Abstract
Introduction
Patients with advanced non-small cell lung cancer (NSCLC) benefit from treatment with immune checkpoint inhibitors (ICIs). Biomarkers such as programmed death-ligand 1 (PD-L1), the tumor mutational burden (TMB) and the mismatch repair (MMR) status are used to predict the prognosis of ICIs therapy. Nevertheless, novel biomarkers need to be further investigated, and a systematic prognostic model is needed for the evaluation of the survival risks of ICIs treatment.
Methods
A cohort of 240 patients who received ICIs from the cBioPortal for Cancer Genomics was evaluated in this research. Clinical information and targeted sequencing data were acquired for analyses. The Kaplan-Meier plot method was used to perform survival analyses, and selected variables were then confirmed by a novel nomogram constructed by the “rms” package of R software.
Results
Seven percent of the NSCLC patients harbored ARID1A mutations, while 4% of the NSCLC patients harbored ARID1B mutations. Mutations in ARID1A and ARID1B were confirmed to be associated with sensitivity to ICIs. Patients harboring these mutations were found to have a better response to treatment (ARID1A: P = 0.045; ARID1B: P = 0.034) and prolonged progression-free survival (ARID1B: P = 0.032). Here, a novel nomogram was constructed to predict the prognosis of ICIs treatment. Elevation of the TMB, enhanced expression of PD-L1 and activation of the antigen presentation process and cellular immunity were found to be correlated with ARID1A and ARID1B mutations.
Conclusion
ARID1A and ARID1B could serve as novel biomarkers for the prognosis and sensitivity to ICIs of advanced NSCLC.
Keywords: ARID1A, ARID1B, NSCLC, Immune checkpoint inhibitors, Prognosis
Introduction
Immune checkpoint inhibitors (ICIs) for cancer treatment have proven to be a great breakthrough in the past decade. In non-small cell lung cancer (NSCLC), ICIs have achieved convincing efficacy and tolerable safety in single-agent therapies or combined treatments and significantly prolonged the overall survival (OS) of patients (Leighl et al. 2019; Reck et al. 2019; Mok et al. 2019; Ready et al. 2019; Horn et al. 2017). During trials evaluating ICIs, several biomarkers have been found to be related to sensitivity to ICIs treatment, including high programmed death-ligand 1 (PD-L1) expression (Mok et al. 2019; Ready et al. 2019; Ott et al. 2019), a high tumor mutational burden (TMB) (Ready et al. 2019; Ott et al. 2019) and mismatch repair (MMR) deficiency (Mandal et al. 2019). In contrast, NSCLC patients harboring mutations of epidermal growth factor receptor (EGFR) and/or anaplastic lymphoma kinase (ALK) might not benefit from ICIs treatment (Gainor et al. 2016; Haratani et al. 2017). However, the reliability of these biomarkers in predicting the prognosis of ICIs treatment in NSCLC, especially the progression-free survival (PFS) of NSCLC patients, remains unclear, and at the same time, multivariate analyses of the sensitivity to ICIs involving the biomarkers described above and the genomic signatures confirmation of patients are necessary because of the heterogeneity and high somatic mutation rate of NSCLC cells. A hazard model for the prognosis of ICIs treatment is urgently needed.
The roles of switch/sucrose nonfermenting (SWI/SNF) chromatin remodeling complexes in a variety of biological processes during cell growth and development, including DNA replication, gene expression and cell differentiation, are essential (Wang et al. 2004; Zhang et al. 2014). These complexes are dysregulated in various cancer types (Huang et al. 2015). Canonical BRG1/BRM-associated factor (BAF), which is one of the three assembled SWI/SNF chromatin remodeling complexes (Naito et al. 2019), mainly consists of AT-rich interactive domain 1A/1B (ARID1A/1B) and DPF2 subunits (Michel et al. 2018; Mashtalir et al. 2018). According to the latest studies, ARID1A deficiency may be a novel biomarker for cancer immunotherapy (Shen et al. 2018; Jiang et al. 2020), and patients with ARID1A deficiency could benefit from ICIs treatment (Okamura et al. 2020). Nevertheless, the role of ARID1B, which serves as the other subunit of ARID1 together with ARID1A, in the sensitivity to ICIs treatment is unknown. Therefore, we performed this study of a previously published cohort of patients (Rizvi et al. 2018) to clarify the role of the ARID1 subunits in the prognosis of ICIs treatment among advanced NSCLC patients and the relationships between the ARID1 subunits with the tumor immune microenvironment (TIME) or other factors related to ICIs sensitivity and to establish a prognostic model for ICIs treatment of advanced NSCLC.
