Abstract
Background
—Maintenance of tight controls on circulating blood metabolites is crucial to normal, healthy tissue and organismal function. A number of single nucleotide polymorphisms (SNPs) have been associated with changes in the levels of blood metabolites. However, the impacts of the metabolite-associated SNPs are largely unknown because they fall within non-coding regions of the genome.
Objective
—We aimed to identify genes and tissues that are linked to changes in circulating blood metabolites by characterizing genome-wide spatial regulatory interactions involving blood metabolite-associated SNPs.
Method
—We systematically integrated chromatin interaction (Hi-C), expression quantitative trait loci (eQTL), gene ontology, drug interaction, and literature-supported connections to deconvolute the genetic regulatory influences of 145 blood metabolite-associated SNPs.
Findings
—We identified 577 genes that are regulated by 130 distal and proximal metabolite-associated SNPs across 48 different human tissues. The affected genes are enriched in categories that include metabolism, enzymes, plasma proteins, disease development, and potential drug targets. Our results suggest that regulatory interactions in other tissues contribute to the modulation of blood metabolites.
Conclusions
—The spatial SNP-gene-metabolite associations identified in this study expand on the list of genes and tissues that are influenced by metabolic-associated SNPs and improves our understanding of the molecular mechanisms underlying pathologic blood metabolite levels.
Keywords: Expression quantitative trait loci, GWAS, Metabolite regulation, 3D genome
1. Introduction
The interaction between genetic variation, environment and lifestyle affects metabolite levels in humans [1]. The levels of these metabolites are central to the development of diseases including cholesterolaemia, cancer, coronary artery disease, angina pectoris, and type 2 diabetes [[2], [3], [4], [5], [6], [7], [8]]. Despite decades of work targeting metabolites of interest, the genetic networks and genes that modify metabolic potential remain poorly characterized and their therapeutic and diagnostic utility remains unrealized.
Whole genome or exome sequencing, in combination with metabolomic techniques, have been used to identify the genetic influencers of the human blood metabolome [[9], [10], [11], [12], [13], [14]]. In arguably the most comprehensive analysis to date, Shin and colleagues [9] performed a genome-wide association study (GWAS) and identified 145 genetic loci (SNPs) that correlate with the levels of approximately 400 blood metabolites. As is typical of all GWAS studies, >65% of the SNPs identified by Shin et al. were in non-coding regions of the genome. Shin et al. predicted causal genes of the genetic loci by scanning 500 kilobase (Kb) regions flanking the SNPs for genes whose functions linked with the corresponding metabolite. While it is commonly accepted that GWAS SNPs are enriched in regulatory elements, identifying the genes affected by the SNPs using a nearest-relevant-gene approach is problematic because the accumulating evidence supports long-range gene regulatory interactions, including inter-chromosomal, between regulatory elements and genes [[15], [16], [17], [18]]. Moreover, recent studies suggest that long-range regulatory interactions may play a more significant role in modifying disease outcomes than proximal interactions [[19], [20], [21]]. Long-range gene regulatory interactions can occur by several mechanisms: 1) epistatic interactions between gene products; 2) widespread action of a transcription factor or non-coding RNA; 3) scanning (e.g. the spreading of silencing complexes along a chromosome); or 4) chromatin looping (e.g. direct physical contact between enhancers and gene promoters) [22]. We, and others, have previously used genome structural data captured by proximity ligation techniques (e.g. Hi-C [23]) and eQTL data (e.g. GTEx [24]) to detect chromatin loops that bring long-range regulatory loci in spatial proximity with their target genes [19,[25], [26], [27], [28], [29]].
Here, we set out to use the 3D genome organization to identify target genes for the 145 blood metabolite-associated SNPs reported by Shin et al. [9]. To achieve this, we interrogated chromatin interaction (Hi-C) data from eight human cell lines [30] (Supplementary Table 1) to identify genes that are in physical contact with the SNPs [29]. Next, we designated functional SNP-gene associations by interrogating expression quantitative trait loci (eQTL) in 48 human tissues from the Genotype-Tissue Expression (GTEx [24], www.gtexportal.org) database [29]. Finally, we employed an array of approaches including gene ontology (KEGG, www.genome.jp), protein classification (Protein Atlas, www.proteinatlas.org), drug target analysis (DGIdb, www.dgidb.org), and literature text mining (PubMed®, www.ncbi.nlm.nih.gov/pubmed) to annotate the genes that are involved in modulating intermediate metabolites in human blood.
2. Methods
2.1. Data sources
The 145 metabolite-associated Single Nucleotide Polymorphisms (SNPs) investigated in this study are the genome-wide significant hits from Shin et al. [9]. Genomic positions of SNPs were obtained from the human hg19 genome build chromosome bed files downloaded from NCBI (See Data Availability). Gene synonyms and full names were obtained from NCBI’s gene information dataset (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/GENE_INFO/Mammalia/Homo_sapiens.gene_info.gz). We used the GENCODE transcript model (See Data Availability) as reference for gene annotations, which is the same reference used in GTEx. All isoforms of a gene were collapsed into a single gene. The human genome used in this study is the hg19 (GRChr37) build of the human genome release 75 (See Data Availability).
