Abstract
Common neurodegenerative diseases are thought to arise from a combination of environmental and genetic exposures. Mendelian randomization is a powerful way to leverage existing genetic data to investigate causal relationships between risk factors and disease. In recent years, Mendelian randomization has gathered considerable traction in neurodegenerative disease research, providing valuable insights into the aetiology of these conditions. This review aims to evaluate the impact of Mendelian randomization studies on translational medicine for neurodegenerative diseases, highlighting the advances made and challenges faced. We will first describe the fundamental principles and limitations of Mendelian randomization and then discuss the lessons from Mendelian randomization studies of environmental risk factors for neurodegeneration. We will illustrate how Mendelian randomization projects have used novel resources to study molecular pathways of neurodegenerative disease and discuss the emerging role of Mendelian randomization in drug development. Finally, we will conclude with our view of the future of Mendelian randomization in these conditions, underscoring unanswered questions in this field.
Keywords: Mendelian randomization, neurodegeneration, genetics
Mendelian randomization uses genetic data to examine causal relationships. Storm et al. review the insights gained from Mendelian randomization studies about environmental risk factors, molecular mechanisms and drug development for neurodegenerative diseases. The authors critically evaluate novel approaches in this field and explore the challenges going forward.
Graphical Abstract
Graphical Abstract.
Introduction
Constructing a thorough understanding of neurodegenerative disease aetiology is a multidisciplinary enterprise. Observational studies have uncovered many environmental risk factors, yet spurious associations may arise if confounding variables influence both exposure and disease. There may also be reverse causation, where the disease causes the exposure. Randomized controlled trials (RCTs) are less affected by such issues; however, large-scale RCTs are prohibitively expensive.
The genetic determinants of neurodegenerative disease have been explored in genome-wide association studies (GWASs) (IMSGC and WTCCC2, 2011; IMSGC, 2013, 2019; GeM-HD Consortium, 2015; van Rheenen et al., 2016; Chang et al., 2017; Jansen et al., 2019; Nalls et al., 2019). GWASs are a tremendous resource for studying molecular pathways and drug targets, and medications with genetic support may be twice as likely to proceed from Phase I to approval (Nelson et al., 2015).
Mendelian randomization (MR) is a powerful method to investigate the interplay between genetic and environmental disease risks, enabling a more robust understanding of pathogenesis. MR has recently gathered considerable traction in neurodegenerative disease research, and this review aims to evaluate the impact of MR studies on translational medicine for these conditions. We will
explain the principles and limitations of MR,
discuss lessons from MR studies of environmental risk factors for neurodegeneration,
describe how MR can provide insight about molecular disease pathways,
address the emerging role of MR in drug development and
conclude with the future directions and limitations of MR in neurodegenerative diseases.
Search strategy
The PubMed and EMBASE databases were searched for key words such as ‘Mendelian randomization’, ‘neuro*’, ‘Parkinson*’, ‘Alzheimer*’, ‘dementia’, ‘Huntington*’ and ‘multiple sclerosis’ to identify MR studies published before 23 July 2019, with no language restrictions. Further articles were located through the references of these articles. Studies were assessed for relevance based on titles, abstracts and full texts.
What is Mendelian randomization?
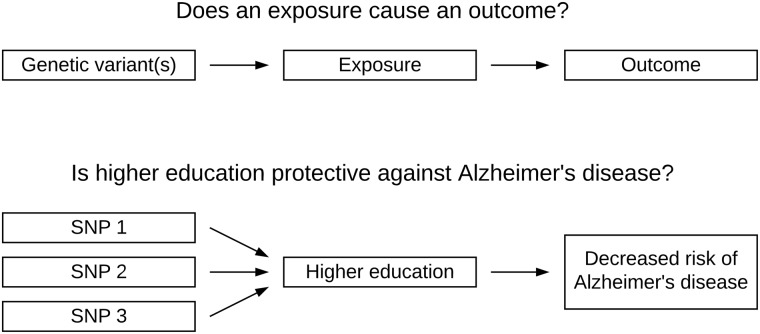
MR builds on the principle that genetic variants mimic exposure to environmental risk factors (Katan, 1986; Davey Smith and Ebrahim, 2003). Since GWASs have identified single-nucleotide polymorphisms (SNPs) associated with many phenotypic traits, MR can be used to interrogate questions such as: is higher education protective against Alzheimer’s disease?
Methodologically, SNPs that are associated with an exposure of interest (e.g. education) are used as the so-called ‘instrumental variable’ or ‘genetic instrument’. The association between the same genetic variants and an outcome (e.g. Alzheimer’s disease) is then calculated (Fig. 1). The SNP-exposure and SNP-outcome associations are combined to infer whether the exposure causes the outcome.
Figure 1.
Mendelian randomization as a concept. Mendelian randomization investigates if an exposure causes an outcome by using genetic variants that are strongly associated with the risk factor of interest. For example, genetic variants linked to education can be used to infer whether education has an effect Alzheimer’s disease risk.
MR overcomes many limitations of observational studies and RCTs. Since the inheritance of a genetic variant is independent of other traits (Mendel’s law of independent assortment), any confounders should be equally common in people with different genotypes. MR is less susceptible to reverse causation, because genotypes are determined at conception and not modifiable by disease. Thanks to openly available GWAS data, MR is much cheaper and quicker than a large-scale observational study or RCT. There are nonetheless key limitations of MR, which are explained in Box 1.
Box 1 MR considerations and limitations
MR operates under three core assumptions, stating that
the genetic variant(s) must be associated with the exposure;
the genetic variant(s) must ‘not’ be associated with any confounders; and
the genetic variant(s) must ‘not’ be associated directly with the outcome.