Methods
Study patients
Using the data derived from the cBioPortal for Cancer Genomics, 240 advanced NSCLC patients who received ICIs treatment and were described in previously published work (Rizvi et al. 2018) were involved in this research. The involved patients were treated with either anti-PD-(L)1 therapy (pembrolizumab) alone or in combination with anti-cytotoxic T-cell lymphocyte-4 (anti-CTLA-4) therapy (pembrolizumab+ipilimumab). Basic clinical information was collected from this cohort of patients, including age, sex and smoking history. The investigators used Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 to assess the efficacy of ICIs treatment, and PFS was assessed as the time that patients remained responsive to treatment (exhibited a complete response [CR], a partial response [PR] or stable disease [SD]). All involved patients gave their consent for the examination of targeted sequencing data, and tumor tissue samples from 86 patients were sent for PD-L1 expression assessment.
Survival analyses and nomogram construction
The Kaplan-Meier plot (KM plot) method was used for univariate analyses of the involved patients, and the log-rank tests were used to detect significant differences. In addition, an online tool was used to perform survival analyses for all NSCLC patients (Gyorffy et al. 2013). Variables selected by the univariate analyses were used for the construction of a nomogram. The “rms” package of R software version 3.1.2 (The R Foundation for Statistical Computing, Vienna, Austria) was used to construct the nomogram. Harrell’s C-indexes ranging from 0.5 (no discrimination) to 1 (perfect discrimination) were used for the verification of discrimination (Bandos et al. 2009), and a visual calibration plot was used for the verification of calibration (Jin et al. 2017). Bootstrap analyses with 1000 resamples were used for these analyses.
Bioinformatic and statistical analyses
TMB assessment was conducted during the published clinical trial (Rizvi et al. 2018), and the exploration of the relationships between ARID1 subunits and immunocytes derived from the TIME was carried out with an online tool (Ru et al. 2019). All statistical analyses were conducted by GraphPad Prism 8.0 software (GraphPad, La Jolla, CA), and Student’s t tests were used to determine statistical significance. P values were determined by two-tailed tests, and P < 0.05 was considered statistically significant.
Results
The prevalence of mutant ARID1 and its role in the prognosis of NSCLC
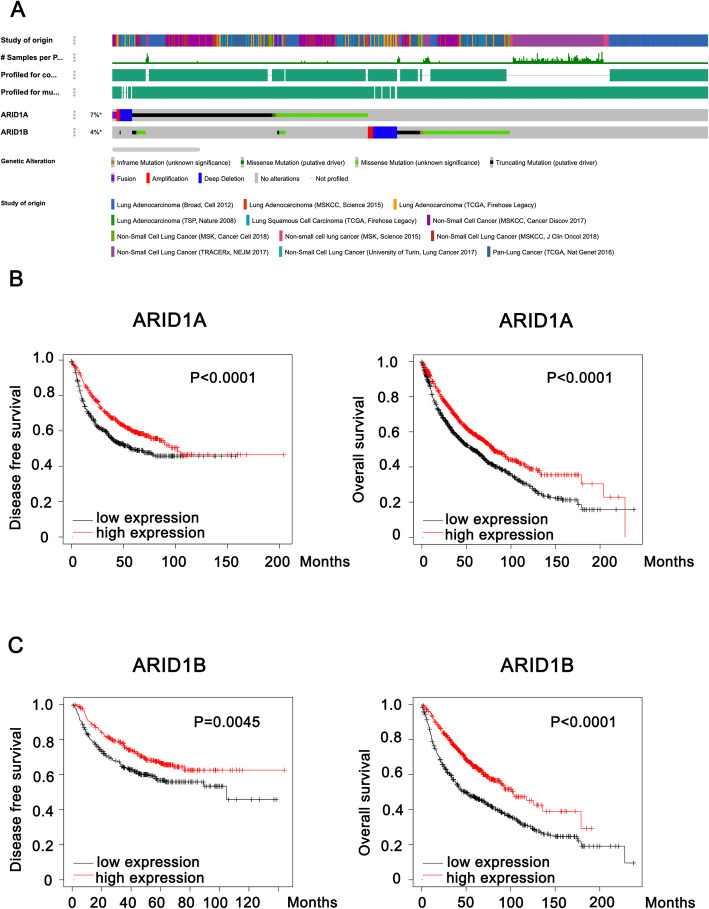
According to the datasets acquired from the cBioPortal for Cancer Genomics, ARID1 mutation is common among NSCLC patients. As shown in Fig. 1a, the mutation frequencies of the subunits of ARID1 including ARID1A and ARID1B were 7 and 4% in NSCLC patients, respectively. As far as we are concerned, gene mutations or hypermethylation lead to low ARID1 protein expression (Guan et al. 2011; Zhang et al. 2013), therefore, we further investigated the relationship between the survival of NSCLC patients and the expression of the ARID1 protein. Figure 1b and c describe the relationships between the disease-free survival (DFS) or OS of NSCLC patients and the expression of ARID1A or ARID1B. As shown in the figure, ARID1A and ARID1B both are convincing biomarkers for NSCLC prognosis with compelling efficiency, and ARID1A or ARID1B deficiency was significantly related to the poor prognosis of NSCLC (ARID1A [DFS: P < 0.0001; OS: P < 0.0001]; ARID1B [DFS: P = 0.0045; OS: P < 0.0001]).