2.2. Identification of SNP target genes
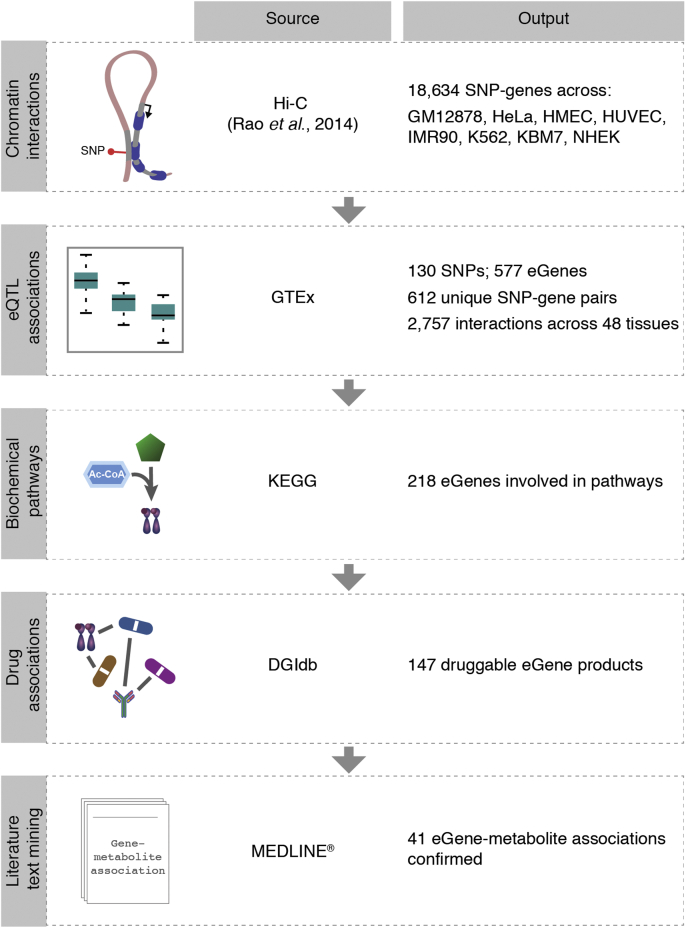
The CoDeS3D [29] pipeline was used to identify target genes of SNPs (Fig. 1). In summary, we downloaded raw Hi-C data from eight cell lines (i.e. GM12878, HeLa, HMEC, HUVEC, IMR90, K562, KBM7 and NHEK; Supplementary Table 1) [30] and processed them to generate chromatin interaction (Hi-C) libraries according to the methods outlined in Rao et al. [30]. For quality assurance, only Hi-C reads with MAPQ score >30 were retained. We digested the hg19 human genome with MboI (the same endonuclease used to prepare the Hi-C libraries) to identify the restriction fragments that harbor the metabolite-associated SNPs. The Hi-C libraries were queried to identify gene fragments that are captured as spatially interacting with the SNP-containing fragments. We eliminated SNP-gene spatial interactions that were captured in only one replicate of one cell line. The resulting spatial SNP-gene pairs were used to query GTEx V7 tissue eQTL analysis databases for cis eSNP-eGene pairs. eSNPs are the SNPs that correlate with a change in the expression of genes in at least one of the 48 human tissues available in the GTEx project. Trans eQTL associations were calculated using the GTEx eQTL calculator API. A Benjamini-Hochberg correction was performed on eSNP-eGene-associations from all tissues and those with adjusted (Benjamini-Hochberg) p-values ≤ 0.05 were deemed significant.
Fig. 1.
Methods workflow
Genes overlapping restriction fragments that spatially interact with fragments containing metabolite-associated SNPs were identified using Hi-C libraries. The resulting spatial SNP-gene pairs were tested for tissue eQTL interactions in the GTEx database. Only significant (FDR ≤ 0.05) tissue eSNP-eGene associations were further analyzed for pathways, drug associations and literature text mining. The Python scripts are available on Github (https://github.com/Genome3d/blood-metabolites-regulation.git).
2.3. Annotation of genes involved in metabolism
The Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY [55] (https://www.kegg.jp/kegg/pathway.html, accessed on 01/07/2018) database was queried with a list of the eGenes to identify their associated pathways (Supplementary 1). The retrieved results were analyzed to identify their first level pathway maps (e.g. metabolism, genetic information processing) and second level maps (e.g. carbohydrate metabolism, transcription). See Code Availability for the Python scripts used for this analysis.
