Abstract
Background
Surface layers (S-layers) are two-dimensional crystalline arrays of repeating proteinaceous subunits that form the outermost layer of many bacterial cell envelopes. Within the Lactobacillus genus, S-layer presence is frequently associated with probiotic-relevant properties such as improved adherence to host epithelial cells and modulation of the immune response. However, recent studies have demonstrated that certain S-layer functions may be supplemented by a novel subset of proteins embedded within its lattice, termed S-layer associated proteins (SLAPs). In the following study, four Lactobacillus acidophilus NCFM SLAPs (LBA0046, LBA0864, LBA1426, and LBA1539) were selected for in silico and phenotypic assessment.
Results
Despite lacking any sequence similarity or catalytic domains that may indicate function, the genes encoding the four proteins of interest were shown to be unique to S-layer-forming, host-adapted lactobacilli species. Likewise, their corresponding deletion mutants exhibited broad, host-relevant phenotypes including decreased inflammatory profiles and reduced adherence to Caco-2 intestinal cells, extracellular matrices, and mucin in vitro.
Conclusions
Overall, the data presented in this study collectively links several previously uncharacterized extracellular proteins to roles in the underlying host adaptive mechanisms of L. acidophilus.
Keywords: Lactobacillus, S-layer, Probiotics, Cell surface
Background
Lactobacillus are Gram-positive, non-sporulating, anaerobic or microaerophilic bacteria, with complex nutritional requirements [1]. These versatile microorganisms inhabit diverse environments including dairy, meat and plant fermentations, as well as the gastrointestinal and urogenital tracts of humans and animals [2]. Although renowned for their applications in food and feed fermentations, several Lactobacillus species are also touted for their health-promoting, probiotic properties [3]. Probiotics are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” [4]. Efficacy tends to correlate with underlying host adaptive mechanisms such as bile and acid tolerance [5, 6], adhesion to mucus and epithelial cells [7, 8], and modulation of the immune response [9, 10] -- characteristics that can oftentimes be linked to the presence of specific extracellular proteins [2, 11, 12].
In general, Lactobacillus exoproteomes are composed of two main groups: secreted proteins that are released from the cell and surface-associated proteins, such as surface (S-)layers [2]. S-layers are two-dimensional crystalline arrays composed of numerous repeating subunits, S-layer proteins (Slps), that are inherently driven to self-assemble on the cell exterior [13, 14]. S-layers have been detected on several Lactobacillus species [1] and shown to play critical roles in adherence to intestinal cells [7, 15, 16] and host immunomodulation [10, 17]. However, recent studies have demonstrated the supplementation of these functions by various proteins embedded within the S-layer lattice. S-layer associated proteins, or SLAPs, first classified in industry-relevant strain Lactobacillus acidophilus NCFM [18], have since been identified on the surfaces of several species within the L. acidophilus homology group including Lactobacillus helveticus, Lactobacillus crispatus, Lactobacillus amylovorus, and Lactobacillus gallinarum [19]. To date, characterized SLAPs have exhibited a broad range of functions encompassing roles in cell maintenance processes such as cell division [20] and autolysin activity [21], as well as host adhesion [22] and immunomodulation [18, 23] mechanisms. Historically, selection of SLAP targets for deletion characterization was predominately based on their intriguing domain architecture or predicted function, but what about the extensive catalog of remaining proteins?
In the present study, four putative uncharacterized L. acidophilus SLAPs, routinely isolated from the cell surface [18, 19] and shown to exhibit either high relative abundance (LBA0864 and LBA1426) or significant induction in stationary growth phase (LBA0046 and LBA1539) [24], were selected for in silico and phenotypic analyses. Remarkably, despite having no sequence homology or predicted catalytic domains, all four genes were detected only in S-layer-forming, host-adapted lactobacilli species. Likewise, their corresponding deletion mutants demonstrated host-relevant phenotypes including modified immunogenicity profiles and reduced adherence to Caco-2 intestinal cells, extracellular matrices (ECMs) and mucin in vitro. Collectively, these results suggest that our four SLAPs of interest contribute to the survival and persistence L. acidophilus within the confines of the host gastrointestinal tract.
Results
Genetic analysis of SLAP loci
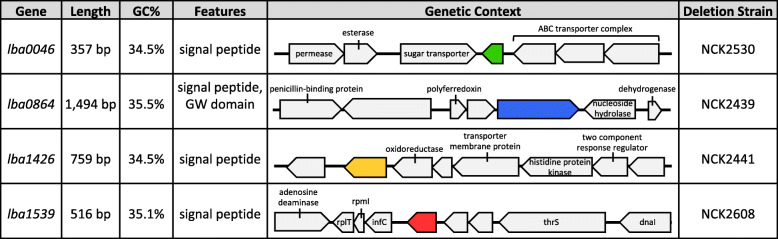
Selected genes were shown to encode four previously uncharacterized surface proteins with no predicted function or COG. Their genetic layouts were analyzed on both a nucleotide and amino acid level (Fig. 1). Gene sizes ranged from 357 to 1494 bp, exhibited low GC content (34.5–35.5%), and were distributed randomly throughout the L. acidophilus chromosome. Though each possessed an N-terminal signal sequence, indicative of secretion and/or incorporation into cell wall/cell membrane components, only lba0864 contained a GW (Gly-Tryp) dipeptide surface anchor. Genetic context was used to gain insight into the potential function of these proteins. The smallest of the genes analyzed, lba0046, was flanked by two transporters, namely a sugar transporter and an ABC transporter of unknown substrate specificity. The largest gene, lba0864, was located upstream of a nucleoside hydrolase and downstream of a hypothetical protein and polyferredoxin gene. The locus encoding LBA1539 was bordered by a 50S ribosomal protein complex and two hypothetical proteins, while lba1426 was situated upstream of a two-component regulatory system. None of the analyzed genes were predicted to be part of an operon.
Fig. 1.
