Abstract
Chemical modifications of small interfering (si)RNAs are used to enhance their stability and potency, and to reduce possible off-target effects, including immunogenicity. We have earlier introduced highly effective antiviral siRNA swarms against herpes simplex virus (HSV), targeting 653 bp of the essential UL29 viral gene. Here, we report a method for enzymatic production and antiviral use of 2′-fluoro-modified siRNA swarms. Utilizing the RNA-dependent RNA polymerase from bacteriophage phi6, we produced 2′-F-siRNA swarms containing either all or a fraction of modified adenosine, cytidine or uridine residues in the antisense strand of the UL29 target. The siRNA containing modified pyrimidines demonstrated high resistance to RNase A and the antiviral potency of all the UL29-specific 2′-F-siRNA swarms was 100-fold in comparison with the unmodified counterpart, without additional cytotoxicity. Modest stimulation of innate immunity signaling, including induced expression of both type I and type III interferons, as well as interferon-stimulated gene 54, by 2′-F-cytidine and 2′-F-uridine modified siRNA swarms occurred at early time points after transfection while the 2′-F-adenosine-containing siRNA was similar to the unmodified antiviral siRNA swarm in this respect. The antiviral efficacy of the 2′-F-siRNA swarms and the elicited cellular innate responses did not correlate suggesting that innate immunity pathways do not significantly contribute to the observed enhanced antiviral activity of the modified siRNAs. The results support further applications of enzymatically produced siRNA molecules with incorporated adenosine nucleotides, carrying fluoro-modification on ribose C2′ position, for further antiviral studies in vitro and in vivo.
Keywords: Antiviral, Herpes simplex virus, siRNA, Innate immunity, Fluoro-modification, Bacteriophage phi6 RNA-dependent RNA polymerase
Graphical abstract
Highlights
-
•
Phage phi6 polymerase can use 2′-F-dNTP substrates to produce 2′-F-modified dsRNA.
-
•
SiRNAs containing 2′-F-modified pyrimidine nucleotides demonstrate resistance to RNase A.
-
•
Enzymatically produced 2′-F-siRNA swarms display low cytotoxicity.
-
•
Antiviral activity of 2′-F-siRNAs against HSV exceeds that of the unmodified siRNAs.
-
•
Innate immunity induction by 2′-F-siRNAs is similar to that of unmodified siRNAs.
1. Introduction
RNA interference (RNAi) is a natural antiviral defense mechanism in plants, fungi, invertebrates, and, under specific conditions, in mammals (Guo et al., 2019; Schuster et al., 2019). Antiviral RNAi comes about via a sequence-specific binding of small interfering RNAs (siRNAs) to viral mRNAs or genomic RNAs, resulting in their degradation or translational repression. In general, siRNA is prone to fast degradation with RNases, and the delivery to the target cells might be difficult. A number of chemical modifications introduced to different positions in the siRNA (sense, antisense, or both strands) have been tested to improve siRNA stability and reduce off-target activities (Braasch et al., 2003; Choung et al., 2006; Czauderna et al., 2003; Fedorov et al., 2006; Jackson et al., 2006; Watts et al., 2008; Hassler et al., 2018).
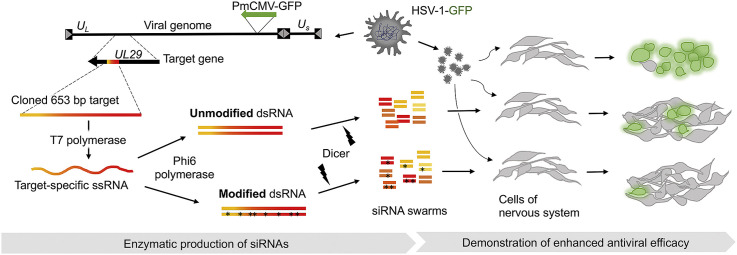
Instead of chemical synthesis and post-synthesis hybridization of RNA oligonucleotides, we produce high-quality molecules for RNAi using viral RNA polymerases and recombinant Giardia Dicer enzyme (Levanova and Poranen, 2018). This results in a swarm of 25-bp-long Dicer-substrate siRNAs (DsiRNAs) (Macrae et al., 2006; Romanovskaya et al., 2012), which can cover several kbp sequence of the target (Nygardas et al., 2009; Niehl et al., 2018; Jiang et al., 2019). When introduced into a cell, such siRNAs are processed by the endogenous Dicer, which enhances the RNAi potency and efficacy (Kim et al., 2005). In the enzymatically synthesized siRNA swarms, each individual siRNA species is present only in low concentrations, diluting out the potential off-target effects (Levanova and Poranen, 2018). The siRNA swarm contains predominantly 5′-monophosphates as only the single siRNAs originating from the very ends of the produced dsRNA molecules contain the 5′-triphosphate moiety, which could induce innate immunity responses. The swarm, comprising of numerous different siRNAs against the target, also widens the antiviral spectrum (Paavilainen et al., 2016; Jiang et al., 2019) and minimizes the potential of emergence of viral variants resistant to a single siRNA.
We have previously generated three antiviral siRNA swarms against herpes simplex virus type 1 (HSV-1) mRNAs encoding essential viral proteins, including glycoprotein B (UL27), the infected cell protein 8 (ICP8; UL29), and ICP27 (UL54) (Romanovskaya et al., 2012; Paavilainen et al., 2016). The siRNA swarm targeting mRNA of HSV single-stranded DNA binding protein ICP8, encoded by the UL29 gene, harbors most pronounced antiviral effect (Paavilainen et al., 2016) and induces only minimal non-specific cellular responses (Romanovskaya et al., 2012). The HSV UL29-derived siRNA swarm elicits modest IFN-λ1 (IL-29), IFN–λ2/3, ISG54, and TLR3 expression (Paavilainen et al., 2015). However, the observed slight activation of innate immune responses did not cause a decrease in cell viability (Romanovskaya et al., 2012; Paavilainen et al., 2015). The anti-HSV-UL29 siRNA swarm also controlled HSV infection in mice and inhibited local virus replication in corneal epithelia in vivo (Paavilainen et al., 2017).