2.4. Drug associations and protein classification
Data from The Human Protein Atlas [56] (https://www.proteinatlas.org/, downloaded on 25/03/19) version 18.1 was queried to obtain the protein classes of eGenes. The Drug Gene Interaction database [57] (DGIdb, http://dgidb.org) was interrogated for information on drugs that target the gene products (Fig. 1). The mechanisms of action of the drugs were also obtained. See Code Availability for the Python scripts used for this analysis.
2.5. Literature support for associations
To find evidence for eSNP-eGene, eSNP-metabolite, or eGene-metabolite associations in literature, we performed a text mining algorithm as follows. We employed the Bio. Entrez python API (see Code Availability) to search the underlying MEDLINE database to retrieve the PubMed IDs for articles that contain, in their titles or abstracts, exact matches of 1) eSNPs (rsIDs) and their linked eGenes (or gene synonyms or full names); 2) eSNPs (rsIDs) and at least one of the metabolites associated to the eSNP by Shin et al. [9]; or 3) eGenes (or their synonyms or full names) and at least one of the metabolites associated to the corresponding eSNP. We then identified intersecting articles for corresponding eSNP-eGene, eSNP-metabolite, and eGene-metabolite associations. Articles supporting gene-metabolite associations not reported in Shin et al. were manually curated by at least two persons.
2.6. Urls
CoDeS3D pipeline: https://github.com/Genome3d/codes3d-v1.
The Drug Gene Interaction database: http://dgidb.org.
GTEx portal: https://www.gtexportal.org/home/
The KEGG PATHWAY database: https://www.kegg.jp/kegg/pathway.html.
The Human Protein Atlas: https://www.proteinatlas.org/
2.7. Data and Code Availability
Supplementary tables are available at https://doi.org/10.17608/k6.auckland.8116097.
Scripts used for data curation, analysis, and visualisation are available at https://github.com/Genome3d/blood-metabolites-regulation.git.
Human genome build hg19 (GRChr37) was downloaded from ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/
SNP annotations (human genome, build hg19) were obtained from ftp.ncbi.nih.gov/snp/organisms/human_9606_b150_GRCh37p13/BED.
Gene annotations (Transcript model from GENCODE) were downloaded from https://storage.googleapis.com/gtex_analysis_v6p/reference/gencode.v19.genes.v6p_model.patched_contigs.gtf.gz.
3. Results
3.1. Metabolite-associated SNPs mark eQTLs
Shin et al. reported 145 metabolite-associated loci that are marked by SNPs with genome-wide significance. 102 of these loci were assigned predicted causal genes based on proximity to the SNPs and an established association between the gene and metabolite [9]. Notably, only 57 (∼39%) of the loci were reported as eQTLs by Shin et al. (Fig. 2a). Integrating genomic organization into the analysis identified eQTLs for 130 (∼90%) of the 145 metabolite-associated SNPs (hereafter eSNPs), more than double that reported by Shin et al. (Fig. 2a, Supplementary Table 2). None of the 15 non-eQTL SNPs (Supplementary Table 3) in our study was reported as being involved in an eQTL by Shin et al.
Fig. 2.
Regulatory interactions involving metabolite-associated SNPs
a) 90% of metabolite-associated SNPs mark eQTLs involved in long-range regulatory interactions. b) 71 of 102 of the causal SNP-gene pairs predicted in Shin et al. are among the 612 eSNP-eGene pairs we identified. c) Bar plot of number of genes regulated per eSNP (mode = 3). d) Violin plots of the normalized effect sizes (beta) of the eQTL interactions in different tissues.
The 130 eSNPs we identified were associated with the expression of 577 genes (i.e. eGenes) through 612 unique eSNP-eGene pairs (Fig. 2b). The eSNP-eGene pairs included 68.6% of the causal SNP-gene pairs predicted by Shin et al. (Fig. 2b, Supplementary Table 4). Notably, the eSNPs were associated with between 1 and 23 eGenes, with 3 eGenes as the mode (Fig. 2c), in a tissue specific manner. This is consistent with previous reports on shared gene regulatory sites [29,31,32]. Altogether, we identified 2757 eSNP-eGene interactions in 48 different human tissues (Supplementary Table 5) of which 621 overlapped Shin et al.‘s predictions (normalized effect size range, −1.69778 to 1.05834), while 2136 are novel (normalized effect size range, −1.15093 to 1.65858; Fig. 2d).
