Abstract
Wastewater-based epidemiology (WBE) has been used to analyze markers in wastewater treatment plant (WWTP) influent to characterize emerging chemicals, drug use patterns, or disease spread within communities. This approach can be particularly helpful in understanding outbreaks of disease like the novel Coronavirus disease-19 (COVID-19) when combined with clinical datasets. In this study, three RT-ddPCR assays (N1, N2, N3) were used to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in weekly samples from nine WWTPs in southeastern Virginia. In the first several weeks of sampling, SARS-CoV-2 detections were sporadic. Frequency of detections and overall concentrations of RNA within samples increased from mid March into late July. During the twenty-one week study, SARS-CoV-2 concentrations ranged from 101 to 104 copies 100 mL−1 in samples where viral RNA was detected. Fluctuations in population normalized loading rates in several of the WWTP service areas agreed with known outbreaks during the study. Here we propose several ways that data can be presented spatially and temporally to be of greatest use to public health officials. As the COVID-19 pandemic wanes, it is likely that communities will see increased incidence of small, localized outbreaks. In these instances, WBE could be used as a pre-screening tool to better target clinical testing needs in communities with limited resources.
Keywords: Wastewater-based epidemiology, COVID-19, SARS-CoV-2, RT-ddPCR
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19), was first documented in late 2019 and declared a global pandemic on March 11, 2020 by the World Health Organization (WHO). The virus responsible for COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is an enveloped, single-stranded RNA virus that has been characterized by high infectivity, relatively high asymptomatic ratio in the population, and potential to result in serious health complications (Bai et al., 2020; Gerrityet al., 2020; Zhou et al., 2020).
Although COVID-19 clinical tests were developed rapidly, production and distribution did not keep up with high demand. Thus, testing was often reserved only for individuals who met strict requirements including symptomology and recent travel to high risk areas (CDC 2020). With these limitations on clinical testing, it is likely that many individuals, both with and without symptoms, were not included in the COVID-19 case estimates being used to make public health decisions (Murakami et al., 2020). Seropositive testing shows promise for retrospective understanding of asymptomatic rates, disease spread within a population, and reinfection risks (Yongchenet al., 2020). However, an additional method for real time or near real time tracking of disease spread at a population level that can inform public health decisions without being invasive is needed.
Wastewater-based epidemiology (WBE) can be used to observe community-level trends through analysis of various markers in wastewater to make inferences about the population (Choi et al., 2018). Although recent WBE studies have primarily focused on pharmaceutical and illicit drug use (Choi et al., 2018; Causanilleset al., 2017; Baz-Lombaet al., 2016; van Nuijis et al., 2011), this approach has promise for better understanding the spread of infectious disease within a population. In fact, some studies looking at various pathogens with WBE were published prior to the COVID-19 pandemic (Hoviet al., 2012; Hellmeret al., 2014; Bisseuxet al., 2018; Brouweret al., 2018). Because wastewater sampling captures the aggregated community signal, it can potentially be used to identify regions where disease incidence is increasing, but remains undetected via individual clinical testing (Peccia et al 2020). In addition, WBE has the potential to identify both symptomatic and asymptomatic individuals (Bivinset al., 2020). This not only results in a less biased dataset, particularly when individual test kits are limited (Murakami et al., 2020), but can also incorporate the asymptomatic population into the crucial assessment of the true population prevalence for epidemiological response and modeling. Lack of a reliable SARS-CoV-2 stool shedding rate is the current limitation in the use of WBE to estimate total infection within a community. Thus, when used in concert with clinical testing data, WBE has the potential to be a powerful tool for officials to use when making public health decisions.
Use of WBE for COVID-19 detection shows much promise. Whilst the routes of infection for people to develop COVID-19 are via exposure to respiratory tract bioaerosol droplets, SARS-CoV-2 RNA has been detected in stool samples from both symptomatic and asymptomatic infected individuals (Holshueet al., 2020; Caiet al., 2020; Tang et al., 2020; Wölfelet al., 2020; Xiao et al., 2020; Zanget al., 2020; Zhang et al., 2020a; Zhang et al., 2020b). Viral shedding in stool samples is likely due to infection of gastrointestinal cells in patients and can continue even after the individual no longer tests positive based on respiratory tract assays (Wölfelet al., 2020; Xiao et al., 2020; Zanget al., 2020a). Although there is indication that virus shed in stool are no longer viable (Wölfelet al., 2020; Zanget al., 2020a), there is not yet consensus regarding whether SARS-CoV-2 should be considered a fecal-oral virus. Nevertheless, shedding of SARS-CoV-2 RNA by infected individuals into wastewater supports the use of WBE as an indicator of COVID-19 presence in communities.
