences (CDS) was carried out. As shown in Figure 5F, CDSs of cytosolic RPS6 and mitochondrial RP MRPL51 were detected not only in glioma cells but also in glioma-derived MVs and Exos, with the residual levels also detected in RNP fractions. OXPHOS genes COX7B and COX7C, encoding the subunits of mitochondrial Cytochrome c oxidase, were also present as full-CDSs in GSC EV fractions (Figure 5G). The transcript for Enolase 1 (ENO1), the key glycolytic enzyme, was detected in both GSC MVs and Exosomes, whereas Phosphoglycerate mutase 1 (PGAM1), which coordinates glycolysis and biosynthesis to support tumor growth, was primarily associated with MVs (Figure 5H). Finally, the MVs also contained mRNAs for transcription factor C-MYC and cyclin CCND3 (Figure 5I). Considering the described metabolic and proliferative phenotypes induced by GBM EVs (Figures 1 and 4), we propose that GBM could reprogram the metabolism of transformed astrocytes by delivering mRNAs for RPs, glycolytic enzymes, oncogenes, and other tumor-promoting factors via EVs.
Activated Glycolysis Correlates with Glioma Progression and Poor Prognosis in Patients with GBM
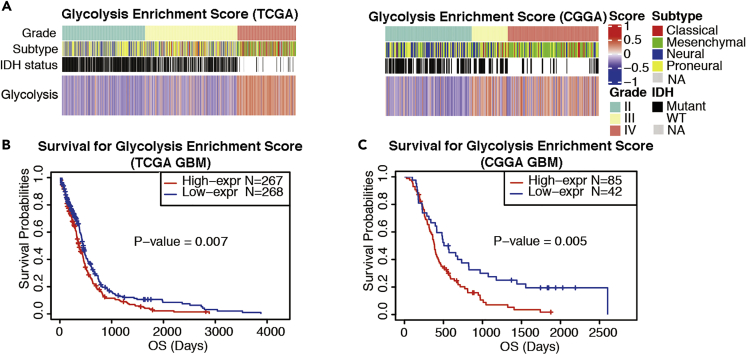
The metabolic shift to aerobic glycolysis has been associated with a survival advantage in rapidly proliferating GBM cells (Strickland and Stoll, 2017; Wolf et al., 2010). To investigate the association between activated glycolysis and glioma progression, we employed the Gene Set Variation Analysis (GSVA) and evaluated glycolysis using the hallmark glycolysis gene set in two cohorts of gliomas of grade II–IV, including 609 patients from TCGA dataset and 301 patients from the CGGA dataset. As shown in Figure 6A, GBM tumors had significantly higher glycolysis enrichment scores (GESs) than grade II gliomas in both TCGA (P < 0.0001) and CGGA datasets (P < 0.0001) (Figure 6A). TCGA GBM dataset also exhibited increased GES relative to the grade III gliomas (P < 0.0001). Furthermore, Kaplan-Meier survival analysis demonstrated that patients with GBM with high GES had worse overall survival than those with low GES in TCGA dataset (Figure 6B, P = 0.007). The top 67% GES also correlated with a worse outcome of GBM patients with GBM when compared with the bottom 33% GES in the CGGA dataset (Figure 6C, P = 0.005).
Figure 6.
Upregulated Glycolysis Is Associated with Glioma Progression and Patient Survival
(A) The Gene Set Variation Analysis (GSVA) demonstrates that enrichment scores for glycolysis mRNAs are positively associated with glioma progression from grade II to grade IV in TCGA and CGGA datasets. The cohorts consisted of 609 tumors of various grades in TCGA and 301 tumors in CGGA.
(B) Kaplan-Meier analysis of 535 patients with GBM demonstrates that high glycolysis enrichment scores are associated with shortened overall survival (OS) of patients with GBM, based on TCGA dataset. Median score was used as the threshold. Kaplan-Meier test.
(C) OS of patients with GBM is associated with high glycolysis enrichment scores, based on CGGA dataset. The association is highly significant for the top two-thirds of the patients compared with the bottom one-third. Kaplan-Meier test.
