Abstract
Neuromyelitis optica spectrum disorders lack imaging biomarkers associated with disease course and supporting prognosis. This complex and heterogeneous set of disorders affects many regions of the central nervous system, including the spinal cord and visual pathway. Here, we use graph theory-based multimodal network analysis to investigate hypothesis-free mixed networks and associations between clinical disease with neuroimaging markers in 40 aquaporin-4-immunoglobulin G antibody seropositive patients (age = 48.16 ± 14.3 years, female:male = 36:4) and 31 healthy controls (age = 45.92 ± 13.3 years, female:male = 24:7). Magnetic resonance imaging measures included total brain and deep grey matter volumes, cortical thickness and spinal cord atrophy. Optical coherence tomography measures of the retina and clinical measures comprised of clinical attack types and expanded disability status scale were also utilized. For multimodal network analysis, all measures were introduced as nodes and tested for directed connectivity from clinical attack types and disease duration to systematic imaging and clinical disability measures. Analysis of variance, with group interactions, gave weights and significance for each nodal association (hyperedges). Connectivity matrices from 80% and 95% F-distribution networks were analyzed and revealed the number of combined attack types and disease duration as the most connected nodes, directly affecting changes in several regions of the central nervous system. Subsequent multivariable regression models, including interaction effects with clinical parameters, identified associations between decreased nucleus accumbens (β = −0.85, P = 0.021) and caudate nucleus (β = −0.61, P = 0.011) volumes with higher combined attack type count and longer disease duration, respectively. We also confirmed previously reported associations between spinal cord atrophy with increased number of clinical myelitis attacks. Age was the most important factor associated with normalized brain volume, pallidum volume, cortical thickness and the expanded disability status scale score. The identified imaging biomarker candidates warrant further investigation in larger-scale studies. Graph theory-based multimodal networks allow for connectivity and interaction analysis, where this method may be applied in other complex heterogeneous disease investigations with different outcome measures.
Keywords: graph theory, multimodal network analysis, aquaporin-4-immunoglobulin G seropositive, neuromyelitis optica, magnetic resonance imaging, optical coherence tomography
Neuromyelitis optica spectrum disorders lack imaging biomarkers associated with disease course. Chien et al. report that multimodal network analysis identified associations of the nucleus accumbens and caudate nucleus volumes with clinical attacks and disease duration. Evaluation of connectivity between different types of data can elucidate damage in complex heterogeneous diseases.
Graphical Abstract
Graphical Abstract.
Introduction
Neuromyelitis optica spectrum disorders (NMOSD) are inflammatory diseases of the central nervous system defined by a clinical spectrum of optic neuritis (ON), myelitis, and more rarely, brainstem and cerebral attacks (Wingerchuk et al., 2015). The majority of patients (about 70%) are seropositive for immunoglobulin (Ig)G antibodies against the astrocytic water channel aquaporin-4 (AQP4-IgG+) (Mori et al., 2018; Cook et al., 2019). Magnetic resonance imaging (MRI) identification of T2-hyperintense brain lesions is useful for initial differential diagnosis and during acute attacks, but brain lesions are often non-specific for disability or prognosis in NMOSD (Kremer et al., 2015). Several imaging markers have been proposed for quantifying direct attack-related damage, including spinal cord atrophy after myelitis (Chien et al., 2018b), neuro-axonal damage in the retina assessed by optical coherence tomography (OCT) after ON (Schmidt et al., 2017; Oberwahrenbrock et al., 2018; Oertel et al., 2018) and T2-hyperintense lesions in symptomatic brainstem syndromes (Kim et al., 2015).
However, it is unclear if the NMOSD disease course also leads to covert or diffuse and clinically relevant central nervous system damage, which can be systematically assessed by imaging biomarkers, for example, in the grey matter (Pache et al., 2016) or the retina (Oertel et al., 2018). Normal MRI volumetric deep grey matter (DGM) measures have been reported by some (Finke et al., 2016), and it has been difficult to attribute clinical disability in NMOSD to systematic measures (Mealy et al., 2019). Others reported cortical grey matter atrophy (Kim et al., 2016) or DGM affection with clinical relevance, i.e. for cognition (Kim et al., 2017; Oertel et al., 2019).