Schematic representation of the four S-layer associated proteins of interest. Table includes the gene length, GC percentage, interesting features, and genetic context of the four S-layer associated protein targets as well as their corresponding deletion strains. Genes are colored as follows, lba0046 green, lba0864 blue, lba1426 yellow, and lba1539 red. Unlabeled gray arrows represent genes of unknown function
Mapping target SLAPs to a phylogenetic tree
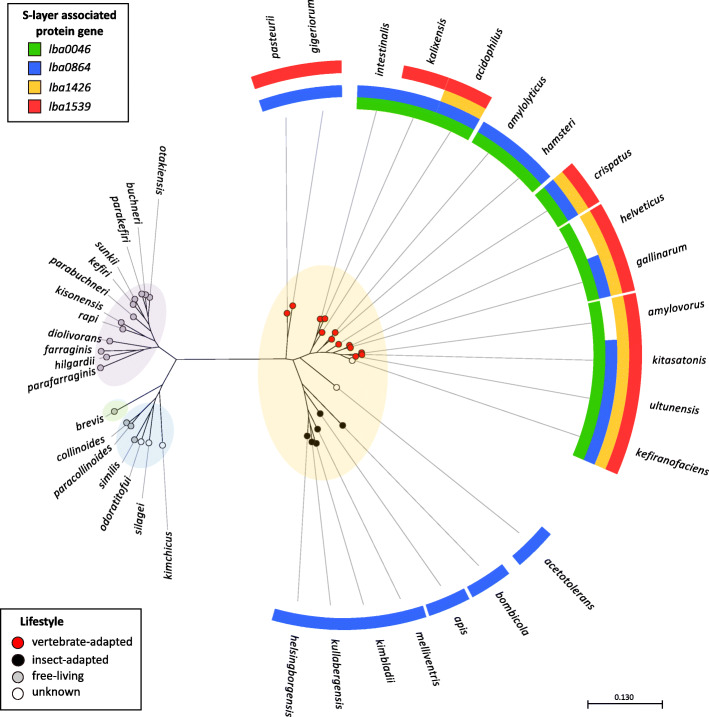
The nucleotide sequences of lba0046, lba0864, lba1426, and lba1539 were searched against a previously curated database of 170 Lactobacillus genomes updated to incorporate the 25 recently classified genera emended within the genus [25]. Orthologs with > 40% nucleotide sequence identity were mapped to a phylogenetic tree based on the pyruvate kinase (Pyk) enzyme sequence (Fig. 2). The strains used to generate this figure are further detailed in Supplementary Table 1. All identified genes had comparable sizes to their query and consistently exhibited low GC content (< 40%). Several genomes possessed more than one copy of a gene, most often lba0046. Figure 2 only depicts S-layer forming strains as none of the investigated genes were present in non-S-layer-formers. Although strains tended to cluster by lifestyle and genera, a clear divide emerged based on the mapped SLAPs (Fig. 2). While only lba0864 was detected in insect-adapted species, vertebrate adapted species possessed at least two SLAPs in every strain, and several contained all four, including L. acidophilus, L. crispatus, L. gallinarum, L. kitasatonis, and L. ultunensis. These results are in stark contrast to free-living strains, which encompass three recently identified genera (Lentilactobacillus, Levilactobacillus, and Secundilactobacillus), and were completely devoid of any target SLAPs.
Fig. 2.
Presence/absence of lba0046, lba0864, lba1426, and lba1539 mapped to a phylogenetic tree. Gene targets were mapped to a phylogenetic tree constructed based on the pyruvate kinase gene sequence. Strain groupings are based on the recent Zheng et al., Lactobacillus reclassification [25] and colored as follows, Lentilactobacillus purple, Levilactobacillus green, Secundilactobacillus blue, and Lactobacillus yellow. Node colors designate lifestyle as described by Duar et al. [26]. Note: Branches shorter than 0.0127 are shown as having length of 0.127
Chromosomal gene deletion and growth curve analysis
A pORI-based upp counterselective gene replacement system [27] was used to generate four separate in-frame deletions within the L. acidophilus NCFM chromosome. The mutant strains are listed in Fig. 1 and detailed in Table 1. Deletions were detected by PCR (Supplementary Fig. 1) and sequenced to confirm the absence of both the target SLAP and any unintended mutations within the flanking regions. All SLAP deletion strains were subjected to growth curve analyses. The OD600 of the parent and mutant strains was measured over the course of 30 h in MRS, as well as MRS containing 2.5% NaCl, 0.2% porcine bile, and 0.5% oxgall. No differences were detected between any of the strains under these conditions (data not shown).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or Plasmid | Genotype or characteristics | Reference |
|---|---|---|
| Strains | ||
| Lactobacillus acidophilus | ||
| NCK56 (NCFM) | Human intestinal isolate | [28] |
| NCK1909 (Δupp) | NCFM with a 315 bp in-frame deletion within the upp gene (lba0770); background/parent strain for NCFM deletion mutants | [27] |
| NCK1910 | NCK1909 harboring the repA helper plasmid pTRK669; host for pORI-based counterselective integration vector | [27] |
| NCK2439 (Δlba0864) | NCK1909 with a 1425 bp in-frame deletion within the lba0864 gene | This study |
| NCK2441 (Δlba1426) | NCK1909 with a 717 bp in-frame deletion within the lba1426 gene | This study |
| NCK2530 (Δlba0046) | NCK1909 with a 342 bp in-frame deletion within the lba0046 gene | This study |
| NCK2608 (Δlba1539) | NCK1909 with a 498 bp in-frame deletion within the lba1539gene | This study |
| Escherichia coli | ||
| EC101 | RepA+ JM101; Kmr; repA from pWV01 integrated in chromosome; cloning host for pORI-based plasmids | [29] |
| NCK1391 | E. coli DH10B harboring pTRK669; cloning host for pORI-based plasmids | [27] |
| NCK1911 | EC101 host harboring pTRK935 integration vector | [27] |
| NCK2438 | EC101 host harboring pTRK1118 recombinant plasmid | This study |
| NCK2440 | EC101 host harboring pTRK1119 recombinant plasmid | This study |
| NCK2529 | EC101 host harboring pTRK1126 recombinant plasmid | This study |
| NCK2607 | DH10B host harboring pTRK1170 recombinant plasmid | This study |
| Plasmids | ||
| pTRK669 | Ori (pWV01), Cmr, RepA+, thermosensitive | [30] |
| pTRK935 | pORI upp-based counterselective integration vector, Emr | [27] |
| pTRK1118 | pTRK935 harboring a mutated copy of lba0864 gene cloned into HindIII/SacI site | This study |
| pTRK1119 | pTRK935 harboring a mutated copy of lba1426 gene cloned into HindIII/SacI site | This study |
| pTRK1126 | pTRK935 harboring a mutated copy of lba0046 gene cloned into HindIII/SacI site | This study |
| pTRK1170 | pTRK935 harboring a mutated copy of lba1426 gene cloned into BamHI/SacI site | This study |
Examination of SLAP mutant cellular morphologies
Flow cytometry and scanning electron microscopy (SEM) were used to detect morphological changes in log phase (6 h) and early stationary phase (12 h) SLAP mutants compared to the parent. Results showed relatively little difference between the strains at either time point (Supplementary Fig. 2). Although NCK2608 (Δlba1539) cells appeared slightly longer in stationary phase flow data, this difference was not discernable from SEM images (Supplementary Fig. 2B). Overall, the strains seemed unencumbered, with no obvious deletion-dependent morphological alterations.
