Abstract
Background
The endosymbiont bacterium Wolbachia is maternally inherited and naturally infects some filarial nematodes and a diverse range of arthropods, including mosquito vectors responsible for disease transmission in humans. Previously, it has been found infecting most mosquito species but absent in Anopheles and Aedes aegypti. However, recently these two mosquito species were found to be naturally infected with Wolbachia. We report here the extent of Wolbachia infections in field-collected mosquitoes from Malaysia based on PCR amplification of the Wolbachia wsp and 16S rRNA genes.
Methods
The prevalence of Wolbachia in Culicinae mosquitoes was assessed via PCR with wsp primers. For some of the mosquitoes, in which the wsp primers failed to amplify a product, Wolbachia screening was performed using nested PCR targeting the 16S rRNA gene. Wolbachia sequences were aligned using Geneious 9.1.6 software, analyzed with BLAST, and the most similar sequences were downloaded. Phylogenetic analyses were carried out with MEGA 7.0 software. Graphs were drawn with GraphPad Prism 8.0 software.
Results
A total of 217 adult mosquitoes representing 26 mosquito species were screened. Of these, infections with Wolbachia were detected in 4 and 15 mosquito species using wsp and 16S rRNA primers, respectively. To our knowledge, this is the first time Wolbachia was detected using 16S rRNA gene amplification, in some Anopheles species (some infected with Plasmodium), Culex sinensis, Culex vishnui, Culex pseudovishnui, Mansonia bonneae and Mansonia annulifera. Phylogenetic analysis based on wsp revealed Wolbachia from most of the mosquitoes belonged to Wolbachia Supergroup B. Based on 16S rRNA phylogenetic analysis, the Wolbachia strain from Anopheles mosquitoes were more closely related to Wolbachia infecting Anopheles from Africa than from Myanmar.
Conclusions
Wolbachia was found infecting Anopheles and other important disease vectors such as Mansonia. Since Wolbachia can affect its host by reducing the life span and provide resistance to pathogen infection, several studies have suggested it as a potential innovative tool for vector/vector-borne disease control. Therefore, it is important to carry out further studies on natural Wolbachia infection in vector mosquitoes’ populations as well as their long-term effects in new hosts and pathogen suppression. 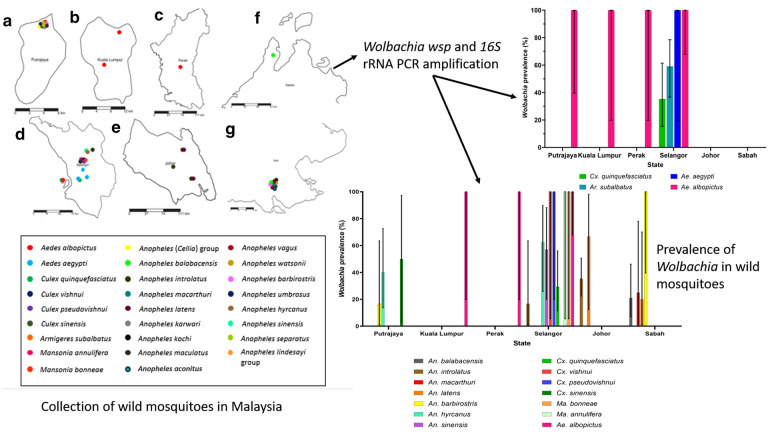
Keywords: Wolbachia, Mosquitoes, wsp gene, 16S rRNA, Anopheles, Vectors
Background
The endosymbiotic bacterium Wolbachia is maternally inherited and naturally infects some filarial nematodes and a diverse range of arthropods, including mosquito vector species which are responsible for disease transmission to humans [1, 2]. Wolbachia is found naturally in various mosquitoes, including Aedes, Culex, Armigeres, Mansonia, Coquillettidia, Culiseta, Hodgesia, Ochlerotatus, Tripteroides and Uranotaenia [3–8] but was believed to be absent in Aedes aegypti and Anopheles mosquitoes [3]. To enhance their spread across the population, Wolbachia has a wide host range as well as wide tissue distributions [9]. Besides, Wolbachia can possibly be transferred horizontally between mosquito populations though the mechanism involved is not fully understood [10].
Wolbachia infection in the host causes a broad range of abnormal reproductive phenotypes which include male-killing, parthenogenesis, feminization and cytoplasmic incompatibility (CI) [11, 12]. Cytoplasmic incompatibility caused by Wolbachia infection is the most well-known and common phenotypic effect and is considered a potential vector control alternative since crosses between individuals with different Wolbachia infection status will result in few or no offspring [13]. This effect confers indirect reproductive advantages to the infected females as infected females can produce viable progenies, but uninfected females when mated with infected males produce fewer viable progenies, if any [14]. Therefore, CI, along with maternal transmission of Wolbachia, allows Wolbachia to spread rapidly through a population [14, 15]. Several factors affect the CI in the host, namely strain of Wolbachia, host species, temperature and rearing density [16–19].
Furthermore, some studies have shown that Wolbachia can inhibit pathogen replication within mosquitoes [20, 21] such as dengue virus (DENV) [22–28], chikungunya virus (CHIKV) [25, 29, 30], yellow fever virus (YFV) [30], Zika virus (ZIKV) [31–33], Plasmodium parasites [25, 34] and filarial nematodes [35]. In studying the impact of Wolbachia infection on Plasmodium infection in Anopheles, transiently and somatically infected Anopheles gambiae with wMelPop or wAlbB Wolbachia strains was found to have significant reductions in Plasmodium berghei or Plasmodium falciparum oocyst infections [36, 37]. Following the increasing findings of natural Wolbachia infections in Anopheles [38], it was also found that natural populations of Wolbachia-infected An. coluzzii females have a lower frequency of Plasmodium infections than Wolbachia-negative individuals [39]. Gomes et al. [40] further showed that naturally infected field An. gambiae, has significantly lower Plasmodium sporozoite infections. These findings support the potential use of Wolbachia in controlling vector-borne disease transmission. Currently, in Malaysia, studies on the release of Wolbachia-infected Ae. aegypti are ongoing to curb the spread of dengue [41].
It was believed that Wolbachia is absent in Ae. aegypti and Anopheles mosquitoes [3] until the year 2014, when the wAnga Wolbachia strain was detected in An. gambiae collected from West Africa [38]. This was followed by several other discoveries of Wolbachia in other Anopheles vectors in Africa [40, 42–45] and Southeast Asia (Myanmar) [46]. Moreover, recent studies have also found Wolbachia naturally infecting Ae. aegypti in the Philippines [47], Thailand [48], Malaysia [49], India [50], Florida [51], New Mexico [52], Texas [53] and Panama [54].
Amplification of Wolbachia genes such as Wolbachia surface protein (wsp), filamenting temperature-sensitive mutant Z (ftsZ) and 16S rDNA using polymerase chain reaction (PCR) has been used to detect Wolbachia in the vector. Previous failure in detecting Wolbachia in Anopheles mosquitoes may be due to limitations in the detection system used such as non-optimal DNA amplification [38]. This has led to the development of a nested PCR amplification of 16S rDNA region for Wolbachia detection in Anopheles mosquitoes [39]. A study by Marcon et al. [55] demonstrated that the combination of 16S rDNA and wsp PCR detection increased the detection efficiency and supergroup differentiation whereas detection with ftsZ was less sensitive which can lead to false-negative for Wolbachia infection mosquitoes.
Thus, this study aimed to determine natural Wolbachia infections in different field-collected mosquitoes from several states in Peninsular Malaysia and East Malaysia (Sabah) using Wolbachia 16S rRNA and wsp gene amplifications, to determine the extent of natural infection of Wolbachia in some of these disease vectors and to understand its role in the dynamics of disease transmission.