Here, we set up an enzymatic production method for modified DsiRNAs using T7 DNA-dependent RNA polymerase (DdRp), phi6 RNA-dependent RNA polymerase (RdRp) and Giardia Dicer. Using this method, we produced UL29-specific siRNA swarms harboring 2′-fluoro-modifications in the ribose moieties of uridine, adenosine or cytidine residues. The incorporated fluoro-modifications generally improved siRNA stability. The 2′-F-siRNA swarms were well tolerated by cells and had enhanced HSV-specific antiviral activity compared to unmodified siRNA swarms.
2. Materials and methods
2.1. Enzymatic synthesis of siRNA swarms and their purification
The 653 bp sequence of the UL29 gene from HSV-1 prototype strain (17+) (GenBank JN555585.1, nucleotides 59,302 to 59,954) was PCR amplified to produce a DNA template for ssRNA production (Romanovskaya et al., 2012) using primers UL29_T7_Rev (5′-TAATACGACTCACTATAGGGCGCAACTTTCGCAATCAAT-3′) and UL29_phi6_Fwd (5′-GGAAAAAAAATGATGGCCGTAAGGGTGT-3′); underlined italics are recognition sequences of T7 DdRp and phi6 RdRp, respectively. The PCR product was converted into ssRNA by incubation with four canonical nucleoside triphosphates (NTPs; Thermo Fisher Scientific, #R0481) and T7 DdRp. The produced ssRNA (1 μg) was subsequently incubated for 6 h at 30 °C with 0.8 μg Phi6 RdRp and a 1 mM NTP mixture, including both canonical and selected sugar-modified NTPs (TriLink Biotechnologies, #N-1007, 2′-fluoro-2′-dATP; #N-1008, 2′-fluoro-2′-dCTP; #N-1009, 2′-fluoro-2′-dGTP; #N-1010, 2′-fluoro-2′-dUTP; #N-1017, 2′-O-Methyl-GTP; or #N-1018, 2′-O-Methyl-UTP) to produce dsRNAs. The generated dsRNA molecules were processed to swarms of 25-bp-long duplexes with in-house produced Giardia intestinalis Dicer (Romanovskaya et al., 2012; Paavilainen et al., 2017). The siRNA molecules were purified by anion-exchange chromatography on a CIMac QA column (BIA Separations, Slovenia, 110.5113–1.3) using an ÄKTApurifier FPLC system (GE Healthcare) (Romanovskaya et al., 2013) at the Instruct-HiLIFE Biocomplex unit (University of Helsinki). The same protocol was used to produce non-specific unmodified control siRNA swarms from a 413 bp long sequence of Escherichia coli lacI gene in pET32b vector using primers: Fwd_pET32b_1581: 5′-TAATACGACTCACTATAGGGCTGCCTGCACTAATGTTCC-3′ and Rev_pET32b_1983: 5′-GGAAAAAAATCGTCGTATCCCACTACC-3’. As confirmed with BLASTn, the selected control sequence has no significant similarities with human, herpesviral or mouse transcriptomes. The 88 bp dsRNA was produced as described previously (Jiang et al., 2011).
2.2. RNA stability
Generated siRNAs were treated with RNase A (#EN0531; Thermo Fisher) under the conditions suitable for dsRNA degradation: 1 μg siRNA was incubated with 0.1 μg of RNase A in 20 μl of 0.1 × SSC buffer (20 × SSC: 3 M sodium chloride + 0.3 M sodium citrate in water, pH 7.0) at room temperature for 15 min. The reactions were stopped by addition of 2.5 μl of 10 × loading dye [50% glycerol, 40 mM EDTA pH 8.0, 0.1% bromphenol blue (w/v), 0.1% xylene cyanol] and immediately frozen at −70 °C. The reaction products and equivalent amounts of undegraded siRNA were analyzed by electrophoresis on a 2.5% (w/v) agarose gel containing 0.2 μg/ml EtBr in Tris-acetate-EDTA (TAE) buffer at 90 V, 200 mA, 1 h. The RNAs were visualized with ChemiDoc Touch Imaging System (Bio-Rad) and the band intensities measured using Fiji (Schindelin et al., 2012). The relative amount of undegraded siRNA was calculated according to the formula: a%=(SRNaseA × 100)/Suntreated, where SRNaseA is a density of RNaseA-treated siRNA and Suntreated is a density of the equivalent amount of untreated sample.
2.3. Cells and virus
An astrocytoma-glioma cell line (U373MG) obtained originally from ATCC (Manassas, VA) was used. The line has been subsequently reclassified as U251 human glioblastoma, but referred here as U373MG to ensure continuity with our previous studies (Paavilainen et al., 2015, 2016; Romanovskaya et al., 2012). U373MG cells were maintained in DMEM with 10% heat inactivated fetal bovine serum (FBS) and 2 mM L-glutamine at 37 °C, 5% CO2. Vero cells (ATCC), used for plaque titration, were maintained in M199 medium with 5% FBS.
Green fluorescent protein (GFP)-expressing HSV-1 strain, HSV-1 (17+)Lox-PmCMVGFP (abbreviated here as HSV-1-GFP), originally received from prof. Beate Sodeik (MHH Hannover Medical School, Germany), was used for antiviral studies (Snijder et al., 2012; Mattila et al., 2015) and propagated as previously described (Romanovskaya et al., 2012).
2.4. Transfection
The U373MG cells were transfected on 96-well plates with either modified or unmodified siRNA swarms, 88 bp dsRNA or water (mock transfection) using Lipofectamine RNAiMAX (#13778-150; Invitrogen, Carlsbad, CA) according to the manufacturer's forward transfection protocol (Romanovskaya et al., 2012). Depending on the experiment, the cells were transfected with 1–10 pmols of RNA per well, which is equivalent to a concentration of 10–100 nM. All experiments were repeated at least twice with three or more biological replicates each.
2.5. Cellular viability
Cellular viabilities in siRNA treatments were assessed 48 h post transfection (hpt) with CellTiter-Glo (#G7571; Promega, Madison, WI) luminescent assay (Romanovskaya et al., 2012; Turunen et al., 2016). The luminescence was quantified with VICTOR Nivo Multimode Plate Reader (PerkinElmer, Waltham, MA). Viability of more than 80% compared to mock- and untreated cells was considered acceptable.
2.6. Quantitation of innate immunity responses by RT-qPCR
The total cellular RNA was extracted from siRNA-treated U373MG cells at 8, 24, and 48 hpt with TRI Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The RNA was processed to complementary DNA using RevertAid H Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA) with random hexamer primers (Thermo Fisher Scientific) after treatment with DNAse (Thermo Fisher Scientific) (Romanovskaya et al., 2012; Paavilainen et al., 2015, 2016).