To date, several studies documenting SARS-CoV-2 RNA in wastewater samples around the world have been published (e.g. Ahmed et al., 2020; Kumar et al., 2020; Medemaet al., 2020; Randazzoet al., 2020; Sherchanet al., 2020). While these early publications were intended to quickly establish a proof of concept for WBE of COVID-19, a large-scale study would be helpful to further validate this approach.
Here we present a regional study of SARS-CoV-2 RNA in wastewater during the rise of COVID-19 cases in southeastern Virginia, USA over the course of a twenty-one week period. Within this study we observed that wastewater measurements of SARS-CoV-2 RNA were a viable means to describe the occurrence and trends (onset) in SARS-CoV-2 infection. Our results indicate the production and sharing of WBE datasets with local health agencies will provide an additional source of reliable information that can be used by governments to inform public health responses to future health crises.
2. Methods
2.1. Hampton Roads Sanitation District
Hampton Roads Sanitation District (HRSD) is a political subdivision of the Commonwealth of Virginia, with a service area of approximately 3,100 square miles that includes 18 cities and counties of southeast Virginia, and serves a population of 1.7 million. A combined capacity of 249 million gallons per day includes nine major (design flow 15-54 MGD) and seven smaller (design flow 0.025-0.1 MGD) wastewater treatment plants (WWTPs).
2.2. Sample collection
Weekly 1L raw wastewater influent samples were aseptically collected at HRSD's nine major plants (Atlantic (AT), Army Base (AB), Boat Harbor (BH), Chesapeake-Elizabeth (CE), James River (JR), Nansemond (NP), Virginia Initiative Plant (VIP), Williamsburg (WB), York River (YR)) beginning the week of March 9th. Flow-weighted composite samples were collected over the course of 24-hours at AT, JR, and VIP plants, while at the remaining plants, grab samples were collected. Samples were gathered mid-morning (between 800 - 1100) and then brought back to HRSD's Central Environmental Laboratory on ice within 6 hours. Samples were immediately concentrated upon receipt, followed by molecular processing within the same week, as described below.
2.3. SARS-CoV-2 concentration, RNA extraction, and quantification
Reverse transcription droplet digital PCR (RT-ddPCR) was used to enumerate SARS-CoV-2 RNA copies using three CDC diagnostic panel assays (Lu et al., 2020). Primer and probe information used in this study are summarized in Supplemental Table S1.
Wastewater concentration was done using an InnovaPrep Concentrating Pipette Select (InnovaPrep, Drexel, MO, USA) for the first 13 weeks, then using electronegative filtration for the remaining 8 weeks. Total recovery for the 2 concentration method workflows were determined by spiking in bovine coronavirus (CALF-GUARD; Zoetis, Parsippany, NJ) and bovine respiratory syncytial virus (Inforce 3 Cattle Vaccine; Zoetis) into 12 wastewater samples from different WWTPs. Recovered concentrations (see Supplemental Table S1 for primers and probes) were converted to percent recovery by dividing by the total spiked concentration (2.34 × 108 copies of bovine coronavirus and 1.14 × 109 copies of bovine respiratory syncytial virus).
For the InnovaPrep (InnovaPrep, Drexel, MO, USA) concentration, raw wastewater samples (125 mL) were centrifuged using an Eppendorf 5804 R (Eppendorf, Hamburg, Germany) for 10 minutes at 10,000 g. Supernatant (100 mL) was then concentrated using a 0.05 μm PS Hollow Fiber concentrating pipette tip on the InnovaPrep Concentrating Pipette Select (InnovaPrep, Drexel, MO, USA). Immediately after filtration, the retentate was eluted with 250-500 μL of Elution Fluid-Tris (InnovaPrep, Drexel, MO, USA). For electronegative concentration, mixed cellulose ester HA filters (HAWP04700; Millipore, Billerica, MA, USA) were used to concentrate SARS-CoV-2 in 100 ml water samples. MgCl2 was added to a final concentration of 25 mM prior to filtration, then the samples were acidified to a pH of 3.5 with 20% HCl. Immediately after InnovaPrep elution or HA filtration, eluate or HA filters were stored in a ‐80°C freezer until total nucleic extraction using NucliSENS easyMag (bioMerieux, Inc., Durham, NC, USA) was completed.
Prior to extraction, 10 μL of 1 × 106 copies/μL Hep G Armored RNA (Asuragen, Austin, TX, USA) was spiked in the lysis buffer with all samples and controls to quantify matrix inhibition. All extractions were performed according to the manufacturer's protocol B 2.0.1 with modifications. The protocol was modified with a 30‐min off board lysis using 2 mL of lysis buffer and 100 μL of magnetic silica beads to minimize inhibition. Using the modified protocol, the samples (the entire concentration volume), standard, and negative extraction control (NEC) were extracted and eluted to a 100 μL final volume. The positive circular RNA plasmid standard was 2019-nCoV_N from Integrated DNA Technologies (IDT, Coralville, IA, USA).