Complex dependencies of clinical observations and central nervous system measures can be captured using graph theory models relying on a network of interactions rather than individual pairwise analyses (Lambiotte et al., 2019). Graph theory models distinct anatomical brain regions as nodes with connections represented by edges, which can be exploited to calculate the strength of interaction between multiple regions (Fleischer et al., 2017). Graph theory-based network analysis is typically applied in a single modality network, e.g. magnetic resonance imaging measures. Here, we extend this approach to evaluate multimodal connectivity (Heath and Sioson, 2009b) between distinct systematic MRI, OCT and clinical measures to identify imaging correlates of disease activity in AQP4-IgG+ NMOSD patients.
Materials and methods
Participants
Cross-sectional data from 65 participants from an on-going observational study at the Experimental and Clinical Research Center and the NeuroCure Clinical Research Center, Charité-Universitätsmedizin Berlin, were screened for inclusion in this retrospective study. Inclusion criteria were a minimum age of 18 years, a diagnosis of NMOSD according to the 2015 International Consensus Diagnostic Criteria (Wingerchuk et al., 2015) with seropositivity for AQP4-IgG in a cell-based assay (Euroimmune, Lübeck, Germany) at any time during the course of the disease, and availability of MRI, OCT images and clinical data. No restrictions were made as to type of attack preventing therapy. Ophthalmologic comorbidities (e.g. glaucoma, myopia > 5dpt) were excluded in OCT analysis only. We included data from 40 patients with NMOSD and 31 age- and sex-matched healthy controls (HC) from our research database (Table 1). Disease duration was calculated from the first clinical symptom to the date of the MRI examination. Clinical assessments consisted of the expanded disability status scale (EDSS), number of ON attacks, myelitis attacks, other NMOSD-related clinical attacks (i.e. area postrema or brainstem syndrome) and combined attacks (i.e. simultaneous myelitis and ON or myelitis and brainstem syndrome). The combined attacks were counted also as separate types of attacks, for example, a patient with a simultaneous myelitis and brainstem attack would have a count of one myelitis and one other attack; however, the total number of attacks would remain as one for that patient. The study was approved by the ethics committee at the Charité-Universitätsmedizin Berlin, Germany (EA1/131/09, EA1/163/12 and EA1/077/11) and was conducted in accordance to the Declaration of Helsinki in its current applicable version. All participants provided written informed consent.
Table 1.
Participant demographics
| HC | AQP4-IgG+ NMOSD | Test statistics | |
|---|---|---|---|
| Number of subjects [n] (total = 71) | 31 | 40 | NA |
| Age [years] (mean ± SD) | 45.92 ± 13.3 | 48.16 ± 14.3 |
|
| Female:male [n] (%female) | 24:7 (77%) | 36:4 (92%) |
|
| Disease duration [years] [mean (range)] | NA | 4.6 (0.47–28.06) | NA |
| Attack types [n] [median (range)] | NA | ON: 1 (0–14) | NA |
| Myelitis: 1 (0–11) | |||
| Other: 0 (0–2) | |||
| Combined: 0 (0–2) | |||
| Total: 3 (1–14) | |||
| EDSS [median (range)] | NA | 4 (0–6.5) | NA |
| Attack preventing therapies [n] (%) | NA | AZA: 8 (20) | NA |
| BLM: 1(2.5) | |||
| GALA: 1 (2.5) | |||
| MTX: 1 (2.5) | |||
| MMF: 1 (2.5) | |||
| RIX: 17 (42.5) | |||
| N: 11 (27.5) |
Clinical demographics of study cohort, where attack preventing therapy counts for all patients were included prior to the MRI date.
AQP4-IgG+ NMOSD = aquaporin-4 immunoglobulin-G seropositive neuromyelitis optica spectrum disorders; AZA = azathioprine; BLM = belimumab; EDSS = expanded disability status scale; GALA = glatiramer acetate; HC = healthy control; MMF = mycophenolate mofetil; MTX = methotrexate; N = none/unknown; ON = optic neuritis; RIX = rituximab.