Discussion
Impressive advances in the studies of EVs and their functions provide fundamental novel insights into intercellular communication and its role in physiology and pathology, such as cancer. Nevertheless, several technical shortcomings still hamper research in the field. Although ultracentrifugation is widely employed for the EV isolation, the disadvantages of this procedure include a compromised yield of EVs and exRNA, their inevitable damage due to the high-gravity force, and co-sedimentation of other non-EV contaminants such as RNPs and lipoproteins, impeding the downstream analysis (Wei et al., 2017; Laurent et al., 2015; Greening et al., 2015). Quantitative analysis of EVs is also restricted by currently deficient methodologies, resulting in limited data reproducibility (Bachurski et al., 2019; Das et al., 2019; Laurent et al., 2015). Furthermore, the comparison of EVs isolated from cancer versus normal cells is challenging since these cell types are typically propagated in vastly different culture conditions, exhibit diverse proliferation and growth rates, and release different amounts of EVs. Based on these considerations, we adopted a stringent sequential filtration protocol (Wei et al., 2017) for a study of GSC-derived EV functions in astrocyte transformation. The protocol relies on the comparison of EV-containing versus EV-depleted GSC CM and produces much lower pressure on EVs than ultracentrifugation (75 versus 14,500 psi, correspondingly). Since the filtration through 20-nm pores efficiently eliminates EVs from the CM (Wei et al., 2017), the differential effects observed in our experiments are caused mainly by the secreted EVs. It should be noted, however, that the 20-nm filters may still retain some large protein, lipoprotein, and RNP complexes, such as ApoE and alpha-2M, ferritin oligomers, or IgM pentamers, and thus contribute to the observed effects (Hiramoto et al., 2018; Krimbou et al., 1998; Kaddis et al., 2007). Non-membranous exomeres, a recently discovered subset of nanoparticles with a diameter close to 35 nm and distinct biophysical and molecular properties (Zhang et al., 2018), may present an additional functional component. Our current experimental design enables the study of diverse EV and non-EV components of CM, ranging in size from 20 to 800 nm as a pooled population. With the currently lacking knowledge of non-EV components present in the GSC-derived EV(+) CM, additional studies dissecting their impact, as well as the contribution of specific subtypes of vesicles (e.g., MVs versus exosomes) would be warranted. Such studies will require an increased amount of input material and further adaptation of the protocol. In addition to exosomes and MVs, very large EVs or oncosomes (1–10 μm) may also carry RNA (Kowal et al., 2016; Conley et al., 2017). However, our data suggest relatively low amounts of exRNA associated with such EVs released by GSC cultures, as compared with exosomes and MVs (Wei et al., 2017), leaving them beyond the scope of the current work.
The horizontal EV transfer is a novel route of intercellular communication that operates over both short and long distances (Broekman et al., 2018). EVs can deliver complex multi-molecular biological messages from one cell to another and may, thereby, spread a pathogenic signal across the tissue microenvironment (Nakano et al., 2015). A number of studies demonstrated that EVs released by GBM are taken up by various cells of the brain microenvironment, including endothelial cells and microglia, and alter their phenotypes, often toward those more supportive for tumor progression and invasion (Gao et al., 2020; Abels et al., 2019; Lucero et al., 2020). Here we demonstrate that glioma-derived EVs are efficiently delivered to astrocytes, the cells possessing high transformation capacity to glioma, which provide a pool of glioma-initiating cells in the adult brain. Notably, our in vitro and in vivo experiments suggest that glioma-derived EVs are by themselves unable to transform astrocytes or accelerate tumor growth in nude mice, in the utilized experimental conditions. However, in the genetic context of an oncogene-induced pre-transformed environment, glioma EVs enhance the transformation in vitro and tumor growth in vivo. EVs potentiate the self-renewal, proliferation, and anchorage-independent growth of human astrocytes triggered by activated Ras, telomerase, or simultaneously inactivated p53 and pRb pathways, the signaling aberrations commonly observed in GBM (Cancer Genome Atlas Research Network, 2008; Brennan et al., 2013). Interestingly, constitutively active EGFRvIII, in combination with Ink4A/Arf (CDKN2A gene) deletion, was insufficient to confer the susceptibility to EV transformation, unless PTEN tumor suppressor was additionally ablated. In triple-mutant EGFRvIII; CDKN2A-/-; PTEN-/- cells, however, the EVs promoted proliferation, colony formation, and tumorigenic conversion. Therefore, the protective growth-restraining effects of PTEN, probably via PI3K/Akt signaling pathway for senescence and apoptosis, appear to abrogate the signaling associated with the EV uptake and cargo release. These data suggest that (1) the effects induced by EVs and their cargo depend on the genetic or mutational status of the recipient cells, (2) specific tumor-suppressive pathways exist to block the spread of EV-associated cancer signals in normal astrocytes, and (3) EVs are only able to promote limited steps of the progression, possibly due to the low copy number of mRNAs and other non-rRNA cargo transcripts. It has also been suggested that EVs may elicit additional effects on astrocytes, such as promote their migration (Oushy et al., 2018). We have not detected the enhanced migration in our experiments; nevertheless, the effects of glioma EVs on migration and invasion of normal and pre-transformed astrocytes in various genetic contexts in vivo would warrant further investigation.