Deletion of SLAP genes reduces the adhesive capacity of L. acidophilus
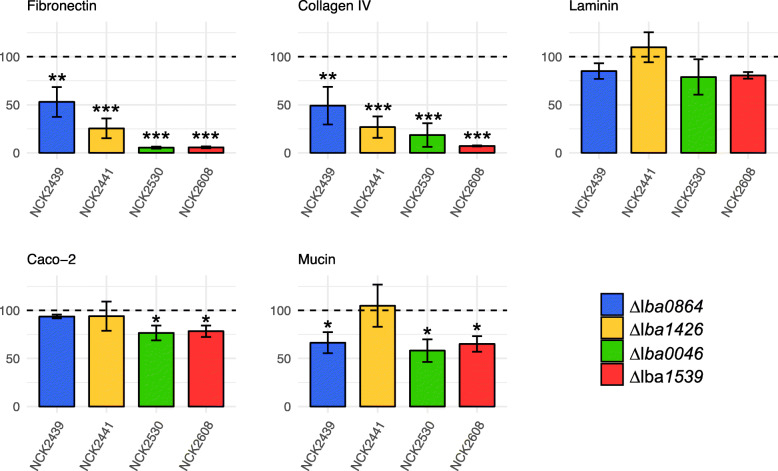
The impact of the SLAP gene deletions on the adhesive capacity of the mutant strains was evaluated using an in vitro adhesion assay. Tested substrates included major ECMs, mucin, and Caco-2 intestinal cells. Assay results were substrate-dependent (Fig. 3). All four mutants demonstrated extensive reductions in adherence to fibronectin and collagen, particularly NCK2608 (Δlba1539) which underwent a > 90% drop relative to the parent strain. Conversely, there were no statistically significant reductions in laminin binding. Despite the relative magnitude of some of the ECM results, Caco-2 adherence deficiencies were only apparent for two of the mutant strains, NCK2530 (Δlba0046) and NCK2608 (Δlba1539), both with relative reductions just over 20%. NCK2441 (Δlba1426) was the only strain to not present some form of mucin-binding deficiency, while the other three mutants demonstrated reductions between 33 and 42%. All strains exhibited similar susceptibility to diluted Triton X-100, eliminating it as a potential source of variability (data not shown).
Fig. 3.
Relative adherence of the SLAP mutants to major extracellular matrices, Caco-2 epithelial cells, and mucin. The L. acidophilus NCK1909 parent strain was standardized to 100% (dotted line). The data represent means of independent biological replicates. Error bars are standard error of the means. Asterisks indicate statistical significance calculated using a Student’s t-test (*** p < 0.001, ** p < 0.01, * p < 0.05)
SLAP deletion-induced alterations to murine dendritic cell (DC) cytokine profiles
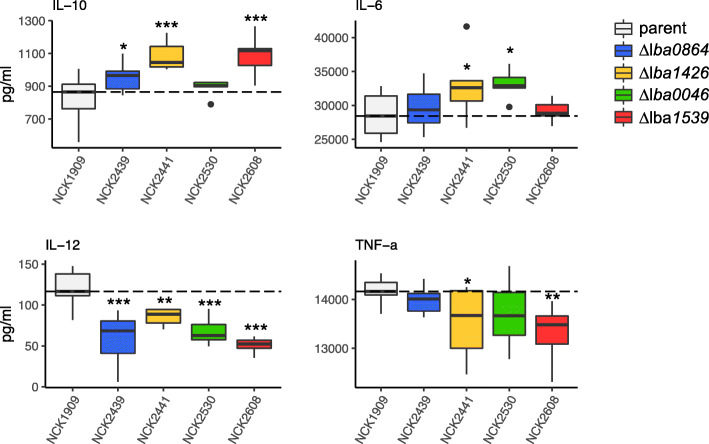
A bacterial/DC co-incubation assay was used to assess the immunomodulatory potential of LBA0046, LBA0864, LBA1426, and LBA1539. Mutant L. acidophilus strains and the parent were exposed to murine DCs for 24 h followed by the measurement of relevant cytokines. To minimize random error, biological replicates were treated as a blocking factor. The block centered data is plotted in Fig. 4. Results indicate that the absence of the four genes produced notably different, predominantly anti-inflammatory, cytokine profiles in comparison to the parent strain. Anti-inflammatory molecule IL-10 was significantly induced by three of the four strains, particularly NCK2441 (Δlba1426) and NCK 2608 (Δlba1539). Likewise, inflammatory molecule IL-12 was repressed by all four strains, while NCK2441 (Δlba1426) and NCK2608 (Δlba1539) reduced TNF-α production. Interestingly, two of the strains, NCK2441 (Δlba1426) and NCK2530 (Δlba0046), slightly but significantly induced pro-inflammatory molecule IL-6.
Fig. 4.