3.2. Metabolite regulatory interactions present in tissues other than blood
We estimated the contribution of Hi-C cell lines and GTEx tissues to the identification of the spatial eSNP-eGene interactions. Cis-eQTL interactions are more likely to occur across multiple cell lines but trans interactions are more cell line specific (Fig. 3a). The contribution of Hi-C cell lines in the identification of eSNP-eGene pairs was estimated by a Hi-C score—defined as the mean interactions across replicates of a cell line. The myelogenous leukemia (K562) cell line contributed the most (total Hi-C score, 1023) to the identification of spatial eSNP-eGene pairs while HMEC (a mammary epithelial cell line) contributed the least (total Hi-C score, 402) (Fig. 3b). The thyroid had the greatest number (137) of eSNP-eGene pairs while the amygdala and C1 vertebrae had the least (11) (Fig. 3c). Although there is a strong correlation (Pearson’s product-moment, 0.94, p-value < 2.2e-16) between the number of eQTLs identified and the number of GTEx RNA-Seq samples per tissue, we also observed variations in the proportions of tissue eQTL interactions and RNA-Seq sample sizes. For example, the cerebellum, for which there were 173 RNA-Seq samples, has the highest proportion (0.34) of eQTL interactions identified per RNA-Seq sample. This is almost 2x the proportion for skeletal muscle (i.e. 0.19), which has the greatest number (564) of RNA-Seq samples. Altogether, these observations reveal the cell line and tissue specificity of metabolic regulatory interactions.
Fig. 3.
Tissue and cell line specificity of metabolic regulatory interactions
a. Trans-eQTL interactions are more cell line specific than cis interactions, which tend to be present across multiple cell lines. b. K562 cell line contributes most to the identification of metabolism-associated eQTL interactions. c. The cerebellum has the most eQTL interactions identified per RNA-Seq sample.
3.3. Metabolite-associated SNPs target genes in metabolic pathways
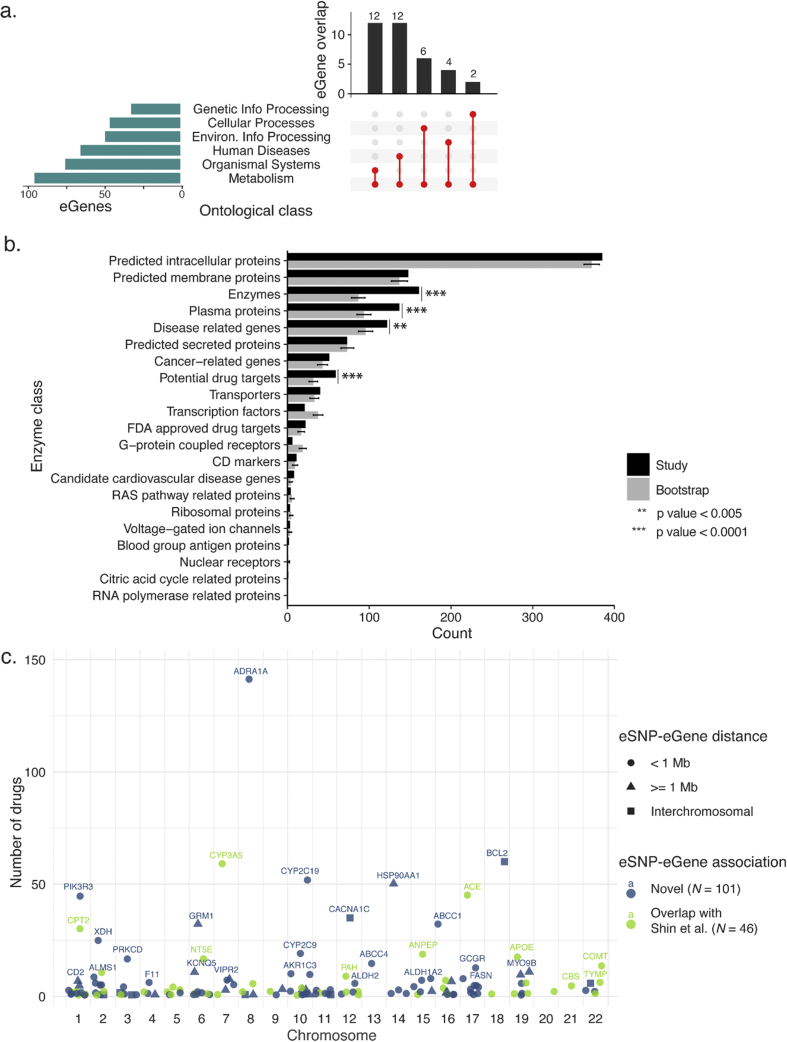
We annotated the eGenes’ biochemical functions using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database. 218 (37.8%) of the eGenes are annotated as being involved in ≥1 biochemical pathway (Supplementary Table 4). As expected, the largest single set of annotations for the eGenes is metabolism (n = 95, Fig. 4a). The next most represented categories i.e. organismal systems (n = 75) and human diseases (n = 65) each share 12 genes with the metabolism category (Fig. 4a). Classification of the eGenes using The Human Protein Atlas identified significant enrichment (via bootstrapping of null sets) of enzymes, plasma proteins, disease related genes and potential drug targets within 490 of the eGenes (Fig. 4b, Supplementary Table 4). The observed enrichment in the metabolic pathway and protein classification analyses is consistent with the eGene products acting as modifiers of metabolic activity.
Fig. 4.