The first assumption can be addressed by selecting genetic variants strongly associated with the exposure, e.g. at genome-wide significance (P < 5 × 10−8). Nevertheless, GWAS-identified SNPs typically have small effect sizes; for example 97 genetic loci account for ∼2.7% of variability in BMI (Locke et al., 2015). Weak instruments limit statistical power of an MR study and can bias the final result (Pierce and Burgess, 2013).
Several SNPs can be combined to mimic one exposure (Haycock et al., 2016; Hemani et al., 2018); e.g. two SNPs represent 2.7% of plasma urate, whereas 26 SNPs explain 7% of urate variance (Kia et al., 2018; Kobylecki et al., 2018). Large study population sizes can also improve statistical power (Haycock et al., 2016), and the exposure and outcome do not have to be measured in the same population. Data from two independent GWASs can therefore be combined using ‘two-sample MR’, as long as the two populations are of the same ancestry.
The latter two core assumptions are violated by genetic pleiotropy, where a genetic locus influences more than one trait. This means that an SNP may affect the outcome through a pathway that does not involve the exposure. Many different methods have been developed to allow for some pleiotropy (Burgess et al., 2015, 2019; Holmes et al., 2017; Hemani et al., 2018).
Some limitations particularly apply to MR in drug development. Most GWASs pertain to disease risk, rather than progression, and MR instruments mimic lifelong, low-dose exposure to a risk factor or drug. This is useful when studying preventative interventions or public health policies, but perhaps less so when predicting the outcome of clinical trials, which typically last a few years only and measure progression. Indeed, there is evidence that MR may overestimate the effect seen in clinical trials (Burgess et al., 2012; Ference et al., 2012).
In addition, there may not be genetic data available to represent a drug. One could use a proxy measure, such as blood lipids to mimic statins rather than levels of the molecular target (Walker et al., 2017). Such proxies cannot detect effects through unknown mechanisms, and SNPs associated with specific gene expression or protein levels may be more suitable (Gamazon et al., 2018; Sun et al., 2018; Võsa et al., 2018; Yao et al., 2018; Zhernakova et al., 2018).
For further reading on MR methods and limitations, we direct readers to the references (Evans and Davey Smith, 2015; Holmes et al., 2017; Davies et al., 2018, Bandres-Ciga et al., 2019b).
Lessons from MR studies of environmental risk factors for neurodegeneration
Confirming findings from epidemiology—Alzheimer’s disease and education
Observational data suggest that one in five Alzheimer’s disease cases may be attributable to low educational attainment (Norton et al., 2014). This association may be confounded by, e.g. socioeconomic status (Raghavan et al., 2019), providing an opportunity for MR studies to shed light.
An MR study by Østergaard et al. found no link between genetically predicted university completion or more years of education and Alzheimer’s disease risk (Østergaard et al., 2015), whereas another MR project suggested that these risk factors may be protective (Larsson et al., 2017). These conflicting findings can be explained by understanding that the power of an MR study depends on how well the genetic variants predict the exposure. The SNP used by Østergaard and colleagues explained 0.022% of variation in years in education (Rietveld et al., 2013), and recent GWASs capture up to 13% and 5.2% of variability in educational attainment and intelligence, respectively (Lee et al., 2018; Savage et al., 2018).
Using the most recent GWASs, an MR study found a decreased Alzheimer’s disease risk with more years in education (Raghavan et al., 2019). There was a small population overlap between the GWASs for education and Alzheimer’s disease, which can bias the MR result (Pierce and Burgess, 2013). Nevertheless, three similarly powered MR analyses found similar results (Anderson et al., 2020; Savage et al., 2018; Jansen et al., 2019). Taken together, genetic evidence indicates that low educational attainment plays a causal role in Alzheimer’s disease.
Relationships between diseases—Alzheimer’s disease, Parkinson’s disease and rheumatoid arthritis
Alzheimer’s disease and Parkinson’s disease share clinical and pathological features (Hamilton, 2000; Hanagasi and Emre, 2005; Kalia and Lang, 2015); however, there is little overlap in their genetic risk (The Brainstorm Consortium, 2018). An MR study has provided further evidence, showing that genetically predicted Parkinson’s disease does not affect Alzheimer’s disease risk (Han et al., 2018). The authors suggested that an SNP in the alpha-synuclein gene SNCA (a risk locus for Parkinson's Disease) was protective for Alzheimer’s disease, but only when using one statistical method. There are many methods to calculate the MR effect, and it is advised to use several approaches with different assumptions to assess the robustness of results (Haycock et al., 2016; Burgess et al., 2017; Slob and Burgess, 2020). If only one method yields a significant result, this should be interpreted with caution.
Furthermore, observational studies suggest that rheumatoid arthritis (an auto-immune inflammatory condition of joints) is inversely associated with Alzheimer’s disease (Smolen et al., 2016). A 2017 MR study found no causal link between these conditions (Policicchio et al., 2017). A different MR study directly probed reverse causation, showing that genetic risk of Alzheimer’s disease does not affect rheumatoid arthritis risk either (Cai et al., 2018). A third MR study found that genetically predicted rheumatoid arthritis raised Alzheimer’s disease risk (Bae and Lee, 2018), but the result lost significance when one SNP was removed from the analysis. This suggests that the MR result is driven or biased by this SNP and that this outlier may affect disease risk through an alternative mechanism, rather than through rheumatoid arthritis (Burgess et al., 2017).