Magnetic resonance imaging
All MRI scans were performed on a 3-T Siemens MAGNETOM Trio Tim (Erlangen, Germany) scanner at the Berlin Center for Advanced Neuroimaging. The MRI protocol for this study included: (i) a T1-weighted 3D magnetization-prepared rapid gradient echo brain MRI (1 × 1 × 1 mm resolution, repetition time (TR) = 1900 ms, echo time (TE) = 3.03 ms), including the upper cervical cord; and (ii) a T2-weighted 3D fluid-attenuated inversion recovery brain MRI (1 × 1 × 1 mm resolution, TR = 6000 ms, TE = 388 ms). All mean upper cervical cord area (MUCCA) measurements were performed at the C2/C3 intervertebral space level using JIM7.0 (Xinapse Systems Ltd, UK) software as described in a previous publication (Chien et al., 2018a). Brain lesion segmentation was performed using the automated lesion segmentation toolbox on fluid-attenuated inversion recovery images (Schmidt et al., 2012) and manually corrected by experienced radiology technicians at our centre. Normalized brain volume (NBV), DGM and brainstem volumes and cortical thickness were obtained from lesion-filled magnetization-prepared rapid gradient echo scans using FSL SIENAX (Smith et al., 2002), FSL FIRST (Patenaude et al., 2011) and the CAT12 toolbox (Dahnke et al., 2013) for MATLAB SPM12. NBV and the total volume of DGM and brainstem structures were normalized by multiplication with the V-Scaling factor, an approximation of head-size given by FSL SIENAX.
Optical coherence tomography
Retinal examinations were performed using a Heidelberg Engineering Spectralis SD-OCT (Heidelberg Engineering, Heidelberg, Germany) with automatic real-time (ART) function. Scan quality was checked using the OSCAR-IB Criteria (Schippling et al., 2015) with the Advised Protocol for OCT Study Terminology and Elements APOSTEL recommendations (Cruz-Herranz et al., 2016). 3.4 mm ring scans around the optic nerve head were used to measure the peripapillary retinal nerve fibre layer (12°, 1536 A-scans 16 ≤automated real time tracking (ART) ≤ 100). The ganglion cell inner plexiform (GCIP) layer volume was measured using a 6 mm diameter cylinder around the fovea from macular volume scans (25° × 30°, 61 vertical B-scans, 768 A-scans per B-scan, ART = 15). Layer segmentation was performed using Eye Explorer 1.9.10.0, viewing module 6.3.4.0 (Heidelberg Engineering, Heidelberg, Germany).
Multimodal network analysis
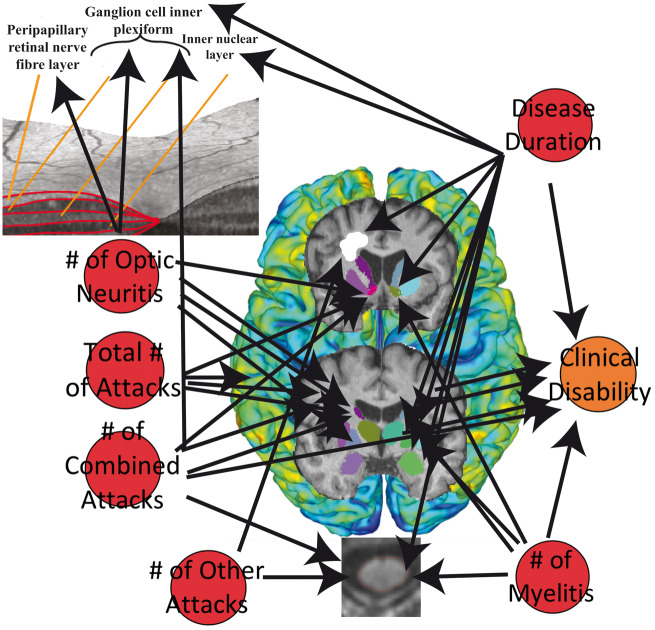
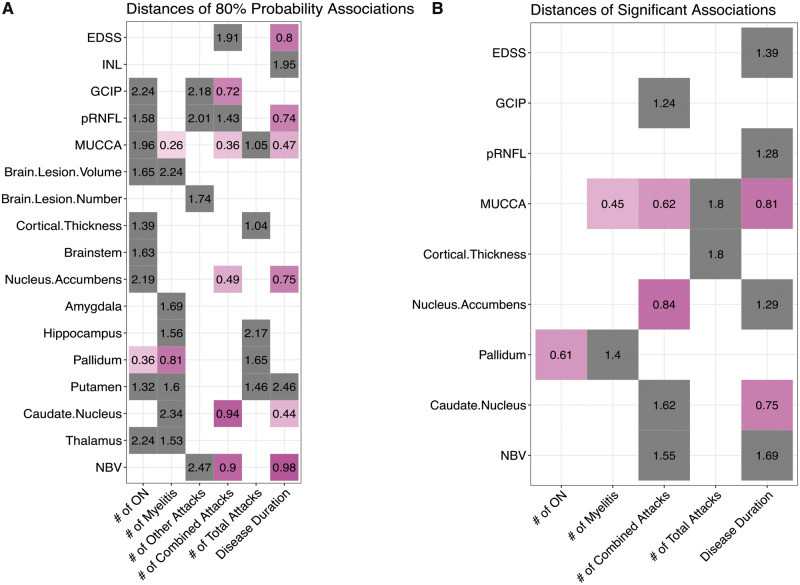