Methods
Mosquito collection
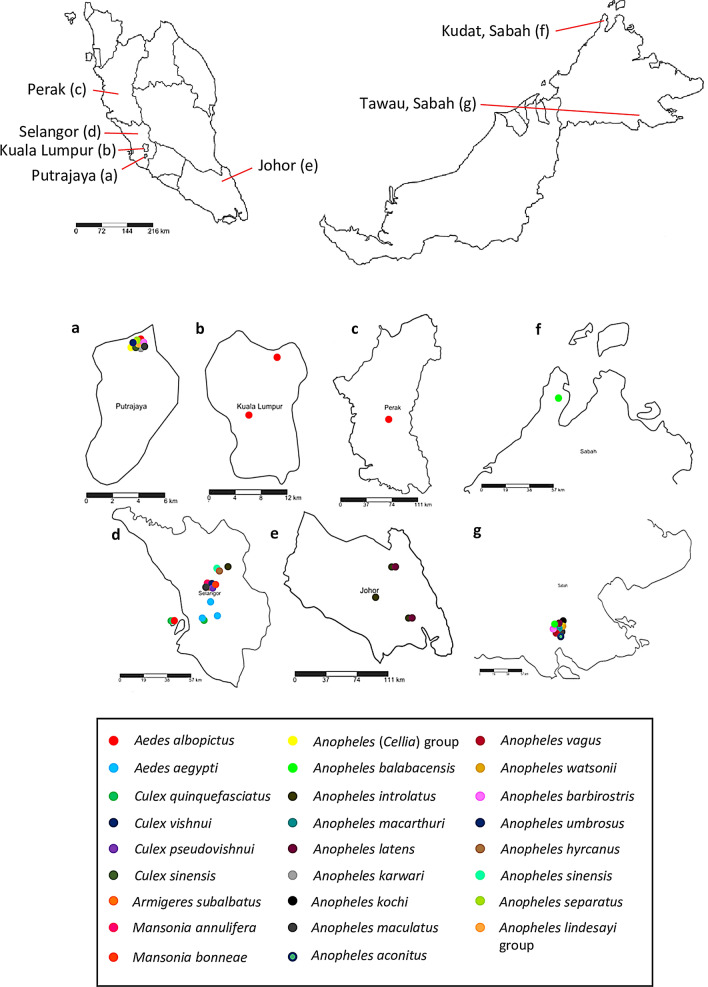
Adult Anopheles spp. and Culicinae mosquitoes collected from 2013 to 2019 in Selangor, Kuala Lumpur, Putrajaya, Johor, Perak and Sabah (Fig. 1) using human landing catch, mosquito magnet trap and sticky trap, were randomly selected and screened for Wolbachia (Additional file 1: Table S1). All specimens were identified morphologically using the taxonomic keys of Reid [56], Sallum et al. [57] (Leucosphyrus group of Anopheles mosquitoes) and Jeffery et al. [58] prior to molecular detection of Wolbachia. Specimens which could not be reliably distinguished based on morphology, such as those of the Leucosphyrus group of Anopheles or those which were not well-preserved, were all molecularly identified by PCR and sequencing of the internal transcribed spacer 2 (ITS2) gene [59]. If specimens could not be identified morphologically or molecularly, they were classified to the lowest level of taxonomic certainty, for example the genus level.
Fig. 1.
Map showing mosquitoes collected from five different states in Malaysia. a Putrajaya. b Kuala Lumpur. c Perak. d Selangor. e Johor. f Kudat, Sabah. g Tawau, Sabah
Additionally, Anopheles samples positive for Plasmodium infections were also included in the study (Additional file 1: Table S2). These 34 samples originating from other vector studies had already been screened for simian Plasmodium parasites (i.e. Plasmodium knowlesi, Plasmodium cynomolgi, Plasmodium inui, Plasmodium fieldi and Plasmodium coatneyi) infection by dissection and observation of the midgut for oocysts and salivary glands for sporozoites followed by PCR [60] in well-preserved samples; or by PCR of whole mosquitoes, head/thorax or abdomen in all other samples. The midguts were stained with 1% merchurochrome for visualization of oocysts, while the salivary glands were squashed in saline and viewed under a light compound microscope at 400× magnification for visualization of sporozoites.
Genomic DNA extraction and molecular detection of Wolbachia
Genomic DNA was extracted from the whole mosquito using InstaGene matrix (Bio-Rad, Hercules, California, USA) according to the manufacturer’s protocol. The extracted DNA was run on a 1% agarose gel electrophoresis to confirm its presence. Detection of Wolbachia was undertaken targeting two conserved Wolbachia genes, wsp and 16S rRNA. Wolbachia in Culicinae mosquitoes was screened using general wsp primers (wsp81F: 5′-TGG TCC AAT AAG TGA TGA AGA AAC-3′; wsp691R: 5′-AAA AAT TAA ACG CTA CTC CA-3′), amplified in a 30 µl reaction volume with 5 µl DNA as template according to the standard PCR protocol [61]. These primers amplify a DNA fragment ranging from 590 to 632 bp. Nested PCR amplifying the 16S rRNA gene was used to detect Wolbachia in Anopheles and Culicinae mosquitoes whose results were negative in the wsp PCR. The initial PCR employed 16S Wolbachia-specific primers (W-Specf: 5′-CAT ACC TAT TCG AAG GGA TAG-3′; W-Specr: 5′-AGC TTC GAG TGA AAC CAA TTC-3′) and was performed in 30 µl reaction volume using 5 µl DNA as template according to standard protocol in Werren & Windsor [62]. Then, two microliters of the initial PCR products were amplified in a 30-µl PCR reaction using specific internal primers (16SNF: 5′-GAA GGG ATA GGG TCG GTT CG-3′; 16SNR: 5′-CAA TTC CCA TGG CGT GAC G-3′) as described in Shaw et al. [39]. The nested 16S rRNA Wolbachia PCR generates a 412-bp DNA fragment. Aedes albopictus (Skuse) was used as positive control for all Wolbachia screening along with negative control (double-distilled water as template). All PCR products were run on 1.5% agarose gel electrophoresis and viewed under UV light prior to Sanger-sequencing by a commercial laboratory (MyTACG, Kuala Lumpur, Malaysia).
Phylogenetic analysis
The nucleotide sequences obtained were aligned using Geneious 9.1.6 software (http://www.geneious.com). All aligned Wolbachia sequences were compared with other sequences available in the GenBank database to determine the percentage identity using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the most similar sequences were downloaded for phylogenetic analysis. The alignment of all available sequences was examined visually and only those residues where variations were observed in sequences obtained from independent PCR amplifications from the same sample or from another independently amplified sample were considered reliable and therefore included in the phylogenetic analysis. All sequences (wsp = 606 nucleotides; 16S rRNA = 374 nucleotides) were exported to MEGA 7.0 software for further alignment and analysis using Clustal W algorithm [63]. Phylogenetic tree for the wsp gene was constructed using the Maximum Likelihood (best-fit substitution model) method. The phylogenetic tree model with lowest Bayesian information criterion (BIC) is considered to describe the substitution pattern the best. Hence, Tamura 3-parameter model was chosen for wsp gene analysis. Wsp gene sequence from Brugia pahangi was incorporated as the outgroup to confirm the outcome of the phylogenetic tree. For 16S rRNA gene, the evolutionary history was inferred using the Maximum Likelihood method (best-fit substitution model). Based on the lowest BIC value, Jukes-Cantor model was chosen to infer the phylogenetic relationship of Wolbachia using the 16S rRNA gene. The 16S rRNA gene sequence of Rickettsia japonica was included as the outgroup. Both phylogeny tests were performed by bootstrap method with 1000 replications. All the evolutionary analyses were performed in MEGA 7.0 software. All sequences used in the phylogenetic analyses were submitted in the GenBank database under the accession numbers MN893348-MN893366 (wsp gene) and MN887537-MN887588 (16S rRNA).
Results
Detection of Wolbachia in mosquitoes
A total of 217 adult mosquitoes representing 26 mosquito species belonging to 5 genera (Anopheles, Culex, Aedes, Mansonia and Armigeres) (Additional file 1: Table S1) were screened for Wolbachia. These mosquitoes were collected from a wide range of habitats such as urban, island and forest (Additional file 1: Table S2). Polymerase chain reaction amplifying the Wolbachia wsp gene was successful in most of the Culicinae mosquito samples except for Culex vishnui, Culex pseudovishnui, Mansonia mosquitoes and some samples of Culex quinquefasciatus. Wolbachia infection was detected in these samples as well as in Anopheles spp. mosquitoes using the 16S rRNA primers.
The overall Wolbachia infection rate for all mosquitoes tested in this study was 46.1%. Wolbachia was detected in 4 species of mosquitoes (40/217) using wsp gene amplification and 15 species of mosquitoes (60/217) using 16S rRNA gene amplification (Table 1). The prevalence of Wolbachia infection within mosquito species ranged from 35.3% (6/17) in Cx. quinquefasciatus to 100% (19/19) in Ae. albopictus for wsp gene. Wolbachia infection rate by detection of Wolbachia 16S rRNA gene ranged between 50–100% for Cx. sinensis, Cx. vishnui, Cx. pseudovishnui and Mansonia mosquitoes. For Anopheles mosquitoes, the infection rate of Wolbachia among each species ranged between 21–57.1%.
Table 1.
