Abstract
Purpose
To test the feasibility of using deep learning for optical coherence tomography angiography (OCTA) detection of diabetic retinopathy.
Methods
A deep-learning convolutional neural network (CNN) architecture, VGG16, was employed for this study. A transfer learning process was implemented to retrain the CNN for robust OCTA classification. One dataset, consisting of images of 32 healthy eyes, 75 eyes with diabetic retinopathy (DR), and 24 eyes with diabetes but no DR (NoDR), was used for training and cross-validation. A second dataset consisting of 20 NoDR and 26 DR eyes was used for external validation. To demonstrate the feasibility of using artificial intelligence (AI) screening of DR in clinical environments, the CNN was incorporated into a graphical user interface (GUI) platform.
Results
With the last nine layers retrained, the CNN architecture achieved the best performance for automated OCTA classification. The cross-validation accuracy of the retrained classifier for differentiating among healthy, NoDR, and DR eyes was 87.27%, with 83.76% sensitivity and 90.82% specificity. The AUC metrics for binary classification of healthy, NoDR, and DR eyes were 0.97, 0.98, and 0.97, respectively. The GUI platform enabled easy validation of the method for AI screening of DR in a clinical environment.
Conclusions
With a transfer learning process for retraining, a CNN can be used for robust OCTA classification of healthy, NoDR, and DR eyes. The AI-based OCTA classification platform may provide a practical solution to reducing the burden of experienced ophthalmologists with regard to mass screening of DR patients.
Translational Relevance
Deep-learning-based OCTA classification can alleviate the need for manual graders and improve DR screening efficiency.
Keywords: deep learning, diabetic retinopathy, artificial intelligence, detection, screening
Introduction
As the leading cause of preventable blindness in working-age adults, diabetic retinopathy (DR) affects 40% to 45% of diabetic patients.1 In the United States alone, the number of DR patients is estimated to increase from 7.7 million in 2010 to 14.6 million by 2050.1 Early detection, prompt intervention, and reliable assessment of treatment outcomes are essential to preventing irreversible visual loss from DR. With early detection and adequate treatment, more than 95% of DR-related vision losses can be prevented.2 Retinal vascular abnormalities, such as microaneurysms, hard exudates, retinal edema, venous beading, intraretinal microvascular anomalies, and retinal hemorrhages, are common DR findings.3 Therefore, imaging examination of retinal vasculature is important for DR diagnosis and treatment evaluation. Traditional fundus photography provides limited sensitivity to reveal subtle abnormality correlated with early DR.4–7 Fluorescein angiography (FA) can be used to improve imaging sensitivity of retinal vascular distortions in DR8,9; however, FA requires intravenous dye injections, which may produce side effects and require additional monitoring and careful management. Optical coherence tomography angiography (OCTA) is a noninvasive method for better visualization of retinal vasculatures.10 OCTA allows visualization of multiple retinal layers with high resolution; thus, it is more sensitive than FA in detecting subtle vascular distortions correlated with early eye conditions.11–13
The recent development of quantitative OCTA offers a unique opportunity to utilize computer-aided disease detection and artificial intelligence (AI) classification of eye conditions. Quantitative OCTA analysis has been explored for objective assessment of, for example, DR,14–17 age-related macular degeneration (AMD),18,19 vein occlusion,20–23 and sickle cell retinopathy (SCR).24 Supervised machine learning has also recently been validated for multiple-task classification to differentiate among control, DR, and SCR eyes.25 In principle, deep learning may provide a simple solution to fostering clinical deployment of AI classification of OCTA images. Deep learning generally refers to the convolutional neural network (CNN) algorithm, which was inspired by the human brain and visual information processing. CNNs contain millions of artificial neurons (also referred to as parameters) to process image features in a feed-forward process by extracting and processing simple features in early layers and complex features in later layers.26 To train a CNN for a specific classification task requires millions of images to optimize the network parameters.27 However, for the relatively new imaging modality OCTA, the limitation of currently available images poses an obstacle for practical implementation of deep learning.