Fig. 1.
ARID1 subunits are tightly associated with the prognosis of NSCLC. a. The prevalence of ARID1 subunits mutations in NSCLC patients according to the cBioPortal for Cancer Genomics; b. The relationship between ARID1A expression and the prognosis of NSCLC patients; c. The relationship between ARID1B expression and the prognosis of NSCLC patients
ARID1A or ARID1B mutation correlates with an improved outcome for ICIs treatment
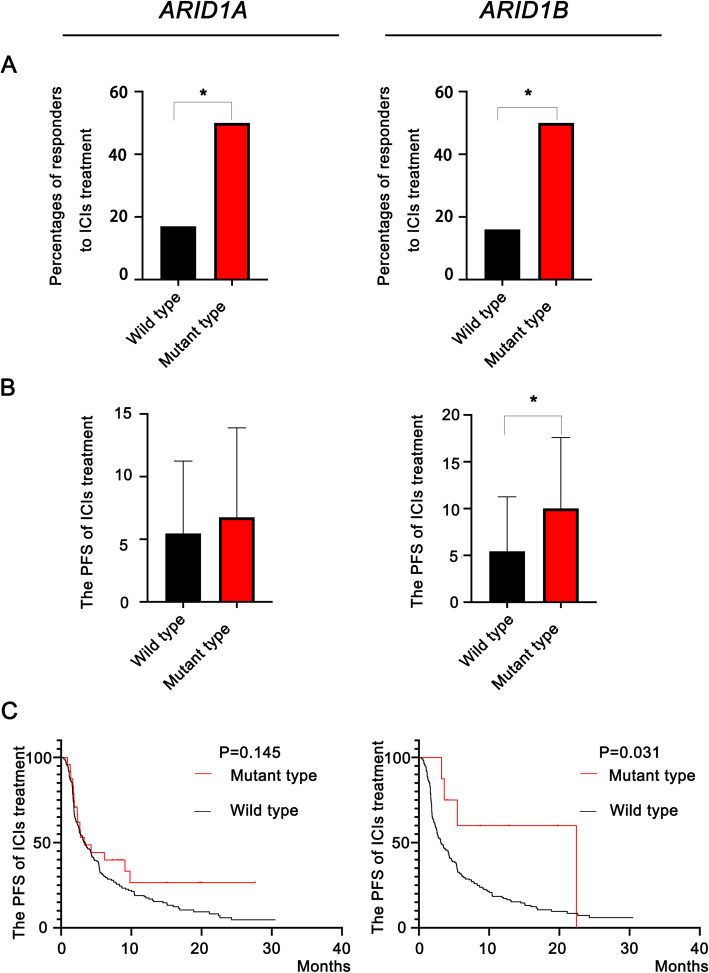
The relationship between ARID1A or ARID1B mutation and the outcome of ICIs treatment was then studied. Through systematic analyses of the datasets from the cBioPortal for Cancer Genomics, we found that both ARID1A and ARID1B mutations were associated with an improved outcome for ICIs treatment in advanced NSCLC patients. As shown in Fig. 2a, more responders (CR + PR + SD) were confirmed in the mutant-type (MT) group than in the wild-type (WT) group for patients harboring ARID1A (50% versus 19%, P = 0.045) or ARID1B (50% versus 16%, P = 0.034) mutations. Figure 2b displays the median PFS (mPFS) with ICIs treatment for the two groups. Patients harboring ARID1A (6.8 months versus 5.5 months, P = 0.313) or ARID1B (10.0 months versus 5.4 months, P = 0.032) mutations benefited more from treatment and achieved a longer PFS time than those in the WT group. Survival analyses for ICIs treatment were then performed as shown in Fig. 2c. Compared with those in the WT group, patients harboring mutant ARID1B achieved significant survival benefits with treatment (P = 0.031). However, although a trend toward a difference existed for the survival curve of patients harboring mutant ARID1A, no statistical significance was found (P = 0.145).
