Introduction:
Neonates are protected from colonizing bacteria by antibodies secreted into maternal milk. Necrotizing Enterocolitis (NEC) is a disease of neonatal preterm infants with high morbidity and mortality that is associated with intestinal inflammation driven by the microbiota1-3. The incidence of NEC is significantly lower in infants fed with maternal milk, though the mechanisms underlying this benefit are not clear4-6. Here, we show that maternal Immunoglobulin A (IgA) is an important factor in protection against NEC. Analysis of IgA-binding of fecal bacteria from preterm infants indicated that maternal milk was the predominant source of IgA in the first month of life and that a relative decrease in IgA-bound bacteria is associated with the development of NEC. Sequencing of IgA-bound and unbound bacteria revealed that prior to disease onset, NEC was associated with increasing domination of the IgA-unbound microbiota by Enterobacteriaceae. Further, we confirmed that IgA is critical in preventing NEC in a murine model, where pups reared by IgA deficient mothers are susceptible to disease despite exposure to maternal milk. Our findings show that maternal IgA shapes the host-microbiota relationship of preterm neonates and that IgA is a critical and necessary factor in maternal milk for the prevention of NEC.
Main
NEC is associated with an intestinal microbiota of decreased diversity and increased Enterobacteriaceae, but this association is not sufficient for disease7,8. Bioactive components of maternal milk, including IgA antibodies, shape the neonatal microbiota9-12. It is not known how the anti-bacterial IgA repertoire of maternal milk varies between women, but mammary gland IgA-producing B cells traffic from the intestine and thus may differ between mothers as a result of individualized microbiomes and infectious histories13-15. We hypothesized that differential binding of the preterm microbiota by maternal IgA is a central feature of NEC pathogenesis.
To analyze immunoglobulin (Ig) binding of gut bacteria in preterm infants we stained fecal samples (Table 1a) with anti-human IgA, IgM and IgG antibodies and measured the Ig-bound populations with flow cytometry16,17. This initial sample set contained 30 samples collected at the time of NEC diagnosis and 39 samples from age-matched controls. Surveyed across all samples, the percentage of IgA-bound bacteria was far greater than the percentages of IgM- and IgG-bound bacteria and samples from maternal milk-fed infants contained a far greater abundance of IgA positive bacteria compared to formula-fed infants (Figure 1a,b and Extended Data 1a,b). Although a majority (11/19) of formula-fed infants had <1% of their intestinal bacteria bound by IgA,some samples from formula-fed infants contained high amounts of IgA positive bacteria (Figure 1b). Because B cells generally do not populate the intestine until about 4 weeks of age18, we hypothesized that fecal samples from formula-fed infants collected before this time point would not contain IgA-bound bacteria. Indeed, we found a significant temporal relationship between age and IgA binding in formula-fed infants that was not observed among maternal milk-fed infants (Figure 1c). A dedicated analysis of samples from a single formula-fed preterm infant revealed no IgA positive bacteria in the first 4 weeks of life, strongly implicating maternal milk as the primary source of perinatal IgA (Extended Data 1c). Limiting our analysis of this data set to 4 weeks post-delivery, we found that samples from infants with NEC contained less IgA-bound bacteria than samples from age-matched controls (Figure 1d). However, NEC infants in this cohort were more likely to be formula fed; additionally, their fecal samples were collected after NEC was diagnosed and treatment had been initiated with antibiotics and cessation of feeding. To eliminate the impact of these confounding variables, we selected and analyzed a prospectively collected longitudinal series of samples from 23 milk-fed preterm infants, of which 43.4% subsequently developed NEC (Table 1b). Critically, we found that the fraction of IgA positive bacteria decreased with time among infants that developed NEC, whereas IgA binding of fecal bacteria showed no relationship in controls. (Figure 1e and Extended Data 2a,b). Thus, it appears that in infants that will develop NEC, a change occurs in either the intestinal microbiota or the maternal IgA repertoire that leads to the ‘escape’ of intestinal bacteria from binding.
Table 1.