In order to overcome the limitation of data size, a transfer learning approach has been demonstrated for implementing deep learning. Transfer learning is a training method to adopt some weights of a pretrained CNN and appropriately retrain certain layers of that CNN to optimize the weights for a specific task (i.e., AI classification of retinal images).28 In fundus photography, transfer learning has been explored to conduct artery–vein segmentation,29 glaucoma detection,30,31 and diabetic macular thinning assessment.32 Recently, transfer learning has also been explored in OCT for detecting choroidal neovascularization, diabetic macular edema,28 and AMD.33
In principle, transfer learning can involve a single layer or multiple layers, because each layer has weights that can be retrained. For example, the specific number of layers required for retraining in a 16-layer CNN (Fig. 1) may vary, depending on the available dataset and specific task interested. Moreover, compared to traditional fundus photography and OCT, deep learning in OCTA classification is still unexplored due to the limited size of publicly available datasets. In this study, we demonstrate the first use of OCTA for automated classification using deep learning. By leveraging transfer learning, we aim to train a small dataset to achieve reliable DR classification. Furthermore, an easy-to-use graphical user interface (GUI) platform was also developed to foster deep-leaning-based DR classification for adoption in a clinical setting.
Figure 1.
The deep learning CNN used for OCTA DR detection is VGG16, a network that contains 16 trainable layers: convolution (Conv) and fully connected (FC) layers. The corresponding output layer dimensions of each layer is shown below each block. All convolution and fully connected layers are followed by a ReLU activation function. The softmax layer is a fully connected layer that is followed by a softmax activation function. Maxpool and Flatten layers are operational layers with no tunable parameters.
Methods
This study adhered to the tenets of the Declaration of Helsinki and was approved by the institutional review board of the University of Illinois at Chicago (UIC).
Data Acquisition
The 6 × 6-mm2 field of view OCTA data were acquired using an AngioVue spectral-domain OCTA system (Optovue, Fremont, CA, USA) with a 70-kHz A-scan rate, a lateral resolution of ∼15 um, and an axial resolution of ∼5 um. The inclusion criterion was an OCTA acquisition quality of 6 or greater. All OCTA images were qualitatively examined for severe motion or shadow artifacts. Images with significant artifacts were excluded for this study. The OCTA data were exported using ReVue (Optovue) software, and custom-developed Python procedures were used for image processing.
The study involved two separate datasets collected with the same recording parameters. The first dataset, consisting of 131 images, was collected at UIC. For external validation, a second dataset, consisting of 46 images, was provided by National Taiwan University.
Classification Model Implementation
The CNN architecture chosen for this study was VGG16.34 The network specifications and design are illustrated in Figure 1. The pretrained weights were obtained from the ImageNet dataset.35 The CNN classifier was trained and evaluated using Python 3.7.1 software with the Keras 2.24 application and TensorFlow 1.13.1 open-source platform backend. Training was performed on a Windows 10 computer (Microsoft, Redmond, WA, USA) using the NVIDIA GeForce RTX 2080 Ti graphics processing unit (Santa Clara, CA, USA). To prevent overfitting, each classifier was trained with early stopping, and experimentally the model (i.e., retrained classifier) converges within ∼70 epochs. During each iteration, data augmentation in the form of random rotation, horizontal and vertical flips, and zoom was performed. In this study, manual classifications were used as reference for determining the receiver operating characteristic (ROC) curves and area under the curve (AUC). AUC was used as an index of performance of the model along with sensitivity (SE), specificity (SP), diagnostic accuracy (ACC). To evaluate the performance of each model, fivefold cross-validation was implemented.
Transfer Learning and Model Selection
In practice, it takes hundreds of thousands of data samples (i.e., images) to optimize the millions of parameters of a CNN. Training CNNs on smaller datasets will often lead to overfitting, such that the CNN has memorized the dataset, resulting in high performance when predicting on the same dataset and but failure to perform well on new data.36 Transfer learning, which leverages the weights in a pretrained network, has been established to overcome the overfitting problem. Transfer learning is well suited for CNNs because CNNs extract features in a bottom-up hierarchical structure. This bottom-up process is analogous to the human visual pathway system.37 Due to the great dissimilarity between the ImageNet dataset used to pretrain the CNN and our own OCTA dataset, we conducted a transfer learning process to determine the appropriate number of retrained layers required in a CNN to achieve robust performance of OCTA classification. For this study, misclassification error was used for quantitative assessment of the CNN performance, and the one standard deviation rule was used for model selection. This rule states that a model selected must be within one positive standard deviation from the misclassification error of the best performing model.38 The model requiring the least number of retrained layers for performance comparable to that of the best trained model was selected.