The immunomodulatory profiles of the SLAP mutants compared to the parent strain were evaluated using a murine dendritic cell co-incubation assay. Cytokines IL-10, IL-6, IL-12 and TNF-α, were measured using enzyme-linked immunosorbent assays. Co-incubations were performed in biological triplicate; bars on the box-whisker plots were constructed using block centered data. The dotted line is used to indicate the median of the control strain. Asterisks indicate statistical significance calculated using a Student’s t-test (*** p < 0.001, ** p < 0.01, * p < 0.05)
Transcriptomic analysis of SLAP deletion mutants
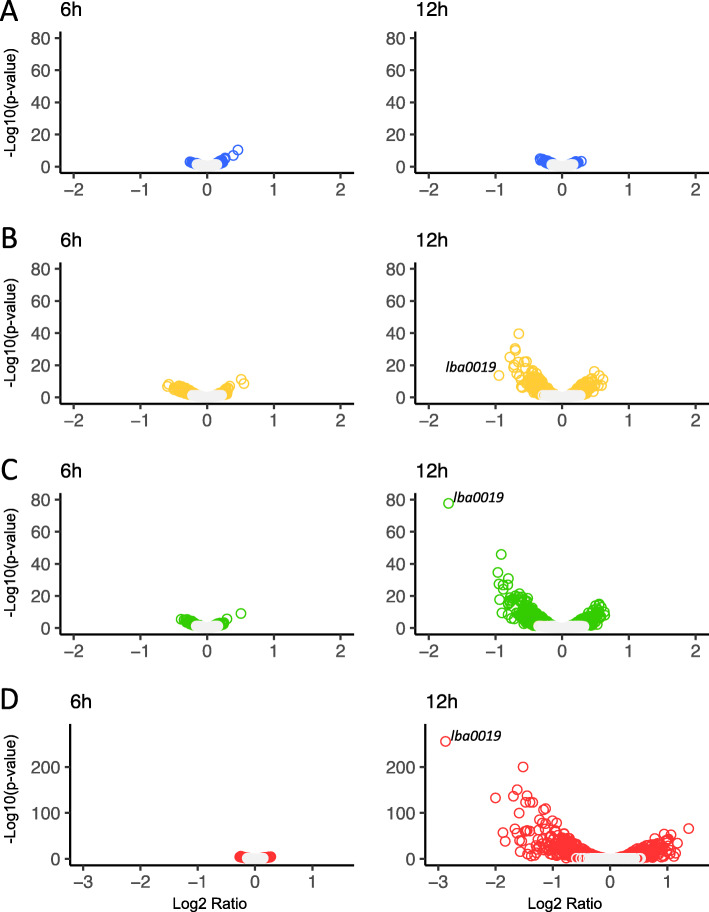
The global transcriptomes of the four SLAP mutant strains relative to the NCK1909 parent were examined in log (6 h) and early stationary (12 h) growth phases. For both conditions, the Log2 ratio was plotted against statistical significance (Fig. 5). The gray circles represent insignificant values, while colored circles are indicative of a p-value < 0.05. At 6 h, all four mutants demonstrated relatively little difference in expression, but by 12 h only the transcriptome of NCK2439 (Δlba0864) still resembled that of the parent strain (Fig. 5A). Although the transcriptomes of the other three mutants appear quite dissimilar, only NCK2530 (Δlba0046) and NCK2608 (Δlba1539) had differentially expressed genes with Log2 ratios > 1 (Fig. 5B-D). Indeed, NCK2608 (Δlba1539) was most influenced by growth phase (Fig. 5D) with 65 differentially expressed genes (Log2 fold change > 1, p-value < 0.0001) which are listed in Table 3 along with their corresponding COGs. Of those genes, 56 were repressed, including six related to cell wall/membrane structure and biogenesis, and 11 predicted to play roles in nucleotide/amino acid transport and metabolism. The vast majority of the remaining genes are poorly characterized or possess unknown functions. Upregulated genes were fewer and encompassed transcription anti-terminator licT, transcriptional regulator lysR, a multiple sugar metabolism (msm) operon regulator, bifunctional protein pyrR, phosphoribosylformylglycinamidine synthase subunit purS and four hypothetical proteins. Remarkably, the most notably downregulated gene, lba0019, was the same for NCK2441, NCK2530, and NCK2608 with Log2 ratios of − 0.9, − 1.7, and − 2.9, respectively. This gene encodes an uncharacterized protein predicted to contain an alpha/beta hydrolase fold and exhibited 50% amino acid identity to several Lactobacillus esterases. It is located in an operon with putative membrane protein, lba0018.
Fig. 5.
Transcriptome profiles of SLAP mutants compared to the parent strain. The transcriptomes of NCK2439 (Δlba0864, A), NCK2441 (Δlba1426, B), NCK2530 (Δlba0046, C) and NCK2608 (Δlba1539, D) relative to the parent strain (NCK1909). Strains were grown in MRS broth for 6 h (left) and 12 h (right). Volcano plots depict Log2 fold change in expression plotted against significance. The gray circles represent insignificant values, while colored circles are indicative of a p-value < 0.05
Table 3.
NCK2608 (Δlba1539) differentially expressed genes in stationary growth phase
| Locus ID | Name | Ratio | COG* |
|---|---|---|---|
| CELLULAR PROCESSES AND SIGNALING | |||
| LBA0833 | ftsW | −1.06 | D |
| LBA1883 | NLP-P60 secreted protein | −2.00 | M |
| LBA1140 | lysin | −1.58 | M |
| LBA0520 | galactosyltransferase | −1.48 | M |
| LBA1743 | cell wall-associated hydrolase | −1.16 | M |
| LBA1744 | glycosidase | −1.13 | M |
| LBA1736 | epsB | −1.09 | M |
| LBA0165 | pepO | −1.40 | O |
| LBA1901 | thioredoxin | −1.38 | O |
| LBA1208 | msrA | −1.25 | O |
| LBA1107 | glutathione reductase | −1.13 | O |
| LBA0096 | htpX | −1.12 | O |
| LBA1659 | response regulator | −1.63 | T |
| LBA0544 | transcriptional regulator | −1.45 | T |
| LBA1132 | ABC transporter component | −1.18 | V |
| LBA0560 | ABC transporter component | −1.07 | V |
| LBA1680 | ABC transporter component | −1.01 | V |
| INFORMATION STORAGE AND PROCESSING | |||
| LBA1519 | pheS | −1.03 | J |
| LBA1899 | transcriptional regulator | −1.44 | K |
| LBA0835 | hypothetical protein | −1.29 | L |
| LBA0797 | radC | −1.04 | L |
| LBA0545 | hypothetical protein | −1.00 | L |
| METABOLISM | |||
| LBA1109 | hypothetical protein | −1.47 | C |
| LBA1220 | pyridine mercuric reductase | −1.45 | C |
| LBA0538 | Na + −H + -exchanging protein | −1.09 | C |
| LBA1896 | asnA | −1.54 | E |
| LBA1961 | oppA | −1.28 | E |
| LBA1177 | iron-sulfur cofactor synthesis | −1.26 | E |
| LBA1045 | ABC transporter component | −1.15 | E |
| LBA1292 | aa transporter | −1.14 | E |
| LBA1658 | prolyl aminopeptidase | −1.04 | E |
| LBA0240 | xanthine phosphoribosyltransferase | −1.40 | F |
| LBA0041 | rtpR | −1.35 | F |
| LBA1631 | deoxyribosyltransferase | −1.24 | F |
| LBA0591 | iunH | −1.18 | F |
| LBA0131 | ribose-p pyrokinase | −1.06 | F |
| LBA0836 | coaD | −1.05 | H |
| LBA0542 | heavy-metal-transporting ATPase | −1.87 | P |
| LBA1771 | ABC transporter component | −1.69 | P |
| LBA0541 | cadA | −1.47 | P |
| LBA0200 | oppB | −1.06 | P |
| POORLY CHARACTERIZED | |||
| LBA0019 | hypothetical protein | −2.87 | S |
| LBA0543 | hypothetical protein | −1.66 | S |
| LBA0493 | aggregation promoting protein | −1.62 | S |
| LBA0834 | hypothetical protein | −1.57 | S |
| LBA1769 | hypothetical protein | −1.52 | S |
| LBA1943 | lipoprotein | −1.40 | S |
| LBA1850 | lysM | −1.23 | S |
| LBA0387 | hypothetical protein | −1.18 | S |
| LBA0208 | hypothetical protein | −1.14 | S |
| LBA1010 | hypothetical protein | −1.03 | S |
| LBA1738 | hflX | −1.01 | S |
| LBA0018 | membrane protein | −1.83 | None |
| LBA1108 | hypothetical protein | −1.59 | None |
| LBA0017 | general stress response | −1.32 | None |
| LBA1221 | hypothetical protein | −1.29 | None |
| LBA0616 | hypothetical protein | 1.01 | M |
| LBA0724 | licT | 1.37 | K |
| LBA1410 | lysR | 1.18 | K |
| LBA1443 | msm operon regulator | 1.06 | K |
| LBA0563 | pyrR | 1.14 | F |
| LBA1558 | purS | 1.11 | F |
| LBA0644 | hypothetical protein | 1.06 | Q |
| LBA0402 | hypothetical protein | 1.02 | None |
| LBA1889 | hypothetical protein | 1.05 | None |
*Clusters of Orthologous Groups (COG) were assigned to significant genes using the EggNOG Database. The categories are as follows: C, Energy production and conversion; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelope biogenesis; O, post-translational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; V, defense mechanisms