RT-ddPCR assays (including the hepatitis G Armored RNA assay, see Supplemental Table S1) were analyzed on a Bio‐Rad QX200 (Bio‐Rad, Hercules, CA, USA). For one‐step RT-ddPCR, a 20 μL final reaction volume comprised 5 μL 1 × one‐step RT‐ddPCR Supermix (Bio‐Rad), 2 μL reverse transcriptase (Bio‐Rad), 1 μL 300 mM DTT, 3 μL forward and reverse primers and probes (final concentrations were 900 and 250 nM, respectively), 5 μL RNase‐free water, and 4 μL RNA (diluted 2x). The reaction mixture was then mixed with 70 μL droplet generation oil in the droplet generator. The resulting droplets were transferred to a 96‐well plate for PCR amplification using the following conditions: 60‐min reverse transcription at 50°C (1 cycle), 10‐min enzyme activation at 95°C (1 cycle), 30‐s denaturation at 94°C (40 cycles), 1‐min annealing/extension cycle at 55°C (40 cycles; ramp rate of ~2–3°C/s), 10‐min enzyme deactivation at 98°C (1 cycle). Finally, droplet reading occurred on the Bio-Rad droplet reader.
Limits of detection (LOD) were calculated by running serial dilutions of the 2019-nCoV_N RNA plasmid standard in 7 replicates over 6 orders of magnitude. The LOD was the concentration at which over 60% of the technical replicates were positive.
2.4. Population normalized SARS-CoV-2 loading estimates
Instantaneous population normalized viral loading to WWTPs during each sampling event was calculated using Eq. (1). Only the N2 assay was used as the CWWTP value in Eq. (1), since it was determined to be the most sensitive. Half the N2 assay LOD was used as the CWWTP concentration when a sample was non-detect.
| (1) |
where;
LWWTP = Population normalized SARS-CoV-2 loading to WWTP (copies per person in the catchment)
CWWTP= N2 assay concentration in samples (copies 100 mL−1)
V =Volume of wastewater entering WWTP during sampling event (MG)
f = Conversion factor between 100 mL and MG
P = Population within WWTP service area
2.5. Data visualization and statistics
All figures were created using R Statistical Computing Software version 3.6.3 (R Core Team, 2020), relying primarily on the dplyr package (Wickham et al., 2015) for data manipulation and the ggplot2 package (Wickham 2016) for plotting. The code used to create each figure can be found at https://github.com/mkc9953/SARS-CoV-2-WW-EPI/tree/master. Clinical testing data were gathered from a repository curated by The New York Times (https://github.com/nytimes/covid-19-data) that reports confirmed COVID-19 cases by city/county.
Kruskall-Wallis analysis was used to examine differences in total number of SARS-CoV-2 assay detections. Dunn's tests were then used to look at pair-wise comparisons between individual assays.
3. Results and discussion
3.1. Confirmed COVID-19 cases in southeastern Virginia, USA
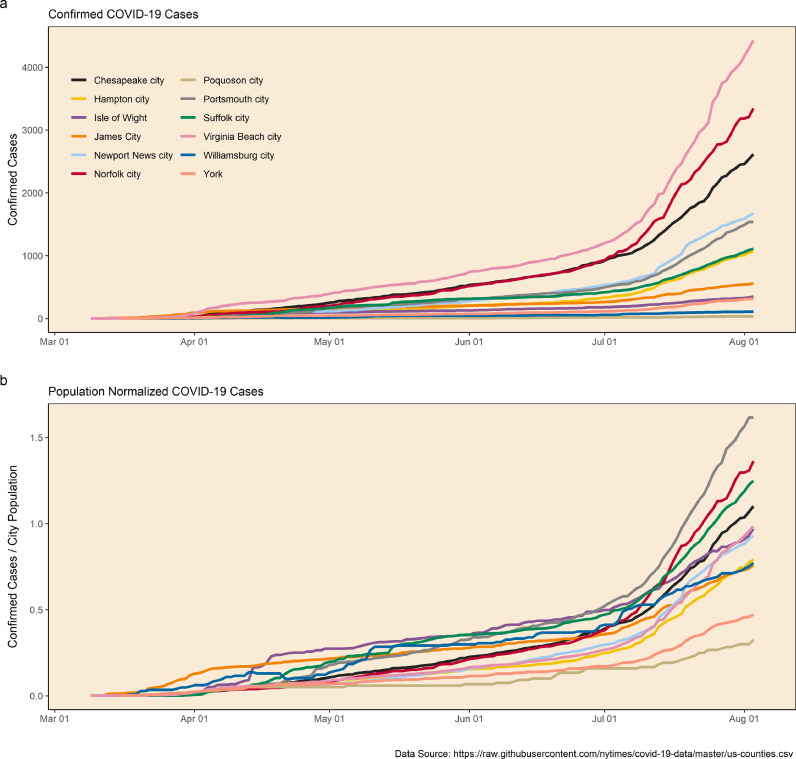
The first confirmed case of COVID-19 in southeastern Virginia was reported on March 9, 2020 in the city of Virginia Beach. Between that time and July 28, 2020, the total number of clinically confirmed cases within the Hampton Roads region grew to 14,904; the total number of cases within the state of Virginia grew to 86,994 during the same time period. Overall, Virginia Beach saw the greatest number of clinical cases (3,788), while Suffolk had the greatest amount of deaths (46). Most other cities within the service area reported total cases in the hundreds while smaller communities had totals ranging from 34 (Poquoson) to 518 (James City) (Fig. 1 a). However, when normalized to population size, there is less of a discrepancy in confirmed case data amongst all the cities and counties (Fig. 1b).