Results
Patient Demographics
Subjects and diabetes mellitus (DM) patients with and without DR were recruited from the UIC retina clinic. The patients presented in this study are representative of a university population of DM patients who require clinical diagnosis and management of DR. Two board-certified retina specialists classified the patients based on the severity of DR according to the Early Treatment Diabetic Retinopathy Study staging system. All patients underwent complete anterior and dilated posterior segment examination (JIL, RVPC). All control OCTA images were obtained from healthy volunteers that provided informed consent for OCT and OCTA imaging. All subjects underwent OCT and OCTA imaging of both eyes (OD and OS). The images used in this study did not include images of eyes with other ocular diseases or any other pathological features in their retina such as epiretinal membranes and macular edema. Additional exclusion criteria included eyes with a prior history of intravitreal injections, vitreoretinal surgery, or significant (greater than a typical blot hemorrhage) macular hemorrhages. Subject and patient characteristics, including sex, age, duration of diabetes, diabetes type, hemoglobin A1c (HbA1c) status, and hypertension prevalence, are summarized in Table 1.
Table 1.
Demographics of OCTA Dataset
| Demographic | Control | No DR | Mild DR | Moderate DR | Severe DR |
|---|---|---|---|---|---|
| Subjects (n) | 20 | 17 | 20 | 20 | 20 |
| Sex, male/female (n) | 12/8 | 6/11 | 11 | 12 | 11 |
| Age (y), mean ± SD | 42 ± 9.8 | 66.4 ± 10.14 | 50.10 ± 12.61 | 50.80 ± 8.39 | 57.84 ± 10.37 |
| Age range (y) | 25–71 | 49–86 | 24–74 | 32–68 | 41–73 |
| Duration of diabetes (y), mean ± SD | — | — | 19.64 ± 13.27 | 16.13 ± 10.58 | 23.40 ± 11.95 |
| Diabetes type II (%) | — | 100 | 100 | 100 | 100 |
| Insulin dependent, Y/N (n) | — | 14/3 | 7/13 | 12/8 | 15/5 |
| Hba1c (%), mean ± SD | — | 5.9 ± 0.7 | 6.5 ± 0.6 | 7.3 ± 0.9 | 7.8 ± 1.3 |
| Hypertension (%) | 10 | 17 | 45 | 80 | 80 |
This study included 24 eyes from 17 patients with diabetes but no DR (NoDR), 75 eyes from 60 patients with DR, and 32 healthy eyes from 20 control subjects. For all subjects and patients, OCTA features were quantified and are summarized in Table 2. In this study, this dataset is referred to as the cross-validation dataset.
Table 2.