a) Post-diagnosis preterm/NEC cohort, single fecal samples taken from NEC patients and controls within 36 hours of NEC diagnosis. Samples analyzed from day of life (DOL) 4 to DOL 70. b) Prospective/longitudinal preterm/NEC cohort. Longitudinally collected samples from infants (gestation age 24 weeks to 31 weeks) captured prospectively (Day-of-life 7 to 40) and assigned to groups after the diagnosis of NEC or discharge from the neonatal intensive care unit. Feeding mode abbreviations Maternal milk only = MMO; Combination of maternal milk and formula = Comb; Formula fed = FF
| A | NEC | Control | B | NEC | Control |
|---|---|---|---|---|---|
| No. of patients | 30 | 39 | 10 | 13 | |
| Avg. gestational age at birth (in weeks) | 29 2/7 | 28 5/7 | 27 1/7 | 27 3/7 | |
| Mean age at NEC diagnosis (Day of life) | 22.2 | NA | 27.4 | NA | |
| Sex | Females- 11 | Females- 15 | Females- 5 | Females- 8 | |
| Males- 19 | Males- 24 | Males- 5 | Males- 5 | ||
| Mean birth weight (grams) | 1176.52 | 1202.26 | 873.1 | 932.92 | |
| Mode of Delivery | Vaginal- 11 | Vaginal- 9 | Vaginal- 0 | Vaginal- 8 | |
| Caesarean- 19 | Caesarean- 30 | Caesarean- 10 | Caesarean- 5 | ||
| Feeding mode | MMO- 6 Comb- 13 |
MMO- 5 Comb- 26 |
MMO- 5 Comb- 5 |
MMO- 5 Comb- 8 |
|
| FF- 11 | FF- 8 | FF- 0 | FF- 0 |
Figure 1. IgA binding to the intestinal bacteria of preterm infants is positively correlated to maternal milk feeding and negatively correlated to the development of NEC.
Flow cytometric analysis of bacterial IgA binding on fecal samples from preterm infants. a) Example of IgA staining on preterm infants. (b-d) Maternal milk fed infants are indicated by black circles, formula fed infants by open red squares. b) Percent IgA-bound bacteria from maternal milk fed (n=50) vs. formula fed (n=19) infants. The box represents the number of samples with <1% IgA binding of intestinal bacteria, two-sided Mann-Whitney test, Mean±SEM. c) Percent IgA binding was correlated by linear regression with time post-delivery in maternal milk fed (n=50) and formula fed (n=19) infants; Pearson’s correlation coefficient. d) Percent IgA bound bacteria from controls (n=28) or infants diagnosed with NEC (n=23), from samples collected <DOL40; red squares indicate formula fed infants; two-sided Mann-Whitney test, Mean±SEM. e) Percent IgA-bound intestinal bacteria from prospectively collected longitudinal samples of NEC patients (Patient number=10, sample size=39) and Controls (Patient number=13, sample size=59), graphed against DOL, each patient is graphed in a different color; linear regression and Pearson’s correlation coefficient.
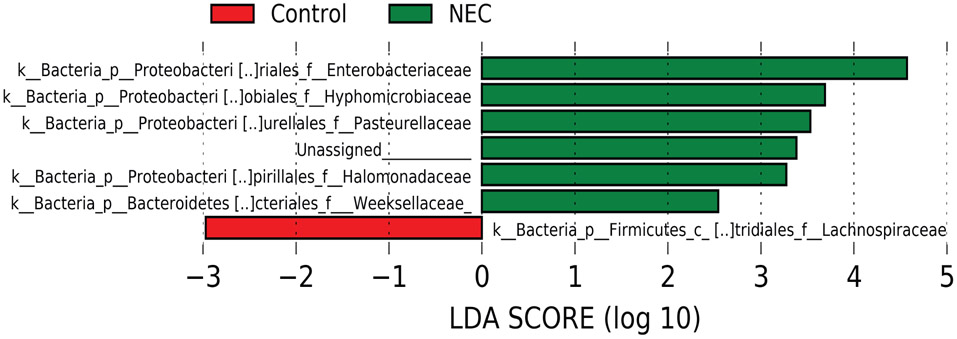
We next sought to identify shifts in the microbiota that might explain this drop in IgA positive bacteria. Analyzing all samples together, we observed a modest enrichment for Enterobacteriaceae and reduction in Gram positive anaerobes such as Lachnospiraceae in infants who will go on to develop NEC (Figure 2a and Extended Data 3). This result aligns well with published reports describing dysbiosis prior to onset of NEC7. We therefore hypothesized that loss of IgA binding of Enterobacteriaceae may be important to the decrease in IgA-bound bacteria in NEC. In support of this hypothesis, we found that the abundance of IgA-bound bacteria varied inversely with the abundance of Enterobacteriaceae among infants that would later develop NEC, but not controls (Figure 2b).
Figure 2. Reduced intestinal bacterial diversity driven by increased IgA unbound Enterobacteriaceae precedes the development of NEC.