Challenges to interpreting MR studies—Parkinson’s disease and body mass index
The clinical implications of MR results are not always straightforward, as is illustrated by an MR study about body mass index (BMI) and Parkinson’s disease. Observational evidence has been conflicting (Wang et al., 2015; Wills et al., 2016; Roos et al., 2018), and an MR study found that genetically higher BMI is protective against Parkinson’s disease (Noyce et al., 2017). Current MR methods detect whether there is a causal relationship, rather than the magnitude thereof, so it is not clear how much of a BMI change would be needed to influence Parkinson’s disease risk. Given that a raised BMI is a well-established risk factor for cardiovascular disease (Benjamin et al., 2019), promoting a raised BMI for the sake of Parkinson’s disease protection produces a challenge.
Environmental risk factors and ethnicity—amyotrophic lateral sclerosis and blood lipids
MR can also be used to explore how disease aetiology differs between ethnic groups. The incidence of amyotrophic lateral sclerosis (ALS) varies between continents such as Europe and Asia but seems homogenous between Europe, North America and New Zealand (Marin et al., 2017). Such patterns may reflect differences in genetic and/or environmental risks.
Reviews suggest that hyperlipidaemia may be protective for ALS prognosis and risk (Jawaid et al., 2018). An MR study assessed whether low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, total cholesterol and triglyceride levels affect ALS risk, using different GWAS data for Europeans and East Asians (Zeng and Zhou, 2018). The authors found that raised low-density lipoprotein cholesterol was significantly associated with increased ALS risk in Europeans, which is in line with another MR study in Europeans (Bandres-Ciga et al., 2019a).
Zeng and Zhou found similar results in an East Asian cohort, but at nominal significance only (Zeng and Zhou, 2018). This population was smaller than the European cohort, so it is unclear whether there is a true biological difference or a lack of power in the East Asian group. Moreover, it has been proposed that observational studies may be confounded by raised BMI, which has been linked to slower ALS progression (Jawaid et al., 2018). Using MR, Zeng et al. showed that genetically predicted BMI is not causally associated with ALS in Europeans nor East Asians (Zeng et al., 2019).
Well-powered GWASs are key to a successful MR, and large GWASs remain mostly limited to European populations (Popejoy and Fullerton, 2016; Sirugo et al., 2019). As such, ethnically diverse GWASs and MR studies are crucial before we can generalize MR results to people of different ancestries.
Using MR to study molecular mechanisms of neurodegenerative disease
MR has recently been extended to molecular disease mechanisms using ‘summary-based MR’, which combines GWAS and gene expression data (Zhu et al., 2016). This is the same as classical MR, except the exposure is the expression of a gene. Expression quantitative trait loci (QTL) are used, which are genetic variants associated with different expression levels of a gene. SNPs associated with gene methylation QTL or protein QTL levels can also be studied using this method.
This approach can identify new genes that may be relevant to disease aetiology. One summary-based MR study used expression QTL associated with the expression of 5366 genes in peripheral blood and data from an ALS GWAS, finding five genes whose expression levels predict ALS risk, none of which were detected by the GWAS (Du et al., 2018). Summary-based MR can also be used to interrogate disease pathways. One study used genes related to endosomal membrane trafficking pathways and showed that expression and methylation of these genes predicted Parkinson’s disease risk (Bandres-Ciga et al., 2019c).
By using the MR approach, these studies provide insight into the ‘causal’ molecular biology of neurodegeneration, and summary-based MR and a derivative thereof (Zhu et al., 2018) have now formed part of several GWASs for neurodegenerative disease (Benyamin et al., 2017; Jansen et al., 2019; Nalls et al., 2019).
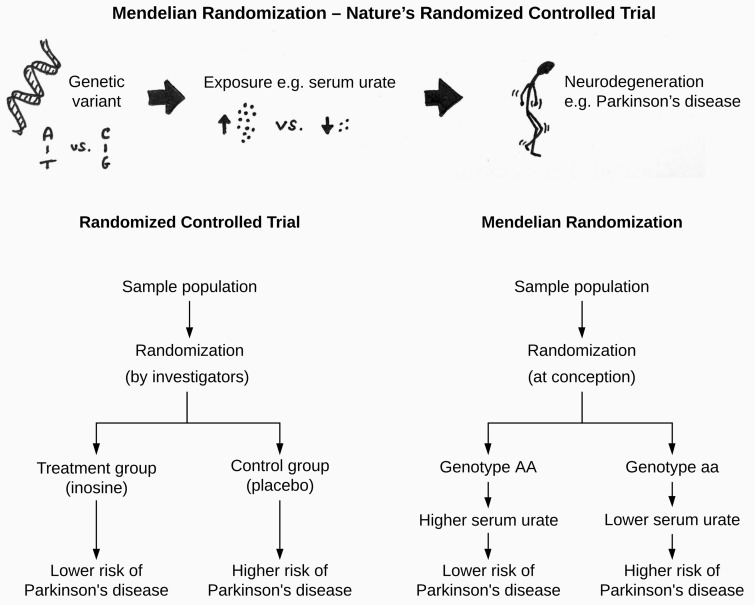
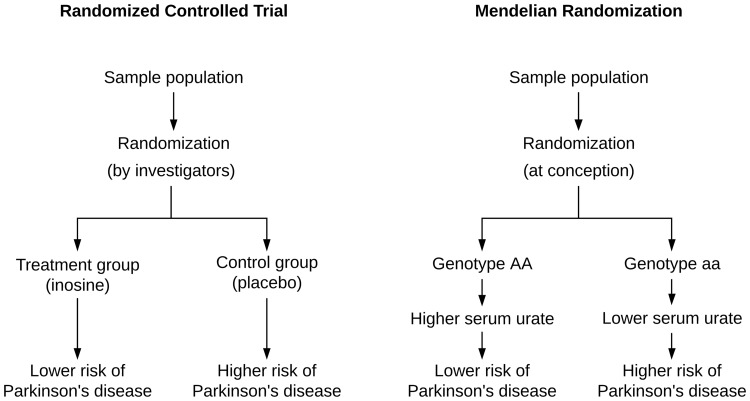
MR in drug development—nature’s randomized controlled trial
It currently takes approximately a decade and $2.6 billion for one drug to proceed from initial testing in humans to licencing (DiMasi et al., 2016), and ∼90% of drugs in Phase I clinical trials are never launched, mostly due to insufficient efficacy and safety (Harrison, 2016; Smietana et al., 2016).