Fig. 1.
Documented cases of COVID-19 by city/county in southeastern Virginia for the study period. Panel ‘a’ presents total confirmed cases. Panel ‘b’ represents total cases normalized by each city's population and plotted as a percent.
Clinical COVID-19 testing for the state of Virginia began on March 5, 2020 and increased dramatically during the time of this study. On the first wastewater sampling date in the present study, a total of 69 patients in all of Virginia had been tested. This increased to 1,180,000 as of July 28th. Although it is likely that the observed increases documented the active spread of the SARS-CoV-2 virus, the inherent confounding influence of increased testing on clinical data should be acknowledged (Murakami et al., 2020).
3.2. Viral assay performance & total surrogate recovery
The theoretical limits of detection (LOD) for assays N1, N2, and N3 were 14.6, 2, and 2.18 copies per reaction, respectively. The N2 assay proved to be the most sensitive for our RT-ddPCR workflow, which is why the N2 assay results were used in subsequent loading analyses and visualizations. This was in contrast to others (e.g. Lu et al., 2020, Vogels et al., 2020) but is likely due to the matrix and specific RT-ddPCR workflows.
Bovine coronavirus (BCoV) and bovine respiratory syncytial virus (BRSV) were used to assess recoveries without concentration as well as with concentration using InnovaPrep and electronegative filtration. Recoveries of the surrogates without concentration (direct extraction of 2 mL samples) were 59% (± 14%) and 75% (± 13%) for BCoV and BRSV, respectively. InnovaPrep (with centrifugation) workflow total recoveries for BCoV and BRSV were 5.5% (± 2.1%) and 7.6% (± 3.0%), respectively. Electronegative filtration workflow total recoveries for BCoV and BRSV were 4.8% (± 2.8%) and 6.6% (± 3.8%), respectively. Although concentration steps used in both workflows during this study likely resulted in reductions of virus signal, concentration was ultimately necessary in order to detect the low viral concentrations documented in the region at the beginning of the study.
Total recoveries were similar across surrogates and workflows, therefore results from the entire 21-week study were reported together and without adjustment. Matrix inhibition of the RT-ddPCR assay, expressed as recovered hepatitis G spike, averaged 50% (± 19%) and 9.4% (± 9.4%) for InnovaPrep and electronegative filtration workflows, respectively. While the total surrogate recoveries were similar for the 2 workflows, the InnovaPrep workflow was less affected by inhibition, as seen in the hepatitis G recoveries. It is likely that the centrifugation step in the InnovaPrep workflow removed solids from suspension, which resulted in less matrix inhibition, but also a lower SARS-CoV-2 signal from particle-attached virus losses. In contrast, the electronegative filtration workflow retained a high percentage of wastewater solids, which likely retained particle attached viruses, but resulted in greater matrix inhibition, as documented by hepatitis G recoveries. Further dilution of samples was not done to alleviate inhibition seen in some samples to maximize low detections.
3.3. Assessment of N1, N2, and N3 assays in wastewater
At the start of this study, the CDC recommended three different assays for SARS-CoV-2 detection: N1, N2, and N3. While N1 and N2 were designed specific to SARS-COV-2, N3 was designed as a more universal assay for the clade 2 and 3 viruses of the Sarbecovirus subgenus (Lu et al., 2020). Typically, environmental microbiology studies use one assay to determine pathogen concentrations in samples (e.g., Worley-Morse et al., 2019; Rose, 2005). To date, several SARS-CoV-2 WBE studies have incorporated multiple assays, including all three CDC assays (e.g. Ahmed et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Wu et al., 2020), while others have incorporated alternative assays (La Rosa et al., 2020; Kocamemi et al., 2020; Wurtzer et al., 2020a, 2020b). To better understand if future WBE studies should run all three CDC assays, we compared detection rates and concentrations among all three throughout the 21-week long study.