Quantitative PCR (qPCR) was performed with Rotor-Gene Q, as previously described (Nygardas et al., 2011). The GAPDH housekeeping mRNA values of the corresponding sample were used for normalization of mRNA expression levels. The mRNA expression of interferon beta (IFN-β) (Peri et al., 2008), lambda 1 (IL-29; IFN-λ1) (Paavilainen et al., 2016), interferon stimulated gene 54 (ISG54) (Romanovskaya et al., 2012), and GAPDH (Nygardas et al., 2009) was quantified using previously published primer sequences.
2.7. Quantitation of antiviral efficacy
The RNA- or control-treated cells were challenged with a clinically relevant infectious dose of HSV-1-GFP at 1000 plaque forming units (pfu) in 100 μl per well [multiplicity of infection (MOI) ≈ 0.06] at 4 hpt in 96-well plates as before (Paavilainen et al., 2016). At 44 h post infection (hpi), the culture supernatant was collected, and the viral titer was determined by plaque titration on Vero cells in 96-well plates.
2.8. Live-cell imaging
The HSV-1-GFP strain enabled fluorescent imaging of the virus infection and thus also of the antiviral effect. The live cell imaging was performed 48 hpt with EVOS Auto FL (Thermo Fisher Scientific) with the whole plate scan program, keeping the imaging parameters equal throughout the scan.
2.9. Statistical analysis
Statistical analysis was conducted with SPSS Statistics 26.0.0.0. (IBM, Armonk, NY). Statistical significances were calculated with Mann-Whitney's non-parametric U test comparing two independent groups.
3. Results
3.1. Phi6 RdRp can use 2′-F-dNTPs as substrate to produce 2′-F-dsRNA for dicer reaction
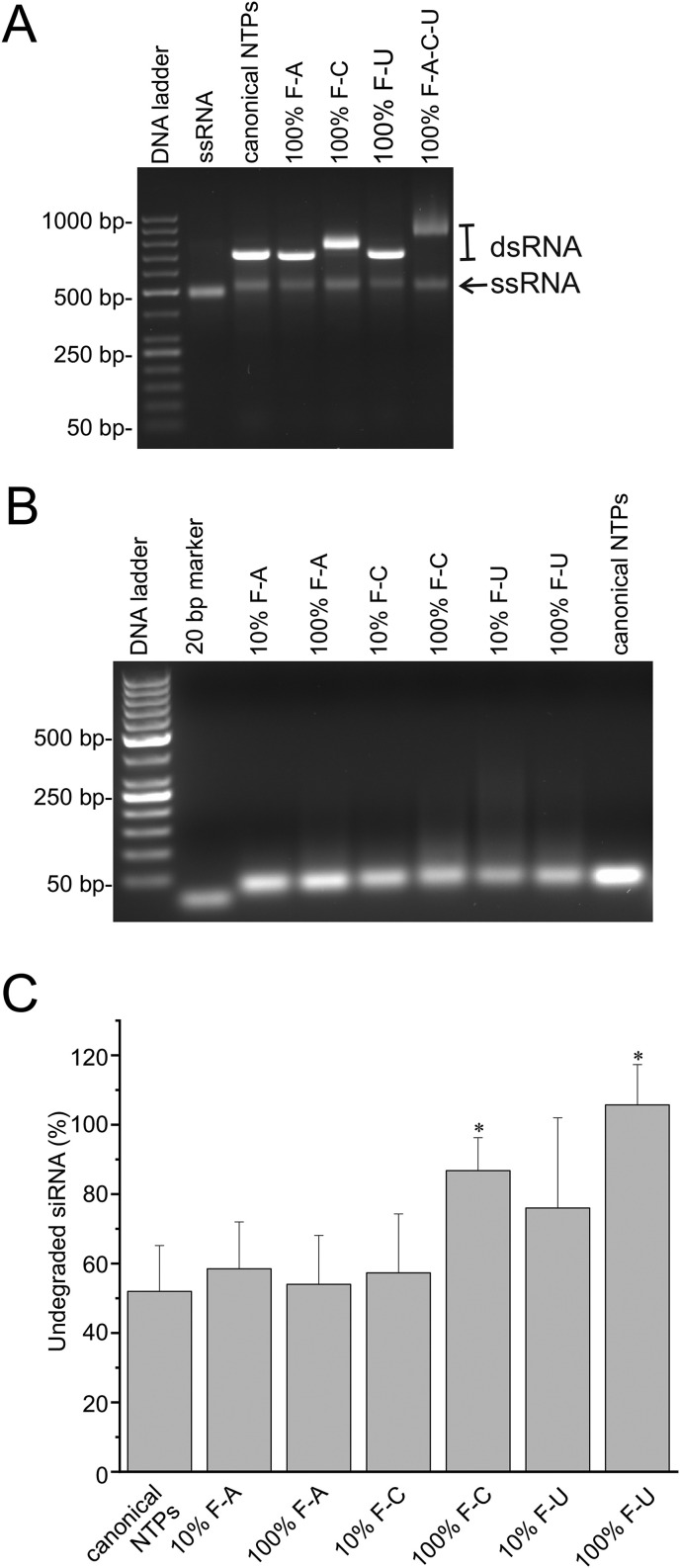
We tested the capability of phi6 RdRp to utilize 2′-F-dNTPs and 2′-OMe-NTPs in the dsRNA synthesis. HSV UL29 gene-specific ssRNA, produced using T7 DdRp, was used as a template for the phi6 RdRp-catalyzed dsRNA synthesis. Initially, the reactions were done by replacing one of the canonical NTPs in the reaction mixture with the corresponding modified NTP. Although dsRNA synthesis was not effective in the presence of 2′-OMe-modified NTPs (Fig. S1) and the quality of the dsRNA product was somewhat compromized in the presence of 2′-F-dGTP (Fig. S2A), phi6 RdRp was able to produce full-length dsRNA products using 2′-F-dATP, 2′-F-dUTP, and 2′-F-dCTP substrates (Fig. 1 A), suggesting that the 113 adenosines, 128 uridines, and 185 cytidines in the antisense strand of the HSV UL29 target sequence can potentially be replaced with their fluoro-modified counterparts. Wild-type phi6 RdRp and a mutant RdRp containing substitution K→A at position 147 (K147A) were initially tested for dsRNA production. The K147A RdRp consistently provided higher dsRNA yields (Fig. S2B) and was therefore used for the subsequent siRNA production. Complete replacement of a canonical rNTP with the corresponding 2′-F-dNTP resulted in slightly decreased yields, which was especially pronounced when all three NTPs (ATP, UTP and CTP) carried the modification (Fig. 1A). Therefore, to produce siRNA swarms for the cellular experiments we used 2′-F-dATP, 2′-F-dCTP, or 2′-F-dUTP, but not their mixture. Furthermore, we set up a dsRNA synthesis reaction in which only a fraction (10%) of a given NTP was fluoro-modified (Fig. S2). The mass spectrometry analysis confirmed that the “10% modified” dsRNA products contained 2′-fluoro-modified nucleotides (Fig. S3). Furthermore, the Giardia Dicer could process the produced UL29-2′-F-dsRNA into a swarm of Dicer-substrate 2′-F-siRNAs (Fig. 1B).