Wolbachia infection prevalence rates in adult female mosquitoes using two conserved Wolbachia genes (16S rRNA and wsp)
| Mosquito species | n | PCR-positive | Prevalence (95% CI) (%)a | ||
|---|---|---|---|---|---|
| wsp | 16S rRNA | wsp | 16S rRNA | ||
| Anopheles (Cellia) group | 1 | 0 | 0 | 0 | 0 |
| An. balabacensis | 19 | 0 | 4 | 0 | 21.1 (7.0–46.1) |
| An. introlatus | 54 | 0 | 17 | 0 | 31.5 (20.0–45.7) |
| An. macarthuri | 4 | 0 | 1 | 0 | 25.0 (1.3–78.1) |
| An. latens | 8 | 0 | 4 | 0 | 50.0 (17.5–82.6) |
| An. karwari | 2 | 0 | 0 | 0 | 0 |
| An. kochi | 1 | 0 | 0 | 0 | 0 |
| An. maculatus | 9 | 0 | 2 | 0 | 22.2 (3.9–59.8) |
| An. vagus | 3 | 0 | 0 | 0 | 0 |
| An. watsonii | 5 | 0 | 0 | 0 | 0 |
| An. barbirostris | 10 | 0 | 5 | 0 | 50.0 (20.1–79.9) |
| An. umbrosus | 6 | 0 | 0 | 0 | 0 |
| An. hyrcanus | 18 | 0 | 9 | 0 | 50.0 (26.8–73.2) |
| An. sinensis | 7 | 0 | 4 | 0 | 57.1 (20.2–88.2) |
| An. lindesayi species group | 1 | 0 | 0 | 0 | 0 |
| An. separatus | 1 | 0 | 0 | 0 | 0 |
| An. aconitus | 1 | 0 | 0 | 0 | 0 |
| Cx. vishnui | 1 | 0 | 1 | 0 | 100 (5.5–100) |
| Cx. pseudovishnui | 2 | 0 | 2 | 0 | 100 (19.8–100) |
| Cx. quinquefasciatus | 17 | 6 | 5 | 35.3 (15.3–61.4) | 29.4 (11.4–56.0) |
| Cx. sinensis | 2 | 0 | 1 | 0 | 50.0 (2.7–97.3) |
| Ma. annulifera | 1 | 0 | 1 | 0 | 100 (5.5–100) |
| Ma. bonneae | 1 | 0 | 1 | 0 | 100 (5.5–100) |
| Ar. subalbatus | 22 | 13 | 0 | 59.1 (36.7–78.5) | 0 |
| Ae. aegypti | 2 | 2 | 0 | 100 (19.8–100) | 0 |
| Ae. albopictus | 19 | 19 | 3 | 100 (79.1–100) | 15.8 (4.2–40.5) |
Note: An individual was considered Wolbachia-infected when any one of the two Wolbachia gene fragments were amplified
Abbreviation: CI, confidence interval
aWolbachia prevalence rate (%) = [No. of Wolbachia positive mosquitoes/Total no. of mosquitoes] × 100
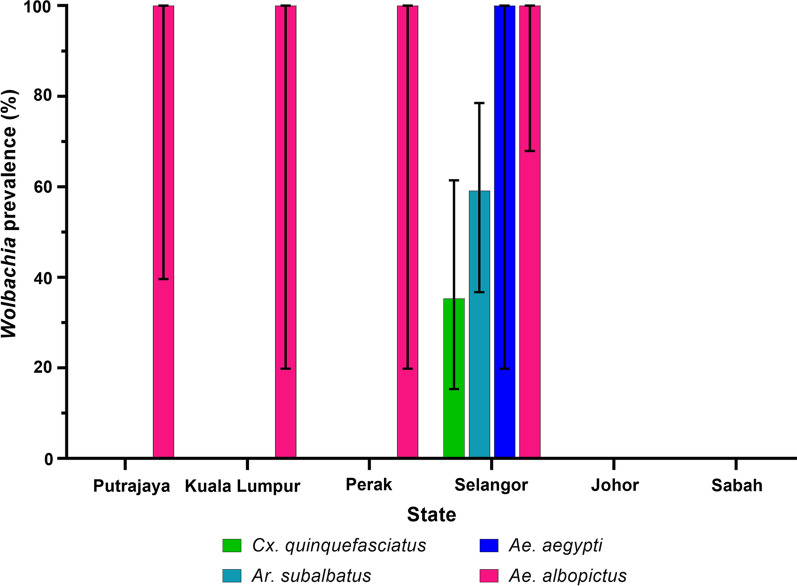
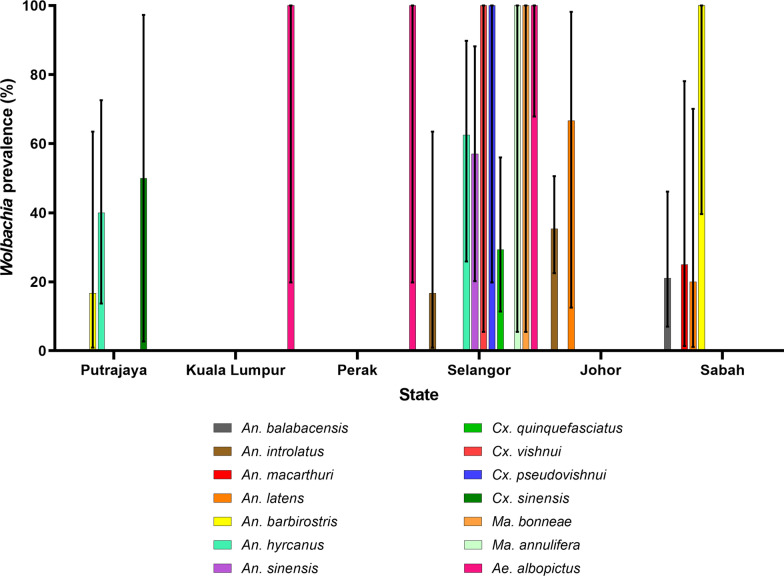
Presence of Wolbachia in different mosquito species from each state is given in Fig. 2 (wsp gene) and Fig. 3 (16S rRNA). While the prevalence appeared to be higher in certain states such as Selangor, this bias could be caused by the prevalence of Wolbachia in Ae. albopictus or higher number of mosquito species collected from Selangor (Fig. 3). Thus, it would be more valid if comparisons were made among mosquito species found in more than one site and are available in similar numbers, without taking Ae. albopictus into account. For example, the prevalence of Wolbachia was higher in An. barbirostris from Sabah than from Putrajaya, and the prevalence of Wolbachia was also higher in An. hyrcanus from Selangor than from Putrajaya. Similarly, the same scenario applies to the prevalence of Wolbachia in the mosquitoes according to ecological types (Table 2), as most mosquitoes were collected from forested areas. Nonetheless, none of the An. balabacensis from the Banggi Island was found to be infected with Wolbachia, compared to infections found in the same species from mainland forested area. As for An. hyrcanus, 40% of the mosquitoes from the wetland were infected compared to 50–75% from the forested area. Thus, mosquitoes from forested areas, may have higher prevalence of Wolbachia compared to those from the wetland (Putrajaya) or island (Banggi Island) areas.
Fig. 2.
Prevalence of Wolbachia-infected mosquitoes collected from different states in Malaysia using wsp primers. Error bar denotes 95% confidence interval (CI)
Fig. 3.
Prevalence of Wolbachia-infected mosquitoes collected from different states in Malaysia using 16S rRNA primers. Error bar denotes 95% confidence interval (CI)
Table 2.