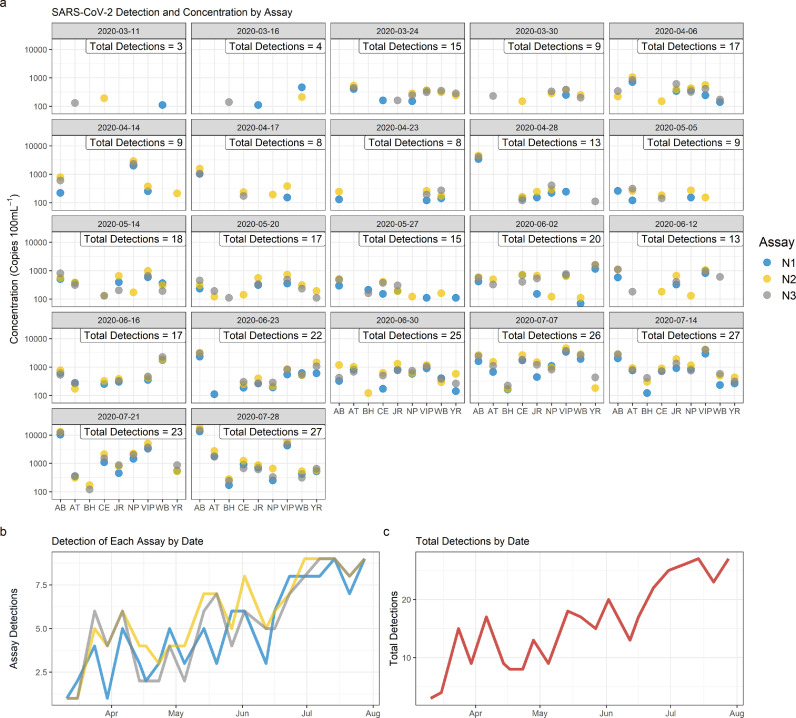
SARS-CoV-2 RNA was detected in raw wastewater influent from at least one WWTP on all sample dates, even during the first week of sampling when there were only two clinical detections in the region (Fig. 2 a). Detections were sporadic in the first several weeks of sampling, with inconsistent detections among the three assays when estimated raw wastewater concentrations of viral RNA were low. For all samples (N=198), 98 had detections for all 3 assays, 22 had detections for 2 of the 3 assays, and 30 had detections for only 1 assay. As total detections increased between March 24th and July 28th, agreement among the three assays improved and detections became more consistent across treatment facilities (Fig. 2a). The N2 assay proved dominant in detection frequency (N=198), with cumulative detections over the 21-week period totaling 107, 125, and 113 for assays N1, N2, and N3, respectively. There was a statistical difference (Kruskal-Wallis chi-squared= 32.49, p<0.001) in the number of N2 and N3 detections from N1 (p<0.001) but no difference between N2 and N3 (p=0.26). It should be noted that N1 had fewer detections for all but two of the sample dates, most likely due to the higher LOD established for this assay (Fig. 2b). Future work will use a different standard to determine the theoretical LOD.
Fig. 2.
SARS-CoV-2 detections for each assay (N1, N2, N3) by sample date. Treatment facilities are noted on the x axis of panel ‘a’. Panel ‘b’ shows total detection by date for each assay. Panel ‘c’ represents total detection of all assays for each sample date.
3.4. SARS-CoV-2 trends in wastewater
Over the course of the entire 21-week study, SARS-CoV-2 concentrations in positive samples were between 101 and 104 copies 100 mL−1. These concentrations are in line with those documented in Australia and Turkey (Ahmed et al., 2020; Kocamemi et al., 2020; Sherchan et al., 2020; Wu et al., 2020). Studies in Spain and France, however, have documented concentrations that were at least two orders of magnitude higher than the concentrations measured in the present study (Randazzo et al., 2020, Wurtzer et al., 2020b). Multiple factors could account for these differences, including disease prevalence in the study regions, efficiency of concentration methods used, and variability in PCR-based workflows.
Aggregated detection trends for all three assays show both increasing and decreasing occurrence over the course of the 21-week sample period (Fig. 2c). Samples collected on the first two sample dates, March 11th and 16th, showed detections at low concentrations at three WWTPs (all less than 300 copies 100mL−1). A sharp increase in detections was documented between March 16th and March 24th, after which a sustained increase in detections was documented over the course of three weeks, through April 14th (Fig. 2c). During this time, between five and seven treatment plants had positive detections, with the maximum number of detections on April 6th (Fig. 2c). Following the peak on April 6th, there was a gradual decline in the total number of detections for the subsequent three sampling dates, with the smallest number of detections since the peak documented on April 23rd (8 detections, Fig. 2a). Starting April 28th through the remaining sample dates, there was an increase in detections documented, most notably in the AB service area, where the highest concentrations seen to date were recorded.