Longitudinal fecal samples from preterm infants prior to the onset of NEC were selected from NEC infants (n=10; 39 samples combined) and Controls (n=13, 59 samples combined) a) Samples were analyzed for the relative abundance of different bacterial OTUs by targeted sequencing of 16S rRNA genes. Shown is the mean relative abundance of different taxa between NEC patients and controls (pooled from all timepoints and infants). b) Relative abundance of Enterobacteriaceae in preterm fecal samples compared to percent IgA-bound bacteria from controls and prospective NEC patients (c-f) Fecal samples were separated into IgA positive and negative pools prior to 16S rRNA sequencing and deconvolution based on post-sort analysis. c) Shannon diversity scores of IgA positive and IgA negative samples from prospective NEC patients and controls, graphed against DOL. d) Relative abundance of Enterobacteriaceae from IgA positive and IgA negative samples from prospective NEC patients and controls, graphed against DOL. e) Relative abundance of combined anaerobic bacterial OTUs from IgA positive and IgA negative samples from prospective NEC patients and controls, graphed against DOL. f) Ratio of reads (IgA negative/IgA positive; Log2 transformed) from paired IgA positive and negative samples, graphed against DOL. Shown are the ratio of total reads, Enterobacteriaceae reads and combined anaerobes. For b-f, each patient is graphed in a different color and R-squared value is based on a linear regression and Pearson’s correlation coefficient.
To directly identify which bacterial taxa might be more or less bound by maternal antibodies in our longitudinal prospective cohort (Table 1b), we physically separated the two fractions and measured microbial diversity by sequencing the V4 region of bacterial 16S rRNA genes (IgSeq)19. In accord with published reports16,20, we did not achieve purity (>99%) in our IgA separations, complicating our ability to discriminate IgA positive and negative bacteria (Extended Data 4a). Post-sort flow cytometric analysis of each sample allowed us to deconvolve the contamination, significantly increasing the correlation between the percent IgA positive bacteria in the unsorted sample and the ratio of IgA positive and negative bacteria reads from IgSeq (Extended Data 4b-d). Deconvolution is particularly important in samples with low levels of IgA positive bacteria, which both validates that our technique is correcting for contamination and improves analysis of the samples most critical to explaining our observation of reduced IgA positive bacteria in NEC (Extended Data 4c and d).
Longitudinal IgSeq analysis of deconvolved samples revealed that control infants showed a significant increase in bacterial diversity amongst IgA negative bacteria over time, while no significant changes were detected in the IgA positive sample (Figure 2c). Conversely, among infants that progressed to NEC, the diversity of both IgA positive and negative fecal bacteria significantly decreased over time (Figure 2c)21. In accord with our analysis of the total microbiome (Figure 2b), IgSeq also revealed that over time the IgA negative intestinal microbiota of NEC infants became dominated by Enterobacteriaceae, while anaerobes (notably Clostridiales and Bifidobacteriales) were virtually undetectable (Figure 2d,e and Extended Data 5). Conversely, amongst controls, the relative abundance of IgA negative anaerobes increased and the relative abundance of IgA negative Enterobacteriaceae decreased, confirming that Enterobacteriaceae are bound at relatively higher frequencies over time in infants that do not develop NEC (Figure 2b,d,e and Extended Data 5). While the IgA positive and negative fractions from controls differed in both diversity and the relative abundance of Enterobacteriaceae, in NEC patients these fractions were not discernibly different (Figure 2c and d). The lack of differences between IgA positive and negative fractions of NEC infants may be explained by very low diversity and domination by Enterobacteriaceae, so essentially there are few taxa available for IgA to bind (Extended Data 5).
An advantage of IgSeq deconvolution is that it allows for a meaningful comparison of taxon abundance between IgA positive and IgA negative samples, as their ratio corresponds to the abundance of IgA-bound bacteria in the unsorted sample (Extended Data 4). When we calculated the ratio of the paired IgA negative and positive reads, we observe a unique increase over time in IgA negative total and Enterobacteriaceae reads in infants who will go on to develop NEC, that correlates well with IgA binding data from flow cytometry (Figure 1e and 2f). In contrast, control infants show no significant shifts in the relative abundances of IgA positive and negative total bacteria, Enterobacteriaceae or anaerobes and only show significant shifts in less abundant OTUs (Figure 2f and Extended Data 6). As Enterobacteriaceae is the most abundant OTU in preterm infants and the only taxa uniquely increasing in the IgA negative fraction of the microbiota prior to disease, the increase in IgA negative Enterobactericeae is the most likely driver of the reduced IgA bound bacteria seen preceding NEC (Figure 1e, 2a, 2f and Extended Data 6). Thus, we have identified that NEC infants uniquely fail to diversify their intestinal microbiota with anaerobic bacteria and instead remain dominated by IgA-unbound Enterobacteriaceae.
To test the possibility that the increase in Enterobacteriaceae was driven by a rapid bloom we measured the number of each bacterial taxa per unit mass in fecal samples22,23. Although we saw increased Enterobacteriaceae amongst some infants developing NEC, inter-individual variation was high and there were no statistically significant differences between cases and controls (Extended Data 7). However, our analysis may not adequately represent bacteria in the small intestine so we cannot rule out the possibility of focal expansions of Enterobacteriaceae associated with NEC24. Nonetheless, we favor a model where loss of IgA binding of the microbiota is induced either by mutation or by transcriptional modifications that allow sub-populations of Enterobacteriaceae to escape maternal IgA but do not lead to increases in their total number.