All systematic MRI, OCT and clinical measures were considered nodes in a network to test for strength of connection between the number of clinical attacks and disease duration with EDSS, brain, spinal cord and retinal measures. Directed correlational analyses between clinical attack counts and disease duration with all other nodes were performed using ANOVA with Type III sum of squares analyses, adjusted for group, thus accounting for only diseased associations of imaging measures with clinical measures. This type of multimodal network approach was proposed by Heath and Siosan (Heath and Sioson, 2009a, b), where multiple networks can be combined with interactions to represent complex biological processes. We calculated the union of three different networks (MRI measures, OCT measures and clinical measures) to give a subnetwork with ‘hyperedges’ derived from the weights calculated from the directed ANOVA F-statistics (Fig. 1A). A threshold F-statistic value of 1.65 or greater was used (giving a probability of 80% observed difference with a numerator degree of freedom of 2 and denominator degree of freedom of 60) to define hyperedges between nodes in the network. Significant network associations (95% F-distribution) were then investigated by further thresholding the P-value from each ANOVA to be <0.05. Connectivity matrices for each network were constructed using the node pairs with F-statistics and P-values within the thresholds set for 80% and 95% probability. Both networks based on an 80% F-distribution and 95% F-distribution were evaluated for connectivity, which depended on the sum of the degree, output weight, closeness to nodes, and betweenness of nodes, as well as the distance between nodes (Rubinov and Sporns, 2010). These particular graph theory measures were chosen since we employed a multimodal network analysis, different than simple conventional directed network analysis, thus we endeavoured to evaluate the component measures which can lead to analysis of centrality, network motifs and network comparison. Shortest distances between nodes are dependent on and calculated using the edge list with corresponding weights (Opsahl et al., 2010), which are related to betweenness in the nodes. Thus, the higher the sum value for connectivity, the more connected the node is with others in the network, suggesting stronger directed associations of that node with clinical measures. The two nodes showing the highest sum value for connectivity were deemed as the most connected nodes in these multimodal networks. Shortest distances were analysed in a matrix format to visualize the closeness of each type of attack to each imaging and clinical measure. This enabled a straightforward and interpretable post hoc analysis with covariates based on these matrices.
Figure 1.