Fig. 1.
Enzymatic production and stability of fluoro-modified dsRNA and siRNA. (A) DsRNA synthesis reactions using 2′-F-dNTPs. Identical dsRNA synthesis reactions were carried out in parallel, and equal amounts from the reactions were analyzed by agarose gel electrophoresis. In the phi6 RdRp-catalyzed dsRNA synthesis reaction, unmodified canonical ATPs, CTPs or UTPs were replaced with 2′-F-dATPs (100% F-A), 2′-F-dCTPs (100% F-C), or 2′-F-dUTPs (100% F-U), respectively, or all the three NTPs were replaced with the corresponding 2′-F-dNTPs (100% F-A-C-U). Input ssRNA was loaded as a control (second well). Since the number of CMPs in the sequence is significantly higher than that for AMPs or UMPs (185 vs 113 and 128, respectively), the dsRNA species containing 2′-F-CMPs (or all the three 2′-F-NMPs) demonstrated lower electrophoretic mobility than those containing 2′-F-AMP or 2′-F-UMP. (B) Production of 2′-F-siRNAs from the 2′-F-dsRNA. UL29-specific 2′-F-dsRNA was digested with Giardia Dicer, and after chromatographic purification the samples were analyzed by agarose gel electrophoresis. 10% indicates that only a fraction of a given NTP in the original dsRNA synthesis reaction was modified and the resulting siRNA swarms are designated 10% F-A, 10% F–C, or 10% F–U. (C) RNase A sensitivity of 2′-F-siRNAs. Samples of unmodified and modified siRNAs were treated with RNase A for 15 min followed by the reaction termination and agarose gel electrophoresis. An equivalent amount of the untreated samples were also loaded on the gel. Densitometric analysis of agarose gels from five independent RNase A treatments was carried out and percentages of undegraded siRNAs from total input siRNA amounts were calculated. The data are presented as mean ± S.D. Asterisks indicate a statistically significant difference from the unmodified control, p ≤ 0.05.
3.2. Stability, cytotoxicity and antiviral efficacy of the 2′-F-siRNA swarms
3.2.1. Stability of the 2′-F-siRNA swarms
RNase A treatment in low salinity conditions was performed to assess sensitivity of the 2′-F-siRNA swarms to RNase. Under the conditions used, about 50% of the unmodified siRNA was degraded (Fig. 1C), whereas fluoro-modifications in the uridine and cytidine residues significantly enhanced the stability of the siRNAs.
3.2.2. Modified 2′-F-siRNA swarms are well tolerated in vitro
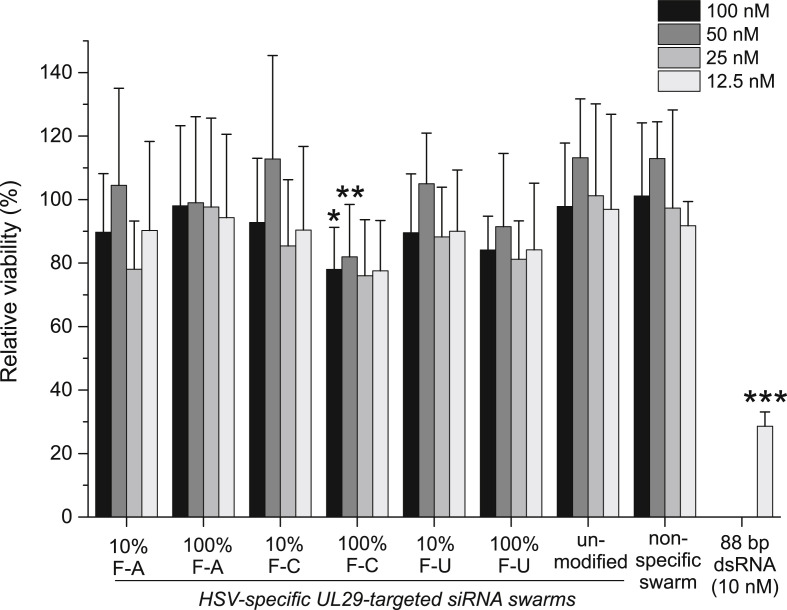
Cellular toxicity of the 2′-F-siRNA swarms was assessed using U373MG cells in 96-well plates. Cells were transfected with each of the HSV-specific UL29-gene derived-2′-F-siRNA swarms, an unmodified UL29-siRNA swarm, a non-specific siRNA swarm derived from E. coli lacI gene, a cytotoxic 88-bp-long control dsRNA, or water. The viability of the cells was assessed 48 hpt with luminescent assay (Fig. 2 ). Transfections with the 2′-F-siRNA swarms resulted in similar levels of cellular viability to that with unmodified UL29-siRNA swarms, with the exception of the fully 2′-F-dCTP modified swarm, which was significantly less tolerated than the unmodified siRNA swarm at 50 and 100 nM. Nevertheless, all the 2′-F-siRNA swarms were well tolerated, with relative cellular viability consistently higher than 80% at doses relevant for antiviral applications. Hence, 2′-F-modifications had no effect on tolerability of the siRNA swarms in the studied concentration range. The UL29-targeting siRNA swarms did not display increased cytotoxicity in comparison to the non-specific siRNA swarm, targeting a non-relevant bacterial sequence. The 88 bp dsRNA used as a toxic control resulted in a clear decrease in cell viability at 10 nM concentration (Fig. 2). The cytotoxicity profile of the modified siRNA swarms was confirmed with non-cancerous human corneal epithelial cells, in which only the fully 2′-F-dCTP modified swarm was significantly less tolerated than the unmodified siRNA swarm (Fig. S4).