Wolbachia infection prevalence rates in adult female mosquitoes in different ecological types across the states in Malaysia
| Ecological type/study site | Mosquito species | Prevalence (95% CI) (%) | |
|---|---|---|---|
| wsp | 16S rRNA | ||
| Urban | |||
| Bangsar, Kuala Lumpur | Ae. albopictus | 100 (5.5–100) | – |
| Jalan Genting, Kuala Lumpur | 100 (5.5–100) | 100 (5.5–100) | |
| Pasir Puteh, Perak | 100 (19.8–100 | 100 (19.8–100) | |
| Damansara Damai, Selangor | Ae. aegypti | 100 (19.8–100) | – |
| Persanda, Selangor | Cx. quinquefasciatus | 42.9 (11.8–79.8) | 28.6 (5.1–69.8) |
| Klang, Selangor | 30.0 (8.1–64.6) | 30.0 (8.1–64.6) | |
| Island | |||
| Pulau Ketam, Selangor | Ae. albopictus | 100 (39.6–100) | – |
| Pulau Banggi, Sabah | An. balabacensis | 0 | 0 |
| Wetland | |||
| Putrajaya | An. hyrcanus | 0 | 40.0 (13.7–72.6) |
| An. (Cellia) group | 0 | 0 | |
| An. barbirostris | 0 | 20.0 (1.1–70.1) | |
| An. umbrosus | 0 | 0 | |
| An. maculatus | 0 | 0 | |
| An. karwari | 0 | 0 | |
| An. separatus | 0 | 0 | |
| An. lindesayi group | 0 | 0 | |
| Cx. sinensis | 0 | 50.0 (2.7–97.3) | |
| Ae. albopictus | 100 | – | |
| Forest | |||
| Ulu Kalong, Selangor | An. introlatus | 0 | 16.7 (0.9–63.5) |
| An. maculatus | 0 | 100 (19.8–100) | |
| Ar. subalbatus | 59.1 (36.7–78.5) | – | |
| Ulu Kalong, Selangor | Ma. bonneae | 0 | 100 (5.5–100) |
| Ae. albopictus | 100 (56.1–100) | – | |
| Culex. sp. | 100 (5.5–100) | – | |
| Bukit Lagong, Selangor | An. maculatus | 0 | 0 |
| An. hyrcanus | 0 | 75.0 (21.9–98.7) | |
| Ma. annulifera | 0 | 100 (5.5–100) | |
| Cx. vishnui | 0 | 100 (5.5–100) | |
| Cx. pseudovishnui | 0 | 100 (5.5–100) | |
| Sg. Sendat, Selangor | An. sinensis | 0 | 57.1 (20.2–88.2) |
| An. hyrcanus | 0 | 50.0 (9.2–90.8) | |
| Tawau, Sabah | An. vagus | 0 | 0 |
| An. maculatus | 0 | 0 | |
| An. watsonii | 0 | 0 | |
| An. funestus | 0 | 0 | |
| An. kochi | 0 | 0 | |
| An. macarthuri | 0 | 25.0 (1.3–78.1) | |
| An. latens | 0 | 20.0 (1.1–70.1) | |
| An. balabacensis | 0 | 23.5 (7.8–50.2) | |
| An. barbirostris | 0 | 100 (39.6–100) | |
| Kluang, Johor | An. introlatus | 0 | 55.6 (22.7–84.7) |
| Mersing, Johor | An. introlatus | 0 | 24.2 (11.7–42.6) |
| An. latens | 0 | 100 (5.5–100) | |
| Kota Tinggi, Johor | An. introlatus | 0 | 50.0 (14.0–86.1) |
| An. latens | 0 | 50.0 (2.7–97.3) | |
| An. umbrosus | 0 | 0 | |
Abbreviation: CI, confidence interval
Plasmodium-positive Anopheles with Wolbachia infection
Out of the 34 Plasmodium-positive Anopheles samples, 14 (38.24%) were also found to have Wolbachia infection (Table 3). Interestingly, only 1 out of 9 (11.11%) samples with oocysts/Plasmodium DNA-positive abdomens were positive for Wolbachia, as compared to 4 out of 8 (50%) samples with sporozoites/Plasmodium DNA-positive head/thorax which were also Wolbachia-infected. However, this difference was not significant (Fisher’s exact test, two-tailed, P = 0.131). There was also no particular difference in Wolbachia prevalence between the small number of Plasmodium-positive Anopheles species investigated in this study.
Table 3.
Plasmodium-positive Anopheles with Wolbachia infection
| Species | No. of samples | No. of samples: stage/type of infection | No. of Wolbachia-positive samples/No. of samples | No. of samples positive for Wolbachia/Total no. of samples |
|---|---|---|---|---|
| An. introlatus | 28 |
17: Plasmodium DNA-positive whole mosquito; 4: oocysts/Plasmodium DNA-positive abdomen; 7: sporozoites/Plasmodium DNA-positive head/thorax |
8/17 whole mosquito samples; 0/4 samples with oocysts/Plasmodium DNA-positive abdomen; 4/7 samples with sporozoites/Plasmodium DNA-positive head/thorax |
12/28 |
| An. latens | 2 |
1: oocysts; 1: Plasmodium DNA-positive abdomen |
1/1 sample with oocysts; 0/1 sample with Plasmodium DNA-positive abdomen |
1/2 |
| An. balabacensis | 2 |
1: sporozoites; 1: oocysts |
0 | 0/2 |
| An. umbrosus | 2 | 2: oocysts | 0 | 0/2 |
| Total | 34 |
17: whole mosquitoes; 9: oocysts/Plasmodium DNA-positive abdomen 8: sporozoites/Plasmodium DNA-positive head/thorax |
8/17 whole mosquito samples; 1/9 samples with oocysts/Plasmodium DNA-positive abdomen; 4/8 samples with sporozoites/Plasmodium DNA-positive head/thorax |
13/34 |
Phylogenetic analysis of Wolbachia in mosquitoes
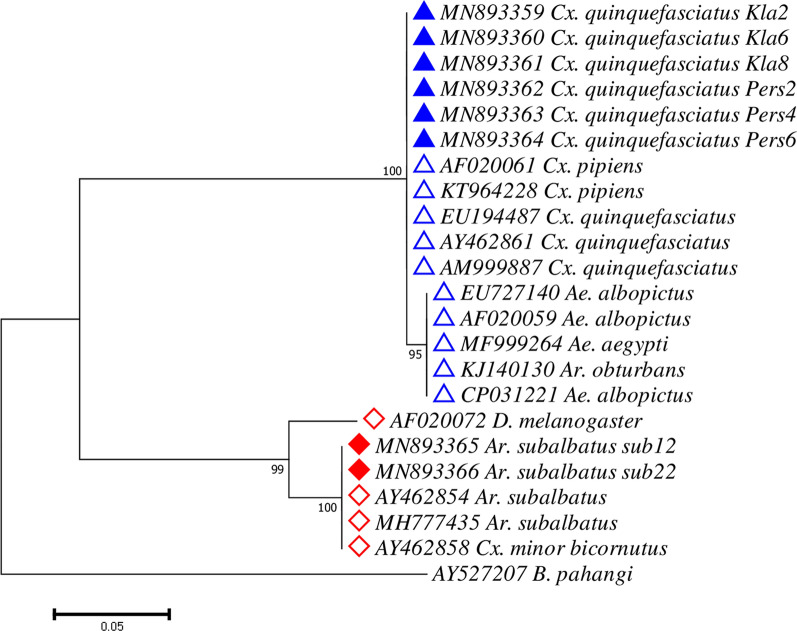
Eight wsp Wolbachia sequences from this study were deemed suitable for phylogenetic analysis after analysis of the sequencing results. We retrieved an additional 14 wsp sequences and 1 outgroup sequence (B. pahangi) from GenBank for phylogenetic analysis. In total, 23 sequences were used for construction of the phylogenetic tree. The alignments for different species of mosquitoes are shown in Additional file 2: Figure S1 (Supergroup A) and Additional file 3: Figure S2 (Supergroup B). Phylogenetic tree of wsp gene inferred using ML method based on Tamura 3-parameter model (lowest BIC value = 2502.136) resulted in two major clades belonging to Supergroups A and B with bootstrap values of 99% and 100%, respectively (Fig. 4). Within the Wolbachia Supergroup B, two sub-clades branched out, one comprised of wAlbB sequences from Ae. albopictus and Ae. aegypti (bootstrap value = 95%) whereas Wolbachia from Cx. quinquefasciatus were grouped with wPip from Cx. pipiens (bootstrap value = 100%).
Fig. 4.
Maximum Likelihood phylogenetic analysis for Wolbachia using the wsp gene. The tree with the lowest Bayesian Information Criterion (BIC) (2725.733879) is shown, incorporating 33 nucleotide sequences (8 sequences from this study; 14 sequences from GenBank) and one outgroup (B. pahangi). Wolbachia sequences of Supergroup A are denoted with red diamonds while those of Supergroup B are denoted with blue triangles. Sequence retrieved from GenBank is denoted with hollow diamond or triangle along with its respective accession number while sequence obtained from this study are denoted with a filled diamond or triangle
A total of 52 16S rRNA Wolbachia sequences were deemed suitable for use in phylogenetic analysis after analysing the sequencing results. Confirmation of Wolbachia supergroup for each sequence was executed using BLAST. All the sequences showed 99–100% nucleotide identities. Additionally, 26 sequences from GenBank were retrieved for phylogenetic analysis including 1 outgroup sequence (R. japonica). Overall, 84 sequences were used for construction of Wolbachia 16S rRNA phylogenetic tree. The alignments of 16S rRNA sequences are shown in Additional file 4: Figure S3 (Supergroup A) and Additional file 5: Figure S4 (Supergroup B). The phylogenetic tree inferred using Maximum Likelihood method based on Jukes-Cantor model (lowest BIC value = 1897.45) resulted in two major clades with high bootstrap value: Supergroup A (bootstrap value = 91%) and Supergroup B (bootstrap value = 85%) (Fig. 5). Within the Supergroup B clade, three sub-clades branched out. The first sub-clade consists of the majority of the Wolbachia sequences obtained from Anopheles mosquitoes (82.9%) from this study and Wolbachia sequences from An. gambiae, Ae. albopictus and Ae. aegypti retrieved from GenBank (Fig. 5). Wolbachia 16S rRNA gene amplified from adults of Cx. quinquefasciatus were clustered within the second sub-clade of Wolbachia Supergroup B, confirming the phylogenetic relationship inferred using wsp gene. A single sample of each Ma. annulifera and Ma. bonneae was found infected with Wolbachia using 16S rRNA gene. Only Wolbachia-infected Ma. annulifera sequence was obtained in this study and it was grouped together in the Supergroup B with wUnif-Mad isolated from Ma. uniformis, as well as Wolbachia from Cx. quinquefasciatus and Anopheles mosquitoes from Kayin state, Myanmar. Repeated attempts to sequence Wolbachia 16s rRNA gene from Wolbachia-infected Ma. bonneae failed to provide reliable sequences and thus were not available. The third sub-clade comprised Wolbachia sequences from Anopheles mosquitoes found in Myanmar. On the other hand, Wolbachia from two Culicinae and seven Anopheles spp. mosquitoes were grouped with Wolbachia Supergroup A.