Fig. 2.
Mutations in ARID1 subunits predict an improved prognosis for cancer immunotherapy. a The different percentages of patients who responded (complete response+partial response+stable disease) to immune checkpoint inhibitors (ICIs) grouped by the genomic signatures of ARID1 subunits; b. The different progression-free survival (PFS) times grouped by the genomic signatures of ARID1 subunits; c. The survival curves for ICIs-treated patients based on the genomic signatures of ARID1 subunits. (*: P<0.05.)
Establishment of a prognostic nomogram for the prognosis of ICIs treatment in NSCLC
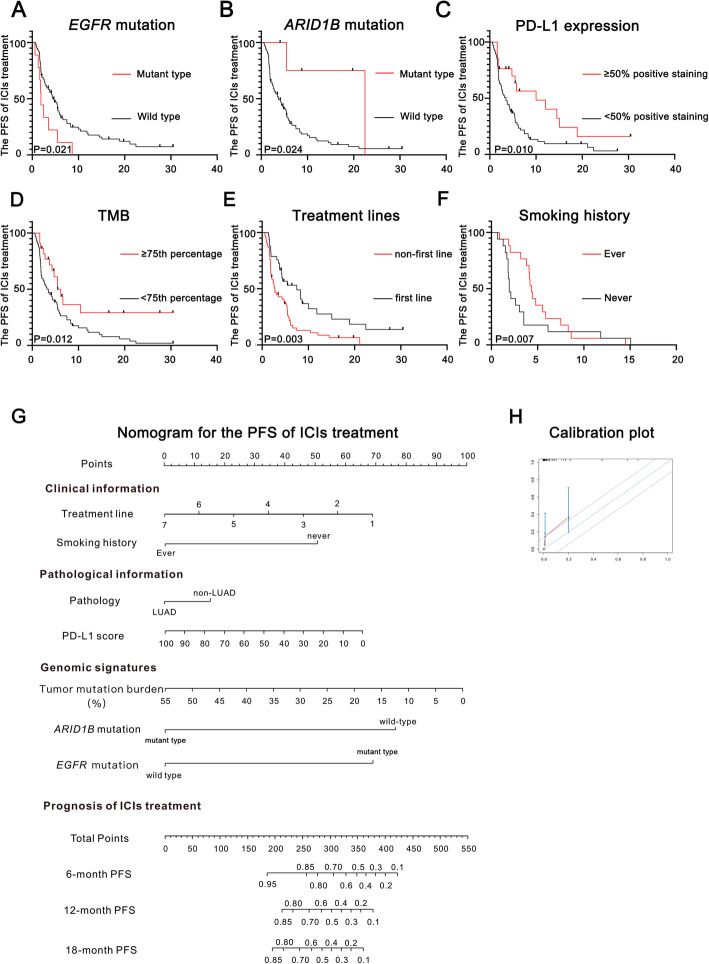
Eighty-six patients of the selected NSCLC cohort with integrated information on clinical features, targeted sequencing and PD-L1 expression evaluated by immunohistochemistry (IHC) were involved in the construction of the novel nomogram. First, univariate analyses were performed to identify variables to include in nomogram construction. As shown in Fig. 3a to f, multiple variables were confirmed to be significantly associated with the prognosis of ICI treatment, including EGFR mutation (P = 0.021), ARID1B mutation (P = 0.024), PD-L1 expression (P = 0.010), TMB (P = 0.012), treatment lines (P = 0.003) and smoking history (P = 0.007). Through the univariate analyses, we found that patients with mutant ARID1B, elevated PD-L1 expression (≥50% percentage positive staining), a high TMB value (≥75th percentage) or a history of smoking could benefit from ICIs treatment, while patients harboring mutant EGFR might not derive survival benefits from ICIs treatment. In addition, first-line administration of ICIs in advanced NSCLC patients might be a better choice than later administration. The nomogram based on these variables was then established as shown in Fig. 3g. In total, 3 types of patient information, including the clinical information, pathological information and genomic signatures, were included in the nomogram. Through this novel nomogram, physicians could easily obtain a score based on the Cox regression model for each variable listed in the graph, and then the total number would be assessed as the sum of all variable scores. Therefore, the survival risks of ICIs treatment for advanced NSCLC patients could be quantified before treatment. The C-index for this prognostic model was 0.71, which suggests that the model has a relatively robust ability to predict the PFS of advanced NSCLC patients treated with ICIs. The calibration plots shown in Fig. 3h indicated that the probabilities of our prognostic model agreed with the accuracy probabilities on acceptable scales (the dashed lines in the calibration plots correspond to a 10% margin of error).