MR provides an exciting opportunity, because it can imitate an RCT. The underlying principle is that genetic variants associated with different concentrations of a drug target act in the same way the corresponding drug does (Evans and Davey Smith, 2015). The SNPs essentially mimic lifelong exposure to low levels of a drug, and the random allocation of genes at birth is similar to randomization in an RCT. In addition, patients do not typically know their genotype, so MR studies are effectively blinded. Figure 2 visualizes the analogy between MR and an RCT, using serum urate and Parkinson’s disease as an example.
Figure 2.
Mendelian randomization is analogous to a placebo-controlled randomized controlled trial. For example, genetic variants associated with higher serum urate levels mimic lifelong exposure to a urate-raising drug.
MR offers a chance to prioritize drug targets using human evidence early in the drug development pipeline. We can also explore repurposing opportunities for already-licenced drugs, which have passed safety assessment and could reach patients sooner and at a lower cost (Evans and Davey Smith, 2015). MR has already been applied in drug development for neurodegenerative disease, and below we discuss several promising examples.
Parkinson’s disease and urate
Epidemiological evidence indicates that higher serum urate may lower Parkinson’s disease risk (Gao et al., 2016; Wen et al., 2017). Four small MR studies have found a protective role of raised serum urate on Parkinson’s disease risk (Gao et al., 2013; González-Aramburu et al., 2013), age of onset (Facheris et al., 2011) and progression to disability requiring dopaminergic treatment at 1 year (Simon et al., 2014). More recently, two considerably larger MR studies found no significant association between plasma urate and Parkinson’s disease risk (Kia et al., 2018; Kobylecki et al., 2018).
This discrepancy may lie in the chosen instrumental variable and sample sizes. Genetically determined urate levels are largely defined by variation in SLC2A9 and ABCG2, which together explain ∼3.4% of serum urate concentration (Köttgen et al., 2013). Most of the early MR studies of urate used SNPs in SLC2A9 only (Facheris et al., 2011; Gao et al., 2013, Simon et al., 2014). Kia et al. used GWAS-identified SNPs that explain 7% of urate variability and a two-sample MR design combining two large-scale GWAS studies, both of which improve statistical power (Kia et al., 2018).
In 2018, a clinical trial studying the urate-raising drug inosine as a treatment for Parkinson’s disease was terminated early, because it was deemed ‘unable to show that inosine slows Parkinson’s progression’ (https://www.michaeljfox.org/news/parkinsons-inosine-trial-ending-early, last accessed 9 April 2020). This potently shows how well-powered MR studies can predict the likely efficacy of a drug.
Alzheimer’s disease and blood pressure
Observational studies suggest that hypertension is a risk factor for Alzheimer’s disease (Norton et al., 2014; Livingston et al., 2017) and that antihypertensive use may be protective (Larsson and Markus, 2018), creating a potential drug repurposing opportunity.
One MR study opposed this evidence, finding that genetically predicted higher systolic blood pressure may decrease Alzheimer’s disease risk (Østergaard et al., 2015). A more recent MR study used SNPs associated with the expression of 12 antihypertensive drug targets and systolic blood pressure (Walker et al., 2019). Only the proxy for angiotensin-converting enzyme inhibitors was associated with Alzheimer’s disease risk.
Furthermore, another MR study has shown that acetylcholinesterase expression may be causally associated with raised blood pressure (Richardson et al., 2020). ACHE encodes the target for acetylcholinesterase inhibitors such as donepezil and galantamine, which are licenced treatments for Alzheimer’s disease (Lane et al., 2018).
MR can also be used to untangle on-target and off-target drug effects. Walker et al. postulated that angiotensin converting enzyme inhibitors may act through a different mechanism, since no link was found between Alzheimer’s disease risk and systolic blood pressure or other antihypertensives (Walker et al., 2019). This principle has been illustrated in cardiometabolic disease, building on observations that statin therapy is associated with an increased risk of type 2 diabetes mellitus (Sattar et al., 2009). Using SNPs in the HMG-CoA reductase gene HMGCR, which encodes the target for statins, an MR study showed that the raised diabetes risk is an on-target effect (Swerdlow et al., 2015). Likewise, if MR evidence suggests that a side effect is an off-target effect, this could encourage the development of a drug with more specificity (Evans and Davey Smith, 2015).
Multiple sclerosis and vitamin D
Multiple sclerosis prevalence rises with latitude on both sides of the equator (Ascherio and Schwarzschild, 2016; Sintzel et al., 2018), and migration studies suggest that multiple sclerosis risk is heavily influenced by environmental factors (Berg-Hansen and Celius, 2015; Ascherio and Munger, 2016; Dobson and Giovannoni, 2019). This is thought to occur because ultraviolet light stimulates vitamin D production in the skin, and vitamin D levels are inversely correlated with multiple sclerosis risk (Sintzel et al., 2018).