3.5. Service area and regional loading estimates
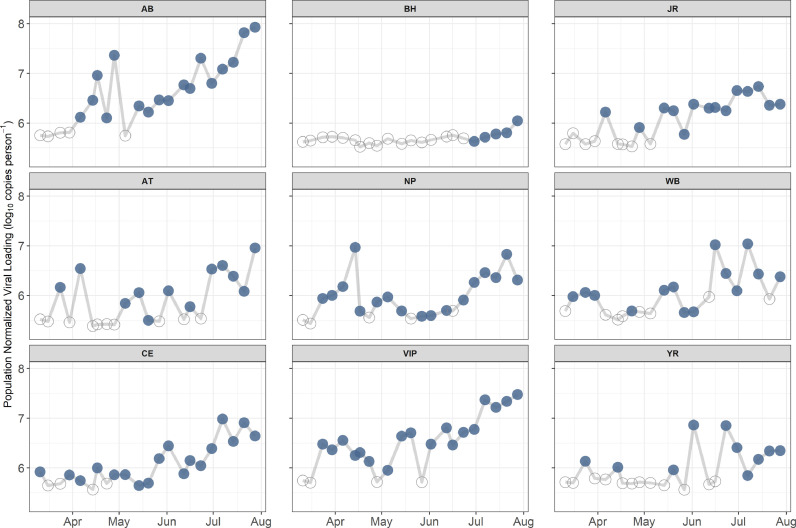
Fig. 3 shows the range of population normalized loading by date for each WWTP. This allows end users to easily visualize specific outbreaks and major trends. Overall, trends increased across all catchments during the study period; in addition, notable shifts in trends can be seen at several of the WWTPs. For example, in the BH service area there were no detections for the first several months until the last five weeks when loading increased, indicating a notable rise in the total number of infected people. Another example is in the CE service area. Loadings were consistent from March through mid-June, after which loading began to trend upwards.
Fig. 3.
Population normalized SARS-CoV-2 loading for each facility. Filled dots indicated samples greater than the limit of detection, hollow dots indicate samples below the limit of detection.
Data presented in Fig. 3 can also be compared to known outbreaks that were documented by the health department during this study, e.g. one in the WB service area and one in the NP service area. Increases in detection in the WB service area that were documented in mid-March were likely caused by an outbreak in James City County, which saw a total of 34 people infected (within 7 days of sampling) based on clinical data. The NP service area spike in mid-April was associated with an Isle of Wight outbreak that infected a total of 55 people (within 7 days of sampling) based on clinical data. Much in the same way, these data can be used to identify potential areas where signs of elevated infection are not documented based on clinical data or areas where there may be evidence of reduced infection rates over time. For example, VIP showed increased evidence of infection within the catchment starting in late March, followed by a steady decline on the following six sample dates. This indicates an increase, followed by a drop-in cases in the VIP service area. The capability to observe declining numbers of cases at the sub-city scale could prove particularly useful as this information is likely obscured in the clinical testing data. Future monitoring of the Hampton Roads wastewater will hopefully provide more evidence of the usefulness of WBE during major declines in the infected population. The combination of clinical testing results and WBE data can provide a more complete picture of how the virus and disease are transmitting in the population.