Quantification of Individual OCTA Features
| Mean ± SD | |||||
|---|---|---|---|---|---|
| Feature | Control | No DR | Mild DR | Moderate DR | Severe DR |
| SCP | |||||
| BVT | 1.11 ± 0.07 | 1.09 ± 0.01 | 1.14 ± 0.05 | 1.17 ± 0.06 | 1.23 ± 0.04 |
| BVC (µm) | 40.16 ± 0.51 | 41.28 ± 0.63 | 41.17 ± 1.35 | 40.95 ± 0.58 | 41.29 ± 1.09 |
| VPI | 29. 77 ± 1.52 | 31.04 ± 1.58 | 28.30 ± 2.09 | 28.91 ± 2.02 | 27.33 ± 3.30 |
| BVD (%) | |||||
| SCP | |||||
| C1, 2 mm | 56.93 ± 4.07 | 42.33 ± 7.48 | 50.44 ± 8.74 | 52.34 ± 5.71 | 44.40 ± 7.67 |
| C2, 4 mm | 56.49 ± 2.69 | 55.27 ± 4.00 | 54.09 ± 4.90 | 54.93 ± 4.06 | 52.47 ± 4.84 |
| C3, 6 mm | 54.45 ± 2.45 | 55.36 ± 3.14 | 52.77 ± 3.55 | 53.71 ± 3.94 | 52.54 ± 5.04 |
| DCP | |||||
| C1, 2 mm | 75.52 ± 3.70 | 63.03 ± 6.95 | 64.97 ± 8.60 | 67.22 ± 5.52 | 57.36 ± 8.46 |
| C2, 4 mm | 78.37 ± 3.87 | 71.52 ± 5.59 | 70.25 ± 6.45 | 70.17 ± 5.12 | 62.50 ± 7.62 |
| C3, 6 mm | 76.70 ± 4.93 | 71.45 ± 6.02 | 68.08 ± 6.62 | 67.11 ± 5.23 | 60.77 ± 7.72 |
| FAZ-A | |||||
| SCP (mm2) | 0.30 ± 0.06 | 0.37 ± 0.16 | 0.33 ± 0.05 | 0.38 ± 0.07 | 0.46 ± 0.06 |
| DCP (mm2) | 0.39 ± 0.08 | 0.40 ± 0.14 | 0.46 ± 0.07 | 0.53 ± 0.12 | 0.58 ± 0.09 |
| FAZ-CI | |||||
| SCP | 1.14 ± 0.11 | 1.14 ± 0.04 | 1.29 ± 0.14 | 1.38 ± 0.14 | 1.46 ± 0.18 |
| DCP | 1.18 ± 0.12 | 1.09 ± 0.02 | 1.31 ± 0.21 | 1.42 ± 0.19 | 1.49 ± 0.17 |
SCP, superficial capillary plexus; BVT, blood vessel tortuosity; BVC, blood vessel caliber; VPI, vessel perimeter index; BVD, blood vessel density; C1, C2, and C3, three circular zones; DCP, deep capillary plexus; FAZ-A, foveal avascular zone area; FAZ-CI, foveal avascular zone contour irregularity.
Model Selection
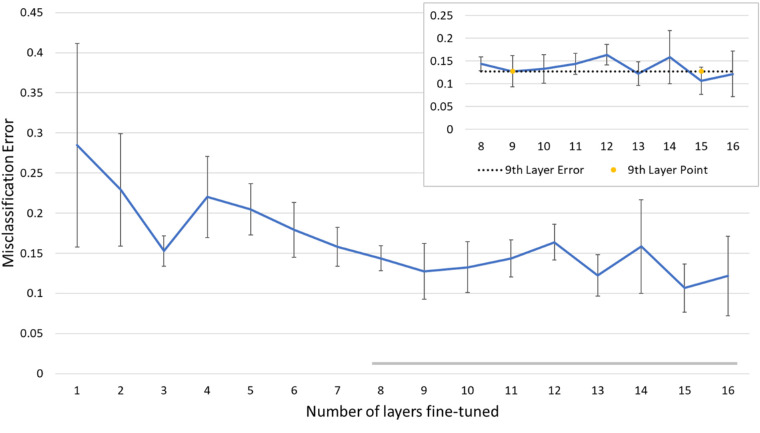
A model selection process was used to identify the required number of retrained layers for the best performance of OCTA classification. The layers in the VGG16 CNN were sequentially retrained starting from the last to the beginning layers. A quantitative comparison of misclassification errors for individual models (i.e., variable number of retrained layers) is provided in Figure 2. In this study, we evaluated each model using the cross-validation dataset. We applied the one standard deviation rule, which recommends choosing a less complex model that is within one standard deviation of the misclassification error of the best performing model. The model retrained with nine layers conformed with the one standard deviation rule and was further examined in a cross-validation study.
Figure 2.
A transfer learning performance study was conducted to determine how many layers are necessary for effective transfer learning in OCTA images. Our model consisted of 16 retrainable layers. The additional graph in the right-hand corner shows that retraining nine layers satisfies the criteria of the one positive standard deviation rule.