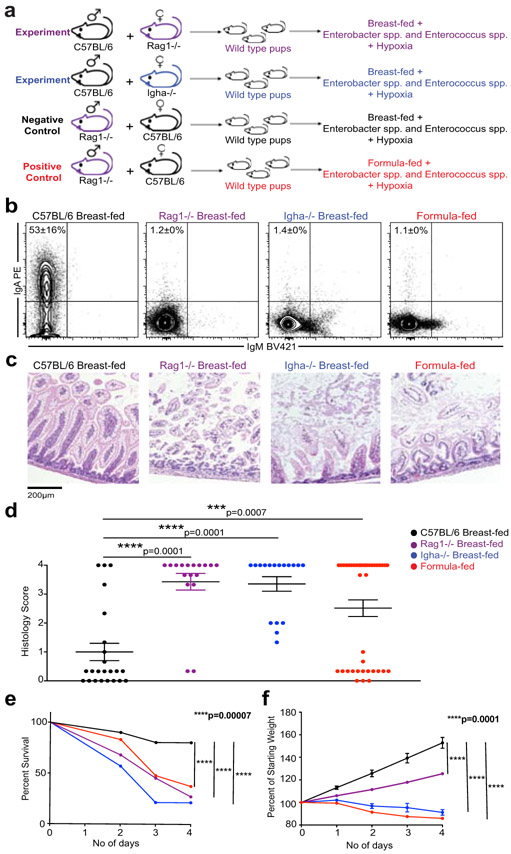
To further define the contribution of maternal IgA to disease pathogenesis, we turned to an experimental murine model of NEC25,26. We bred mice so that heterozygote wild-type pups were fed by mothers that either can (C57BL/6) or cannot produce IgA (Rag1−/− or Igha−/−), and compared them to formula-fed positive controls (Figure 3a). We confirmed that mice, like humans, produce little IgA during their first two weeks of life and that dams are the primary source of neonatal IgA18,27,28 (Figure 3b). We also determined that Enterobacter spp. gavaged into pups (C57BL/6 dams) to induce NEC was enriched in the IgA positive fraction, indicating that murine dams may produce protective IgA without being vaccinated (Extended Data 8). Strikingly, pups undergoing the NEC protocol that were breast-fed by mothers lacking IgA (Rag1−/− or Igha−/−) showed a phenotype that was indistinguishable from formula-fed controls. Specifically, they exhibited increased mortality, and severe intestinal damage characterized by shortened necrotic villi and mucosal sloughing (Figure 3c-e). Furthermore, pups fed by Igha−/− mothers exhibited a significant reduction in weight gain compared to pups fed by wild-type mothers (Figure 3f). We have thus shown, using an experimental model of NEC, that maternal milk only protects against NEC when it contains IgA.
Figure 3. IgA is a necessary component of breast milk to prevent the development of experimental NEC.
a) Experimental NEC mouse model where wild-type pups are fed by dams that either can (C57BL/6; n=31) or cannot produce IgA (Rag1−/−; n=22 or Igha−/−; n=14). Formula fed mice (n=48) are used as a positive control b) Representative IgA staining of the fecal matter of 8 day old pups from a), shows absence of IgA-bound bacteria in dam breast-fed and formula-fed pups. C57BL/6 (n=19), Rag1−/− (n=9), Igha−/− (n=8) and formula fed (n=23) c) Representative images of H&E staining of small intestine of pups undergoing murine NEC protocol; pups fed by C57BL/6 (n=23), Rag1−/− (n=17), Igha−/− (n=17) and formula fed (n=40) d) Histology scores of the small intestines of pups from c), one-way ANOVA with multiple comparisons; Mean±SEM. e) Percent number of pups from a) surviving the NEC protocol at different timepoints; statistics determined by Log-rank (Mantel-Cox) test. f) Weights of pups from a), statistical difference calculated by one-way ANOVA with multiple comparisons on weights at experimental completion point (day 4); Mean±SEM. Data shown in panel b-f is grouped together from 3 individual experiments with minimum n=5 pups in each group for every experiment.
Discussion
Previous studies have shown associations between the abundance of Enterobacteriaceae and NEC7. We now show that IgA-unbound Enterobacteriaceae is more closely linked to NEC development than total Enterobacteriaceae abundance. Animal studies have indicated that both host and maternal IgA is important in controlling Enterobacteriaceae and establishing a mature microbiota characterized by fastidious anaerobic bacteria29,30. Our results indicate that binding of bacteria by maternally-derived IgA may promote diversity in the microbiome and the acquisition of anaerobic bacteria during the critical window when infants make little or no IgA of their own, perhaps by limiting inflammation driven by Enterobacteriaceae31-33.