MR studies show that genetically predicted lower vitamin D may increase the risk of multiple sclerosis (Mokry et al., 2015; Rhead et al., 2016) and paediatric-onset multiple sclerosis (Gianfrancesco et al., 2017). The paediatric study used SNPs from a GWAS in adults, so these SNPs may not reflect vitamin D status in children. In addition, most control subjects were adults. Control subjects are traditionally age-matched, yet it is possible that paediatric controls may develop multiple sclerosis later in life, whereas adult controls are definite ‘negatives’ for paediatric multiple sclerosis. Furthermore, present-day MR assumes a linear relationship, so it is unclear if vitamin D is always protective, or for example only in clinically deficient individuals.
Disease risk as an outcome—Huntington’s disease and telomeres
An important limitation of MR in drug development is that most GWAS datasets pertain to disease risk rather than progression. In other words, most MR studies to date predict whether an intervention could prevent disease, rather than slow or hinder its progression. For example, although MR studies show an inverse relationship between vitamin D levels and multiple sclerosis risk, no significant association has been found with multiple sclerosis severity nor age at onset (Rhead et al., 2016).
An exception to this trend pertains to Huntington’s disease, a neurodegenerative condition caused by a trinucleotide (cytosine-adenine-guanine) repeat expansion in the huntingtin gene (Ross et al., 2014). The trinucleotide repeat length determines some but not all variability in age at motor symptom onset (GeM-HD Consortium, 2015), and an MR study suggests that longer telomeres (repetitive DNA sequences at the end of chromosomes) may delay Huntington’s disease onset (Aziz and Weydt, 2018). These SNPs are associated with telomere length in leukocytes, which are correlated with that in neurons (Zhan et al., 2015; Aziz and Weydt, 2018). Generally, an MR instrument and exposure do not have to be functionally linked and they must only be strongly correlated, so that the SNP(s) reliably represents the exposure.
Future directions and outstanding questions
The expanding popularity of MR has facilitated its critical evaluation and steady improvement (Burgess et al., 2015, 2018; Holmes et al., 2017; Hemani et al., 2018), and initiatives such as the UK Biobank, Million Veterans Program and 23andMe (https://research.23andme.com/, last accessed 9 April 2020) provide genetic data for a wealth of phenotypic traits (Gaziano et al., 2016; Bycroft et al., 2018). In addition, guidelines have been produced to standardize the reporting quality of MR studies (Burgess et al., 2019, Davey Smith et al., 2019).
These developments make MR increasingly accessible, enabling ‘high-throughput’ projects such as an online platform collating over 5000 GWASs to facilitate MR for Parkinson’s disease (Noyce et al., 2019). Nonetheless, it remains crucial to perform MR studies with a reasoned approach, carefully tailored to the research question (Burgess and Davey Smith, 2019). Together with ethnically diverse data and GWASs for progression (Blauwendraat et al., 2019; Iwaki et al., 2019), these resources will expedite MR for neurodegenerative disease prevention and drug development.
With regard to molecular-level MR, it remains unclear whether eQTLs are the best tool. Since most drugs act on proteins, it is perhaps more suitable to use pQTLs, i.e. SNPs associated with different protein levels rather than gene expression. pQTL studies to date have measured protein levels in blood only (Emilsson et al., 2018; Sun et al., 2018; Yao et al., 2018), which may differ from the central nervous system. On the other hand, if MR evidence shows that blood protein levels affect neurodegenerative disease, a corresponding drug may not need to cross the blood–brain barrier to exert a therapeutic effect.
Tissue diversity has been more thoroughly explored for eQTL data (Gamazon et al., 2018; Taylor et al., 2019; Richardson et al., 2020). For example, an MR study found that FBN2 expression in heart tissue was linked to diastolic blood pressure, whereas FBN2 expression in lung tissue was linked to forced vital capacity (Richardson et al., 2020). Such evidence suggests that biologically relevant tissues may be crucial for molecular-level MR projects. The sample sizes of tissue-diverse studies are small (Gamazon et al., 2018; Võsa et al., 2018), so the importance of tissue specificity remains uncertain. Finally, QTL data are notably not functional and these SNPs may not accurately reflect protein activity.
Conclusion
If we view disease aetiology as a combination of genetics and environment, MR provides an invaluable meeting point. MR has yielded many fruitful results and lessons for neurodegeneration, propelling our understanding about environmental risk factors and molecular pathophysiology. This engenders an advantageous evidence base for public health interventions to prevent disease. We eagerly anticipate further GWASs with progression outcomes and ethnically diverse populations, as well as tissue-specific QTL data, which may mitigate the challenges to translating MR results to the clinic. As these resources continue to grow, MR becomes an ideal tool for drug development, able to prioritize medications at an early stage and ascertain molecular mechanisms of action. We encourage carefully designed, standardized use of this exciting technique, boosting the number of success stories.
Funding
C.S.S. is funded by Rosetrees Trust, John Black Charitable Foundation and the University College London MB PhD Programme. D.A.K. is supported by an MB PhD Award from the International Journal of Experimental Pathology. M.A. is funded by the Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia. N.W.W. is a National Institute for Health Research senior investigator and receives support from the European Union Joint Programme—Neurodegenerative Disease Research Medical Research Council Comprehensive Unbiased Risk factor Assessment for Genetics and Environment in Parkinson’s disease. N.W.W. is also supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Competing interests
The authors report no competing interests.