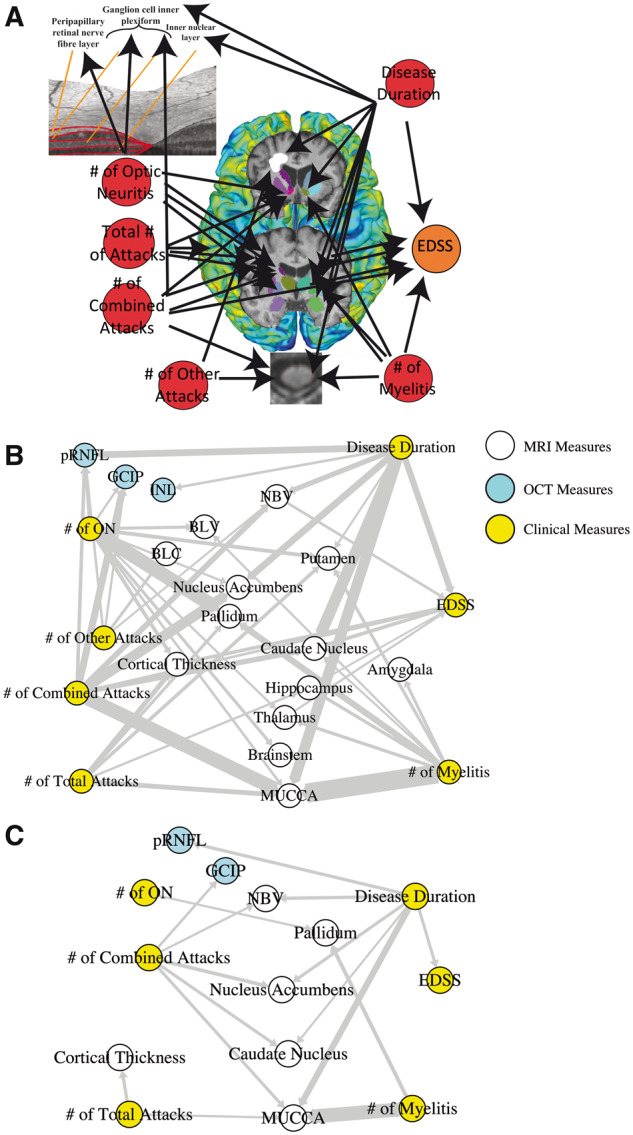
Application of graph theory-based multimodal network analysis on AQP4-IgG+ NMOSD MRI, OCT and clinical data. (A) Representation of the nodes in the multimodal network analysis employed in this study, where clinical attacks and disease duration were investigated for their effects on retinal measures, cortical thickness, DGM volumes, MUCCA, brain lesions and EDSS; (B) Multimodal network analysis of 80% probability F-distribution in AQP4-IgG+ NMOSD patient MRI, OCT and clinical measures, where width of arrows indicate F-statistic value (wider = higher value); (C) Multimodal network analysis of 95% probability F-distribution in AQP4-IgG+ NMOSD patient MRI, OCT and clinical measures, where width of arrows indicate F-statistic value (wider = higher value). BLC = brain lesion count; BLV = brain lesion volume; EDSS = expanded disability status scale; GCIP = ganglion cell inner plexiform; INL = inner nuclear layer; MUCCA = mean upper cervical cord area; NBV = normalized brain volume; ON = optic neuritis; pRNFL = peripapillary retinal nerve fibre layer.
Statistical analysis
Proportional group differences of patient demographics in age and sex were calculated using a Chi-squared test. Group differences between NMOSD and HC for all MRI and OCT measures were tested using a Mann–Whitney U-test, without correction for multiple testing. Network connections of MRI, OCT and EDSS measures within a 95% F-distribution that showed close distances (shortest weighted path length) with attack types and disease duration in the multimodal network analysis were further investigated for effect sizes and changes in the brain, spinal cord and retina of AQP4-IgG+ NMOSD patients, using multivariable linear modelling corrected for by age. Sex was not corrected for, due to the low number of males in the cohort (4 out of 40). For connections with multiple close distances with attack types and/or disease duration, the attack types and/or disease duration were used as interacting covariates. Leave-one-out-cross-validation was used to evaluate prediction error of multivariable linear models, where low overall mean squared error (mean squared error < 20% of mean) over several fold tests would indicate low prediction error. All statistical analyses and plots were produced using R version 3.4.0. Statistical significance in all tests was set to a P-value < 0.05, reflecting true test statistics (confidence intervals) and correlation coefficients (effect sizes).
Data availability
All data from this study can be shared at the request of other investigators for purposes of replicating procedures and results.
Results
Group differences in magnetic resonance imaging and optical coherence tomography measures
Participant clinical demographics are given in Table 1. Patient imaging measures were lower than HC (in order of decreasing effect size) for MUCCA, GCIP, pallidum volume, NBV and caudate nucleus volume in Mann–Whitney U group comparisons (Table 2), but higher than HC for T2-brain lesion count and volume. This is in line with an earlier study using in part, overlapping data from the same cohort study (HC: n = 12, NMOSD: n = 32), showing none or minimal volumetric changes in the DGM (Finke et al., 2016).
Table 2.