To investigate the EV-mediated mechanisms underlying alterations in the transformation and proliferative capacity of the recipient astrocytes, we performed their transcriptomic analysis. This analysis suggested global changes in the levels of metabolic genes, particularly factors of glycolysis, upon the EV uptake. Metabolic assays revealed the activation of both mitochondrial respiration and glycolysis in these cells. Interestingly, the interplay between the activated glycolysis and OXPHOS facilitates the growth, invasion, and drug resistance of tumor cells and contributes to the tumorigenic adaptation to varying microenvironments. Cancer cells may switch their metabolic phenotypes between OXPHOS and glycolysis during tumor progression and metastasis and in response to external perturbations (Yu et al., 2017; Jia et al., 2019, Deberardinis et al., 2008). They may also enter a hybrid metabolic state, not usually present in normal cells, with both glycolysis and OXPHOS highly activated. Such metabolic plasticity generates the energy and the intermediates for biosynthetic processes necessary for tumor growth (Paudel and Quaranta, 2019). Notably, although our data along with prior reports suggest that high expression of glycolytic signature genes is generally unfavorable for survival of patients with glioma (Strickland and Stoll, 2017; Chen et al., 2017; Wolf et al., 2010), the factors such as sex and genetics, as well as overall heterogeneity of a tumor and its microenvironment, contribute to the relationship between glycolysis and OXPHOS pathways and the association of these pathways with patients survival (Ippolito et al., 2017; Stadlbauer et al., 2018). Here, we demonstrated that GBM EVs may reprogram the metabolism of the recipient pre-transformed astrocytes by activating both glycolysis and OXPHOS. This finding provides additional perspectives of the dynamic conversation between cancer cells and associated cells of the tumor microenvironment modulated by EVs. Notably, several key glycolytic enzymes, such as GAPDH, enolase, pyruvate kinase, and phosphoglycerate kinase, have been found by proteomic studies in exosomes (Graner et al., 2009; Goran Ronquist, 2019), and ExoCarta lists them among the top exosomal proteins. Our study further indicates that some of the observed metabolic alterations might be associated with the direct transfer of mRNAs encoding metabolic factors from GBM to the induced astrocytes.
ExRNA functions, exRNA biomarkers, and exRNA/EV-based therapeutics are the topics of multiple studies relying on various models of cancer development and progression. The RNA encapsulated in EVs can elicit the tumorigenic transformation of immortalized rodent fibroblasts, epithelial cells, and stem cells (Wang et al., 2019). Since the majority of exRNA is relatively short (<200 nt), most reports focused on small non-coding RNA species, such as microRNA. Our quantitative analysis of GSC exRNA indicated that mRNA reads account for at least 20% of longer RNA species (>200 nt) present in the rRNA-depleted libraries of EVs-encapsulated exRNA (Wei et al., 2017). Both fragmented and full-length mRNAs were found in all extracellular fractions, including vesicular and extravesicular, although there appears to be size limitation to the cargo RNA in EVs (Wei et al., 2017). A possibility of protein synthesis from a horizontally transferred mRNA template has been demonstrated (Lai et al., 2015; Ridder et al., 2014, 2015); nevertheless, the impact of endogenous mRNA transfer between the cells remains poorly investigated, likely due to the low magnitude of this process and technical challenges associated with its research (Wei et al., 2017). We identified several classes of mRNAs highly enriched in GBM EVs that can be taken up by the recipient astrocytes and induce the downstream functional effects. Most remarkably, transcripts for RP, OXPHOS, and glycolytic factors account for more than 50% of the EV-abundant mRNA species. Representative mRNAs with the complete ORFs and protein-coding potential were detected in glioma EVs. Upon the horizontal transfer and translation, these factors may contribute to the proliferative and metabolic phenotypes observed in the brain astrocytes and thus have a substantial impact on their physiology and progression toward the cancerous state. mRNAs encoding RPs represent the most enriched and abundant class of mRNAs found in EVs, including both MVs and exosomes. RPs are the immediate factors whose levels are dynamically regulated and quickly adjusted based on the cell requirements in protein synthesis. Translational machinery is exceedingly active in proliferative tumor cells to keep up with increasing needs in newly synthesized proteins (Sulima et al., 2017), operating in these cells at much higher levels than in normal slowly dividing or largely postmitotic (e.g., glial) cells. Dysregulated ribosome biogenesis is known to be involved in the development and progression of most human cancers (Sulima et al., 2017; Pelletier et al., 2018). Overall, balancing the levels of RP mRNAs with translational capacity during cell transformation and carcinogenesis is one of the hallmarks of cancer.