Glossary
- ALS =
amyotrophic lateral sclerosis
- BMI =
body mass index
- GWAS =
genome-wide association study
- MR =
Mendelian randomization
- QTL =
quantitative trait locus
- RCT =
randomized controlled trial
- SNP =
single-nucleotide polymorphism
References
- Anderson EL, Howe LD, Wade KH, Ben-Shlomo Y, Hill WD, Deary IJ, et al. Education, intelligence and Alzheimer’s disease: evidence from a multivariable two-sample Mendelian randomization study. International Journal of Epidemiology 2020; 1–10. [DOI] [PMC free article] [PubMed]
- Ascherio A, Munger KL.. Epidemiology of multiple sclerosis: from risk factors to prevention—an update. Semin Neurol 2016; 36: 103–14. [DOI] [PubMed] [Google Scholar]
- Ascherio A, Schwarzschild MA.. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol 2016; 15: 1257–72. [DOI] [PubMed] [Google Scholar]
- Aziz NA, Weydt P.. Telomere length as a modifier of age-at-onset in Huntington disease: a two-sample Mendelian randomization study. J Neurol 2018; 265: 2149–51. [DOI] [PubMed] [Google Scholar]
- Bae S-C, Lee YH.. Causal association between rheumatoid arthritis and a decreased risk of Alzheimer’s disease A Mendelian randomization study. Z Rheumatol 2018; 78: 359–64. [DOI] [PubMed] [Google Scholar]
- Bandres-Ciga S, Noyce AJ, Hemani G, Nicolas A, Calvo A, Mora G, et al. ; The ITALSGEN Consortium. Shared polygenic risk and causal inferences in amyotrophic lateral sclerosis. Ann Neurol 2019. a; 85: 470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres-Ciga S, Noyce AJ, Traynor BJ.. Mendelian randomization—a journey from obscurity to center stage with a few potholes along the way. JAMA Neurol 2019. b; 77: 7. [DOI] [PubMed] [Google Scholar]
- Bandres-Ciga S, Saez-Atienzar S, Bonet-Ponce L, Billingsley K, Vitale D, Blauwendraat C, et al. ; The International Parkinson's Disease Genomics Consortium (IPDGC). The Endocytic membrane trafficking pathway plays a major role in the risk of Parkinson’s disease. Mov Disord 2019. c; 34: 460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. ; On behalf of the American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation 2019; 139: e56–66. [DOI] [PubMed] [Google Scholar]
- Benyamin B, He J, Zhao Q, Gratten J, Garton F, Leo PJ, et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun 2017; 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg-Hansen P, Celius EG.. Socio-economic factors and immigrant population studies of multiple sclerosis. Acta Neurol Scand 2015; 132: 37–41. [DOI] [PubMed] [Google Scholar]
- Blauwendraat C, Heilbron K, Vallerga CL, Bandres-Ciga S, Von Coelln R, Pihlstrøm L, et al. Disease age at onset genome-wide association study: defining heritability, genetic loci, and α-synuclein mechanisms. Mov Disord 2019; 34: 866–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG.. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 2017; 28: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Butterworth A, Malarstig A, Thompson SG.. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ 2012; 345: e7325. [DOI] [PubMed] [Google Scholar]
- Burgess S, Davey Smith G.. How humans can contribute to Mendelian randomization analyses. Int J Epidemiol 2019; 48: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res 2019; 4: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Foley CN, Zuber V.. Inferring causal relationships between risk factors and outcomes from genome-wide association study data. Annu Rev Genom Hum Genet 2018; 196: 1–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Scott RA, Timpson NJ, Smith GD, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015; 30: 543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562: 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Xin Z, Zuo L, Li F, Liu B.. Alzheimer’s disease and rheumatoid arthritis: a Mendelian randomization study. Frontiers in Neuroscience 2018; 12: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, Van Der BM, Kerchner GA, et al. ; International Parkinson's Disease Genomics Consortium. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet 2017; 49: 1511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Dimou N, Egger M, Gallo V, Higgins JPT, Langenberg C, et al. STROBE-MR: guidelines for strengthening the reporting of Mendelian randomization studies. PeerJ Prepr 2019; 7: 1–7. [Google Scholar]
- Davey Smith G, Ebrahim S.. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003; 32: 1–22. [DOI] [PubMed] [Google Scholar]
- Davies NM, Holmes MV, Smith GD.. Reading Mendelian randomisation studies: a guide, glossary and checklist for clinicians. BMJ 2018; 362: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi JA, Grabowski HG, Hansen RW.. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ 2016; 47: 20–33. [DOI] [PubMed] [Google Scholar]
- Dobson R, Giovannoni G.. Multiple sclerosis—a review. Eur J Neurol 2019; 26: 27–40. [DOI] [PubMed] [Google Scholar]
- Du Y, Wen Y, Guo X, Hao J, Wang W, He A, et al. A genome-wide expression association analysis identifies genes and pathways associated with amyotrophic lateral sclerosis. Cell Mol Neurobiol 2018; 38: 635–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018; 361: 769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Davey Smith G.. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genom Hum Genet 2015; 16: 327–50. [DOI] [PubMed] [Google Scholar]
- Facheris MF, Hicks AA, Minelli C, Hagenah JM, Kostic V, Campbell S, et al. Variation in the uric acid transporter gene SLC2A9 and its association with AAO of Parkinson’s disease. J Mol Neurosci 2011; 43: 246–50. [DOI] [PubMed] [Google Scholar]
- Ference BA, Yoo W, Alesh I, Mahajan N, Mirowska KK, Mewada A, et al. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol 2012; 60: 2631–9. [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Segrè AV, Van De Bunt M, Wen X, Xi HS, Hormozdiari F, et al. ; GTEx Consortium. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 2018; 50: 956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xu H, Huang X, Chen H.. Short communication: Genetic variations of SLC2A9 in relation to Parkinson’s disease. Transl Neurodegener 2013; 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, O'Reilly ÉJ, Schwarzschild MA, Ascherio A.. Prospective study of plasma urate and risk of Parkinson disease in men and women. Neurology 2016; 86: 520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016; 70: 214–23. [DOI] [PubMed] [Google Scholar]