Fig. 2.
Viability of U373MG cells treated with 2′-F-siRNA swarms. U373MG cells were transfected with the indicated siRNA swarms at the concentration range of 12.5–100 nM. The antisense strand of the modified siRNA swarms was fully (referred to as 100%) or partially (referred to as 10%) modified containing either 2′-F-dAMP, 2′-F-dCMP or 2′-F-dUMP as the modified nucleotide (10% or 100% F-A, F–C or F–U, respectively). 88-bp-long phi6-derived dsRNA with known cytotoxicity was used as a control RNA, at a concentration of 10 nM. The viability of the cells was quantified at 48 hpt and shown as relative viability compared to mock- and untreated samples. The error bars represent the standard deviation of the mean from two separate experiments with 3–5 biological replicates each. Significant difference to the unmodified UL29-specific swarm is marked with asterisks (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001).
3.2.3. Fluoro-modifications increase antiviral efficacy of enzymatically produced UL29-siRNA swarms
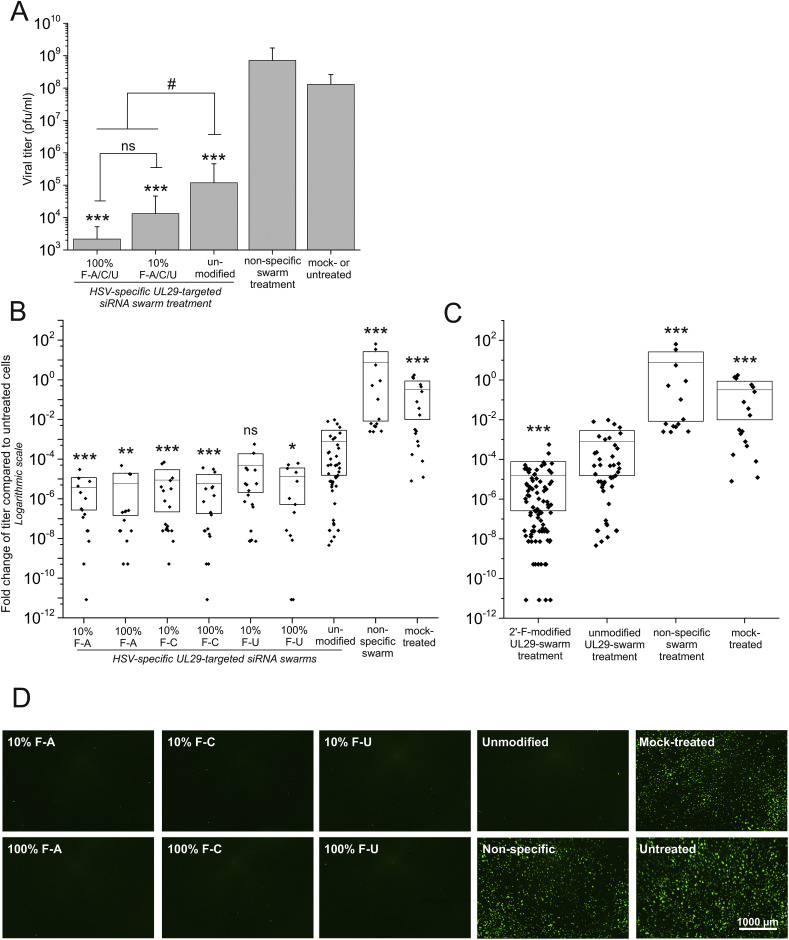
The antiviral efficacy of the UL29-2′-F-siRNA swarms was quantified at 48 hpt by plaque titration from culture supernatants of HSV-1-GFP-infected U373MG cells. Before infection, cells were treated with HSV-targeted modified or unmodified UL29-siRNA swarms, a non-specific siRNA swarm, water (mock-treated), or left untreated (Fig. 3 ). After treatment with the unmodified UL29 siRNA swarm, the viral titer was reduced at least three orders of magnitude compared to the untreated samples (Fig. 3A). The antiviral effect of the UL29-2′-F-siRNA swarms was even more pronounced, resulting in a five orders of magnitude reduction in HSV titer (Fig. 3B). Compared to treatment with the non-specific siRNA swarm, treatment with the modified and unmodified UL29 siRNA swarms resulted in a significant decrease of titer (p = 0.00000000165 and p = 0.000000178, respectively), confirming the sequence specificity of the antiviral effects (Fig. 3A). Furthermore, treatment with the UL29-2′-F-siRNA swarms in comparison with the unmodified counterparts resulted in significantly lower titers (p = 0.018) (Fig. 3A) and an extensive decrease of viral shedding (p = 0.000001) (Fig. 3C). The difference in antiviral efficacy between the 10% and 100% modified swarms was non-significant (Fig. 3A). Nonetheless, the 2′-F-siRNA swarms harboring modifications in cytidine or adenosine residues, i.e. 2′-F-dCMP-siRNA and 2′-F-dAMP-siRNA swarms respectively, yielded significantly higher antiviral potency compared to those having uridine modifications (2′-F-dUMP-siRNA), when using the unmodified UL29-siRNA swarm as a reference (Fig. 3B). The 10% modified 2′-F-dUMP-siRNA swarm was the only UL29-2′-F-siRNA swarm that did not show improved antiviral efficacy in comparison with the unmodified UL29-swarm (Fig. 3B).
Fig. 3.