In accordance with these ideas, the RP mRNAs form a distinct group of highly expressed transcripts characterized by relatively small length (median length 1,260 bp) and 5′ Terminal Oligopyrimidine Tract (TOP) motif that regulates translation of these mRNAs. This cis-regulatory element coordinates the synchronous and stoichiometric expression of the protein components of the translation machinery and operates downstream of mTOR signaling (Thoreen et al., 2012; Gentilella et al., 2015). Its dysregulation enables uncontrolled ribosome synthesis, a critical requirement for unrestrained cellular proliferation (Strimpakos et al., 2009). Notably, PTEN loss leads to an activation of mTOR (Hollander et al., 2011). We demonstrate here that the levels of RP mRNAs are globally elevated in GBM when compared with the control brains. Our work suggests that EV-mediated release and horizontal transfer of RP mRNAs may have a dual role in carcinogenesis, first as a mechanism for the cancer cell adaptation to environmental cues by adjusting its translational machinery via the secretion of RP mRNA, and second, via the synergistic transfer of this class of mRNAs to pre-transformed astrocytes to boost their translational capacity. We hypothesize that both the size and structural properties of these mRNAs, as well as their interactions with specific binding partners, may contribute to their preferential loading to EVs. Further investigation of RP mRNAs may advance our limited understanding of the mRNA sorting and EV-loading mechanisms.
Recent studies demonstrated that ribosome activity is not only a critical regulator of growth but also that of metabolism. For instance, ribosomal availability affects glycolysis and mitochondrial function (Sousa et al., 2016). Notably, in addition to RP mRNAs, GBM EVs transport mRNAs encoding key glycolytic enzymes and factors of OXPHOS. For instance, ENO1 mRNA is encapsulated in both MVs and exosomes. This mRNA encodes alpha-enolase, a glycolytic enzyme that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate and is frequently overexpressed in glioma and multiple other cancers (Song et al., 2014). Along with its direct role in glycolysis, ENO1 promotes cell proliferation by regulating the PI3K/AKT signaling pathway and is prognostic for glioma progression (Song et al., 2014). Another factor PGAM1, phosphoglycerate mutase, whose mRNA was mostly associated with MVs, releases energy during glycolysis by catalyzing the conversion of 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG). PGAM1 is, too, associated with the proliferation of glioma cells and disease progression (Xu et al., 2016; Gao et al., 2013). Interestingly, knockdown of each ENO1 and PGAM1 had similar tumor-inhibitory effects and extended survival of mice with orthotopic GBM xenografts (Xu et al., 2016; Song et al., 2014).
A rapidly growing body of evidence has demonstrated that aerobic glycolysis does not entail a complete shutdown of OXPHOS in tumors. Mitochondrial respiration occurring in cancer cells might provide a primary source of energy for the low-cycling tumor cells, such as tumor stem cells that are responsible for tumor relapse (Viale et al., 2015). Five complexes across the mitochondrial membrane can transfer electrons from donors generated by the TCA cycle and fatty acid oxidation and result in ATP production. The potential vulnerability of the electron transport chain in GBM cells through inhibiting complexes have been indicated as promising therapeutics (Molina et al., 2018; Naguib et al., 2018). Interestingly, we found that several Cytochrome c oxidase (Complex IV)-coding mRNAs were remarkably enriched in GBM MVs and demonstrated a protein-coding potential for some of them, e.g., COX7B and COX7C mRNAs. Altogether, these data indicated that mRNAs encoding glycolytic enzymes and OXPHOS factors secreted by glioma cells in EVs might work in an orchestrated way to reprogram the metabolism of the GBM microenvironment.
Our work also uncovered that full CDSs of c-Myc and CCND3 mRNAs were encapsulated in glioma MVs. The MYC oncogene encodes a transcription factor, c-Myc, which is often genetically deregulated in cancer and correlates with the grade of glioma malignancy (Wang et al., 2008). c-Myc modulates metabolic reprogramming in the pathogenesis of glioma and is also critically important for proliferation, growth, and survival of glioma cancer stem cells (Wang et al., 2008). The drastic growth of gliomas is also partially driven by frequent mutation and transcriptional dysregulation of cell cycle factors (Touat et al., 2017), especially the ones involved in the control of G1/S phase transition, such as cyclin D3 encoded by CCND3 (Buschges et al., 1999). The EV-mediated transfer of these established oncogenes may contribute to the proliferative and metabolic phenotypes observed in the astrocytes.