- GeM-HD Consortium. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 2015; 162: 516–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfrancesco MA, Rhead B, Xu E, Graves JS, Waldman A, Lotze T, et al. ; For the Network of Pediatric Multiple Sclerosis Centers. Evidence for a causal relationship between low vitamin D, high BMI, and pediatric-onset MS. Neurology 2017; 88: 1623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aramburu I, Sánchez-Juan P, Jesús S, Gorostidi A, Fernández-Juan E, Carrillo F, et al. Genetic variability related to serum uric acid concentration and risk of Parkinson’s disease. Mov Disord 2013; 28: 1737–40. [DOI] [PubMed] [Google Scholar]
- Hamilton RL. Lewy bodies in Alzheimer’s disease: a neuropathological review of 145 cases using alpha-synuclein immunohistochemistry. Brain Pathol 2000; 10: 378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Tian R, Ren P, Zhou W, Wang P, Luo M, et al. Parkinson’s disease and Alzheimer’s disease: a Mendelian randomization study. 2018; 19: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanagasi H. A, Emre M.. Treatment of behavioural symptoms and dementia in Parkinson’s disease. Fundam Clin Pharmacol 2005; 19: 133–46. [DOI] [PubMed] [Google Scholar]
- Harrison RK. Phase II and phase III failures: 2013–2015. Nat Rev Drug Discov 2016; 15: 817–8. [DOI] [PubMed] [Google Scholar]
- Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Smith GD.. Statistical commentary Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies 1. Am J Clin Nutr 2016; 103: 965–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018; 7: e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MV, Ala-Korpela M, Davey Smith G.. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol 2017; 14: 577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMSGC. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013; 45: 1353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMSGC. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019; 365: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMSGC, WTCCC2. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011; 476: 214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki H, Blauwendraat C, Leonard HL, Liu G, Maple-Grødem J, Corvol JC, et al. Genetic risk of Parkinson disease and progression: an analysis of 13 longitudinal cohorts. Neurol Genet 2019; 5: e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen I, Savage J, Watanabe K, Bryois J, Williams D, Steinberg S, et al. Genetic meta-analysis identifies 10 novel loci and functional pathways for Alzheimer’s disease risk. Nat Genet 2019; 258533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawaid A, Khan R, Polymenidou M, Schulz PE.. Disease-modifying effects of metabolic perturbations in ALS/FTLD. Mol Neurodegener 2018; 13: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Lang AE.. Parkinson’s disease. Lancet 2015; 386: 896–912. [DOI] [PubMed] [Google Scholar]
- Katan MB. Apoliporotein E isoforms, serum cholesterol, and cancer. Lancet 1986; 327: 507–8. [DOI] [PubMed] [Google Scholar]
- Kia DA, Noyce AJ, White J, Speed D, Nicolas A, Burgess S, et al. Mendelian randomization study shows no causal relationship between circulating urate levels and Parkinson’s disease. Ann Neurol 2018; 84: 191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobylecki CJ, Nordestgaard BG, Afzal S.. Plasma urate and risk of Parkinson’s disease: a Mendelian randomization study. Ann Neurol 2018; 84: 178–90. [DOI] [PubMed] [Google Scholar]
- Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013; 45: 145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane CA, Hardy J, Schott JM.. Alzheimer’s disease. Eur J Neurol 2018; 25: 59–70. [DOI] [PubMed] [Google Scholar]
- Larsson SC, Markus HS.. Does treating vascular risk factors prevent dementia and Alzheimer’s disease? A systematic review and meta-analysis. J Alzheimer’s Dis 2018; 64: 657–68. [DOI] [PubMed] [Google Scholar]
- Larsson SC, Traylor M, Malik R, Dichgans M, Burgess S, Markus HS, et al. Modifiable pathways in Alzheimer’s disease: Mendelian randomisation analysis. BMJ 2017; 359: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 2018; 50: 1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, et al. Dementia prevention, intervention, and care. Lancet 2017; 390: [DOI] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, . et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin B, Boumédiene F, Logroscino G, Couratier P, Babron M-C, Leutenegger AL, et al. Variation in worldwide incidence of amyotrophic lateral sclerosis: a meta-analysis. Int J Epidemiol 2017; 46: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry LE, Ross S, Ahmad OS, Forgetta V, Smith D, Leong A, et al. Vitamin D and risk of multiple sclerosis: a Mendelian randomization study. 2015; 2: 1–20. [DOI] [PMC free article] [PubMed]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 2019; 18: 1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015; 47: 856–60. [DOI] [PubMed] [Google Scholar]
- Norton S, Matthews FE, Barnes DE, Yaff K, Brayne C.. Potential for primary prevention of Alzheimer’s disease: an analysis of population-based data. 2014; 13: 788–94. [DOI] [PubMed] [Google Scholar]