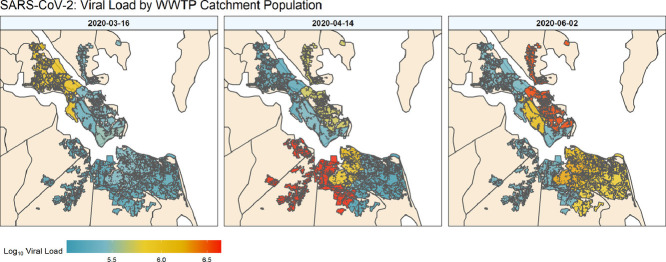
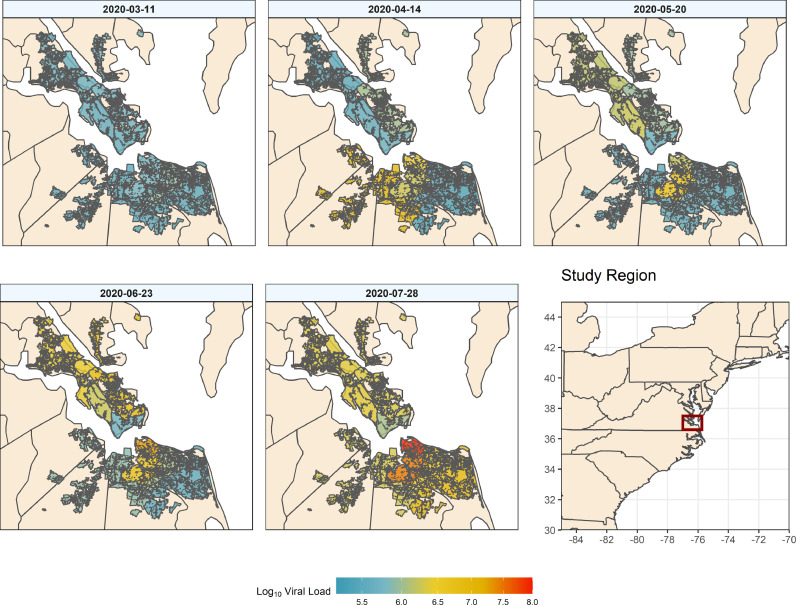
Fig. 4 shows spatial data by week in the form of heat maps representing population normalized loading in sampled WWTP catchments. Only one heat map is shown per month but Supplemental Video S1 shows an animation of all weeks sampled. Maps such as these are helpful to spatially visualize the data and identify likely locations for regional outbreaks. For example, on March 11th, evidence of infection was low throughout the entire region. However, on April 14th there was early evidence of widespread cases in three (WB, VIP, NP) of the nine studied catchments. The May 20th panel shows evidence of increased loading at other service areas (YR, JR), while catchments (i.e. NP) that previously had high loading were reduced. June and July highlight the increase and spread of SARS-COV-2 loading throughout the region, and delineate more densely impacted areas. Spatially displaying population normalized loadings show the irregular outbreak of localized hot spots. This demonstrates that, while clinically confirmed cases uniformly increase for a city as more testing is completed, the actual viral spread is likely more heterogeneous, being heavily influenced by local outbreaks. Thus, WBE has the potential to target where more localized clinical testing might be needed to fully understand sporadic hotspots that are likely to emerge as the COVID-19 pandemic wanes.
Fig. 4.
Population normalized SARS-CoV-2 loading (log10 copies/person) overlaid onto the respective facility catchment. Filled polygons represent discrete catchments for each of the nine sampled treatment plants.
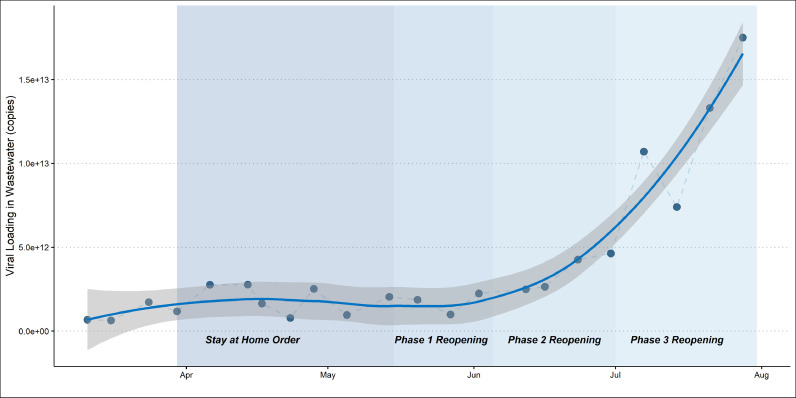
Fig. 5 shows regional loading estimates over time. WBE instantaneous loading data from all 9 WWTP catchments were combined weekly to estimate instantaneous regional loading. The Hampton Roads regional loading estimates mirror the trends in catchment-level population normalized loading. Starting in mid-June there is an obvious, significant inflection upwards in loading corresponding with the Virginia phase reopenings. The late-March to mid-April increase in loading prior to the stay at home was evident, as well as the small decline and plateau in loading before the phase reopenings. The rising limb of regional loading could be incorporated into analyses of clinical testing data to determine the extent to which increases in clinical detection are simply a product of increased testing. Future work will also examine the lead-lag association between Hampton Roads SARS-CoV-2 wastewater data and regional confirmed clinical data.
Fig. 5.
SARS-CoV-2 loading (copies) with LOWESS smoothing for the studied region over 21 weeks.
4. Conclusion
-
•
It is important that public health officials have an array of reliable data sources available to them when making regional decisions
-
•
Clinical datasets can be inherently biased depending on various factors, including patient screening prior to testing, testing supply limitations, and how invasive and/or unpleasant testing is for patients
-
•
WBE methods are often less impacted by these types of sample collection biasbut may incorporate uncertainly associated with temporal and spatial variations in molecular signals within the sewer, decay of nucleic acids, and rainfall impacts on overall load measurements
-
•
Here we propose methods for analyzing and presenting WBE data so that it can be used in concert with clinical results to provide a more complete picture for community officials
Funding
This work was supported by Hampton Roads Sanitation District and the National Science Foundation award 2027752.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Jim Pletl for assistance with conceptualization and editing. We also thank Jonathan Nelson, April Richardson, Jonathan Milisci, Raechel Davis, and Allison Larson for sample collection and analysis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2020.116296.