To conclude, GBM EVs enhance the transformation of astrocytes through metabolic reprogramming. Transformed cells adapt their protein synthesis and metabolism to support tumor growth and recurrence via the mechanisms that include the EV-mediated horizontal mRNA transfer. The concordant transmission of translation-competent messages encoding co-functional protein factors acting along the specific signaling pathways may ensure their balance and maximize the downstream effects. Of note, the hyperactive mTOR signaling, along with the regulation of ribosomal production and protein synthesis, and induction of potent Warburg effect in cancer (Duvel et al., 2010), has been also implicated in the control of exosome release (Zou et al., 2019). It is, therefore, plausible that enhanced protein synthesis and proliferation in gliomas, the switch in tumor metabolism, and RNA secretion and horizontal transfer are under the coordinated control of mTOR, the exciting possibility that remains to be further investigated.
Limitations of the Study
This study has not dissected the functions of the specific subtypes of glioma-derived EVs (e.g., microvesicles versus exosomes, as well as large EVs such as oncosomes) or other large extracellular complexes that could be co-isolated with EVs, but warranted their prospective investigation in glial transformation. The detailed mechanisms of the EVs-mediated facilitation of astrocytic transformation and effects on metabolism also require further exploration. Identification of structural properties and sequence motifs that may facilitate mRNA/RNA sorting to EVs and the in-depth study of the impact of horizontal RNA transfer on neoplastic transformation also necessitate additional work.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Anna M. Krichevsky (akrichevsky@bwh.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
RNAseq fq files and processed data generated during this study are available through GEO, accession code GSE143189.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the U19 CA179563 grant and U19 administrative supplement through the NIH Common Fund and the Office of Strategic Coordination/Office of the NIH Director (A.M.K. and A.C.), R21 NS098051, AG060019, and R01 CA215072 grants (A.M.K.), and a grant from the National Natural Science Foundation of China (No. 81901069, Z.W.). We thank Dr. Xandra Breakefield and members of Krichevsky and Breakefield laboratories for helpful discussions.
Author Contributions
A.M.K. and Z.W. conceived and designed the study; A.Z. performed most experiments; H.J.J. performed soft agar assay; Z.W., R.R., R.E.F., E.D., R.A., and E.J.U. assisted with experiments; Y.Y. and S.Y. assisted with microscopy; Z.W. and W.Y. performed bioinformatics analysis; A.C. and Y.Y. contributed to data analysis; A.Z., Z.W., and A.M.K. wrote the paper. All authors revised and approved the manuscript.
Declaration of Interests
All authors declare no competing financial interests.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101420.
Contributor Information
Zhiyun Wei, Email: zwei@bwh.harvard.edu.
Anna M. Krichevsky, Email: akrichevsky@bwh.harvard.edu.
Supplemental Information
References
- Abels E.R., Maas S.L.N., Nieland L., Wei Z., Cheah P.S., Tai E., Kolsteeg C.J., Dusoswa S.A., Ting D.T., Hickman S. Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. 2019;28:3105–3119.e7. doi: 10.1016/j.celrep.2019.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski D., Schuldner M., Nguyen P.H., Malz A., Reiners K.S., Grenzi P.C., Babatz F., Schauss A.C., Hansen H.P., Hallek M. Extracellular vesicle measurements with nanoparticle tracking analysis - an accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell Vesicles. 2019;8:1596016. doi: 10.1080/20013078.2019.1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian E.B., Chen E.F., Xu Y.D., Yang Z.H., Tang F., Ma C.C., Wang H.L., Zhao B. Exosomal lncRNAATB activates astrocytes that promote glioma cell invasion. Int. J. Oncol. 2019;54:713–721. doi: 10.3892/ijo.2018.4644. [DOI] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G., Mckenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekman M.L., Maas S.L.N., Abels E.R., Mempel T.R., Krichevsky A.M., Breakefield X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018;14:482–495. doi: 10.1038/s41582-018-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschges R., Weber R.G., Actor B., Lichter P., Collins V.P., Reifenberger G. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 1999;9:435–442. doi: 10.1111/j.1750-3639.1999.tb00532.x. discussion 432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D., Turcan S. Origin of gliomas. Semin. Neurol. 