Fig. 3.
Novel nomogram to predict the prognosis of immune checkpoint inhibitor (ICI) treatment. a The survival curves for ICIs-treated patients based on the EGFR mutational status; b. The survival curves for ICIs-treated patients based on the ARID1B mutational status; c. The survival curves for ICIs-treated patients based on PD-L1 expression; d. The survival curves for ICIs-treated patients based on the tumor mutational burden (TMB); e. The survival curves for ICIs-treated patients based on treatment lines; f. The survival curves for ICIs-treated patients based on smoking history; g. The novel nomogram based on patient information to predict the prognosis of ICI treatment; h. The calibration plot for the nomogram
The mutation status of ARID1A or ARID1B is associated with the TMB level, PD-L1 expression and the TIME modulation of NSCLC
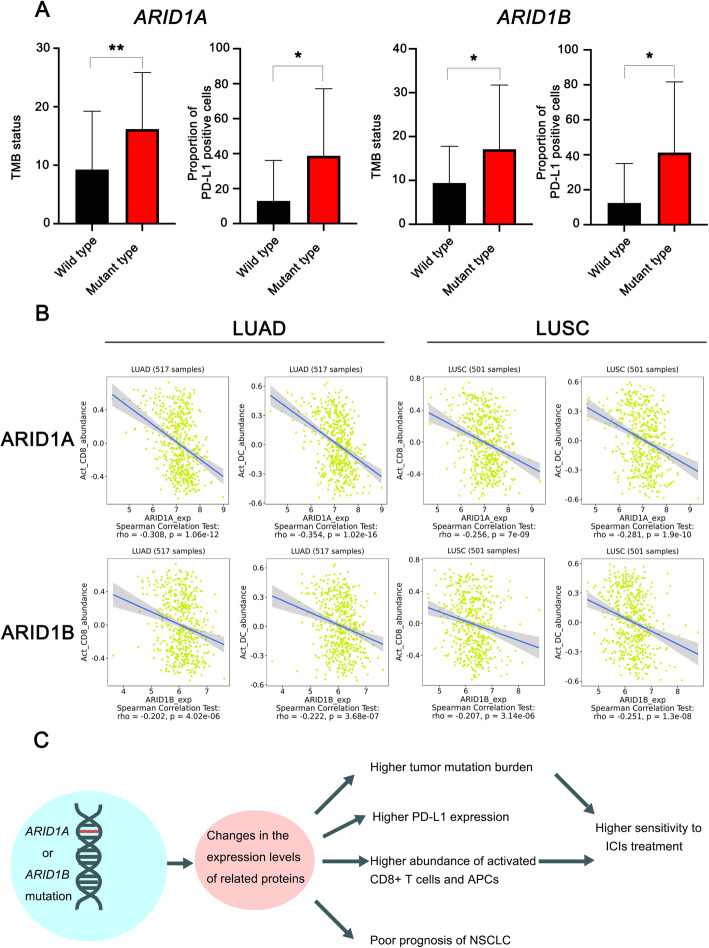
Based on the research above, we reasonably deduced that ARID1A or ARID1B mutation serves as a novel biomarker for ICIs treatment and could have a connection with factors that are proven to be associated with sensitivity to cancer immunotherapy. As shown in Fig. 4a, ARID1A or ARID1B mutations were associated with a higher TMB value (ARID1A: 16.2 versus 9.3, P = 0.001; ARID1B: 17.1 versus 9.4, P = 0.020) and a higher proportion of PD-L1-positive cells (ARID1A: 38.9% versus 12.9%, P = 0.040; ARID1B: 41.3% versus 12.4%, P = 0.020) in advanced NSCLC patients. Figure 4b reveals the relationships between the TIME with the expression of ARID1A or ARID1B. As shown in the figure, ARID1A or ARID1B expression was related to immunosuppression in 517 lung adenocarcinoma (LUAD) samples and 501 lung squamous cell carcinoma (LUSC) samples, which is mainly characterized by significant reductions in the abundances of activated CD8+ T cells and activated dendritic cells (DCs). This result suggested that ARID1A or ARID1B deficiency might modulate the TIME via the activation of the antigen presentation process and cellular immunity and thus contribute to the change in the sensitivity to ICIs treatment.