Functional annotation of eGenes associated with metabolite eSNPs
a) eGenes are enriched for metabolism (horizontal bars) in KEGG pathways analysis. The metabolism genes are also involved in other pathways (vertical bars). b) Protein classification of genes is significantly enriched for enzymes, plasma proteins, potential drug targets, disease related genes. Error bars represent one standard deviation of 10,000 bootstraps. c) 147 eGene products are druggable based on DGIdb analysis. 68.7% (101) of the druggable products are encoded by eGenes that were not previously linked to the metabolite-associated SNPs.
We screened the Drug Gene Interaction database (DGIdb) to identify which of the eGenes encode potentially targetable products. Products of 147 (25.5%) of the eGenes are targets of at least one drug (Fig. 4c, Supplementary Table 6). 68, 119, and 142 of the druggable eGenes are also involved with metabolism, >1 biochemical pathway, and >1 protein class respectively. 101 of the 147 druggable eGenes have not been previously linked to genetic variants associated with metabolism.
There were several notable examples of metabolite associated gene–eSNP networks. For example, APOE, which has been associated with modulation of total cholesterol [33], was reported by Shin et al. as an eGene and predicted as the causal gene of nearby (3 Kb) cholesterol-associated rs445925. In addition to APOE (in suprapubic skin), our study revealed novel eQTL for rs445925 with BCAM (in basal ganglia, 90 Kb away), RYR1 (in skeletal muscle, 6.3 Mb away), and RERE (in suprapubic skin, on chromosome 1) (Fig. 5a).
Fig. 5.
eGene-metabolite associations
a) Cholesterol-associated rs445925 marks an eQTL that correlates with expression levels of APOE, BCAM, RYR1, and RERE. All four genes have literature (PMIDs) [33,[35], [36], [37]] supporting their links with cholesterol. b) ACSM5 pairs as an eGene with rs11647589, rs6497490, and rs1394678, which were associated with 3-phenylpropionate, X-11478 (an unknown metabolite), and indolepropionate respectively. The metabolism of 3-phenylpropionate and indolepropionate requires a Coenzyme A ligase such as ACSM5 [38,39].
We also identified network examples where eGenes linked to multiple regulatory hubs. For example, Acyl-CoA Synthetase Medium Chain Family Member 5 (ACSM5) associates with eQTL SNPs rs11647589 (in 16 tissues, intronic in ACSM5), rs1394678 (in 14 tissues, 38.5 Kb from ACSM5, intronic in ACSM2A), and rs6497490 (in tibial artery, 24.9 Kb from ACSM5, intronic in ACSM2A). rs11647589, rs1394678, and rs6497490 are associated with 3-phenylpropionate, indolepropionate, and X-117478 (an unknown metabolite) respectively (Fig. 5b). rs11647589, rs1394678 and rs6497490 mark distinct eQTLs because they are not in linkage disequilibrium (defined as LD>=0.8 in the EUR population, HaploReg v4.1 [34]).
3.4. Literature text mining supports eGene-metabolite associations
We conducted a stringent semi-automated literature text mining to identify literature that supported links between the eGenes and the metabolites with which the eSNPs are associated. First, we queried the MEDLINE® database using PubMed® APIs for research article titles or abstracts that contain the eSNP-eGene, eSNP-metabolite, or eGene-metabolite pairs (Fig. 6a). We then manually curated articles supporting eGene-metabolite pairs. The manual curation confirmed literature support for 41 eGene-metabolite pairs, 14 of which were not reported by Shin et al. (Supplementary Table 7). 29 of the literature-supported associations include genes that are involved in at least one biochemical pathway and whose products are druggable (Fig. 6b and c).
Fig. 6.
Functional annotation of eGenes associated with metabolite eSNPs
a) There is more literature support for eGene-metabolite associations than for eSNP-eGene or eGene-metabolite associations. b) A schematic of eGenes that are in biochemical pathways, encode druggable products, and whose relationship with their SNP metabolite has literature support. c) Ideogram of 119 druggable eGenes in b. Genes are colored green if the SNP-gene association was reported in Shin et al., or blue if it is not. Numbers in brackets correspond to the locus number (in Supplementary Table 2) of the eSNPs that spatially interact with the eGenes. The red-colored numbers indicate that the relationship between an eGenes and its SNP-associated metabolite (Supplementary Table 4) is supported in literature via text mining. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The literature supported the putative functional outcomes for the rs445925-APOE, BCAM, RYR1, RERE network as follows: 1) BCAM had a suggestive association in a GWAS meta-analysis of LDL cholesterol response to statins [35]; 2) RYR1, together with other nonalcoholic steatohepatitis related genes, has been linked to dietary cholesterol [36]; and 3) RERE was associated with circulating blood CD34+, which positively relates to total cholesterol [37]. Therefore, these eQTL associations do not only link SNPs to genes that are relevant to cholesterol, they also reveal the tissues that might be important for its metabolism.