2018;38:5–10. doi: 10.1055/s-0037-1620238. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Shi Y., Li Y., He Z.C., Zhou K., Zhang X.N., Yang K.D., Wu J.R., Kung H.F., Ping Y.F. A glycolysis-based ten-gene signature correlates with the clinical outcome, molecular subtype and IDH1 mutation in glioblastoma. J. Genet. Genomics. 2017;44:519–530. doi: 10.1016/j.jgg.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang Y., Wang H., Zeng Y.Y., Li Z., Li M.L., Li F.F., You J., Zhang Z.M., Tzeng C.M. WW domain-binding protein 2 acts as an oncogene by modulating the activity of the glycolytic enzyme ENO1 in glioma. Cell Death Dis. 2018;9:347. doi: 10.1038/s41419-018-0376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley A., Minciacchi V.R., Lee D.H., Knudsen B.S., Karlan B.Y., Citrigno L., Viglietto G., Tewari M., Freeman M.R., Demichelis F. High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 2017;14:305–316. doi: 10.1080/15476286.2016.1259061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Extracellular R.N.A.C.C., Ansel K.M., Bitzer M., Breakefield X.O., Charest A., Galas D.J., Gerstein M.B., Gupta M., Milosavljevic A. The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019;177:231–242. doi: 10.1016/j.cell.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Duvel K., Yecies J.L., Menon S., Raman P., Lipovsky A.I., Souza A.L., Triantafellow E., Ma Q., Gorski R., Cleaver S. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Yu B., Yan Y., Shen J., Zhao S., Zhu J., Qin W., Gao Y. Correlation of expression levels of ANXA2, PGAM1, and CALR with glioma grade and prognosis. J. Neurosurg. 2013;118:846–853. doi: 10.3171/2012.9.JNS112134. [DOI] [PubMed] [Google Scholar]
- Gao X., Zhang Z., Mashimo T., Shen B., Nyagilo J., Wang H., Wang Y., Liu Z., Mulgaonkar A., Hu X.L. Gliomas interact with non-glioma brain cells via extracellular vesicles. Cell Rep. 2020;30:2489–2500 e5. doi: 10.1016/j.celrep.2020.01.089. [DOI] [PubMed] [Google Scholar]
- Gentilella A., Kozma S.C., Thomas G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim. Biophys. Acta. 2015;1849:812–820. doi: 10.1016/j.bbagrm.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goran Ronquist K. Extracellular vesicles and energy metabolism. Clin. Chim. Acta. 2019;488:116–121. doi: 10.1016/j.cca.2018.10.044. [DOI] [PubMed] [Google Scholar]
- Graner M.W. Roles of extracellular vesicles in high-grade gliomas: tiny particles with outsized influence. Annu. Rev. Genomics Hum. Genet. 2019;20:331–357. doi: 10.1146/annurev-genom-083118-015324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner M.W., Alzate O., Dechkovskaia A.M., Keene J.D., Sampson J.H., Mitchell D.A., Bigner D.D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- Gyuris A., Navarrete-Perea J., Jo A., Cristea S., Zhou S., Fraser K., Wei Z., Krichevsky A.M., Weissleder R., Lee H. Physical and molecular landscapes of mouse glioma extracellular vesicles define heterogeneity. Cell Rep. 2019;27:3972–3987 e6. doi: 10.1016/j.celrep.2019.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallal S., Mallawaaratchy D.M., Wei H., Ebrahimkhani S., Stringer B.W., Day B.W., Boyd A.W., Guillemin G.J., Buckland M.E., Kaufman K.L. Extracellular vesicles released by glioblastoma cells stimulate normal astrocytes to acquire a tumor-supportive phenotype via p53 and MYC signaling pathways. Mol. Neurobiol. 2019;56:4566–4581. doi: 10.1007/s12035-018-1385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto E., Tsutsumi A., Suzuki R., Matsuoka S., Arai S., Kikkawa M., Miyazaki T. The IgM pentamer is an asymmetric pentagon with an open groove that binds the AIM protein. Sci. Adv. 2018;4:eaau1199. doi: 10.1126/sciadv.aau1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg F.H., Pruitt A. Assumptions in the radiotherapy of glioblastoma. Neurology. 1980;30:907–911. doi: 10.1212/wnl.30.9.907. [DOI] [PubMed] [Google Scholar]
- Hollander M.C., Blumenthal G.M., Dennis P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito J.E., Yim A.K., Luo J., Chinnaiyan P., Rubin J.B. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI Insight. 2017;2:e92142. doi: 10.1172/jci.insight.92142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenjaroenpun P., Kremenska Y., Nair V.M., Kremenskoy M., Joseph B., Kurochkin I.V. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;1:e201. doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Lu M., Jung K.H., Park J.H., Yu L., Onuchic J.N., Kaipparettu B.A., Levine H. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. U S A. 2019;116:3909–3918. doi: 10.1073/pnas.1816391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddis C.S., Lomeli S.H., Yin S., Berhane B., Apostol M.I., Kickhoefer V.A., Rome L.H., Loo J.A. Sizing large proteins and protein complexes by electrospray ionization mass spectrometry and ion mobility. J. Am. Soc. Mass Spectrom. 2007;18:1206–1216. doi: 10.1016/j.jasms.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A., Jr., Friedman A.H., Friedman H., Gallia G.L., Giovanella B.C. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U S A. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbou L., Tremblay M., Davignon J., Cohn J.S. Association of apolipoprotein E with alpha2-macroglobulin in human plasma. J. Lipid Res. 1998;39:2373–2386. [PubMed] [Google Scholar]