Fig. 4.
ARID1A or ARID1B mutations are tightly associated with sensitivity to immune checkpoint inhibitors (ICIs). a. The comparison of tumor mutational burden (TMB) values and PD-L1 expression grouped by the genomic signatures of ARID1 subunits; b. The correlations between ARID1A or ARID1B expression and the abundances of activated CD8+ T cells and activated dendritic cells (DC) in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC); c. The underlying relationships between prognosis and ARID1A or ARID1B mutation deduced from this research. (**: P<0.01; *: P<0.05)
Discussion
As is well known, the PD-L1 expression or TMB level did not disclose satisfied efficiency in selecting patients who might benefit from immunotherapy while plasma NGS circulating-free DNA (cfDNA) analysis in NSCLC might provides new biomarkers for cancer immunotherapy (Rossi et al. 2020). According to the previous studies, the co-mutations of TP53 and STK11 serve as an important role in modulating the TIME of NSCLC and related to the sensitivity to ICIs treatment (Skoulidis and Heymach 2019). Especially for STK11 and KEAP1 mutations, which were found more prevalent altered in KRAS-mutated LUAD, demonstrate their roles in inducing the primary resistance to ICIs treatment (Skoulidis and Heymach 2019; Skoulidis et al. 2018; Papillon-Cavanagh et al. 2020). On the contrary, mutations of ARID1A, which belongs to the ARID1 family member, might be associated with a different outcome among patients received ICIs treatment and participate to the improved ICIs outcome in NSCLC via associating KRAS mutations as reported by Gandara D, et al. Besides, Goswami S, et al. confirmed that ARID1A mutations plus CXCL13 expression as combinatorial biomarkers to predict the sensitive phenotype to ICIs in metastatic urothelial carcinoma (Goswami et al. 2020). In addition, the NGS examination based on MYSTIC trial (Rizvi et al. 2020) revealed that ARID1A mutations are associated with improved outcomes with the PD-1 inhibitor durvalumab plus the CTLA4 inhibitor tremelimumab reported by Rizvi NA, et al. Based on these studies for new biomarkers of cancer immunotherapy especially for ARID1A mutations, we conducted our research in order to further investigate the significance of ARID1 mutations, including ARID1A and ARID1B, in predicting the prognosis of ICIs treatment for advanced NSCLC.
To the best of our knowledge, our research is the first to illustrate the roles of the ARID1 subunits in the prognosis of cancer immunotherapy and to establish a prognostic model for immunotherapy in advanced NSCLC patients. Through this research, the roles of ARID1A and ARID1B in the prognosis of NSCLC were clearly clarified, and both ARID1A deficiency and ARID1B deficiency were related to the poor prognosis of NSCLC patients. However, ARID1A or ARID1B mutations and the resultant functional deficiencies were tightly associated with the sensitive phenotype for cancer immunotherapy. Patients harboring ARID1A or ARID1B mutations were more likely to have a good response to treatment with ICIs, and their PFS time might be prolonged through ICIs treatment. These correlations were also verified by the prognostic nomogram based on the Cox regression model established in this research. Here, we provided patients and physicians with this novel nomogram to estimate the survival risks of ICI treatment and determine an appropriate regimen for treatment and follow-up before treatment is initiated.
The results from the latest studies suggest that ARID1A mutation and the resultant functional deficiency are associated with the elevation in PD-L1 expression and the TMB value in ovarian cancer (Shen et al. 2018) and gastric cancer (Li et al. 2019), which indicates the underlying relationship between ARID1A and potential therapeutic anticancer immunity. Whether this correlation can be verified in NSCLC and the potential role of ARID1B in cancer immunotherapy remain to be further studied. The bioinformatic analyses in this research indicated that ARID1A or ARID1B mutations were associated with instability in the cancer cell genome and cancer mutability, as indicated by the elevated TMB, which is a proven biomarker for cancer immunotherapy. Patients harboring ARID1A or ARID1B mutations are likely to exhibit enhanced expression of PD-L1. In addition, through the activation of the antigen presentation process and anticancer cellular immunity, ARID1A and ARID1B deficiencies could modulate the TIME of NSCLC and initiate a therapeutic immune reaction to tumors. In this research, we confirmed the important role of ARID1A deficiency in elevating cancerous mutability and changing tumors to exhibit an aggressive phenotype in NSCLC, which was proposed by published works (Shen et al. 2018; Jiang et al. 2020; Okamura et al. 2020; Rossi et al. 2020) on other cancer types. Additionally, our research is the first to investigate the role of ARID1B, which is another ARID1 subunit similar to ARID1A, in changing the phenotype of cancer. Given the results reported in this research, we proposed that ARID1A and ARID1B might be equally important in cancer immunotherapy and the prognosis of NSCLC.