Discussion
S-layer associated proteins (SLAPs) of L. acidophilus have been linked to a broad range of functions including some of the critical roles once believed to be the sole responsibility of the S-layer [7, 18]. In the present study, four putative uncharacterized SLAPs, LBA0046, LBA0864, LBA1426, and LBA1539, were selected for functional analysis. The only consistent feature among the target loci was the possession of an N-terminal signal peptide, which denotes transfer across the cytoplasmic membrane via the Sec translocase system, and confirms their extracellular localization [31]. The nucleotide sequences encoding these proteins were searched for in a curated database of 170 genomes [32]. Orthologs were only present in S-layer-forming strains, which is in agreement with previous SLAP studies [19, 32]. Although Lactobacillus species tend to cluster based on lifestyle [26], mapping of SLAP presence added an additional level of granularity. Examined SLAPs were confined to host-adapted Lactobacillus species, while lba0046, lba1426, and lba1539 were unique to vertebrates. Interestingly, no SLAPs were detected in the free-living strains, which represented three newly identified genera (Lentilactobacillus, Levilactobacillus, and Secundilactobacillus). Likewise, their low GC content is consistent with the genomes of host-adapted species which have lost DNA repair genes and undergone subsequent mutational bias toward A and T [26]. The absence of these proteins from free-living Lactobacillus species suggests that SLAPs evolved expressly for the purpose of host adaption.
The genes encoding the four SLAPs of interest were deleted from the L. acidophilus NCFM chromosome via a pORI-based upp counterselective gene replacement system [27]. Our previous proteomic analysis of the L. acidophilus SLAP profile revealed the upregulation of LBA0046, LBA1426, and LBA1539 during the transition from log to stationary growth phase; in fact, LBA0046 and LBA1539 were among the most induced proteins within the dataset [24]. Expression analyses for Δlba0046, Δlba1426, Δlba1539 at 6 h and 12 h supported these findings by only demonstrating differential gene expression during stationary growth phase. These results were most apparent for Δlba1539 which underwent considerable gene repression, most notably for cell wall biogenesis and structural genes, as well as an aggregation promoting factor (apf) protein previously shown to contribute to the survival of L. acidophilus during transit through the digestive tract and predicted to mediate host interactions [33]. Surprisingly, in comparison to the parent, the most significantly downregulated gene, lba0019, was the same for all three strains. This gene likely encodes an esterase, and is co-transcribed with a putative membrane protein. Esterases are a type of hydrolase enzyme which catalyze the cleavage and formation of ester bonds and have wide substrate specificities [34]. Although we were unable to find more information about the specific role of this protein, its orthologs appear to be confined to host-adapted S-layer-forming lactobacilli.
Examination of mutant cellular morphologies via flow cytometry and SEM revealed no obvious differences in comparison to the parent strain. This data, coupled with unaltered growth, even under stressful conditions, suggests that the mutant cell surfaces remained intact. Prior deletion studies of L. acidophilus SLAPs yielded similar results [18, 22], and imply a more direct mechanism for the observed phenotypes, as opposed to SLAPs involved in cell wall maintenance, such as CdpA and IgdA, whose deletion phenotypes were presumed to be pleiotropic responses resulting from disordered cell surfaces [20, 32]. This inferred directness is most apparent for LBA0864, whose deletion had almost no effect on the transcriptome, yet its corresponding mutant strain exhibited significant adherence reductions and immunomodulatory shifts in comparison to the parent. Additionally, LBA0864 was the only examined protein to possess a GW anchor. The GW domain, termed for a conserved Gly-Trp (GW) dipeptide, constitutes a motif for cell-surface anchoring in Listeria and other Gram-positive bacteria [35]. Within Listeria, this domain enables the protein LnlB to re-associate to the bacterial cell, even when added from extracellular medium, and is thought to play a role in cell invasion [36]. These characteristics support the assumption that LBA0864 has a surface localized function likely related to host interaction.
In general, surface proteins of L. acidophilus NCFM are important mediators of adhesion to intestinal epithelial cells, mucus, and extracellular matrices [7, 22, 37, 38]. Among the remaining three mutant strains tested, binding deficiencies were found to be substrate-specific, again consistent with a more direct mechanism of action. In a previous study which mutated the mucus-binding (Mub), fibronectin-binding (FbpA), and SlpA proteins of L. acidophilus, resultant strains established that multiple proteins individually contribute to the organism’s ability to adhere to intestinal cells in vitro [7]. In fact, the authors argue that the severe binding deficiency of the slpA mutant was likely due to the loss of multiple proteins that may have been embedded within the S-layer. Our results support this claim by demonstrating the influence of several uncharacterized surface proteins on adherence to not only Caco-2 intestinal cells, but also specific extracellular matrices and mucin. Bacteria that are only able to adhere to mucus, but unable to associate with the epithelium, may be washed away with degraded mucins, thus adhesion to extracellular matrices is critical to the realization of certain probiotic attributes [2]. Despite the severe reductions in fibronectin and collagen adhesion, examined SLAPs were all devoid of signature binding domains. Nonetheless, when compared to an L. acidophilus mutant with a deleted Type-III fibronectin binding protein [22], our Δlba0046 and Δlba1539 strains demonstrated substantially more dramatic relative reductions in fibronectin adhesion. Although these four SLAPs do not possess any sequence homology, it is possible that they contain novel domains which contribute to the adhesive capacity of L. acidophilus.