Fig. 5.
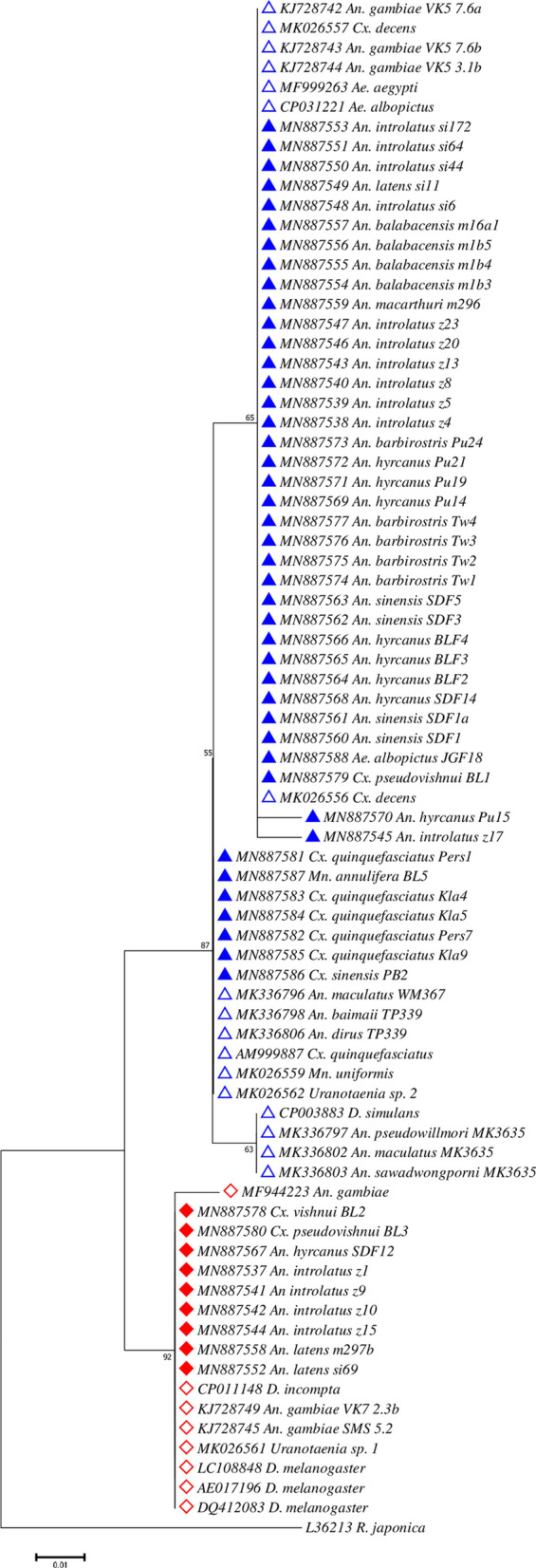
Maximum Likelihood phylogenetic analysis for Wolbachia using the 16S rRNA gene. The tree with the lowest Bayesian Information Criterion (BIC) (2020.770483) is shown, incorporating 83 nucleotide sequences (52 sequences from this study; 31 sequences from GenBank) and one outgroup (R. japonica). Wolbachia sequences of Supergroup A are denoted with red diamonds while those of Supergroup B are denoted with blue triangles. Sequences retrieved from GenBank are denoted with an open diamond or triangle along with its respective accession number while sequences obtained from this study are denoted with a filled diamond or triangle
While investigating if any strains of Wolbachia was associated with a particular species of mosquito, Wolbachia from Armigeres subalbatus and the only one sample from Cx. vishnui were all found to be grouped in Supergroup A, whereas Wolbachia from Cx. quinquefasciatus mosquitoes were of Supergroup B. Other than these, Wolbachia from other mosquito species belonged to either supergroups, although most of them were grouped in Wolbachia Supergroup B.
Discussion
In our study, we assessed the prevalence of Wolbachia in mosquito species collected from Peninsular Malaysia and Sabah. To our knowledge, this study is the first report of natural Wolbachia infections in field adult mosquito populations in Malaysia detected using Wolbachia wsp and 16S rRNA PCR amplifications. Our study further supports the higher sensitivity of the 16S rRNA nested PCR described by Shaw et al. [39] over the standard wsp PCR from Zhou et al. [61] for the detection of Wolbachia in mosquitoes due to genetic divergence or weak infection density [44]. The overall infection rate of all tested mosquitoes in this study was 46.1%. Wolbachia infection rates of 28.1% and 37.8% have been previously reported in the natural mosquito populations from neighboring countries Singapore [3] and Thailand [8], respectively.
The nested PCR utilizing 16S rRNA primers detected Wolbachia in seven species of Anopheles (41/144; 28.5%), i.e. An. balabacensis, An. latens, An. introlatus, An. macarthuri (Leucosphyrus Group), An. barbirostris (Barbirostris Group), An. hyrcanus and An. sinensis (Hyrcanus Group). Of these, An. latens [64], An. introlatus [59] and An. balabacensis [65] of the Leucosphyrus Group were incriminated as vectors of Plasmodium knowlesi in Malaysia. Additionally, An. balabacensis is also a vector of human malaria and Bancroftian filariasis in Sabah, Malaysia [66]. Other Anopheles species such as An. barbirostris [67] and An. sinensis [68] are vectors of human malaria, whereas An. hyrcanus has been postulated to be a vector for malaria due to its simultaneous occurrence with malaria cases [69]. These data along with the report of Wolbachia in natural mosquito populations in Kayin state, Myanmar [46] and Africa [38–40, 42–44], reinforce the occurrence of natural Wolbachia infections in malaria vectors.
Conclusive prevalence of Wolbachia based on different habitats cannot be drawn. This is due to the heterologous distribution of the mosquito species themselves and differences in number of mosquito species available between the sites. The prevalence appeared to be biased in certain states or environments mostly due to more mosquito species collected from the particular site than the others. However, there was a slight indication that Wolbachia infections can be more prevalent in forested areas than wetlands or island. This can be attributed to the richer flora and fauna in the forested areas which may have many more hosts with stable Wolbachia, which may facilitate horizontal transfers to other species.
Molecular phylogeny based on 16S rRNA sequences revealed that most Wolbachia infecting Anopheles (34/41) clustered with wAlbB (Supergroup B) isolated from Ae. albopictus [70], whereas the remaining Wolbachia strains from other Anopheles (7/41) were more closely related to wMel (D. melanogaster) of Supergroup A. Interestingly, all the Wolbachia infecting Anopheles detected in this study were shown to be more closely related with those recently reported in African malaria vectors (wAnga) [38–40, 42–44] than with the ones from Myanmar [46] (Fig. 5). Although recent publications suggest a lack of concrete evidence regarding the presence of Wolbachia in Anopheles and Ae. aegypti [71, 72], our results show that Wolbachia is present in Anopheles. Our sequences were clean and matched those of Wolbachia Supergroup A and B. However, we agree that our limited study shows that the prevalence of Wolbachia in Anopheles is much lower than the natural presence of Wolbachia in Ae. albopictus.