Antiviral activity of siRNA swarms is improved by 2′-F-dNMP modifications. U373MG cells were infected with 1000 pfu of HSV-1-GFP (MOI≈0.06) at 4 h after treatment with each of the UL29-2′-F-siRNA swarms (50 nM), unmodified UL29-siRNA swarm (50 nM) or non-specific swarm (50 nM). The guide strand of modified siRNA swarms was fully (100%) or partially (10%) modified and contained 2′-F-dAMP, 2′-F-dCMP or 2′-F-dUMP as the modified nucleotide (10% or 100% F-A, F-C or F-U, respectively). The controls were transfected with water only (mock-treated) or were left untreated. At 48 hpt, the culture supernatants were collected for quantitation of the virus (A, B, and C), and live cell imaging was performed (D). (A) Viral titer (pfu/ml) was determined from the supernatant samples by plaque titration. The samples treated with partially modified (10%) 2′-F-siRNA swarms (A, C, or U), and those treated with fully (100%) modified 2′-F-siRNA swarms were grouped for statistical analysis. The error bars represent standard deviation of the mean from two separate experiments with at least three replicate samples of all individual treatments and controls (N = 15–33 or N = 6–12, respectively). The significance of the changes is shown against both the non-specific swarm as well as the mock- and untreated samples (***, p ≤ 0.001). The significant changes against a group of comparison are marked with # (#: p ≤ 0.05, ns: non-significant). (B, C) Viral shedding quantitated from the culture supernatants is shown for each individual siRNA swarm (B) or collectively for the modified siRNA swarms (C). The fold change of viral titer is presented in comparison with the untreated samples. The efficacies of the siRNA swarms, with the indicated modified nucleotides, are presented separately (N = 13–18) in panel B and collectively (N = 96) in panel C. The mean and standard deviation (shown with a horizontal line and a box, respectively) for each sample group represent five separate experiments (N = 13–96). The diamonds illustrate the individual samples. Significant changes against the unmodified UL29-specific swarm are marked with asterisks (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, ns: non-significant). (D) Representative fluorescent images of the HSV-1-GFP -infected cells with the indicated siRNA swarm or control treatments were taken at 48 hpt. The scale bar represents 1000 μm.
The efficacy of the HSV-targeted siRNA swarms against HSV-1-GFP infection on U373MG cells was visualized by living cell fluorescent imaging at 48 hpt (at 44 hpi; Fig. 3D). The virus-derived GFP signal was prominent in infected cells, untreated or control-treated, whereas in cells pretreated with unmodified UL29-siRNAs or UL29-2′-F-siRNAs, the GFP signal could not be detected using standard imaging parameters.
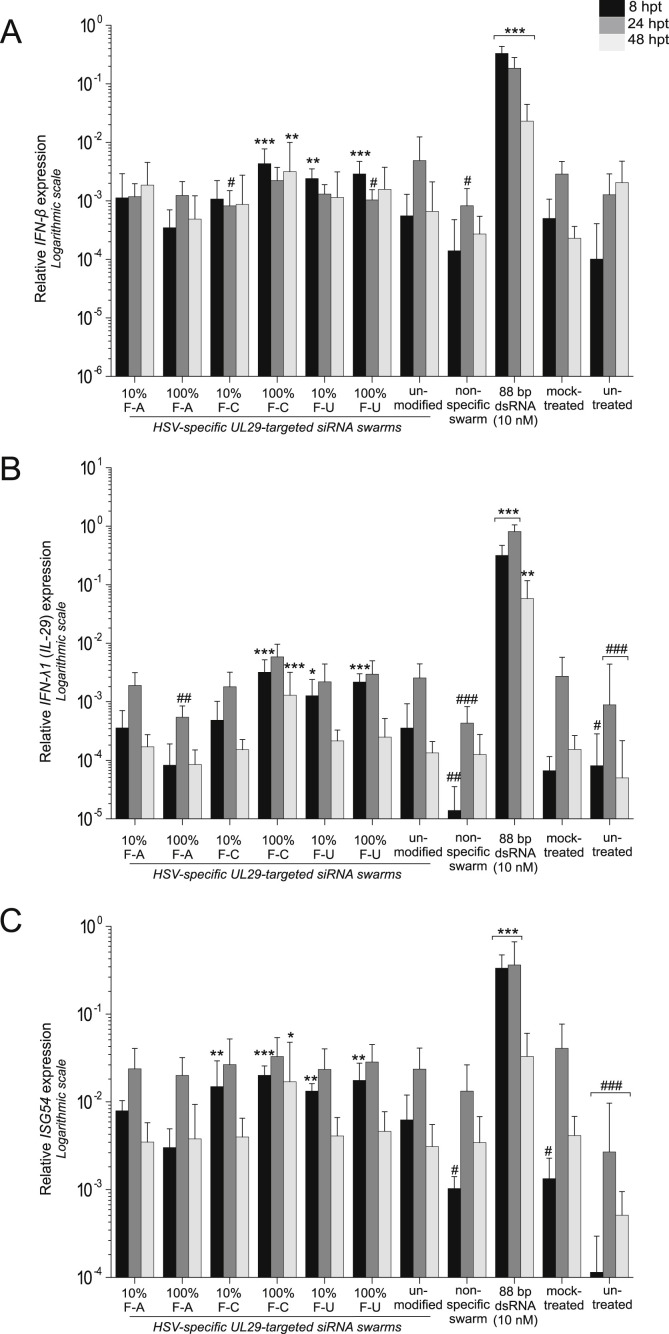
3.3. Induction of innate immunity responses by 2′-F-siRNA swarms
Consistent with earlier research, transfection of 88 bp dsRNA, an immunostimulatory and cytotoxic molecule (Jiang et al., 2011), induced IFN-β, IL-29 and ISG54 expression in the transfected cells (Fig. 4 ) (Romanovskaya et al., 2012; Paavilainen et al., 2015). In comparison with untreated cells, the unmodified UL29 siRNA swarm induced modest ISG54 and IL-29 responses (Fig. 4B and C) but did not significantly alter IFN-β expression (Fig. 4A) at any time point. Notably, only the ISG54 levels at 8 hpt differed significantly between the unmodified UL29 siRNA and transfection reagent alone (mock-treated) (Fig. 4C), indicating that the transfection reagent causes the majority of the responses in comparison with the untreated cells.
Fig. 4.
Innate immunity responses induced by 2′-F-siRNA swarms. U373MG cells were treated with the indicated siRNA swarms (50 nM) or controls. The antisense strand of the modified siRNA swarms used was fully (100%) or partially (10%) modified containing either 2′-F-dAMP, 2′-F-dCMP or 2′-F-dUMP as the modified nucleotide (10% or 100% F-A, F–C or F–U, respectively). The expression levels of interferon beta (IFN-β; panel A), interferon lambda-1 (IFN-λ1, IL-29; panel B) and interferon-stimulated gene 54 (ISG54; panel C) were quantified at 8 hpt (black bars), 24 hpt (dark grey bars) or 48 hpt (light grey bars) by RT-qPCR. The expression levels, normalized to housekeeping gene GAPDH, are presented on a logarithmic scale. The means and standard deviations, from two separate experiments with at least four repeat samples, are presented. Significantly higher induction of IFNs and ISGs in comparison to the unmodified UL29 siRNA swarm treatment is marked with asterisks (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001) and significantly lower induction with # (#: p ≤ 0.05, ##: p ≤ 0.01, ###: p ≤ 0.001). If no p-value is shown, the difference to the unmodified UL29-specific swarm is not significant.