For host-adapted bacteria, extracellular features are critical for not only adhesion, but also immunomodulation. The capacity of lactobacilli to variably induce IL-12 and TNF- α, and to a lesser extent, IL-6 and IL-10, may determine which immune response is favored [9]. In comparison to other lactobacilli, L. acidophilus NCFM tends to exhibit a slight proinflammatory profile with a very low IL-10/IL-12 ratio [39] that has been directly linked to S-layer presence [17]. The deletion mutants in this study were associated with less inflammatory phenotypes based on either the significant induction of IL-10, reduction of IL-12, or both. This was most apparent with NCK2608 (Δlba1539), which also triggered a significant drop in TNF-α. Unexpected was the slight upregulation of pro-inflammatory molecule IL-6 by Δlba0046 and Δlba1426, results which could be attributed to low-level induction of auxiliary surface molecules, though statistical significance was much lower in comparison to other evaluated cytokines. Overall, our results suggest that SLAPs contribute to the slight proinflammatory profile of L. acidophilus, a characteristic that was once attributed to the S-layer.
Conclusions
In summary, the data presented in this study collectively links several previously uncharacterized extracellular proteins to underlying mechanisms contributing to L. acidophilus probiotic activity. Initial in silico findings revealed high conservation of these genes among host-adapted S-layer forming lactobacilli and suggest a probable host-related function. Complementary in vitro assay results supported this claim, particularly in regards to host adhesion and immune stimulation, both critical for probiotic functionality. Mutant transcriptome profiles, although not exceptionally divergent from the parent, did highlight a similarly repressed putative esterase whose future functional characterization may indirectly broaden our understanding of the SLAP gene subset. Overall, deciphering the complex adaptative features that promote both survival and persistence of probiotics in the host intestines is essential for elucidating and augmenting their health benefits. Our results demonstrate both the importance of SLAPs in host adaptation and add to the ever-expanding catalog of probiotic-relevant molecules.
Methods
Sequence analysis and phylogenetic tree mapping
Genetic context was examined by extracting previously annotated sequences encoding LBA0046, LBA0864, LBA1426, LBA1539 and flanking regions from the L. acidophilus NCFM genome (NC_006814) using Geneious software [40]. Protein domains were detected and assigned using UniProt and InterPro databases [41, 42]. Phylogenetic mapping was conducted to gain insight into the presence of these loci within a database of 170 genomes [5] updated to take into account the recently published Lactobacillus genus reclassifications [25]. Orthologs were identified and extracted via Geneious annotation and extraction workflows [40], then imported into CLC Genomics Workbench (Qiagen). S-layer presence was determined using the UniProt annotation tool suite to search for proteomes present within the UniProt database, as previously described [32]. A phylogenetic tree containing only S-layer-forming bacteria was constructed using a published method based on the nucleotide sequence of the Pyk enzyme [5, 43]. SLAP orthologs and Duar et al. [26] species lifestyles were mapped to the tree using the CLC Genomics metadata feature, however L. acidophilus NCFM was re-classified as vertebrate-adapted [32].
Bacterial strains and growth conditions
The bacteria and plasmids used in this study are listed in Table 1, while Tables 2 and 3 details the PCR primers required for generating the mutant strains. Escherichia coli EC101 acted as a host for cloning the deletion constructs for all genes except lba1539, for which NCK1391 (E. coli DH10B harboring pTRK669) was substituted. E. coli EC101 was propagated in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) with aeration at 37 °C in the presence of 40 μg/ml kanamycin (Sigma-Aldrich, St. Louis, MO, USA), while NCK1391 was grown in Luria-Bertani (LB) broth (Difco) with aeration at 32 °C in the presence of 15 μg/ml chloramphenicol. Recombinant E. coli cells containing pTRK935-based plasmids were selected using 150 μg/ml erythromycin. Propagation and selection of Lactobacillus strains was performed as previously described [32]. For growth curve analyses, overnight bacterial cultures were used to inoculate 96-well microplates (Corning Costar, Corning, NY) containing MRS broth or MRS broth supplemented with 2.5% (w/v) NaCl (Fisher Scientific, Hampton, NH, USA), 0.2% (w/v) porcine bile (Sigma) or 0.5% (w/v) oxgall (Difco). Plates were sealed with clear adhesive film then incubated at 37 °C in a Fluostar Optima microplate reader (BMG Labtech, Cary, NC). The OD600 was measured every hour for 30 h.
Table 2.
PCR primers used in this study
| Primer Name | Sequence* |
|---|---|
| Construction of deletion mutants | |
| 0046HindIIIF | GATCTAAAGCTTGCTCAACATTATTAACGGTTC |
| 0046R | TTCTAACATAATGAATACCTCGTA |
| 0046Soe | AGGTATTCATTATGTTAGAAGACTAATCTAGATCAAGATTCATCA |
| 0046SacIR | GATCTAGAGCTCCATACTACTTCTGCGTCTTC |
| 0864HindIIIF | GATCTAAAGCTTCTGCTGATATTGATGCAGTGAGTGG |
| 0864R | CGCACCGGCAATAACTATTCCCTTAAT |
| 0864Soe | AAGGGAATAGTTATTGCCGGTGCGCGTGCAGAATTAACTCAAGGTCGC |
| 0864SacIR | GATCTAGAGCTCCGTGCACTTGACACAGATCCTG |
| 1426HindIIIF | GATCTAAAGCTTTATAGATTAATTGACTGCAGCC |
| 1426R | CGCTGCCATTGAAGTAATTA |
| 1426Soe | TAATTACTTCAATGGCAGCGAACTAATCTATTAATGAAGAAACTCGT |
| 1426SacIR | GATCTAGAGCTCCTTATCGTTCATGCCAAGAA |
| 1539BamHIF | GATCTAGGATCCATCACTTGATCGATCATCTG |
| 1539R | CTTCATCTGAATATCTCCTCT |
| 1539Soe | AGAGGAGATATTCAGATGAAGTTGATAAAATAATCTACTACTTTGTGA |
| 1539SacIR | GATCTAGAGCTCCTCTTAGGTGCAAGCATTAA |
| PCR analysis and DNA sequencing of deletion targets | |
| 0046up | CTATCTGTATGATGCTTCCAC |
| 0046dw | GTACCTCAATCTGTTGTAATCTC |
| 0864up | ACAAGCTAGAGGTATGGCTGG |
| 0864dw | CCACATGAATGGCGTATGGC |
| 1426up | AAGCCGTTGTATTGAATGATGGTAG |
| 1426dw | CGCGAATCATCAATTCACGGTA |
| 1539up | CAGGATAGGGATGCACATGC |
| 1539dw | CGACGTTGACGTGTTACTGT |
*The primers, the 5′-to-3′ sequences are given and restriction enzyme sites are underlined
Chromosomal deletion of SLAP genes targets
DNA manipulations and transformation was performed as previously described [32]. The SLAPs encoded by lba0046, lba0864, lba1426, and lba1539 were deleted from the L. acidophilus NCFM chromosome via a pORI-based upp counterselective gene replacement system [27]. Briefly, in-frame deletions were created by PCR amplifying sequences upstream and downstream of the deletion targets (Table 2). Subsequent purified products were joined by splicing using overlap extension PCR (SOE-PCR), then amplified and cloned into the pTRK935 integration vector. Recombinant plasmids were transformed into E. coli and eventually electroporated into L. acidophilus NCK1910 (Table 1). Recovery of single- and double-crossover recombinants was performed as previously described [27]. Gene deletions were confirmed by sequencing the entirety of both flanking regions. For all subsequent phenotypic assays, NCK1909 (Δupp) served as the control strain (Table 1).