Plasmodium-infected Anopheles mosquitoes were also found to be Wolbachia-infected. These mosquitoes harbored oocysts or sporozoites. Results from this study found that Wolbachia prevalence is higher (though not significant) in Anopheles mosquitoes with sporozoites (50%) than in mosquitoes with oocysts (11.11%). This is in agreement with a study finding significant reductions in P. berghei or P. falciparum oocyst infections in An. gambiae transiently and somatically-infected with wMelPop or wAlbB Wolbachia strains [36, 37]. It was also found that natural populations of Wolbachia-infected An. coluzzii females have lower frequency of Plasmodium infections than Wolbachia-negative individuals. Although the study did not explicitly mention the infection stages of the parasite in these mosquitoes, it is assumed that most of these Plasmodium infections could be at the oocyst stage since Plasmodium DNA was detected from dissected abdomens and thoraces of the mosquitoes 5 days after indoor collection [39]. Another interesting study also showed that Culex pipiens (the natural vector of P. relictum), naturally infected with Wolbachia, had increased sporozoites than those without Wolbachia [73]. Thus, it seems to demonstrate that when Wolbachia is naturally present in the mosquitoes, it may perturb oocysts development but promote sporozoites production. This however, is the opposite to the findings of a study on An. gambiae from Mali, Africa showing that Wolbachia infection was significantly lower in sporozoite-positive mosquitoes compared to the sporozoite-negative mosquitoes. However, Wolbachia-positive mosquitoes had higher oocyst infection compared to Wolbachia negative-mosquitoes [40]. It seems that Wolbachia can suppress the prevalence and intensity of P. falciparum sporozoite infections in wild An. gambiae [40]. On the other hand, An. stephensi experimentally infected with wAlbB strain of Wolbachia was able to suppress both P. falciparum oocysts and sporozoite infection rates [34]. Due to these contrasting results, it is pertinent for us to study the life-cycle and transmission dynamics of Plasmodium in Anopheles vectors infected and uninfected with Wolbachia. It is possible that Wolbachia may affect only certain Plasmodium parasites present in the vector such as P. falciparum as reported by Bian et al. [34] or simian malaria parasites in this study, or it might influence vector competence of certain vector species [73]. As such, effect of Wolbachia on An. balabacensis, An. latens and An. introlatus in the present study remains obscure and hence, there is a need for extensive mosquito collections and thorough investigations.
Naturally, Ae. aegypti does not harbor Wolbachia. Interestingly, in this study the two Ae. aegypti which emerged from larvae collected from sticky traps were positive for Wolbachia. However, Wolbachia has been found in large percentage of Ae. aegypti in Manila [47], and in Kuala Lumpur, Malaysia, Wolbachia of Supergroup B has been reported from Ae. aegypti larvae which were also positive for DENV 2 virus [49]. Similarly, in India [50], Wolbachia has been detected in natural Ae. aegypti populations and was designated as wAegB. Additionally, Wolbachia-infected Ae. aegypti has been reported from Mexico (> 50%) [52], Florida [51], Panama [54] and Texas [53]. It is noteworthy that during this study, no Wolbachia-infected mosquito colony is being reared in our insectarium, neither is there any routine screening nor handling of Wolbachia material.
The Wolbachia infecting Cx. quinquefasciatus was shown to be more closely related to wPip, isolated from Cx. pipiens within the Supergroup B than with wAlbB from Ae. albopictus, similar to previous reports [4, 74]. Our results also revealed the presence of Wolbachia in Ar. subalbatus which was grouped within the Supergroup A. Interestingly, all Cx. vishnui, Cx. pseudovishnui, Cx. sinensis (vector of Japanese encephalitis), Ma. annulifera, Ma. bonneae and some Cx. quinquefasciatus mosquitoes failed to show a positive wsp PCR amplification. Through nested PCR of the 16S rRNA gene, this is the first report of Wolbachia infection detected in Cx. sinensis, Cx. vishnui, Cx. pseudovishnui, Ma. bonneae and Ma. annulifera. However, due to the low number of samples collected, further studies are needed to determine if this is a result of strain variability or low-density infections which resulted in failure of amplification using wsp primers.
While the PCR and sequencing results of the present study are convincing, especially by showing that the Wolbachia strains found infecting the mosquitoes belong to the two supergroups (A and B, which are associated with arthropods), there is a growing call for careful investigations into natural Wolbachia infections in previously unreported mosquitoes. This is because, it has been estimated that over half of terrestrial arthropod species are infected with Wolbachia [75]. Therefore, the environment is a factor that possibly affects the presence or absence of Wolbachia in some mosquitoes [43]. Thus, environmental contamination of the samples from plants, endo- and ectoparasites, Wolbachia-contaminated/infected food in the wild, eggs and larval habitats and other cohabitating insects cannot be ruled out [46, 71]. Furthermore, reports of Wolbachia found with low prevalence in some species and difficulty in detecting them, especially in Anopheles spp. [42–44, 46], indicate that there could be a lack of stable symbiotic relationship between Wolbachia and the hosts [71]. Therefore, to demonstrate evidence of a stable, intraovarially-transmitted Wolbachia symbiont in a host [71], steps that should be taken include: (i) visualizing Wolbachia in different host tissues using fluorescent in situ hybridization (FISH) or electron microscopy; (ii) demonstrating that the infection is and can be maternally transmitted by performing reciprocal crosses; and (iii) showing that Wolbachia can be removed from the mosquitoes by antibiotic or heat treatment [72].
Nonetheless, the growing reports of Wolbachia infections in previously unreported mosquito vectors, open avenues for further investigations into its prevalence and should prompt careful evaluations on the release of Wolbachia-infected mosquitoes for disease/vector control programmes. Additionally, the presence of Asaia, an acetic acid bacterium found in some Anopheles species is postulated for the absence of Wolbachia in Anopheles mosquitoes [76]. Asaia has been shown to impede vertical transmission of Wolbachia in Anopheles mosquitoes experimentally and found negatively related with Wolbachia in mosquito reproductive organs [77]. Therefore, the dynamics of Wolbachia infections with vector capacity, microbial interactions within the host and disease transmission require further in-depth studies.
Conclusions
The present study reports the presence of Wolbachia in Anopheles and Ae. aegypti which were not previously reported, as well as in other important mosquito vector genera such as Culex and Mansonia using 16S rRNA and wsp genes. Given the fact that Wolbachia can reduce the life span of its host, prevent pathogen from completing its life-cycle, as well as reduce susceptibility of host to pathogen infection, Wolbachia is being considered for vector control programmes and is now released on a large scale in many countries. Thus, it is pertinent to carry out large-scale studies on the natural infection of Wolbachia in mosquito vectors using more sensitive and accurate methods of detection as described above. The long-term effect of introduced Wolbachia into new hosts and its effect on pathogen suppression should also be studied.
Supplementary information
Additional file 1: Table S1. Summary of mosquito species used in the study from six states in Malaysia. Collection was carried out using human landing catch (HLC), mosquito magnetic trap and ovitrap. Table S2. Mosquito collection sites. Mosquitoes were collected from different settings: urban, village, island, forest and wetland across several states in Malaysia. Table S3. Anopheles samples with Plasmodium infections.
Additional file 2: Figure S1. Alignment and variable sites of Wolbachia wsp sequences (Supergroup A) with nucleotide positions.
Additional file 3: Figure S2. Alignment and variable sites of Wolbachia wsp sequences (Supergroup B) with nucleotide positions.
Additional file 4: Figure S3. Alignment and variable sites of Wolbachia 16S rRNA (Supergroup A) sequences with nucleotide positions.
Additional file 5: Figure S4. Alignment and variable sites of Wolbachia 16S rRNA (Supergroup B) sequences with nucleotide positions.
Acknowledgements
The authors thank the staff of vector control program for the help they provided during the collection of mosquitoes. The authors would also like to thank other colleagues who have volunteered to collect some of the mosquitoes and Dr. Francesco Baldini for Wolbachia DNA from An. arabiensis.
Abbreviations
- wsp
Wolbachia surface protein
- 16S rRNA
16S ribosomal RNA
Authors’ contributions
IV, WML and JWKL conceived and designed the study. WML, WKW, SP, WYWS, NKJ, CSL, VLL and NMH were responsible for the field work. WML, WKW, SP, NKY and NMH carried out laboratory work. WML, WKW, SP and JWKL carried out the molecular work. ML, VLL, JWKL and IV analysed the data and prepared the manuscript. All other authors were responsible for editorial processing of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by Ministry of Education, Malaysia Long-term Research Grant Scheme LRGS 2018-1(LR002C-2018). The funding body has no role in the design of the study and collection, analysis and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files. The newly generated sequences were deposited in the GenBank database under the accession numbers MN893348-MN893366 (wsp gene) and MN887537-MN887588 (16S rRNA).