Cross-Validation Study
Using a fivefold cross-validation method, we evaluated the performance of the selected nine layers retrained on our cross-validation dataset. The cross-validation performance summarized in Tables 3 and 4 reveals that this model achieved an average of 87.27% ACC, 83.76% SE, and 90.82% SP among the three categories of control, NoDR, and DR. For the individual class predictions, NoDR had the highest accuracy, followed by control and then DR. This performance is reflected in the ROC graphs and AUC in Figure 3. For our cross-validation study, the overall AUC was 0.96, with individual AUC values for control, NoDR, and DR of 0.97, 0.98, and 0.97, respectively.
Table 3.
Cross-Validation Multi-Label Confusion Matrix (n = 131)
| Predicted Label | |||
|---|---|---|---|
| True Label | Control | No DR | DR |
| Control | 25 | 3 | 4 |
| NoDR | 1 | 23 | 0 |
| DR | 9 | 8 | 58 |
Table 4.
Cross-Validation Evaluation Metrics (n = 131)
| Mean ± SD | ||||
|---|---|---|---|---|
| Metric | Control | No DR | DR | Average |
| ACC (%) | 87.022 ± 0.059 | 90.840 ± 0.020 | 83.970 ± 0.050 | 87.277 ± 0.034 |
| SE (%) | 78.123 ± 0.152 | 95.835 ± 0.089 | 77.334 ± 0.076 | 83.764 ± 0.105 |
| SP (%) | 89.899 ± 0.050 | 89.720 ± 0.022 | 92.858 ± 0.041 | 90.825 ± 0.018 |
Figure 3.

ROC curves for the cross-validation performance of the model for individual class performance (control, NoDR, and DR) and the average performance of the model.
External Validation
In addition to cross-validation, we validated our CNN classifier using an external dataset from a cohort of NoDR and DR patients from National Taiwan University Hospital. The dataset was comprised of 13 NoDR patients (eight males), with an average age of 62 ± 8.35 years, and 21 DR patients (12 males), with an average age of 61 ± 12.80 years. Other clinical information regarding the dataset was not available to us. The validation results revealed an average of 70.83 ± 0.021% ACC, 67.23 ± 0.026% SE, and 73.96 ± 0.026% SP. The confusion matrix for the validation dataset is summarized in Table 5.
Table 5.
External Validation Multi-Label Confusion Matrix (n = 46)
| Predicted Label | |||
|---|---|---|---|
| True Label | Control | No DR | DR |
| Control | |||
| NoDR | 1 | 13 | 6 |
| DR | 1 | 7 | 18 |
Evaluation of Clinical Deployment
The model with the best performance was integrated with a custom-designed GUI platform to evaluate the potential of AI classification of DR using OCTA in a clinical environment. The GUI was developed using Java 11.01, an open source programming language. As shown in Figure 4, the GUI adopted an interface that is commonly used in retina clinics to enable easy adoption by ophthalmic personnel. By clicking the “Load image” button, one OCTA image can be selected (Fig. 4A) and displayed (Fig. 4B) for visual examination. By clicking the “Predict” button, automated AI classification can be performed. The output of the DR classification is displayed in the “Results” box. Additional information about classification confidence is also available for clinical reference. The GUI platform has been tested by three ophthalmologists (JIL, RPVC, and DT) to verify the feasibility of applying the deep-learning-based AI classification of DR using OCTA in a clinical environment. For each OCTA image, the GUI-based classification can be completed within 1 minute.
Figure 4.
GUI platform for DR classification using OCTA.
Discussion
In this study, a deep learning model was trained using transfer learning for automated classification of OCTA images for three categories: healthy control, NoDR, and DR. In our study, we utilized a selection processes to determine the optimal number of layers for fine-tuning. This selection process revealed that fine-tuning nine layers is optimal. To evaluate this model, we performed a fivefold cross-validation. The results of cross-validation were promising; however, the cross-validation dataset may contain some inherent biases, such as the inclusion of both eyes from one subject. To verify our classifier, we tested an external dataset to confirm the performance of automated DR classification.