Examination of mutant cellular morphologies
Flow cytometry was used to examine changes to cell size and granularity resulting from SLAP gene deletions. Mutant and parent strains were grown to log (6 h) and early stationary (12 h) growth phases in MRS broth. Cells were centrifuged at 3220×g for 10 min, then washed and resuspended in phosphate buffered saline (PBS, pH 7.4, Thermo Fisher Scientific, Waltham, MA, USA). Data was acquired using a CytoFLEX Flow Cytometer instrument (Beckman Coulter, Brea, CA, USA) and analyzed with CytExpert software (Beckman Coulter). Changes in morphology were also visualized with SEM. Log (6 h) and early stationary phase (12 h) cells, cultured in MRS broth, were fixed in a solution of 3% glutaraldehyde in 0.1 M sodium cacodylate (pH 5.5) and stored at 4 °C. Fixed cells were processed by the CALS Center for Electron Microscopy (CEM) at North Carolina State University. Images were acquired with a JEOL JEM-5900LV SEM (JEOL USA, Peabody, MA) at 15 kV.
Adhesion to mucin, extracellular matrices (ECMs), and Caco-2 intestinal cells
Binding assays were performed as previously described [32]. Adhesion substrates consisted of Mucin (Type III from porcine stomach, Sigma), fibronectin (from human plasma, Sigma), collagen (type IV from human cell culture, Sigma), and laminin (from Engelbreth-Holm-Swarm murine sarcoma/basement membrane; Sigma), as well as Caco-2 intestinal cells purchased from the American Type Culture Collection. Relative adherence percentages were calculated by standardizing the parent strain adherence to 100%. Mucin and ECM adhesion assays were performed in biological triplicate with four technical replicates, while Caco-2 adhesion assays were performed a minimum of three times with two technical replicates. Statistical significance was determined using a Student’s t-test.
Bacteria/DC co-incubation and cytokine quantification
Bone marrow-derived C57BL/6 murine immature dendritic cells (DCs) were purchased from Astarte-Biologics (Bothell, WA) and preserved in liquid nitrogen. Bacterial co-incubation assays were performed as previously described [32]. Cytokine measurements for tumor necrosis factor α (TNF-α) and interleukins IL-6, IL-10 and IL-12 were obtained using Single-Analyte ELISArray kits (Qiagen) as per manufacturer’s instructions. Assays were performed in biological triplicate with two technical replicates. To reduce random error, replicates were treated as a blocking factor. Significance of block centered data was analyzed using a Student’s t-test.
RNA extraction, sequencing, and transcriptional analysis
Total RNA was isolated from the L. acidophilus parent strain and SLAP-deficient mutants propagated in MRS broth for 6 and 12 h. Cells were grown statically under ambient atmospheric conditions, pelleted by centrifugation (3220×g, 5 min, RT), then flash frozen and stored at − 80 °C. RNA isolation and sequencing was performed as previously described by Klotz et al., [32]. Geneious software [40] was used to filter and map reads to the L. acidophilus NCFM reference genome and differential expression levels were calculated using the DESeq2 package [28]. Clusters of Orthologous Groups (COGs) were assigned using EggNOG 5.0 [44]. Transcriptomic datasets generated in this study are available in the National Center for Biotechnology database under BioProject ID PRJNA576881.
Supplementary information
Additional file 1 Supplementary Fig. 1. SLAP deletions visualized using PCR. Confirmation of the S-layer associated protein gene deletions from the L. acidophilus NCFM chromosome using primers that flanked the deletion region. P, NCK1909 parent strain, Δ, SLAP deletion strain.
Additional file 2 Supplementary Fig. 2. Cellular morphologies of the parent strain and SLAP mutants. Cellular morphologies of the parent strain and SLAP mutants in logarithmic (6 h, A) and early stationary (12 h, B) growth phase visualized using flow cytometry and scanning electron microscopy.
Additional file 3 Supplementary Table 1. List of strains used, Zheng et al., reclassification, lifestyles, and isolation sources.
Acknowledgements
We would like to thank Valerie Lapham of the CALS Center for Electron Microscopy, NC State University, for her electron microscopy technical expertise. We would also like to thank Rosemary Sanozky-Dawes and Meichen Pan for technical advice and assistance preparing Caco-2 epithelial cells for in vitro adherence studies.
Abbreviations
- GOG
Clusters of Orthologous Groups
- DC
Dendritic cell
- ECM
Extracellular matrix
- LAB
Lactic acid bacteria
- SEM
Scanning electron microscopy
- S-layer
Surface-layer
- SLP
S-layer protein
- SLAP
S-layer associated protein
Authors’ contributions
CK, YJG, SOF, and RB designed the study; CK carried out the work, analyzed the results, and prepared the manuscript under the advisement of RB, YJG, and SOF. All authors have read and approved the manuscript.
Funding
This research was funded, by the North Carolina Agricultural Foundation and DuPont Nutrition & Health. Funding organizations had no input in experimental design and execution.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available at the National Center for Biotechnology database under BioProject ID PRJNA576881 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA576881).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01908-2.