Ethics approval and consent to participate
This study was approved by the Medical Research and Ethics Committee (NMRR, Reference No. NMRR-12-786-13048 and NMRR 19-962-47606 (IIR)), for the collection of mosquitoes and registered in the National Medical Research Register of the Malaysian Ministry of Health.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Meng Li Wong, Email: mengli.wong0105@gmail.com.
Jonathan Wee Kent Liew, Email: jon_wkent@hotmail.com, Email: jonathanliew@um.edu.my.
Wai Kit Wong, Email: waikit_789@hotmail.com.
Sandthya Pramasivan, Email: sandthya96@gmail.com.
Norzihan Mohamed Hassan, Email: norzihan.m.h@gmail.com.
Wan Yusoff Wan Sulaiman, Email: wanyus@um.edu.my.
Nantha Kumar Jeyaprakasam, Email: j_nanthkumar@hotmail.com.
Cherng Shii Leong, Email: qqshiiqq@gmail.com.
Van Lun Low, Email: lucaslow24@gmail.com.
Indra Vythilingam, Email: indrav@um.edu.my.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-04277-x.
References
- 1.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 2.Zug R, Hammerstein P. Still a host for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PloS ONE. 2012;7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kittayapong P, Baisley KJ, Baimai V, O’Neill SL. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae) J Med Entomol. 2000;37:340–345. doi: 10.1093/jmedent/37.3.340. [DOI] [PubMed] [Google Scholar]
- 4.Nugapola N, De Silva W, Karunaratne S. Distribution and phylogeny of Wolbachia strains in wild mosquito populations in Sri Lanka. Parasit Vectors. 2017;10:230. doi: 10.1186/s13071-017-2174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raharimalala FN, Boukraa S, Bawin T, Boyer S, Francis F. Molecular detection of six (endo-) symbiotic bacteria in Belgian mosquitoes: first step towards the selection of appropriate paratransgenesis candidates. Parasitol Res. 2016;115:1391–1399. doi: 10.1007/s00436-015-4873-5. [DOI] [PubMed] [Google Scholar]
- 6.Rasgon JL, Scott TW. An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae) J Med Entomol. 2004;41:255–257. doi: 10.1603/0022-2585-41.2.255. [DOI] [PubMed] [Google Scholar]
- 7.Soni M, Bhattacharya C, Sharma J, Khan SA, Dutta P. Molecular typing and phylogeny of Wolbachia: a study from Assam, north-eastern part of India. Acta Trop. 2017;176:421–426. doi: 10.1016/j.actatropica.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Wiwatanaratanabutr I. Geographic distribution of wolbachial infections in mosquitoes from Thailand. J Invertebr Pathol. 2013;114:337–340. doi: 10.1016/j.jip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Yamada R, Iturbe-Ormaetxe I, Brownlie JC, O’Neill SL. Functional test of the influence of Wolbachia genes on cytoplasmic incompatibility expression in Drosophila melanogaster. Insect Mol Biol. 2011;20:75–85. doi: 10.1111/j.1365-2583.2010.01042.x. [DOI] [PubMed] [Google Scholar]
- 10.O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill SL, Hoffman A, Werren JH. Influential passengers: inherited microorganisms and arthropod reproduction. Oxford: Oxford University Press; 1997. [Google Scholar]
- 12.Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 13.Brelsfoard CL, Dobson SL. Wolbachia-based strategies to control insect pests and disease vectors. Asia-Pac J Mol Biol Biotechnol. 2009;17:55–63. [Google Scholar]
- 14.Turelli M, Hoffman AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 15.Fraser JE, De Bruyne JT, Iturbe-Ormaetxe I, Stepnell J, Burns RL, Flores HA, et al. Novel Wolbachia-transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLoS Pathog. 2017;13:e1006751. doi: 10.1371/journal.ppat.1006751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy DJ, Hoffmann AA. Environmental effects on cytoplasmic incompatibility and bacterial load in Wolbachia-infected Drosophila simulans. Entomol Exp Appl. 1998;86:13–24. [Google Scholar]
- 17.Panteleev D, Goriacheva II, Andrianov BV, Reznik NL, Lazebnyi OE, Kulikov AM. The endosymbiotic bacterium Wolbachia enhances the nonspecific resistance to insect pathogens and alters behavior of Drosophila melanogaster. Genetika. 2007;43:1277–1280. [PubMed] [Google Scholar]
- 18.Wiwatanaratanabutr I, Kittayapong P. Effects of crowding and temperature on Wolbachia infection density among life cycle stages of Aedes albopictus. J Invertebr Pathol. 2009;102:220–224. doi: 10.1016/j.jip.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Wiwatanaratanabutr S, Kittayapong P. Effects of temephos and temperature on Wolbachia load and life history traits of Aedes albopictus. Med Vet Entomol. 2006;20:300–307. doi: 10.1111/j.1365-2915.2006.00640.x. [DOI] [PubMed] [Google Scholar]
- 20.Iturbe-Ormaetxe I, Walker T, O’Neill SL. Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 2011;12:508–518. doi: 10.1038/embor.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joubert DA, Walker T, Carrington LB, De Bruyne JT, Kien DH, Hoang Nle T, et al. Establishment of a Wolbachia superinfection in Aedes aegypti mosquitoes as a potential approach for future resistance management. PloS Pathog. 2016;12:e1005434. doi: 10.1371/journal.ppat.1005434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PloS Pathog. 2010;6:e1000833. doi: 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frentiu FD, Zakir T, Walker T, Popovici J, Pyke AT, Van den Hurk AF, et al. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PloS Negl Trop Dis. 2014;8:e2688. doi: 10.1371/journal.pntd.0002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joanne S, Vythilingam I, Teoh B-T, Leong C-S, Tan K-K, Wong M-L, et al. Vector competence of Malaysian Aedes albopictus with and without Wolbachia to four dengue virus serotypes. Trop Med Int Health. 2017;22:1154–1165. doi: 10.1111/tmi.12918. [DOI] [PubMed] [Google Scholar]
- 25.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 26.Mousson L, Zouache K, Arias-Goeta C, Raquin V, Mavingui P, Failloux AB. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PloS Negl Trop Dis. 2012;6:e1989. doi: 10.1371/journal.pntd.0001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 28.Xi Z, Khoo CC, Dobson SL. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science. 2005;310:326–328. doi: 10.1126/science.1117607. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad NA, Vythilingam I, Lim YAL, Zabari NZAM, Lee HL. Detection of Wolbachia in Aedes albopictus and their effects on chikungunya virus. Am J Trop Med Hyg. 2017;96:148–156. doi: 10.4269/ajtmh.16-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, et al. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PloS Negl Trop Dis. 2012;6:e1892. doi: 10.1371/journal.pntd.0001892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aliota MT, Peinad SA, Velez ID, Osorio JE. The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci Rep. 2016;6:28792. doi: 10.1038/srep28792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chouin-Carneiro T, Ant TH, Herd C, Louis F, Failloux AB, Sinkins SP. Wolbachia strain wAlbA blocks Zika virus transmission in Aedes aegypti. Med Vet Entomol. 2019;34:116–119. doi: 10.1111/mve.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutra HLC, Rocha MN, Dias FBS, Mansur SB, Caragata EP, Moreira LA. Wolbachia blocks currently circulating Zika virus isolates in Brazillian Aedes aegypti mosquitoes. Cell Host Microbe. 2016;19:771–774. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 35.Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326:134–136. doi: 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7:e1002043. doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kambris Z, Blagborough AM, Pinto SB, Blagrove MSC, Godfray HCJ, Sinden RE, et al. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6:e1001143. doi: 10.1371/journal.ppat.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldini F, Segata N, Pompon J, Marcenac P, Robert SW, Dabiré RK, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5:3985. doi: 10.1038/ncomms4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw WR, Marcenac P, Childs LM, Buckee CO, Baldini F, Sawadogo SP, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7:11772. doi: 10.1038/ncomms11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomes FM, Hixson BL, Tyner MDW, Ramirez JL, Canepa GE, Silva EALT, et al. Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc Natl Acad Sci USA. 2017;114:12566–12571. doi: 10.1073/pnas.1716181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nazni WA, Hoffmann AA, NoorAfizah A, Cheong YL, Mancini MV, Golding N, et al. Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. Curr Biol. 2019;29:4241–4248. doi: 10.1016/j.cub.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldini F, Rouge J, Kreppel K, Mkandawile G, Mapua SA, Sikulu-Lord M, et al. First report of natural Wolbachia infection in the malaria mosquito Anopheles arabiensis in Tanzania. Parasit Vector. 