- Lai C.P., Kim E.Y., Badr C.E., Weissleder R., Mempel T.R., Tannous B.A., Breakefield X.O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L.C., Abdel-Mageed A.B., Adelson P.D., Arango J., Balaj L., Breakefield X., Carlson E., Carter B.S., Majem B., Chen C.C. Meeting report: discussions and preliminary findings on extracellular RNA measurement methods from laboratories in the NIH Extracellular RNA Communication Consortium. J. Extracell Vesicles. 2015;4:26533. doi: 10.3402/jev.v4.26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero R., Zappulli V., Sammarco A., Murillo O.D., Cheah P.S., Srinivasan S., Tai E., Ting D.T., Wei Z., Roth M.E. Glioma-derived miRNA-containing extracellular vesicles induce angiogenesis by reprogramming brain endothelial cells. Cell Rep. 2020;30:2065–2074.e4. doi: 10.1016/j.celrep.2020.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J.R., Sun Y., Protopopova M., Gera S., Bandi M., Bristow C., Mcafoos T., Morlacchi P., Ackroyd J., Agip A.A. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018;24:1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- Naguib A., Mathew G., Reczek C.R., Watrud K., Ambrico A., Herzka T., Salas I.C., Lee M.F., El-Amine N., Zheng W. Mitochondrial complex I inhibitors expose a vulnerability for selective killing of pten-null cells. Cell Rep. 2018;23:58–67. doi: 10.1016/j.celrep.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano I., Garnier D., Minata M., Rak J. Extracellular vesicles in the biology of brain tumour stem cells--Implications for inter-cellular communication, therapy and biomarker development. Semin. Cell Dev. Biol. 2015;40:17–26. doi: 10.1016/j.semcdb.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Oushy S., Hellwinkel J.E., Wang M., Nguyen G.J., Gunaydin D., Harland T.A., Anchordoquy T.J., Graner M.W. Glioblastoma multiforme-derived extracellular vesicles drive normal astrocytes towards a tumour-enhancing phenotype. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20160477. doi: 10.1098/rstb.2016.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel B.B., Quaranta V. Metabolic plasticity meets gene regulation. Proc. Natl. Acad. Sci. U S A. 2019;116:3370–3372. doi: 10.1073/pnas.1900169116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Thomas G., Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- Rich J.N., Guo C., Mclendon R.E., Bigner D.D., Wang X.F., Counter C.M. A genetically tractable model of human glioma formation. Cancer Res. 2001;61:3556–3560. [PubMed] [Google Scholar]
- Ridder K., Keller S., Dams M., Rupp A.K., Schlaudraff J., Del Turco D., Starmann J., Macas J., Karpova D., Devraj K. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12:e1001874. doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder K., Sevko A., Heide J., Dams M., Rupp A.K., Macas J., Starmann J., Tjwa M., Plate K.H., Sultmann H. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology. 2015;4:e1008371. doi: 10.1080/2162402X.2015.1008371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.E., Sneider A., Witwer K.W., Bergese P., Bhattacharyya S.N., Cocks A., Cocucci E., Erdbrugger U., Falcon-Perez J.M., Freeman D.W. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: an ISEV position paper arising from the ISEV membranes and EVs workshop. J. Extracell Vesicles. 2019;8:1684862. doi: 10.1080/20013078.2019.1684862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Wurdinger T., Van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Luo Q., Long H., Hu Z., Que T., Zhang X., Li Z., Wang G., Yi L., Liu Z. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol. Cancer. 2014;13:65. doi: 10.1186/1476-4598-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa F.L., Nelson-Sathi S., Martin W.F. One step beyond a ribosome: the ancient anaerobic core. Biochim. Biophys. Acta. 2016;1857:1027–1038. doi: 10.1016/j.bbabio.2016.04.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer A., Zimmermann M., Doerfler A., Oberndorfer S., Buchfelder M., Coras R., Kitzwogerer M., Roessler K. Intratumoral heterogeneity of oxygen metabolism and neovascularization uncovers 2 survival-relevant subgroups of IDH1 wild-type glioblastoma. Neuro Oncol. 