References
- 1.Hynonen U, Palva A. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol. 2013;97(12):5225–5243. doi: 10.1007/s00253-013-4962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou MM, Siezen RJ, Bron PA. The extracellular biology of the lactobacilli. FEMS Microbiol Rev. 2010;34(2):199–230. doi: 10.1111/j.1574-6976.2009.00208.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnson BR, Klaenhammer TR. Impact of genomics on the field of probiotic research: historical perspectives to modern paradigms. Antonie Van Leeuwenhoek. 2014;106(1):141–156. doi: 10.1007/s10482-014-0171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. The international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastro Hepat. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.O’Flaherty S, Crawley A, Theriot CM, Barrangou R: The Lactobacillus Bile Salt Hydrolase Repertoire Reveals Niche-Specific Adaptation mSphere 2018, 3(3):e00140–00118. [DOI] [PMC free article] [PubMed]
- 6.Pfeiler EA, Azcarate-Peril MA, Klaenhammer TR. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J Bacteriol. 2007;189(13):4624–4634. doi: 10.1128/JB.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck BL, Altermann E, Svingerud T, Klaenhammer TR. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2005;71(12):8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celebioglu HU, Olesen SV, Prehn K, Lahtinen SJ, Brix S, Abou Hachem M, Svensson B. Mucin- and carbohydrate-stimulated adhesion and subproteome changes of the probiotic bacterium Lactobacillus acidophilus NCFM. J Proteome. 2017;163:102–110. doi: 10.1016/j.jprot.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2002;168(1):171–178. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, et al. S layer protein a of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A. 2008;105(49):19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebeer S, Vanderleyden J, De Keersmaecker SC: Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 2008, 72(4):728–764, Table of Contents. [DOI] [PMC free article] [PubMed]
- 12.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8(3):171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 13.Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12(3):211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 14.Sleytr UB, Beveridge TJ. Bacterial S-layers. Trends Microbiol. 1999;7(6):253–260. doi: 10.1016/S0966-842X(99)01513-9. [DOI] [PubMed] [Google Scholar]
- 15.Antikainen J, Anton L, Sillanpaa J, Korhonen TK. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol. 2002;46(2):381–394. doi: 10.1046/j.1365-2958.2002.03180.x. [DOI] [PubMed] [Google Scholar]
- 16.Avall-Jaaskelainen S, Lindholm A, Palva A. Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl Environ Microbiol. 2003;69(4):2230–2236. doi: 10.1128/AEM.69.4.2230-2236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki S, Yokota K, Igimi S, Kajikawa A. Comparative analysis of immunological properties of S-layer proteins isolated from Lactobacillus strains. Microbiology. 2019. [DOI] [PubMed]
- 18.Johnson B, Selle K, O'Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiol-Sgm. 2013;159:2269–2282. doi: 10.1099/mic.0.070755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson BR, Hymes J, Sanozky-Dawes R, Henriksen ED, Barrangou R, Klaenhammer TR. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl Environ Microbiol. 2015;82(1):134–145. doi: 10.1128/AEM.01968-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altermann E, Buck LB, Cano R, Klaenhammer TR. Identification and phenotypic characterization of the cell-division protein CdpA. Gene. 2004;342(1):189–197. doi: 10.1016/j.gene.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Johnson BR, Klaenhammer TR. AcmB is an S-layer-associated beta-N-Acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2016;82(18):5687–5697. doi: 10.1128/AEM.02025-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hymes JP, Johnson BR, Barrangou R, Klaenhammer TR. Functional analysis of an S-layer-associated Fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2016;82(9):2676–2685. doi: 10.1128/AEM.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson BR, O'Flaherty S, Goh YJ, Carroll I, Barrangou R, Klaenhammer TR. The S-layer associated serine protease homolog PrtX impacts cell surface-mediated microbe-host interactions of Lactobacillus acidophilus NCFM. Front Microbiol. 2017;8:1185. doi: 10.3389/fmicb.2017.01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klotz C, O'Flaherty S, Goh YJ, Barrangou R. Investigating the effect of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front Microbiol. 2017;8:2174. doi: 10.3389/fmicb.2017.02174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter J, et al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol. 2020;70(4):2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 26.Duar RM, Lin XXB, Zheng JS, Martino ME, Grenier T, Perez-Munoz ME, Leulier F, Ganzle M, Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017;41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 27.Goh YJ, Azcarate-Peril MA, O'Flaherty S, Durmaz E, Valence F, Jardin J, Lortal S, Klaenhammer TR. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2009;75(10):3093–3105. doi: 10.1128/AEM.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders ME, Klaenhammer TR. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J Dairy Sci. 2001;84(2):319–331. doi: 10.3168/jds.S0022-0302(01)74481-5. [DOI] [PubMed] [Google Scholar]
- 29.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177(24):7011–7018. doi: 10.1128/JB.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell WM, Klaenhammer TR. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl Environ Microb. 2001;67(9):4361–4364. doi: 10.1128/AEM.67.9.4361-4364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natale P, Bruser T, Driessen AJM. Sec- and tat-mediated protein secretion across the bacterial cytoplasmic membrane - distinct translocases and mechanisms. Bba-Biomembranes. 2008;1778(9):1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Klotz C, Goh Y, O’Flaherty S, Johnson B, Barrangou R. Deletion of S-layer associated Ig-like domain protein disrupts the Lactobacillus acidophilus cell surface. Front Microbiol. 2020;11:345. doi: 10.3389/fmicb.2020.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh YJ, Klaenhammer TR. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2010;76(15):5005–5012. doi: 10.1128/AEM.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bornscheuer UT. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev. 2002;26(1):73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- 35.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25(2):285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 36.Bierne H, Cossart P. InIB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. J Cell Sci. 2002;115(17):3357–3367. doi: 10.1242/jcs.115.17.3357. [DOI] [PubMed] [Google Scholar]
- 37.Call EK, Goh YJ, Selle K, Klaenhammer TR, O'Flaherty S. Sortase-deficient lactobacilli: effect on immunomodulation and gut retention. Microbiology. 2015;161(Pt 2):311–321. doi: 10.1099/mic.0.000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Flaherty SJ, Klaenhammer TR. Functional and phenotypic characterization of a protein from Lactobacillus acidophilus involved in cell morphology, stress tolerance and adherence to intestinal cells. Microbiology. 2010;156(Pt 11):3360–3367. doi: 10.1099/mic.0.043158-0. [DOI] [PubMed] [Google Scholar]
- 39.Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, Dewulf J, Brassart D, Mercenier A, Pot B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol. 2007;13(2):236–243. doi: 10.3748/wjg.v13.i2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47(D1):D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The UniProt C UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandt K, Barrangou R: Phylogenetic Analysis of the Bifidobacterium Genus Using Glycolysis Enzyme Sequences. Front Microbiol 2016, 7(657). [DOI] [PMC free article] [PubMed]
- 44.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44(D1):D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Supplementary Fig. 1. SLAP deletions visualized using PCR. Confirmation of the S-layer associated protein gene deletions from the L. acidophilus NCFM chromosome using primers that flanked the deletion region. P, NCK1909 parent strain, Δ, SLAP deletion strain.
Additional file 2 Supplementary Fig. 2. Cellular morphologies of the parent strain and SLAP mutants. Cellular morphologies of the parent strain and SLAP mutants in logarithmic (6 h, A) and early stationary (12 h, B) growth phase visualized using flow cytometry and scanning electron microscopy.
Additional file 3 Supplementary Table 1. List of strains used, Zheng et al., reclassification, lifestyles, and isolation sources.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available at the National Center for Biotechnology database under BioProject ID PRJNA576881 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA576881).