As shown in Fig. 4c, we generated new results in this research and reasonably deduced that mutation of ARID1A or ARID1B leads to changes in the expression levels of related proteins, which results in the poor prognosis of NSCLC on the one hand but enhances the sensitivity to ICI treatment in advanced NSCLC through elevating the TMB, enhancing PD-L1 expression and modulating the anticancer immune reaction on the other hand. Admittedly, our research might have several limitations. Further investigations into the role of ARID1B in cancer immunotherapy are needed, given the small sample size in this research. Although the results suggest that ARID1A and ARID1B may participate in the modulation of the TIME and are associated with the elevations in tumor mutability and PD-L1 expression, no molecular mechanism was explored in this research. Further studies are needed to explore the mechanism underlying these correlations.
Conclusion
ARID1A and ARID1B could serve as novel biomarkers for the prognosis and sensitivity to ICIs treatment of advanced NSCLC. ARID1A or ARID1B mutations and the resultant functional deficiencies are tightly associated with cancer mutability, PD-L1 expression and TIME modulation and are associated with a good prognosis for ICIs treatment. The relationship between the prognosis of ICIs treatment and ARID1B mutation could be verified by the novel nomogram for ICIs treatment.
Acknowledgements
Our work was supported by Special Funding for the Qilu Sanitation and Health Leading Talents Cultivation Project (to Helei Hou); the Chinese Postdoctoral Science Foundation (2017 M622143 to Helei Hou); and the Qingdao Postdoctoral Application Research Funded Project (2016052 to Helei Hou).
Abbreviations
- SWI/SNF
Switch/sucrose nonfermenting
- ARID1A
AT-rich interactive domain 1A
- ARID1B
AT-rich interactive domain 1B
- BAF
BRM-associated factor
- NSCLC
Non-small cell lung cancer
- EGFR
Epidermal growth factor receptor
- ALK
Anaplastic lymphoma kinase
- IHC
Immunohistochemistry
- DFS
Disease-free survival
- ICIs
Immune checkpoint inhibitors
- PD-L1
Programmed death-ligand 1
- TMB
Tumor mutational burden
Authors’ contributions
Conception/design: Helei Hou and Hong Li; Provision of study material or patients: Helei Hou and Dantong Sun; Collection and/or assembly of data: Dantong Sun, Lu Tian, Yang Wo and Qiaoling Liu; Data analysis and interpretation: Dantong Sun, Lu Tian; Manuscript writing: Helei Hou, Dantong Sun and Yan Zhu; Final approval of manuscript: All authors.
Funding
Special Funding for the Qilu Sanitation and Health Leading Talents Cultivation Project (to Helei Hou); the Chinese Postdoctoral Science Foundation (2017 M622143 to Helei Hou); and the Qingdao Postdoctoral Application Research Funded Project (2016052 to Helei Hou).
Availability of data and materials
All data generated during this study are included in this published article. The datasets generated in the current study are available in the cBioportal for Cancer Genomics [Cerami et al. 2012; Gao et al. 2013] (http://www.cbioportal.org/).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dantong Sun, Email: sundantongerik@163.com.
Lu Tian, Email: tianlu@stu.ouc.edu.cn.
Yan Zhu, Email: zhuyan0398@163.com.
Yang Wo, Email: woyang789@gmail.com.
Qiaoling Liu, Email: qiaolingliu1986@163.com.
Shihai Liu, Email: Shiliumed@126.com.
Hong Li, Email: hongli@shsci.org.
Helei Hou, Email: houhelei@qdu.edu.cn.