The absence of literature support for eGene-metabolite associations does not equate to an absence of relationship. For example, the intergenic SNP rs7809615 is associated with ratio of androsterone sulfate and 4-androsten-3beta,17beta-diol disulfate 2 levels in the blood metabolite GWAS (Supplementary Table 2). Our method, as well as that of Shin et al., links rs7809615 to CYP3A5 (Fig. 6c), which encodes a cytochrome P450 known to be active against steroids such as testosterone [40]. We further observed that rs7809615 also links with CYP3A7 (Fig. 6c), which encodes another P450. Although our stringent text mining did not find articles that directly linked the two target eGenes, the STRING database (v10.5, accessed 26/04/2019) reports a binding interaction between CYP3A5 and CYP3A7.
4. Discussion
We systematically integrated evidence from chromatin interactions, eQTL data, gene ontology, protein classifications, drug interactions, and the published literature to identify and characterize genes that are regulated by metabolite-associated SNPs. Our results provide a step-change in our current understanding of the variant-gene regulatory interactions of metabolism, particularly those that rely on chromatin looping.
The integration of chromatin interaction and eQTL data in this study enabled the mapping of long-range regulatory interactions for ∼90% of the metabolite-associated SNPs. This mapping was robust to the different data sources that were used to identify the eQTLs (GTEx [24]) and the blood metabolite associated SNPs (KORA and Twins UK) [9]. The ∼10% of metabolite-associated SNPs that were not identified as eQTLs in our analysis can be explained by the following: a) they tag other functional loci in linkage disequilibrium but do not have any regulatory effect themselves; b) they are eQTLs but their gene associations were missed due to the transient nature of chromatin loops or the specificity in the developmental timing of the linkage of these SNPs to their metabolites; or c) their underlying regulatory mechanism does not require chromatin proximity (e.g. it occurs by diffusion of regulatory factors or non-coding RNA, or the variant impacts on post-transcriptional regulation). While we have reproduced ∼70% of SNP-gene regulatory mapping predicted in Shin et al., we propose that the novel spatial eQTL associations discovered in this study provide a useful resource to unravel the holistic genetic architecture that underlies metabolism.
There are number of caveats to this study. Firstly, the integration of heterogenous biological data has inherent limitations. For example, the chromatin interaction information used in this study is primarily from immortalized cell lines. By contrast, the eQTL mapping used mRNA data obtained from donated tissues. The problem of tissue averaging is well-known in mRNA studies [41]. However, the changes in mRNA levels that were associated with the eQTLs we identified were large enough to affect the overall transcript profile of the tissue. Secondly, eQTL analysis implicitly assumes mRNA levels are predictive of protein levels. There is good correlation between mRNA and protein levels in differentially expressed genes and cells in a steady state [42]. However, the relationship between mRNA and protein levels is affected by factors including modulation of translation rates and protein half-life, protein synthesis delay, and protein transport [43]. To overcome these limitations, future investigations would ideally use chromatin interaction, genotype, and gene expression information that is generated from the same population of cells. This would overcome cell population structural biases and delineate genotype-specific chromatin rewiring and expression profiles.
The tissue SNP-gene mappings identified in this study are consistent with known tissue-metabolite relationships. For example, previous studies support our observation of cholesterol-associated regulatory interactions in the basal ganglia [44,45], skeletal muscle [46,47], and skin [48,49]. Similarly, our observation of disproportionately high metabolism-associated eQTL interactions per RNA-Seq sample in the cerebellum and testis agrees with known metabolic dysfunction in these tissues (e.g. cerebellar metabolism in patients with diaschisis [50], and the glucose sensing mechanism of Sertoli cells that responds to homeostatic conditions in the testis [51]). The detection of a metabolite-associated eQTL interaction in a tissue therefore suggests a modulation in the production or utilisation of the metabolite in that tissue.
We have identified regulatory interactions involving genes that encode for proteins that do not directly act on the associated metabolite. We propose that metabolite-associated eQTLs affect metabolism by modulating the expression of genes that encode the protein components (i.e. enzymes, transcription factors, inhibitors, transporters, receptors etc.) that are collectively responsible for metabolism in a holistic sense, including the environment in which it occurs. For example, spatial regulatory interactions with genes that encode for plasma proteins could be important for metabolism because plasma proteins aid in the transport and regulation of metabolites, ion, lipids and hormones. Further investigation into the metabolic roles of eQTL associations between metabolite-associated SNPs and uncharacterised genes or genes in non-metabolism ontological classes is therefore necessary. Collectively, our observations are consistent with the understanding that homeostatic metabolite levels in the blood are the sum of the rates of import from the environment, production and degradation of metabolites in all of the tissues that contact blood [[52], [53], [54]].
5. Conclusions
In conclusion, we have shown that blood metabolite-associated SNPs mark expression quantitative trait loci that affect the expression of proximal and distal genes in tissues that are relevant to the associated metabolites in particular, and to metabolism in general. The eSNP-eGene-metabolite connections identified in this study reproduce and expand on the list of genes that are likely to be influenced by metabolite-associated genetic variants and thus provide a useful resource for further empirical study of the genetic influences of the human metabolome.