2018;11:635. doi: 10.1186/s13071-018-3249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffries CL, Lawrence GG, Golovko G, Kristan M, Orsborne J, Spence K, et al. Novel Wolbachia strains in Anopheles malaria vectors from sub-Saharan Africa. Wellcome Open Res. 2018;3:113. doi: 10.12688/wellcomeopenres.14765.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niang EHA, Bassene H, Makoundou P, Fenollar F, Weill M, Mediannikov O. First report of natural Wolbachia infection in wild Anopheles funestus population in Senegal. Malar J. 2018;17:408. doi: 10.1186/s12936-018-2559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayala D, Akone-Ella O, Rahola N, Kengne P, Ngangue MF, Mezeme F, et al. Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evol Appl. 2019;12:1583–1594. doi: 10.1111/eva.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawasdichai S, Chaumeau V, Dah T, Kulabkeeree T, Kajeechiwa L, Phanaphaduntham M, et al. Natural Wolbachia infections in malaria vectors in Kayin state, Myanmar. Wellcome Open Res. 2019;4:11. doi: 10.12688/wellcomeopenres.15005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvajal TM, Hashimoto K, Harnandika RK, Amalin DM, Watanabe K. Detection of Wolbachia in field-collected Aedes aegypti mosquitoes in metropolitan Manila, Philippines. Parasit Vectors. 2019;12:361. doi: 10.1186/s13071-019-3629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thongsripong P, Chandler JA, Green AB, Kittayapong P, Wilcox BA, Kapan DD, et al. Mosquito vector-associated microbiota: metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol Evol. 2017;8:1352–1368. doi: 10.1002/ece3.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teo CHJ, Lim PKC, Voon K, Mak JW. Detection of dengue viruses and Wolbachia in Aedes aegypti and Aedes albopictus larvae from four urban local localities in Kuala Lumpur, Malaysia. Trop Biomed. 2017;34:583–597. [PubMed] [Google Scholar]
- 50.Balaji S, Jayachandran S, Prabagaran SR. Evidence for the natural occurrence of Wolbachia in Aedes aegypti mosquitoes. FEMS Microbiol Lett. 2019;366:fnz055. doi: 10.1093/femsle/fnz055. [DOI] [PubMed] [Google Scholar]
- 51.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol Ecol. 2016;25:5806–5826. doi: 10.1111/mec.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulkarni A, Yu W, Jiang J, Sanchez C, Karna AK, Martinez KJL, et al. Wolbachia pipientis occurs in Aedes aegypti populations in New Mexico and Florida, USA. Ecol Evol. 2019;9:6148–6156. doi: 10.1002/ece3.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hegde S, Khanipov K, Albayrak L, Golovko G, Pimenova M, Saldaña MA, et al. Microbiome interaction networks and community structure from laboratory-reared and field-collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus mosquito vectors. Front Microbiol. 2018;9:2160. doi: 10.3389/fmicb.2018.02160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett KL, Gómez-Martínez C, Chin Y, Saltonstall K, McMillan WO, Rovira JR, et al. Dynamics and diversity of bacteria associated with the disease vectors Aedes aegypti and Aedes albopictus. Sci Rep. 2019;9:12160. doi: 10.1038/s41598-019-48414-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marcon HS, Coscrato VE, Selivon D, Perondini AL, Marino CL. Variations in the sensitivity of different primers for detecting Wolbachia in Anastrepha (Diptera: Tephritidae) Braz J Microbiol. 2011;42:778–785. doi: 10.1590/S1517-838220110002000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reid J. Anopheline mosquitoes of Malaya and Borneo. England: Staples Printer Limited; 1968. [Google Scholar]
- 57.Sallum MAM, Peyton EL, Harrison BA, Wilkerson RC. Revision of the Leucosphyrus group of Anopheles (Cellia) (Diptera: Culicidae) Rev Bras Entomol. 2005;49(Suppl. 1):1–152. [Google Scholar]
- 58.Jeffery J, Rohela M, Muslimin M, Abdul Aziz S, Jamaiah I, Kumar S, et al. Illustrated keys: some mosquitoes of Peninsula Malaysia. Kuala Lumpur: University of Malaya Press; 2012. [Google Scholar]
- 59.Vythilingam I, Lim YAL, Venugopalan B, Ngui R, Leong CS, Wong ML, et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor Malaysia (2009–2013): epidemiologic and entomologic analysis. Parasit Vectors. 2014;7:436. doi: 10.1186/1756-3305-7-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PloS Pathog. 2011;7:e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou W, Rousset F, O’Neil S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–1285. doi: 10.1098/rspb.2000.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis Version 70 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan CH, Vythilingam I, Matusop A, Chan ST, Singh B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J. 2008;7:52–59. doi: 10.1186/1475-2875-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong ML, Chua TH, Leong CS, Khaw LT, Fornace K, Wan Yusoff WS, et al. Seasonal and spatial dynamics of the primary vector of Plasmodium knowlesi within a major transmission focus in Sabah, Malaysia. PloS Negl Trop Dis. 2015;9:e0004135. doi: 10.1371/journal.pntd.0004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hii J, Kan S, Pereira M, Parmar S, Campos R, Chan M. Bancroftian filariasis and malaria in island and hinterland populations in Sabah, Malaysia. Trop Geogr Med. 1985;37:93–101. [PubMed] [Google Scholar]
- 67.Amerasinghe PH, Amerasinghe FP, Konradsen F, Fonseka KT, Wirtz RA. Malaria vectors in a traditional dry zone village in Sri Lanka. Am J Trop Med Hyg. 1999;60:421–429. doi: 10.4269/ajtmh.1999.60.421. [DOI] [PubMed] [Google Scholar]
- 68.Feng X, Zhang S, Huang F, Zhang L, Feng J, Xia Z, et al. Biology, bionomics and molecular biology of Anopheles sinensis Wiedemann 1828 (Diptera: Culicidae), main malaria vector in China. Front Microbiol. 2017;8:1473. doi: 10.3389/fmicb.2017.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramsdale CD. Internal taxonomy of the Hyrcanus group of Anopheles (Diptera, Culicidae) and its bearing on the incrimination of vectors of malaria in the west of the Palaearctic region. J Eur Mosq Control Assoc. 2001;10:1–8. [Google Scholar]
- 70.Sinha A, Li Z, Sun L, Carlow CKS. Complete genome sequence of the Wolbachia wAlbB endosymbiont of Aedes albopictus. Genome Biol Evol. 2019;11:706–720. doi: 10.1093/gbe/evz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chrostek E, Gerth M. Is Anopheles gambiae a natural host of Wolbachia. mBio. 2019;10:e00784. doi: 10.1128/mBio.00784-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross PA, Callahan AG, Yang Q, Jasper M, Arif M, Afizah AN, et al. An elusive endosymbiont: does Wolbachia occur naturally in Aedes aegypti? Ecol Evol. 2020;10:1581–1591. doi: 10.1002/ece3.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zélé F, Nicot A, Berthomieu A, Weill M, Duron O, Rivero A. Wolbachia increases susceptibility to Plasmodium infection in a natural system. Proc Biol Sci. 2014;281:20132837. doi: 10.1098/rspb.2013.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osei-Poku J, Han C, Mbogo CM, Jiggins FM. Identification of Wolbachia strains in mosquito disease vectors. PloS ONE. 2012;7:e49922. doi: 10.1371/journal.pone.0049922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc Biol Sci. 2015;282:20150249. doi: 10.1098/rspb.2015.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hughes GL, Dodson BL, Johnson RM, Murdock CC, Tsujimoto H, Suzuki Y, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA. 2014;111:12498. doi: 10.1073/pnas.1408888111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossi P, Ricci I, Cappelli A, Damiani C, Ulissi U, Mancini MV, et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit Vectors. 2015;8:278. doi: 10.1186/s13071-015-0888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Summary of mosquito species used in the study from six states in Malaysia. Collection was carried out using human landing catch (HLC), mosquito magnetic trap and ovitrap. Table S2. Mosquito collection sites. Mosquitoes were collected from different settings: urban, village, island, forest and wetland across several states in Malaysia. Table S3. Anopheles samples with Plasmodium infections.
Additional file 2: Figure S1. Alignment and variable sites of Wolbachia wsp sequences (Supergroup A) with nucleotide positions.
Additional file 3: Figure S2. Alignment and variable sites of Wolbachia wsp sequences (Supergroup B) with nucleotide positions.
Additional file 4: Figure S3. Alignment and variable sites of Wolbachia 16S rRNA (Supergroup A) sequences with nucleotide positions.
Additional file 5: Figure S4. Alignment and variable sites of Wolbachia 16S rRNA (Supergroup B) sequences with nucleotide positions.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additional files. The newly generated sequences were deposited in the GenBank database under the accession numbers MN893348-MN893366 (wsp gene) and MN887537-MN887588 (16S rRNA).