Future studies will be required to elucidate the mechanism by which IgA controls and modifies gut bacterial colonization in newborns. IgA has been shown to modify bacterial surface protein expression and motility which may limit the ability of bacteria to gain access to the intestinal epithelium17,34. IgA may accomplish these tasks by ‘enchaining’ bacterial cells, allowing for easier expulsion and preventing gene transfer35. Importantly, the current study did not discriminate between bacteria at the strain level. Thus, it remains to be determined whether the loss of IgA binding results from the appearance of new organisms not constrained by the existing IgA repertoire, or alternatively from changes to bacterial genomes and/or gene expression that allow early colonizers to escape IgA binding36,37. Temporal changes in bacterial binding could also result from shifts in the maternal IgA repertoire.
Previous attempts to prevent NEC with intravenous immunoglobulins have largely failed to show efficacy38,39. However, the repertoire of intravenous antibodies may differ from that of secretory IgA and bacterial specificity was not accounted for in these studies. Future efforts might allow for precision microbiome-informed strategies that enable augmentation of milk or preterm infant formulas with rationally selected protective antibodies.
Methods
Mice
C57BL/6 mice were purchased from Taconic. Rag1−/− mice were obtained from Jackson Laboratories. Igha−/− mice were obtained from Dr. Yasmine Belkaid (NIH/NIAID). All mice were maintained at and all experiments were performed in an American Association for the Accreditation of Laboratory Animal Care-accredited animal facility at the University of Pittsburgh and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals under an animal study proposal approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. Mice were housed in specific pathogen-free (SPF) conditions.
Human Fecal Samples
The human study protocol was approved by the Institutional Review Board (Protocol Nos. PRO16030078, PRO09110437) of the University of Pittsburgh. Fecal samples were collected fresh or from the diaper of preterm infants at UPMC Magee-Womens Hospital and frozen immediately at −80°C. The samples were later divided into age-matched controls and NEC depending on the incidence of NEC.
Fecal IgA Flow Cytometry and Magnetic Sorting of IgA+ and IgA− Bacteria
Either fecal pellets collected from mice after sacrifice or ~50 mg of frozen human fecal material was placed in 1.5ml Eppendorf tubes and 1ml Phosphate Buffered Saline (PBS) was added. The fecal material was disrupted by a combination of vortexing and pipetting and passed through a 40μm filter to remove food/fibrous material. The fecal material is diluted with PBS to obtain a bacterial OD of ~0.4 to maintain equality between samples and to prevent the magnetic columns from clogging. A volume of 200μl of the suspended bacterial material was then frozen as an ‘unsorted’ control. An additional 200μl of the suspended material was divided equally on a 96-well plate for IgA staining and isotype control for each sample to eliminate non-specific binding. The fractions were washed with twice with staining buffer (1% Bovine Serum Albumin (Sigma) in PBS-filtered through a 2.2μm filter). The bacteria were stained with Syto BC (Green Fluorescent nuclear acid stain, Invitrogen-1:400), APC Anti-Human IgA (Miltenyi Biotec clone IS11-8E10) (1:10)/ Anti-Human IgA APC (Miltenyi Biotec clone REA1014) (1:50), Anti-Human IgM BV421 (BD Biosciences clone G20-127) (1:30)/ BV421 Mouse Anti-Human IgG (BD Biosciences clone G18-145) (1:10) or PE-conjugated Anti-Mouse IgA (eBioscience clone mA-6E1) (1:500), Anti-Mouse Rat IgM BV421 (BD Biosciences clone R6-60.2) (1:30)/ Anti-Mouse Rat IgG2a isotype (BD Biosciences clone R35-95), Anti-Mouse IgG FITC (BioLegend clone Poly4060) (1:30) and blocking buffer of 20% Normal Mouse Serum for human or 20% Normal Rat Serum for mouse samples (ThermoFisher). The isotype control was stained similarly using APC Mouse IgG1 isotype control (Miltenyi Biotec clone-IS5-21F5) (1:10) or PE-conjugated Rat Anti-Mouse IFNγ (eBioscience clone XMG1.2). The stained samples were incubated in the dark for an hour at 4°C. Samples were then washed three times with 200μl of staining buffer before flow-cytometric analysis (LSRFortessa-BD Biosciences).
For magnetic activated cell sorting (MACS), we used 500μl of the suspended fecal material to compensate for the loss of material during sorting and scaled our staining volume accordingly. Anti-IgA stained fecal bacterial pellets were incubated in 1ml per sample of staining buffer containing 45μl of anti-APC or anti-PE MACS Microbeads (Miltenyi Biotec) (20 min at 4°C in the dark), washed twice with 1 ml Staining Buffer (8000 x rpm, 5 min, 4°C), and then sorted using MS columns (Miltenyi Biotec). The flow-through was collected as IgA-unbound (IgA-negative) fraction. Columns were washed with 70% ethanol and sterile PBS between separations. The IgA-bound fraction was added in the column and the steps mentioned above were repeated four times for maximum enrichment. 100μl each of the IgA-bound and IgA-unbound fraction was used for post-sort flow cytometric analysis (along with unsorted sample). Absolute bacterial counts were determined by adding a known number of AccuCheck Counting beads (Life Technologies) to antibody stained fecal samples of a given mass, which allows for the calculation of the total number of SYTO (DNA)+ events in any given sample. This can then be multiplied by the measured abundance of any OTU to represent the number of bacteria of that taxon/mass in any sample.