2018;20:1536–1546. doi: 10.1093/neuonc/noy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland M., Stoll E.A. Metabolic reprogramming in glioma. Front. Cell Dev. Biol. 2017;5:43. doi: 10.3389/fcell.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimpakos A.S., Karapanagiotou E.M., Saif M.W., Syrigos K.N. The role of mTOR in the management of solid tumors: an overview. Cancer Treat. Rev. 2009;35:148–159. doi: 10.1016/j.ctrv.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Sulima S.O., Hofman I.J.F., De Keersmaecker K., Dinman J.D. How ribosomes translate cancer. Cancer Discov. 2017;7:1069–1087. doi: 10.1158/2159-8290.CD-17-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri B., Soleimani M., Aval S.F., Memari F., Zarghami N. C6 glioma-derived microvesicles stimulate the proliferative and metastatic gene expression of normal astrocytes. Neurosci. Lett. 2018;685:173–178. doi: 10.1016/j.neulet.2018.08.034. [DOI] [PubMed] [Google Scholar]
- Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touat M., Idbaih A., Sanson M., Ligon K.L. Glioblastoma targeted therapy: updated approaches from recent biological insights. Ann. Oncol. 2017;28:1457–1472. doi: 10.1093/annonc/mdx106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Drapkina O., Tonevitsky A. Transcriptome of extracellular vesicles: state-of-the-art. Front. Immunol. 2019;10:202. doi: 10.3389/fimmu.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Van Der Vos K.E., Abels E.R., Zhang X., Lai C., Carrizosa E., Oakley D., Prabhakar S., Mardini O., Crommentuijn M.H., Skog J. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol. 2016;18:58–69. doi: 10.1093/neuonc/nov244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S., Thompson C.B. Metabolic reprogramming in brain tumors. Annu. Rev. Pathol. 2017;12:515–545. doi: 10.1146/annurev-pathol-012615-044329. [DOI] [PubMed] [Google Scholar]
- Viale A., Corti D., Draetta G.F. Tumors and mitochondrial respiration: a neglected connection. Cancer Res. 2015;75:3685–3686. doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu J., Sun G., Meng H., Wang J., Guan Y., Yin Y., Zhao Z., Dong X., Yin S. Glioblastoma extracellular vesicles induce the tumour-promoting transformation of neural stem cells. Cancer Lett. 2019;466:1–12. doi: 10.1016/j.canlet.2019.09.004. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang H., Li Z., Wu Q., Lathia J.D., Mclendon R.E., Hjelmeland A.B., Rich J.N. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Batagov A.O., Schinelli S., Wang J., Wang Y., El Fatimy R., Rabinovsky R., Balaj L., Chen C.C., Hochberg F. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017;8:1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen P.Y., Weller M., Lee E.Q., Alexander B.A., Barnholtz-Sloan J.S., Barthel F.P., Batchelor T.T., Bindra R.S., Chang S.M., Chiocca E.A. Glioblastoma in adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro. Oncol. 2020 doi: 10.1093/neuonc/noaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A., Agnihotri S., Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget. 2010;1:552–562. doi: 10.18632/oncotarget.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Gong J., Wang C., Wang Y., Song Y., Xu W., Liu Z., Liu Y. The diagnostic value and functional roles of phosphoglycerate mutase 1 in glioma. Oncol. Rep. 2016;36:2236–2244. doi: 10.3892/or.2016.5046. [DOI] [PubMed] [Google Scholar]
- Yu L., Lu M., Jia D., Ma J., Ben-Jacob E., Levine H., Kaipparettu B.A., Onuchic J.N. Modeling the genetic regulation of cancer metabolism: interplay between glycolysis and oxidative phosphorylation. Cancer Res. 2017;77:1564–1574. doi: 10.1158/0008-5472.CAN-16-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Freitas D., Kim H.S., Fabijanic K., Li Z., Chen H., Mark M.T., Molina H., Martin A.B., Bojmar L. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018;20:332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Acquaviva J., Ramachandran P., Boskovitz A., Woolfenden S., Pfannl R., Bronson R.T., Chen J.W., Weissleder R., Housman D.E. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc. Natl. Acad. Sci. U S A. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Lai M., Zhang Y., Zheng L., Xing Z., Li T., Zou Z., Song Q., Zhao X., Xia L. Exosome release is regulated by mTORC1. Adv. Sci. (Weinh) 2019;6:1801313. doi: 10.1002/advs.201801313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq fq files and processed data generated during this study are available through GEO, accession code GSE143189.