Funding
This work was funded by High Value Nutrition National Science, New Zealand (MBIE/HVN grant #3710040) to JOS & TF, and the New Zealand Ministry of Business, Innovation and Employment (MBIE grant #UOAX1611) to JOS . JOS and WS are funded by a Royal Society of New Zealand Marsden Fund (Grant 16-UOO-072).
CRediT authorship contribution statement
Tayaza Fadason: Writing - original draft, Writing - review & editing, Visualization. William Schierding: Writing - review & editing, Conceptualization. Nikolai Kolbenev: Software, Methodology. Jiamou Liu: Software, Methodology. John R. Ingram: Writing - review & editing. Justin M. O’Sullivan: Conceptualization, Writing - review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metop.2020.100035.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kastenmüller G., Raffler J., Gieger C., Suhre K. Genetics of human metabolism: an update. Hum Mol Genet. 2015;24:R93–R101. doi: 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Outschoorn U.E., Peiris-Pagés M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 3.Sonnenburg J.L., Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature. 2016;531:651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiolase C.A., Kantor P.F., Lucien A., Kozak R., Lopaschuk G.D. 2016. The antianginal drug trimetazidine shifts cardiac energy metabolism from fatty acid oxidation to glucose oxidation by inhibiting mitochondrial long-chain; pp. 580–588. [DOI] [PubMed] [Google Scholar]
- 6.Gilde A.J. Peroxisome proliferator-activated receptor (PPAR) α and PPARβ/δ, but not PPARγ, modulate the expression of genes involved in cardiac lipid metabolism. Circ Res. 2003;92:518–524. doi: 10.1161/01.RES.0000060700.55247.7C. [DOI] [PubMed] [Google Scholar]
- 7.Akbaraly T. Association of circulating metabolites with healthy diet and risk of cardiovascular disease: analysis of two cohort studies. Sci Rep. 2018;8:8620. doi: 10.1038/s41598-018-26441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong Z. Targeted metabolomic analysis of plasma metabolites in patients with coronary heart disease in southern China. Medicine (Baltimore) 2019;98 doi: 10.1097/MD.0000000000014309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin S.-Y. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draisma H.H.M. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee E.P. An exome array study of the plasma metabolome. Nat Commun. 2016;7:12360. doi: 10.1038/ncomms12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettunen J. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long T. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 14.Kordalewska M., Markuszewski M.J. Metabolomics in cardiovascular diseases. J Pharmaceut Biomed Anal. 2015;113:121–136. doi: 10.1016/j.jpba.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 15.Dekker J., Misteli T. Long-range chromatin interactions. Cold Spring Harb. Perspect. Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claussnitzer M. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguet F. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaneau O. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science. 2019;364(80):eaat8266. doi: 10.1126/science.aat8266. [DOI] [PubMed] [Google Scholar]
- 19.Martin P. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat Commun. 2015;6:10069. doi: 10.1038/ncomms10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Võsa U. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv. 2018;18:10. [Google Scholar]
- 21.Liu X., Li Y.I., Pritchard J.K. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177 doi: 10.1016/j.cell.2019.04.014. 1022-1034.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dekker J., Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman-Aiden E. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(80):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ardlie K.G. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(80):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jäger R. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun. 2015;6:6178. doi: 10.1038/ncomms7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schierding W., Antony J., Cutfield W.S., Horsfield J.A.J.A., O’Sullivan J.M. Intergenic GWAS SNPs are key components of the spatial and regulatory network for human growth. Hum Mol Genet. 2016:1–11. doi: 10.1093/hmg/ddw165. 0. [DOI] [PubMed] [Google Scholar]
- 27.Fadason T., Ekblad C., Ingram J.R., Schierding W.S., Justin M. Physical interactions and expression quantitative traits loci identify regulatory connections for obesity and type 2 diabetes associated SNPs. Front Genet. 2017;8:1–12. doi: 10.3389/fgene.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schierding W. GWAS on prolonged gestation (post-term birth): analysis of successive Finnish birth cohorts. J Med Genet. 2018;55:55–63. doi: 10.1136/jmedgenet-2017-104880. [DOI] [PubMed] [Google Scholar]