DNA Extraction
All microbial DNA was extracted using the MO BIO PowerSoil DNA Isolation kit (single tube extractions). The unsorted, IgA-bound and IgA-unbound pellets were resuspended in Solution TD1 by pipetting and vortexing and ~200μl of 0.1mm diameter Zirconia/Silica beads (Biospec) were added and shaken horizontally on a lab mixer for 12-18 min at maximum speed using a MO BIO vortex adaptor. All remaining steps followed the manufacturer’s protocol. The DNA extracted was stored at −20°C for further 16S amplicon PCR and sequencing.
16S Amplicon PCR, Sequencing and Analysis
PCR amplification of the small subunit ribosomal RNA gene (16S rRNA) was performed in triplicate 25μl reactions. Reactions were held at 94°C for 3 min to denature the DNA, with amplification performed for 30 cycles at 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s; followed by a final extension of 10 min at 72°C. Amplicons were produced utilizing primers adapted for the Illumina MiSeq. Amplicons target the V4 region and primers utilized either the Illumina adaptor, primer pad and linker (forward primer) or Illumina adaptor, Golay barcode, primer pad and linker (reverse primer) followed by a sequence targeting a conserved region of the bacterial 16S rRNA gene as described40,41. The only deviation from the protocol was that PCR was run for 30 cycles. Amplicons were cleaned using the Qiagen UltraClean 96 PCR Cleanup Kit. Quantification of individual amplicons was performed with the Invitrogen Quant-iT dsDNA High Sensitivity Assay Kit. Amplicons were then pooled in equimolar ratio. Agarose gel purification was performed to further purify the amplicon pool and remove undesired PCR products prior to submission for paired-end sequencing on the Illumina MiSeq. Read pairing, clustering and core diversity statistics were generated through PEAR, UPARSE and QIIME and R42,43. LEfSe was used to compare family level relative abundances between NEC and control groups44. Raw 16S rRNA data has been uploaded to NCBI BioSample/SRA and is available under accession number PRJNA526906.
Deconvolution and microbiome data analysis:
Flow cytometry was used to determine the percentage of IgA positive and IgA negative bacteria in each sample (unsorted, IgA positive, IgA negative) post magnetic separation. We assumed that contamination affected each OTU equally and that the IgA positive and IgA negative samples are reciprocal (fractions of the same whole). The raw reads from sequencing of 16S rRNA genes were then deconvolved by summing the proportion of IgA bound or IgA unbound (as measured by flow cytometry) across the paired (infant and time point [Day of life]) IgA positive and IgA negative samples for each OTU (Extended Data 3c).
Example to solve for IgA positive from one time point and one OTU ‘X’:
The deconvoluted data was processed through the QIIME2 workflow to create alpha diversity metrics with sampling depth chosen based on alpha rarefaction plotting. Abundance of individual or ‘pooled’ (Anaerobes) OTUs was then calculated using the deconvolved values. The algorithm for deconvolution is available on GitHub (https://github.com/handlab/IgA_Seq_Deconvolution).
If IgSeq is accurate, the ratio of the read numbers between IgA positive and negative (IgA+/IgA++IgA−) samples should roughly correspond to the percent IgA+ bacteria in the unsorted sample from where they were derived. This relationship does not hold for samples with low levels of IgA positive bacteria (Extended Data 4c) prior to deconvolution but is much improved after our method has been applied (Extended Data 4d)
We categorized all of the following OTUs as ‘Anaerobes’: Bifidobacteriaceae, Prevotellaceae, Bacteroidiales_S24-7, Clostridiaceae, Lachnospiraceae, Peptostreptococcaceae, Ruminococcaceae, Veillonellaceae, Tissierellaceae (Figure 2e and Extended Data 6b).
Quantitative PCR for 16S rRNA.
PCR amplification of the small subunit ribosomal RNA gene (16S rRNA) was performed in triplicate 10μl reactions. Reactions were held at 95°C for 3 min to denature the DNA, with amplification performed for 35 cycles (95°C for 10 s and 60°C for 30 s). The forward primer sequence of 16S is ACTCCTACGGGAGGCAGCAGT and the reverse primer sequence of 16S ATTACCGCGGCTGCTGGC.
Quantitative PCR for Enterobacter spp.