Current research for deep learning in ophthalmology has focused on using fundus photography and OCT. Transfer learning has been demonstrated as a means to train CNNs with fundus images to predict the future DR progression.39 Recently, Li et al.40 employed transfer learning to train a CNN for detecting referable DR from non-referable DR in fundus images. Similarly, Lu et al.41 trained a CNN for classification of OCT scans to detect retinal abnormalities, such as serous macular detachment, cystoid macular edema, macular hole, and epiretinal membrane. All of these studies reported good classification (>90% accuracy) when utilizing transfer learning to train a relatively large dataset (>500 images per cohort). One study by Eltanboly et al.42 proposed a deep learning computer-aided design system for the detection of DR in OCT scans. That study utilized a dataset of 52 clinical OCT scans and achieved 92% accuracy in fourfold cross-validation and 100% accuracy on one hold-out validation. Although these results are promising, that study did not use an external database to validate. In comparison to our study, the cross-validation performance revealed 87.65% accuracy and an external validation accuracy of 70.83%. It should be noted that the compromised classification performance could indicate that cross-validation alone is not substantial enough for validating deep learning models, and external validation is highly important for future studies.
Recent work related to the classification of DR in OCTA has primarily used supervised machine learning methods.14,25 However, the disadvantage of supervised machine learning approaches is that the user must extract quantitative features, such as blood vessel density, blood vessel tortuosity, or foveal avascular zone area. On the other hand, deep learning requires minimal user input. A user, such as a clinician or technician, inputs an image, and the CNN can extract features from the input image to output a prediction. With the advantage of being easy to use, deep learning can work well for mass-screening programs. To the best of our knowledge, this study is the first exploration of using deep learning for OCTA classification of DR.
There are some limitations for this current study. The dataset used to train the CNN models was acquired from one device and may contain biases. This is evident in the external validation, which revealed a lower accuracy compared to the cross-validation accuracy. This decreased accuracy may be due to the differences between ethnicities. For example, the cross-validation dataset largely consisted of cohorts from an African and Hispanic American population, whereas the external dataset consisted primarily of cohorts from an East Asian population. The next step to address this issue would be to create a dataset consisting of multiple devices from different populations, but to accomplish this multiple-institution collaboration is required. One limitation of this study (and deep learning in general) is the lack of interpretability. The CNN can make a prediction, but the user, such as the physician, will not know how the CNN inferred its prediction. Based on our work and other published studies, as well as future studies involving AI technologies, researchers could use tools such as occlusion maps43 to help clinicians understand how the CNN made its prediction, in addition to utilizing external validation in order to build trust and confidence.
Based on the results of this study, we incorporated our trained CNN into a custom GUI to evaluate the potential of using automated AI classification in a clinical setting. The design of the GUI was inspired by commonly used software in the UIC retinal clinic and was developed using open source software. By demonstrating the potential of quick deployment of AI technologies and with the promising results in our study and other published studies, we hope to build confidence and foster the use of AI technologies in a clinical setting. Deep learning can help clinicians gain valuable time when screening patients and can reduce the need for manual graders. In the future, AI can potentially be used to assist clinicians in understanding pathological mechanisms and the development of new treatment options.
Acknowledgments
Supported by grants from the National Institutes of Health (R01EY030842, R01EY029673, R01EY030101, R01EY023522, P30 EY001792); by a T32 Institutional Training Grant for a training program in the biology and translational research on Alzheimer's disease and related dementias (T32AG057468); by an unrestricted grant from Research to Prevent Blindness; and by a Richard and Loan Hill endowment.
Disclosure: D. Le, None; M. Alam, None; C.K. Yao, None; J.I. Lim, None; Y.-T. Hsieh, None; R.V.P. Chan, None; D. Toslak, None; X. Yao, None
References
- 1. National Eye Institute. Eye health data and statistics. Available at: https://nei.nih.gov/eyedata/diabetic. Accessed September 1, 2019.