Appendix. Supplementary materials
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B. Firstconfirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baz-Lomba J.A., Salvatore S., Gracia-Lor E., Bade R., Castiglioni S., Castrignanò E., Causanilles A., Hernandez F., Kasprzyk-Hordern B., Kinyua J., McCall A.K. Comparison of pharmaceutical, illicit drug, alcohol, nicotine and caffeine levels in wastewater with sale, seizure and consumption data for 8 European cities. BMC Public Health. 2016;16(1):1035. doi: 10.1186/s12889-016-3686-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.L., Henquell C. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in central France, 2014 to 2015. Eurosurveillance. 2018;23(7) doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins, A., North, D., Ahmad, A., Ahmed, W., Alm, E., Been, F., Bhattacharya, P., Bijlsma, L., Boehm, A.B., Brown, J. and Buttiglieri, G., 2020. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight Against COVID-19. [DOI] [PubMed]
- Brouwer A.F., Eisenberg J.N., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. 2018;115(45):E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Xu J., Lin D., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Wang X., Ge Y., Xia A. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC), 2020. Evaluating and Testing Persons for Coronavirus Disease 2019 (COVID-19). https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html.
- Causanilles A., Ruepert C., Ibáñez M., Emke E., Hernández F., de Voogt P. Occurrence and fate of illicit drugs and pharmaceuticals in wastewater from two wastewater treatment plants in Costa Rica. Sci. Total Environ. 2017;599:98–107. doi: 10.1016/j.scitotenv.2017.04.202. [DOI] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: Past, present and future. TrAC Trends Anal. Chem. 2018;105:453–469. [Google Scholar]
- Gerrity D., Betancourt W., Bibby K., Reyes F., Holden P., Kotlarz N., McLellan S., Papp K. Implications of COVID-19 for Water, Wastewater, and Water Reuse. WaterReuse Fact Sheet. 2020 https://watereuse.org/wp-content/uploads/2020/04/COVID19-Water-Fact-Sheet.pdf [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T., Shulman L.M., Van der Avoort H., Deshpande J., Roivainen M., De Gourville E.M. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140(1):1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. 2020 [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L, Lucentini L. First detection of SARS-COV-2 in untreated wastewaters in italy. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26(8) doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Tech. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Murakami M., Hata A., Honda R., Watanabe T. Letter to the Editor: Wastewater-Based Epidemiology Can Overcome Representativeness and Stigma Issues Related to COVID-19. Environ. Sci. Tech. 2020 doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Weinberger D.M. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
- Rose J.B. IWA Publishing; 2005. Reduction of pathogens, indicator bacteria, and alternative indicators by wastewater treatment and reclamation processes. [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., Wu W.J., Yuan C., Yu M.L., Li P., Yan J.B. Detection of Novel Coronavirus by RT-PCR in Stool Specimen from Asymptomatic Child, China. Emerg. Infect. Dis. 2020;26(6) doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuijs A.L., Castiglioni S., Tarcomnicu I., Postigo C., de Alda M.L., Neels H., Zuccato E., Barcelo D., Covaci A. Illicit drug consumption estimations derived from wastewater analysis: a critical review. Sci. Total Environ. 2011;409(19):3564–3577. doi: 10.1016/j.scitotenv.2010.05.030. [DOI] [PubMed] [Google Scholar]
- Vogels C.B., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Muenker M.C., Moore A.J., Klein J. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020:1–7. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: elegant graphics for data analysis.https://ggplot2.tidyverse.org ISBN 978-3-319-24277-4. [Google Scholar]
- Wickham, H., Francois, R., Henry, L. and Müller, K., 2015. dplyr: A grammar of data manipulation. R package version 0.8.4. https://CRAN.R-project.org/package=dplyr.
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020:1–5. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Worley‐Morse T., Mann M., Khunjar W., Olabode L., Gonzalez R. Evaluating the fate of bacterial indicators, viral indicators, and viruses in water resource recovery facilities. Water Environ. Res. 2019;91(9):830–842. doi: 10.1002/wer.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., Chen Y., Gu B. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microb. Infect. (just-accepted) 2020:1–14. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S. TMPRSS2 and TMPRSS4 mediate SARS-CoV-2 infection of human small intestinal enterocytes. bioRxiv. 2020 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92(6):680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.