PCR amplification of the small subunit ribosomal RNA gene (23S rRNA) was performed in triplicate 10μl reactions. Reactions were held at 95°C for 3 min to denature the DNA, with amplification performed for 35 cycles (95°C for 10 s and 60°C for 30 s). The forward primer sequence of Enterobacter 23S is AGTGGAACGGTCTGGAAAGG and the reverse primer sequence of Enterobacter 23S TCGGTCAGTCAGGAGTATTTAGC45.
Induction of NEC
NEC is induced in 7- to 8-day-old mice by hand-feeding mice formula via gavage 5 times/day (22-gauge needle; 200μl volume; Similac Advance infant formula [Ross Pediatrics, Columbus, Ohio]/ Esbilac canine milk replacer 2:1). The formula is supplemented with 107 CFUs of Enterobacter spp. (99%) and Enterococcus spp. (1%) and mice are rendered hypoxic (5%O2, 95% N2) for 10 minutes in a hypoxic chamber (Billups-Rothenberg, Del Mar, CA) twice daily for 4 days46,47. We used males and females in all experiments. Disease is monitored by weighing mice daily prior to the second feed. The severity of disease was determined on histologic sections of the entire length of the small intestines stained with hematoxylin and eosin by trained personnel who were blinded to the study conditions according to previously published scoring system from 0 (normal) to 4(severe)48.
Statistics
Statistical tests used are indicated in the figure legends. Lines in scatter bar charts represent the mean of that group. Group sizes were determined based on the results of preliminary experiments. Mouse studies were performed in a non-blinded fashion. Statistical significance was determined with the two-tailed unpaired Student’s t-test or non-parametric Mann-Whitney test when comparing two groups and one-way ANOVA with multiple comparisons, when comparing multiple groups. All statistical analyses were calculated using Prism software (GraphPad). Differences were considered to be statistically significant when p < 0.05.
Data Availability Statement
Patient-related data not included in the paper were generated as part of clinical trials and may be subject to patient confidentiality. The human study protocol was approved by the Institutional Review Board (Protocol Nos. PRO16030078, PRO09110437) of the University of Pittsburgh. All raw and analyzed sequencing data can be found at the NCBI Sequence Read Archive (accession number: PRJNA526906). Algorithm for deconvolution of IgSeq data available on GitHub (https://github.com/handlab/IgA_Seq_Deconvolution).
Biological Materials Availability Statement
All materials used in the production of this paper are available upon request (timothy.hand@chp.edu). Some reagents may require a Material Transfer Agreement through the University of Pittsburgh.
Extended Data
Extended Data Fig. 1.
Maternal milk-derived antibodies binding to intestinal bacteria from preterm infants.
Extended Data Fig. 2.
Fraction of intestinal bacteria bound by IgA in preterm infants.
Extended Data Fig. 3.
Linear discriminant analysis of the microbiota of infants that will develop NEC and controls.
Extended Data Fig. 4.
Deconvolution method to decrease the effect of contamination in IgSeq.
Extended Data Fig. 5.
Longitudinal analysis of the intestinal microbiota of preterm infants.
Extended Data Fig. 6.
Ratio of IgA− to IgA+ reads for low-abundance taxa.
Extended Data Fig. 7.
Absolute number of bacteria and number of bacteria associated with the dominant taxa in preterm infants.
Extended Data Fig. 8.

Enterobacter spp. is enriched in the IgA+ fraction of breast-fed mouse pups.
Acknowledgements
The authors would like to thank the Univ. of Pittsburgh In-Situ Hybridization Lab for the preparation of tissue slides, and the Univ. of Pittsburgh Division of Laboratory Animal Research for assistance with animal husbandry. We would like to thank M. Band, C. Wright, T. Akraiko and the Roy J. Carver Biotechnology Center DNA Sequencing Center at the University of Illinois for assistance with NextGen Sequencing. Igha−/− mice were kindly provided by Y. Belkaid (NIH, Bethesda). We would like to thank A. Poholek, L. Konnikova, S. Canna and the members of the Hand and Morowitz labs for discussion and critical reading of the manuscript, N. Palm for advice on IgSeq analysis and D. Kostka for advice on deconvolution of IgSeq samples. The authors declare no competing financial interests. This project was supported in part by the UPMC Children’s Hospital of Pittsburgh. MG is supported by K08DK101608, R03DK111473 and R01DK118568 from the National Institutes of Health, March of Dimes Foundation Grant No. 5-FY17-79, the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital. MJM and sample collection are supported by R01-AI-092531. TWH is supported by the Richard King Mellon Foundation Institute for Pediatric Research.
Footnotes
Competing Interests Statement
The authors have no competing interests to declare.