- 2. Glasson NM, Crossland LJ, Larkins SL. An innovative Australian outreach model of diabetic retinopathy screening in remote communities. J Diabetes Res. 2016; 2016: 1267215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nayak J, Bhat PS, Acharya UR, Lim CM, Kagathi M. Automated identification of diabetic retinopathy stages using digital fundus images. J Med Syst. 2008; 32: 107–115. [DOI] [PubMed] [Google Scholar]
- 4. Zahid S, Dolz-Marco R, Freund KB, et al.. Fractal dimensional analysis of optical coherence tomography angiography in eyes with diabetic retinopathy. Invest Ophthalmol Vis Sci. 2016; 57: 4940–4947. [DOI] [PubMed] [Google Scholar]
- 5. Gramatikov BI. Modern technologies for retinal scanning and imaging: an introduction for the biomedical engineer. Biomed Eng Online. 2014; 13: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mendis KR, Balaratnasingam C, Yu P, et al.. Correlation of histologic and clinical images to determine the diagnostic value of fluorescein angiography for studying retinal capillary detail. Invest Ophthalmol Vis Sci. 2010; 51: 5864–5869. [DOI] [PubMed] [Google Scholar]
- 7. Cheng S-C, Huang Y-M. A novel approach to diagnose diabetes based on the fractal characteristics of retinal images. IEEE Trans Inf Technol Biomed. 2003; 7: 163–170. [DOI] [PubMed] [Google Scholar]
- 8. Gass JD. A fluorescein angiographic study of macular dysfunction secondary to retinal vascular disease. VI. X-ray irradiation, carotid artery occlusion, collagen vascular disease, and vitritis. Arch Ophthalmol. 1968; 80: 606–617. [DOI] [PubMed] [Google Scholar]
- 9. Talu S, Calugaru DM, Lapascu CA. Characterisation of human non-proliferative diabetic retinopathy using the fractal analysis. Int J Ophthalmol. 2015; 8: 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goh JKH, Cheung CY, Sim SS, Tan PC, Tan GSW, Wong TY. Retinal imaging techniques for diabetic retinopathy screening. J Diabetes Sci Technol. 2016; 10: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thompson IA, Durrani AK, Patel S. Optical coherence tomography angiography characteristics in diabetic patients without clinical diabetic retinopathy. Eye. 2019; 33: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Talisa E, Chin AT, Bonini Filho MA, et al.. Detection of microvascular changes in eyes of patients with diabetes but not clinical diabetic retinopathy using optical coherence tomography angiography. Retina. 2015; 35: 2364–2370. [DOI] [PubMed] [Google Scholar]
- 13. Mo S, Krawitz B, Efstathiadis E, et al.. Imaging foveal microvasculature: optical coherence tomography angiography versus adaptive optics scanning light ophthalmoscope fluorescein angiography. Invest Ophthalmol Vis Sci. 2016; 57: OCT130–OCT140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alam M, Zhang Y, Lim JI, Chan RV, Yang M, Yao X. Quantitative optical coherence tomography angiography features for objective classification and staging of diabetic retinopathy. Retina. 2020; 40: 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsieh Y-T, Alam MN, Le D, et al.. Optical coherence tomography angiography biomarkers for predicting visual outcomes after ranibizumab treatment for diabetic macular edema. Ophthalmol Retina. 2019; 3: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alam MN, Son T, Toslak D, Lim JI, Yao X. Quantitative artery-vein analysis in optical coherence tomography angiography of diabetic retinopathy. In: Manns F, Söderberg PG, Ho A, eds. Proceedings of SPIE: Ophthalmic Technologies XXIX, Vol. 10858 Bellingham, WA: SPIE; 2019: 1085802. [Google Scholar]
- 17. Le D, Alam M, Miao BA, Lim JI, Yao X. Fully automated geometric feature analysis in optical coherence tomography angiography for objective classification of diabetic retinopathy. Biomed Opt Express. 2019; 10: 2493–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moult E, Choi W, Waheed NK, et al.. Ultrahigh-speed swept-source OCT angiography in exudative AMD. Ophthalmic Surg Lasers Imaging Retina. 2014; 45: 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng F, Zhang Q, Motulsky EH, et al.. Comparison of neovascular lesion area measurements from different swept-source OCT angiographic scan patterns in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2017; 58: 5098–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu Z, Lin J, Gao C, et al.. Quantitative assessment of the retinal microvasculature using optical coherence tomography angiography. J Biomed Opt. 2016; 21: 066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabral D, Coscas F, Glacet-Bernard A, et al.. Biomarkers of peripheral nonperfusion in retinal venous occlusions using optical coherence tomography angiography. Transl Vis Sci Technol. 2019; 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samara WA, Shahlaee A, Sridhar J, Khan MA, Ho AC, Hsu J. Quantitative optical coherence tomography angiography features and visual function in eyes with branch retinal vein occlusion. Am J Ophthalmol. 2016; 166: 76–83. [DOI] [PubMed] [Google Scholar]