- 29.Fadason T., Schierding W., Lumley T., O’Sullivan J.M. Chromatin interactions and expression quantitative trait loci reveal genetic drivers of multimorbidities. Nat Commun. 2018;9:5198. doi: 10.1038/s41467-018-07692-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao S.S.P. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong P., Monahan J., Prendergast J.G.D. Shared regulatory sites are abundant in the human genome and shed light on genome evolution and disease pleiotropy. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F., Wang J., Pierce B.L., Chen L.S. Identifying cis -mediators for trans -eQTLs across many human tissues using genomic mediation analysis. Genome Res. 2017;27:1859–1871. doi: 10.1101/gr.216754.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low-Kam C. Variants at the APOE/C1/C2/C4 locus modulate cholesterol efflux capacity independently of high-density lipoprotein cholesterol. J. Am. Heart Assoc. 2018;7 doi: 10.1161/JAHA.118.009545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postmus I. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat Commun. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J.Q. Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat Commun. 2018;9:4490. doi: 10.1038/s41467-018-06931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen K.S. Circulating CD34+ progenitor cell frequency is associated with clinical and genetic factors. Blood. 2013;121:e50–e56. doi: 10.1182/blood-2012-05-424846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. Philipps Universität; Marburg.: 2014. R)-Indolelactyl-CoA dehydratase, the key enzyme of tryptophan reduction to indolepropionate in Clostridium sporogenes. [Google Scholar]
- 39.Ebenau-Jehle C. Anaerobic metabolism of indoleacetate. J Bacteriol. 2012;194:2894–2903. doi: 10.1128/JB.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamakoshi Y., Kishimoto T., Sugimura K., Kawashima H. Human prostate CYP3A5: identification of a unique 5′-untranslated sequence and characterization of purified recombinant protein. Biochem Biophys Res Commun. 1999;260:676–681. doi: 10.1006/bbrc.1999.0960. [DOI] [PubMed] [Google Scholar]
- 41.Tang F. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edfors F. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol. 2016;12:883. doi: 10.15252/msb.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., Beyer A., Aebersold R. Review on the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Onyewuenyi I.C., Muldoon M.F., Christie I.C., Erickson K.I., Gianaros P.J. Basal ganglia morphology links the metabolic syndrome and depressive symptoms. Physiol Behav. 2014;123:214–222. doi: 10.1016/j.physbeh.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Brouwer E.J.M. Prevalence and vascular risk factors of basal ganglia calcifications in patients at risk for cerebrovascular disease. J Neuroradiol. 2019:2–7. doi: 10.1016/j.neurad.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Barrientos G., Sánchez-Aguilera P., Jaimovich E., Hidalgo C., Llanos P. Membrane cholesterol in skeletal muscle: a novel player in excitation-contraction coupling and insulin resistance. J. Diabetes Res. 2017;1–8 doi: 10.1155/2017/3941898. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habegger K.M. Fat-induced membrane cholesterol accrual provokes cortical filamentous actin destabilisation and glucose transport dysfunction in skeletal muscle. Diabetologia. 2012;55:457–467. doi: 10.1007/s00125-011-2334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sörensen B.M. Cardiovascular risk factors as determinants of retinal and skin microvascular function: the Maastricht Study. PloS One. 2017;12 doi: 10.1371/journal.pone.0187324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tashakkor A.Y., Mancini G.B.J. The relationship between skin cholesterol testing and parameters of cardiovascular risk: a systematic review. Can J Cardiol. 2013;29:1477–1487. doi: 10.1016/j.cjca.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 50.Shamoto H., Chugani H.T. Glucose metabolism in the human cerebellum: an analysis of crossed cerebellar diaschisis in children with unilateral cerebral inrjury. J Child Neurol. 1997;12:407–414. doi: 10.1177/088307389701200701. [DOI] [PubMed] [Google Scholar]
- 51.Alves M.G., Martins A.D., Cavaco J.E., Socorro S., Oliveira P.F. Diabetes, insulin-mediated glucose metabolism and Sertoli/blood-testis barrier function. Tissue Barriers. 2013;1 doi: 10.4161/tisb.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boulet M.M. Alterations of plasma metabolite profiles related to adipose tissue distribution and cardiometabolic risk. Am. J. Physiol. Metab. 2015;309:E736–E746. doi: 10.1152/ajpendo.00231.2015. [DOI] [PubMed] [Google Scholar]
- 53.Giesbertz P. Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia. 2015;58:2133–2143. doi: 10.1007/s00125-015-3656-y. [DOI] [PubMed] [Google Scholar]
- 54.Torell F. Multi-organ contribution to the metabolic plasma profile using hierarchical modelling. PloS One. 2015;10 doi: 10.1371/journal.pone.0129260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uhlen M. Tissue-based map of the human proteome. Science. 2015;347(80) doi: 10.1126/science.1260419. 1260419–1260419. [DOI] [PubMed] [Google Scholar]
- 57.Cotto K.C. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary tables are available at https://doi.org/10.17608/k6.auckland.8116097.
Scripts used for data curation, analysis, and visualisation are available at https://github.com/Genome3d/blood-metabolites-regulation.git.
Human genome build hg19 (GRChr37) was downloaded from ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/
SNP annotations (human genome, build hg19) were obtained from ftp.ncbi.nih.gov/snp/organisms/human_9606_b150_GRCh37p13/BED.
Gene annotations (Transcript model from GENCODE) were downloaded from https://storage.googleapis.com/gtex_analysis_v6p/reference/gencode.v19.genes.v6p_model.patched_contigs.gtf.gz.