- Noyce AJ, Bandres-Ciga S, Kim J, Heilbron K, Kia D, Hemani G, et al. The Parkinson’s disease Mendelian randomization research portal. Mov Disord 2019; 34: 1864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce AJ, Kia DA, Hemani G, Nicolas A, Price TR, Pablo-fernandez E, et al. ; International Parkinson Disease Genomics Consortium. Estimating the causal influence of body mass index on risk of Parkinson disease: a Mendelian randomisation study. PLoS Med 2017; 14: e1002314–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard SD, Mukherjee S, Sharp SJ, Proitsi P, Lotta LA, Day F, et al. ; Alzheimer’s Disease Genetics Consortium. Associations between potentially modifiable risk factors and Alzheimer’s disease: a Mendelian randomization study. PLoS Med 2015; 12: e1001841–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Burgess S.. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013; 178: 1177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policicchio S, Ahmad AN, Powell JF, Proitsi P.. Rheumatoid arthritis and risk for Alzheimer’s disease: a systematic review and meta-analysis and a Mendelian randomization study. Sci Rep 2017; 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM.. Genomics is failing on diversity. Nature 2016; 538: 161–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan NS, Vardarajan B, Mayeux R.. Genomic variation in educational attainment modifies Alzheimer disease risk. Neurol Genet 2019; 5: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Bäärnhielm M, Gianfrancesco M, Mok A, Quach H, Shen L, et al. Mendelian randomization shows a causal effect of low vitamin D on multiple sclerosis risk. 2016; 2: 1–8. [DOI] [PMC free article] [PubMed]
- Richardson TG, Hemani G, Gaunt TR, Relton CL, Smith GD.. A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat Commun 2020; 11: 563379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 2013; 340: 1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol 2014; 10: 204–16. [DOI] [PubMed] [Google Scholar]
- Roos E, Grotta A, Yang F, Bellocco R, Ye W, Adami H-O, et al. Body mass index, sitting time, and risk of Parkinson disease. Neurology 2018; 90: e1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar N, Preiss D, Murray HM, Welsh P, Buckley BM, Craen Ajm D, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. 2009; 375: 735–42. [DOI] [PubMed]
- Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 2018; 50: 912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon K, Eberly S, Gao X, Oakes D, Tanner CM, Shoulson I, et al. ; on behalf of the Parkinson Study Group. Mendelian randomization of serum urate and Parkinson disease progression. Ann Neurol 2014; 76: 862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sintzel MB, Rametta M, Reder AT.. Vitamin D and multiple sclerosis: a comprehensive review. Neurol Ther 2018; 7: 59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G, Williams SM, Tishkoff SA.. The missing diversity in human genetic studies. Cell 2019; 177: 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slob EAW, Burgess S.. A comparison of robust Mendelian randomization methods using summary data. Genetic Epidemiology 2020; [Epub ahead of print] 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smietana K, Siatkowski M, Møller M.. Trends in clinical success rates. Nat Rev Drug Discov 2016; 15: 379–80. [DOI] [PubMed] [Google Scholar]
- Smolen JS, Aletaha D, Mcinnes IB.. Rheumatoid arthritis. Lancet 2016; 388: 2023–38. [DOI] [PubMed] [Google Scholar]
- Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature 2018; 558: 73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JEL, Shah T, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: Evidence from genetic analysis and randomised trials. Lancet 2015; 385: 351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K, Smith GD, Relton CL, Gaunt TR, Richardson TG.. Prioritizing putative influential genes in cardiovascular disease susceptibility by applying tissue-specific Mendelian randomization. Genome Med 2019; 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Brainstorm Consortium. Analysis of shared heritability in common disorders of the brain. Science 2018; 360: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 2016; 48: 1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Võsa U, Claringbould A, Westra H-J, Bonder MJ, Deelen P, Zeng B, et al. Unraveling the polygenic architecture of complex traits using blood eQTL meta- analysis. bioRxiv 2018;1–57. Preprint accessed on 9 July 2019. [Google Scholar]
- Walker VM, Davey Smith G, Davies NM, Martin RM.. Mendelian randomization: a novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int J Epidemiol 2017; 46: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker VM, Kehoe PG, Martin RM, Davies NM.. Repurposing antihypertensive drugs for the prevention of Alzheimer’s disease: a Mendelian randomization study. Int J Epidemiol 2019; 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L, Wang Y-T, Li J-F, Zhang Y-Z, Yin H-L, Han B.. Body mass index and risk of Parkinson’s disease: a dose-response meta-analysis of prospective studies. PLoS One 2015; 10: e0131778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen M, Zhou B, Chen Y-H, Ma Z-L, Gou Y, Zhang C-L, et al. Serum uric acid levels in patients with Parkinson’s disease: a meta-analysis. PLoS One 2017; 12: e0173731–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AA, Pérez A, Wang J, Su X, Morgan J, Rajan SS, et al. ; for the NINDS Exploratory Trials in Parkinson Disease (NET-PD) Investigators. Association between change in body mass index, unified Parkinson’s disease rating scale scores, and survival among persons with Parkinson disease secondary analysis of longitudinal data from NINDS exploratory trials in Parkinson disease long-term study. JAMA Neurol 2016; 73: 321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T, et al. Genome‐wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun 2018; 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P, Yu X, Xu H.. Association between premorbid body mass index and amyotrophic lateral sclerosis: Causal inference through genetic approaches. Front Neurol 2019; 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P, Zhou X.. Causal effects of blood lipids on amyotrophic lateral sclerosis: a Mendelian randomization study. Hum Mol Genet 2018; 28: 734–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Song C, Karlsson R, Tillander A, Reynolds CA, Pedersen NL, et al. Telomere length shortening and Alzheimer disease—a Mendelian randomization study. JAMA Neurol 2015; 72: 1202–3. [DOI] [PubMed] [Google Scholar]
- Zhernakova DV, Le TH, Kurilshikov A, Atanasovska B, Bonder MJ, Sanna S, et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat Genet 2018; 50: 1524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016; 48: 481–7. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 2018; 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]