References
- 1.Neu J & Walker WA Necrotizing enterocolitis. N Engl J Med 364, 255–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hackam D & Caplan M Necrotizing enterocolitis: Pathophysiology from a historical context in Seminars in pediatric surgery, Vol. 27 11–18 (Elsevier, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hintz SR, et al. Neurodevelopmental and growth outcomes of extremely low birth weight infants after necrotizing enterocolitis. Pediatrics 115, 696–703 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Cortez J, et al. Maternal milk feedings reduce sepsis, necrotizing enterocolitis and improve outcomes of premature infants. J Perinatol 38, 71–74 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Le Doare K, Holder B, Bassett A & Pannaraj PS Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front Immunol 9, 361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas A & Cole TJ Breast milk and neonatal necrotising enterocolitis. Lancet 336, 1519–1523 (1990). [DOI] [PubMed] [Google Scholar]
- 7.Pammi M, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome 5, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brower-Sinning R, et al. Mucosa-associated bacterial diversity in necrotizing enterocolitis. PloS one 9, e105046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good M, et al. The human milk oligosaccharide 2′-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. British Journal of Nutrition 116, 1175–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisgrill L, et al. Human lactoferrin attenuates the proinflammatory response of neonatal monocyte‐derived macrophages. Clinical & Experimental Immunology (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y-M, et al. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. 24, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandtzaeg P The mucosal immune system and its integration with the mammary glands. The Journal of pediatrics 156, S8–S15 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindner C, et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol 16, 880–888 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Wilson E & Butcher EC CCL28 controls immunoglobulin (Ig)A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J Exp Med 200, 805–809 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell host & microbe 14, 571–581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rognum TO, Thrane PS, Stoltenberg L, Vege Å & Brandtzaeg P Development of intestinal mucosal immunity in fetal life and the first postnatal months. Pediatric research 32, 145 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Bunker JJ, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell host & microbe 17, 153–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yee WH, et al. Incidence and timing of presentation of necrotizing enterocolitis in preterm infants. Pediatrics 129, e298–e304 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. science 339, 708–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandeputte D, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Olm MR, Bhattacharya N, Crits-Christoph A, Firek BA, Baker R, Song YS, Morowitz MJ, Banfield JF Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria that may stimulate TLR4 receptors. bioRxiv 10.1101/558676(2019). [DOI] [PMC free article] [PubMed]
- 25.Barlow B, et al. An experimental study of acute neonatal enterocolitis—the importance of breast milk. Journal of pediatric surgery 9, 587–595 (1974). [DOI] [PubMed] [Google Scholar]
- 26.Jilling T, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. The Journal of Immunology 177, 3273–3282 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogier EW, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proceedings of the National Academy of Sciences 111, 3074–3079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris NL, et al. Mechanisms of neonatal mucosal antibody protection. J Immunol 177, 6256–6262 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Planer JD, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirpuri J, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 5, 28–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slack E, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Byndloss MX, et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson DA, McNulty NP, Guruge JL & Gordon JI IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Moor K, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Gauger EJ, et al. Role of motility and the flhDC Operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect Immun 75, 3315–3324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leatham MP, et al. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect Immun 73, 8039–8049 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eibl MM, Wolf HM, Fürnkranz H & Rosenkranz A Prevention of necrotizing enterocolitis in low-birth-weight infants by IgA–IgG feeding. New England Journal of Medicine 319, 1–7 (1988). [DOI] [PubMed] [Google Scholar]
- 39.Foster JP, Seth R & Cole MJ Oral immunoglobulin for preventing necrotizing enterocolitis in preterm and low birth weight neonates. Cochrane Database Syst Rev 4, CD001816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references
- 40.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal 6, 1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences 108, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel CB, Shanker R, Gupta VK & Upadhyay RS Q-PCR Based Culture-Independent Enumeration and Detection of Enterobacter: An Emerging Environmental Human Pathogen in Riverine Systems and Potable Water. Front Microbiol 7, 172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Good M, et al. Lactobacillus rhamnosus HN001 decreases the severity of necrotizing enterocolitis in neonatal mice and preterm piglets: evidence in mice for a role of TLR9. Am J Physiol Gastrointest Liver Physiol 306, G1021–1032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Good M, et al. Breast milk protects against the development of necrotizing enterocolitis through inhibition of Toll-like receptor 4 in the intestinal epithelium via activation of the epidermal growth factor receptor. Mucosal immunology 8, 1166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radulescu A, et al. Heparin-binding epidermal growth factor–like growth factor overexpression in transgenic mice increases resistance to necrotizing enterocolitis. Journal of pediatric surgery 45, 1933–1939 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Patient-related data not included in the paper were generated as part of clinical trials and may be subject to patient confidentiality. The human study protocol was approved by the Institutional Review Board (Protocol Nos. PRO16030078, PRO09110437) of the University of Pittsburgh. All raw and analyzed sequencing data can be found at the NCBI Sequence Read Archive (accession number: PRJNA526906). Algorithm for deconvolution of IgSeq data available on GitHub (https://github.com/handlab/IgA_Seq_Deconvolution).