References
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed]
- Bandos AI, Rockette HE, Song T, et al. Area under the free-response ROC curve (FROC) and a related summary index. Biometrics. 2009;65(1):247–256. doi: 10.1111/j.1541-0420.2008.01049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res. 2016;22(18):4585–4593. doi: 10.1158/1078-0432.CCR-15-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):11. [DOI] [PMC free article] [PubMed]
- Goswami S, Chen Y, Anandhan S, et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci Transl Med. 2020;12(548):eabc4220. doi: 10.1126/scitranslmed.abc4220. [DOI] [PubMed] [Google Scholar]
- Guan B, Mao TL, Panuganti PK, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35(5):625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haratani K, Hayashi H, Tanaka T, et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann Oncol. 2017;28(7):1532–1539. doi: 10.1093/annonc/mdx183. [DOI] [PubMed] [Google Scholar]
- Horn L, Spigel DR, Vokes EE, et al. Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: two-year outcomes from two randomized, open-label, phase III trials (CheckMate 017 and CheckMate 057) J Clin Oncol. 2017;35(35):3924–3933. doi: 10.1200/JCO.2017.74.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HT, Chen SM, Pan LB, et al. Loss of function of SWI/SNF chromatin remodeling genes leads to genome instability of human lung cancer. Oncol Rep. 2015;33(1):283–291. doi: 10.3892/or.2014.3584. [DOI] [PubMed] [Google Scholar]
- Jiang T, Chen X, Su C, et al. Pan-cancer analysis of ARID1A alterations as biomarkers for immunotherapy outcomes. J Cancer. 2020;11(4):776–780. doi: 10.7150/jca.41296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Cao J, Cai Y, et al. A nomogram for predicting the risk of invasive pulmonary adenocarcinoma for patients with solitary peripheral subsolid nodules. J Thorac Cardiovasc Surg. 2017;153(2):462–469. doi: 10.1016/j.jtcvs.2016.10.019. [DOI] [PubMed] [Google Scholar]
- Leighl NB, Hellmann MD, Hui R, et al. Pembrolizumab in patients with advanced non-small-cell lung cancer (KEYNOTE-001): 3-year results from an open-label, phase 1 study. Lancet Respir Med. 2019;7(4):347–357. doi: 10.1016/S2213-2600(18)30500-9. [DOI] [PubMed] [Google Scholar]
- Li L, Li M, Jiang Z, et al. ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells. 2019;8(7). [DOI] [PMC free article] [PubMed]
- Mandal R, Samstein RM, Lee KW, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364(6439):485–491. doi: 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashtalir N, D’Avino AR, Michel BC, et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175(5):1272–1288. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel BC, D’Avino AR, Cassel SH, et al. A non-canonical SWI:SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20(12):1410–1420. doi: 10.1038/s41556-018-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- Naito T, Udagawa H, Umemura S, et al. Non-small cell lung cancer with loss of expression of the SWI/SNF complex is associated with aggressive clinicopathological features, PD-L1-positive status, and high tumor mutation burden. Lung Cancer. 2019;138:35–42. doi: 10.1016/j.lungcan.2019.10.009. [DOI] [PubMed] [Google Scholar]
- Okamura R, Kato S, Lee S, et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J Immunother Cancer. 2020;8(1):e000438. [DOI] [PMC free article] [PubMed]
- Ott PA, Bang YJ, Piha-Paul SA, et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol. 2019;37(4):318–327. doi: 10.1200/JCO.2018.78.2276. [DOI] [PubMed] [Google Scholar]
- Papillon-Cavanagh S, Doshi P, Dobrin R, et al. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020;5(2):e000706. [DOI] [PMC free article] [PubMed]
- Ready N, Hellmann MD, Awad MM, et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol. 2019;37(12):992–1000. doi: 10.1200/JCO.18.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37(7):537–546. doi: 10.1200/JCO.18.00149. [DOI] [PubMed] [Google Scholar]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36(7):633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Cho BC, Reinmuth N, et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the MYSTIC phase 3 randomized clinical trial. JAMA Oncol. 2020;6(5):661–674. doi: 10.1001/jamaoncol.2020.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G, Russo A, Tagliamento M, et al. Precision medicine for NSCLC in the era of immunotherapy: new biomarkers to select the most suitable treatment or the most suitable patient. Cancers (Basel) 2020;12(5):1125. doi: 10.3390/cancers12051125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- Shen J, Ju Z, Zhao W, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24(5):556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8(7):822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19(9):495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nagl NG, Wilsker D, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J. 2004;383(Pt 2):319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun Q, Shan M, et al. Promoter hypermethylation of ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers. PLoS One. 2013;8(1):e53931. doi: 10.1371/journal.pone.0053931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu X, Zhang M, et al. ARID1A is downregulated in non-small cell lung cancer and regulates cell proliferation and apoptosis. Tumour Biol. 2014;35(6):5701–5707. doi: 10.1007/s13277-014-1755-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during this study are included in this published article. The datasets generated in the current study are available in the cBioportal for Cancer Genomics [Cerami et al. 2012; Gao et al. 2013] (http://www.cbioportal.org/).