2′-F-dUMP- and 2′-F-dCMP-siRNAs induced higher IFN-β, IL-29 and ISG54 expression compared to unmodified UL29-siRNA swarm, particularly at the earliest time point (Fig. 4). Innate immunity signaling induced by 2′-F-dAMP-siRNA swarms and that by the unmodified UL29 siRNA were at equal levels, or even lower. Overall, the induction of interferon signaling by modified or unmodified siRNA swarms was not extensive (only ca. 10-fold compared to mock- and untreated cells).
4. Discussion
We have previously reported a novel enzymatic approach to synthetize large pools of siRNA molecules, which we refer to as siRNA swarms. The antiviral siRNA swarms directed aginst the current target of choice, the UL29 gene of HSV-1, have previously demonstrated their efficacy against circulating pathogenic HSV strains in vitro and corneal infection in vivo, unveiling their potential as antiviral drugs. In the current study, we pursued the modification of the RNAs using nucleotides with 2′-modified ribose, in order to further enhance the antiviral potency and stability of the siRNA preparations. Eventually, we are able to report successful enzymatic incorporation of 2′-F-dNTP nucleotides to biologically active dsRNA molecules. All 2′-F-siRNA swarms were well tolerated and highly effective against HSV-1 in vitro. The observed enhanced antiviral efficacy of the 2′-F-siRNA swarms was unrelated to the elicited cellular innate immunity responses, proving the sequence specificity of the modified siRNAs.
We produced siRNA swarms, in which either all or a fraction of adenosines, cytidines or uridines were replaced by 2′-F-dNMPs. Replacement of a 2′-OH group of ribose with a fluorine has been shown to stabilize RNA molecules against nucleases (Monia et al., 1993). SiRNAs are primarily degraded by RNase A (Turner et al., 2007), which recognizes the 2′-OH of pyrimidine nucleosides for cleavage (Cuchillo et al., 2011). Therefore, a common strategy is to modify all pyrimidines (Hassler et al., 2018) to increase the siRNA stability. Accordingly, substitution of the canonical CMPs or UMPs by the corresponding 2′-F-dNMPs resulted in a significant increase in siRNA stability in our experiments (Fig. 1C).
Conventionally, modifications are introduced into RNA molecules using chemical solid phase synthesis. Some wild-type or mutant DdRps of autographiviruses (T7, Syn5) can be used for enzymatic synthesis of single-stranded 2′-F-RNAs (Sousa and Padilla, 1995; Zhu et al., 2015) which may be further hybridized to produce dsRNA. We demonstrate here that bacteriophage phi6 RdRp efficiently utilizes 2′-F-dNTPs allowing enzymatic production of perfectly duplexed dsRNAs without the often error-prone post-synthesis hybridization. Similarly, to other wild-type bacteriophage polymerases (Sousa and Padilla, 1995; Zhu et al., 2015), phi6 RdRp did not tolerate 2′-OMe modifications (Fig. S2).
All of the modified siRNA swarms were non-toxic for U373MG cells at the relevant antiviral concentrations. The findings were reproducible in human corneal epithelial cells (Fig. S4), which represent another potential target tissue for antiviral therapy of diseases caused by HSV-1. Furthermore, modified swarms had higher antiviral activity than the unmodified swarm, as demonstrated by the significant reduction of viral titer in plaque assays. Notably, there was no difference between siRNA swarms containing only a fraction of modified dNMPs and those containing all modified dCMPs, dUMPs, or dAMPs in the antisense strand (Fig. 3). The antiviral effect of siRNAs containing only a fraction of modified dUMPs was not statistically different from the unmodified siRNA swarm. Interestingly, a higher stability to nucleases of siRNA swarms containing 2′-F-dUMPs or 2′-F-dCMPs did not translate into higher antiviral efficacy than those with 2′-F-dAMPs.
The immunostimulatory potential of 2′-F-dAMP-siRNAs resembled that of the unmodified siRNA swarm and was thus minimal. At the early time point (8 hpt), 2′-F-dUMP-siRNA and 2′-F-dCMP-siRNA swarms induced slightly higher expression of IFN-β, IL-29, and ISG54 than the unmodified siRNA, implying that their antiviral activity at earlier time points might be partly non-specific and related to the stimulation of the innate immune response. However, 2′-F-dAMP-siRNA swarms demonstrated enhanced antiviral efficacy in comparison with the unmodified swarm, but similar immunostimulatory activity. Therefore, antiviral 2′-F-dAMP-siRNA swarm is a promising candidate for further in vivo studies with HSV. The elevated potency in combination with the existing benefits of the siRNA swarm approach offers also a new, potential approach for therapy of virus infections lacking treatment so far, including novel emerging viruses, such as SARS-CoV-2.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
Dr. Ian MacRae is thanked for the generous gift of pFB-GiDcr plasmid used for the production of Giardia Dicer. Tanja Westerholm, Ritva Kajander, and Marja-Leena Mattila are thanked for technical assistance. Prof. Arto Urtti (University of Helsinki and University of Eastern Finland, Finland) is acknowledged for providing the corneal epithelial cells.