- 23. Dave VP, Pappuru RR, Gindra R, et al.. OCT angiography fractal analysis-based quantification of macular vascular density in BRVO eyes. Can J Ophthalmol. 2018; 4: 297–300. [DOI] [PubMed] [Google Scholar]
- 24. Alam M, Thapa D, Lim JI, Cao D, Yao X. Quantitative characteristics of sickle cell retinopathy in optical coherence tomography angiography. Biomed Opt Express. 2017; 8: 1741–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alam M, Le D, Lim JI, Chan RV, Yao X. Supervised machine learning based multi-task artificial intelligence classification of retinopathies. J Clin Med. 2019; 8: 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kheradpisheh SR, Ghodrati M, Ganjtabesh M, Masquelier T. Deep networks can resemble human feed-forward vision in invariant object recognition. Sci Rep. 2016; 6: 32672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cox DD, Dean T. Neural networks and neuroscience-inspired computer vision. Curr Biol. 2014; 24: R921–R929. [DOI] [PubMed] [Google Scholar]
- 28. Kermany DS, Goldbaum M, Cai W, et al.. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018; 172: 1122–1131.e9. [DOI] [PubMed] [Google Scholar]
- 29. Hemelings R, Elen B, Stalmans I, Van Keer K, De Boever P, Blaschko MB. Artery-vein segmentation in fundus images using a fully convolutional network. Comput Med Imaging Graph. 2019; 76: 101636. [DOI] [PubMed] [Google Scholar]
- 30. Gómez-Valverde JJ, Antón A, Fatti G, et al.. Automatic glaucoma classification using color fundus images based on convolutional neural networks and transfer learning. Biomed Opt Express. 2019; 10: 892–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ting DSW, Pasquale LR, Peng L, et al.. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019; 103: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arcadu F, Benmansour F, Maunz A, et al.. Deep learning predicts OCT measures of diabetic macular thickening from color fundus photographs. Invest Ophthalmol Vis Sci. 2019; 60: 852–857. [DOI] [PubMed] [Google Scholar]
- 33. Lee CS, Tyring AJ, Wu Y, et al.. Generating retinal flow maps from structural optical coherence tomography with artificial intelligence. Sci Rep. 2019; 9: 5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Available at: https://arxiv.org/pdf/1409.1556.pdf. Accessed June 19, 2020.
- 35. Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2009;248–255. [Google Scholar]
- 36. Salman S, Liu X. Overfitting mechanism and avoidance in deep neural networks. Available at: https://arxiv.org/pdf/1901.06566.pdf. Accessed June 19, 2020.
- 37. Joukal M. Anatomy of the human visual pathway. In: Skorkovská K, ed., Homonymous visual field defects. Cham, Switzerland: Springer International; 2017;1–16. [Google Scholar]
- 38. Hastie T, Tibshirani R, Wainwright M. Statistical learning with sparsity: the lasso and generalizations. Boca Raton, FL: CRC Press: 2015. [Google Scholar]
- 39. Arcadu F, Benmansour F, Maunz A, Willis J, Haskova Z, Prunotto M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit Med. 2019; 2: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li F, Liu Z, Chen H, Jiang M, Zhang X, Wu Z. Automatic detection of diabetic retinopathy in retinal fundus photographs based on deep learning algorithm. Transl Vis Sci Technol. 2019; 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu W, Tong Y, Yu Y, Xing Y, Chen C, Shen Y. Deep learning-based automated classification of multi-categorical abnormalities from optical coherence tomography images. Transl Vis Sci Technol. 2018; 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eltanboly A, Ismail M, Shalaby A, et al.. A computer-aided diagnostic system for detecting diabetic retinopathy in optical coherence tomography images. Med Phys. 2017; 44: 914–923. [DOI] [PubMed] [Google Scholar]
- 43. Lee CS, Baughman DM, Lee AY. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. 2017; 1: 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]