This work benefited from access to the Instruct-HiLIFE Biocomplex unit, a FINStruct and Instruct-ERIC center and a member of Biocenter Finland. The use of the facilities and expertise of Metabo-HiLIFE Viikki Metabolomics Unit, member of Biocenter Finland, is gratefully acknowledged. The work was financially supported by the Jane and Aatos Erkko Foundation, grant #170046, and by Sigrid Jusélius Foundation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104916.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Braasch D.A., Jensen S., Liu Y., Kaur K., Arar K., White M.A., Corey D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Cuchillo C.M., Nogues M.V., Raines R.T. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna F., Fechtner M., Dames S., Aygun H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y., Anderson E.M., Birmingham A., Reynolds A., Karpilow J., Robinson K., Leake D., Marshall W.S., Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Li Y., Ding S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019;19:31–44. doi: 10.1038/s41577-018-0071-x. [DOI] [PubMed] [Google Scholar]
- Hassler M.R., Turanov A.A., Alterman J.F., Haraszti R.A., Coles A.H., Osborn M.F., Echeverria D., Nikan M., Salomon W.E., Roux L., Godinho B., Davis S.M., Morrissey D.V., Zamore P.D., Karumanchi S.A., Moore M.J., Aronin N., Khvorova A. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 2018;46:2185–2196. doi: 10.1093/nar/gky037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., Marshall W., Khvorova A., Linsley P.S. Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Osterlund P., Sarin L.P., Poranen M.M., Bamford D.H., Guo D., Julkunen I. Innate immune responses in human monocyte-derived dendritic cells are highly dependent on the size and the 5' phosphorylation of RNA molecules. J. Immunol. 2011;187:1713–1721. doi: 10.4049/jimmunol.1100361. [DOI] [PubMed] [Google Scholar]
- Jiang M., Osterlund P., Westenius V., Guo D., Poranen M.M., Bamford D.H., Julkunen I. Efficient inhibition of avian and seasonal influenza A viruses by a virus-specific dicer-substrate small interfering RNA swarm in human monocyte-derived macrophages and dendritic cells. J. Virol. 2019;93:e01916–e01918. doi: 10.1128/JVI.01916-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Levanova A., Poranen M.M. RNA interference as a prospective tool for the control of human viral infections. Front. Microbiol. 2018;9:2151. doi: 10.3389/fmicb.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Mattila R.K., Harila K., Kangas S.M., Paavilainen H., Heape A.M., Mohr I.J., Hukkanen V. An investigation of herpes simplex virus type 1 latency in a novel mouse dorsal root ganglion model suggests a role for ICP34.5 in reactivation. J. Gen. Virol. 2015;96:2304–2313. doi: 10.1099/vir.0.000138. [DOI] [PubMed] [Google Scholar]
- Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. Evaluation of 2'-modified oligonucleotides containing 2'-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- Niehl A., Soininen M., Poranen M.M., Heinlein M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018;16:1679–1687. doi: 10.1111/pbi.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygardas M., Aspelin C., Paavilainen H., Roytta M., Waris M., Hukkanen V. Treatment of experimental autoimmune encephalomyelitis in SJL/J mice with a replicative HSV-1 vector expressing interleukin-5. Gene Ther. 2011;18:646–655. doi: 10.1038/gt.2011.4. [DOI] [PubMed] [Google Scholar]
- Nygardas M., Vuorinen T., Aalto A.P., Bamford D.H., Hukkanen V. Inhibition of coxsackievirus B3 and related enteroviruses by antiviral short interfering RNA pools produced using phi6 RNA-dependent RNA polymerase. J. Gen. Virol. 2009;90:2468–2473. doi: 10.1099/vir.0.011338-0. [DOI] [PubMed] [Google Scholar]
- Paavilainen H., Lehtinen J., Romanovskaya A., Nygardas M., Bamford D.H., Poranen M.M., Hukkanen V. Inhibition of clinical pathogenic herpes simplex virus 1 strains with enzymatically created siRNA pools. J. Med. Virol. 2016;88:2196–2205. doi: 10.1002/jmv.24578. [DOI] [PubMed] [Google Scholar]
- Paavilainen H., Romanovskaya A., Nygardas M., Bamford D.H., Poranen M.M., Hukkanen V. Innate responses to small interfering RNA pools inhibiting herpes simplex virus infection in astrocytoid and epithelial cells. Innate Immun. 2015;21:349–357. doi: 10.1177/1753425914537921. [DOI] [PubMed] [Google Scholar]
- Paavilainen H., Lehtinen J., Romanovskaya A., Nygardas M., Bamford D.H., Poranen M.M., Hukkanen V. Topical treatment of herpes simplex virus infection with enzymatically created siRNA swarm. Antivir. Ther. 2017;22:631–637. doi: 10.3851/IMP3153. [DOI] [PubMed] [Google Scholar]
- Peri P., Mattila R.K., Kantola H., Broberg E., Karttunen H.S., Waris M., Vuorinen T., Hukkanen V. Herpes simplex virus type 1 Us3 gene deletion influences toll-like receptor responses in cultured monocytic cells. Virol. J. 2008;5:140. doi: 10.1186/1743-422X-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanovskaya A., Paavilainen H., Nygardas M., Bamford D.H., Hukkanen V., Poranen M.M. Enzymatically produced pools of canonical and Dicer-substrate siRNA molecules display comparable gene silencing and antiviral activities against herpes simplex virus. PloS One. 2012;7 doi: 10.1371/journal.pone.0051019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanovskaya A., Sarin L.P., Bamford D.H., Poranen M.M. High-throughput purification of double-stranded RNA molecules using convective interaction media monolithic anion exchange columns. J. Chromatogr. A. 2013;1278:54–60. doi: 10.1016/j.chroma.2012.12.050. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S., Miesen P., van Rij R.P. Antiviral RNAi in insects and mammals: parallels and differences. Viruses. 2019;11 doi: 10.3390/v11050448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder B., Sacher R., Ramo P., Liberali P., Mench K., Wolfrum N., Burleigh L., Scott C.C., Verheije M.H., Mercer J., Moese S., Heger T., Theusner K., Jurgeit A., Lamparter D., Balistreri G., Schelhaas M., De Haan C.A., Marjomaki V., Hyypia T., Rottier P.J., Sodeik B., Marsh M., Gruenberg J., Amara A., Greber U., Helenius A., Pelkmans L. Single-cell analysis of population context advances RNAi screening at multiple levels. Mol. Syst. Biol. 2012;8:579. doi: 10.1038/msb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.J., Jones S.W., Moschos S.A., Lindsay M.A., Gait M.J. MALDI-TOF mass spectral analysis of siRNA degradation in serum confirms an RNAse A-like activity. Mol. Biosyst. 2007;3:43–50. doi: 10.1039/B611612D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen A., Hukkanen V., Kulmala J., Syrjanen S. HSV-1 infection modulates the radioresponse of a HPV16-positive head and neck cancer cell line. Anticancer Res. 2016;36:565–574. [PubMed] [Google Scholar]
- Watts J.K., Deleavey G.F., Damha M.J. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Zhu B., Hernandez A., Tan M., Wollenhaupt J., Tabor S., Richardson C.C. Synthesis of 2'-fluoro RNA by Syn5 RNA polymerase. Nucleic Acids Res. 2015;43:e94. doi: 10.1093/nar/gkv367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.