Group comparison of MRI and OCT measures
| HC | AQP4-IgG+ NMOSD | Test statistics | |
|---|---|---|---|
| NBV [mL] (mean ± SD) | 1542.5 ± 79.4 | 1500.5 ± 88.5 | W = 441, P = 0.038 |
| Thalamus [mL] (mean ± SD) | 20.8 ± 1.76 | 20.5 ± 1.82 | W = 604, P = 0.858 |
| Caudate nucleus [mL] (mean ± SD) | 9.46 ± 1.12 | 8.96 ± 1.05 | W= 446, P = 0.0439 |
| Putamen [mL] (mean ± SD) | 13.0 ± 1.33 | 12.6 ± 1.53 | W = 498, P = 0.160 |
| Pallidum [mL] (mean ± SD) | 4.82 ± 0.35 | 4.57 ± 0.46 | W = 420, P = 0.020 |
| Hippocampus [mL] (mean ± SD) | 10.3 ± 1.18 | 10.3 ± 1.23 | W = 616, P = 0.969 |
| Amygdala [mL] (mean ± SD) | 3.66 ± 0.52 | 3.86 ± 0.58 | W = 745, P = 0.150 |
| Nucleus accumbens [mL] (mean ± SD) | 1.21 ± 0.22 | 1.11 ± 0.25 | W = 482, P = 0.111 |
| Brainstem [mL] (mean ± SD) | 29.7 ± 2.41 | 29.9 ± 2.51 | W = 653, P = 0.708 |
| Cortical thickness [mm2] (mean ± SD) | 2.68 ± 0.09 | 2.71 ± 0.15 | W = 703, P = 0.341 |
| T2 brain lesion count [n] [(median (range)] | 3 (0–62) | 13.5 (0–139) | W = 890, P = 2.84e−4 |
| T2 brain lesion volume [mL] (mean ± SD) | 0.33 ± 0.57 | 2.86 ± 7.13 | W = 975, P = 3.40e−6 |
| MUCCA [mm2] (mean ± SD) | 75.3 ± 7.67 | 69.2 ± 7.61 | W = 319, P = 3.70e−4 |
| pRNFL [μm] (mean ± SD) | 95.0 ± 9.70 | 82.6 ± 24.1 | W = 276, P = 0.130 |
| GCIP [mm3] (mean ± SD) | 1.90 ± 0.14 | 1.68 ± 0.33 | W = 201, P = 8.14e−3 |
| INL [mm3] (mean ± SD) | 0.94 ± 0.06 | 0.95 ± 0.08 | W = 343, P = 0.908 |
Group comparisons of imaging measures from MRI and OCT, without adjustments or multiple testing correction. Bolded test statistics indicate statistical significance.
AQP4-IgG+ NMOSD = aquaporin-4-immunoglobulin-G seropositive neuromyelitis optica spectrum disorders; GCIP = ganglion cell inner plexiform; HC = healthy controls; INL = inner nuclear layer; MUCCA = mean upper cervical cord area; NBV = normalized brain volume; pRNFL = peripapillary retinal nerve fibre layer.
Multimodal network analysis with 80% probability of observed correlation
We performed a multimodal network analysis with an 80% probability (F-distribution) threshold to minimize false negatives. Analysis of nodal correlations within this network revealed that all MRI, OCT and clinical measures were associated with different attack types and disease duration in AQP4-IgG+ NMOSD (Fig. 1B).
The number of combined attacks and disease duration was shown to be the most connected nodes to NMOSD MRI, OCT and clinical disability (EDSS) nodes. This was calculated using the sum of the degree (number of connections), output (directed weights of hyperedges between nodes), closeness (strength of connections) and betweenness (strength of interaction between two nodes) shown in Table 3.
Table 3.
Network analysis measures with an 80% probability of observed correlation
| Node | Degree | Output | Closeness | Betweenness | Sum |
|---|---|---|---|---|---|
| NBV | NA | NA | NA | NA | NA |
| Thalamus | NA | NA | NA | NA | NA |
| Caudate nucleus | NA | NA | NA | NA | NA |
| Putamen | NA | NA | NA | NA | NA |
| Pallidum | NA | NA | NA | NA | NA |
| Hippocampus | NA | NA | NA | NA | NA |
| Amygdala | NA | NA | NA | NA | NA |
| Nucleus accumbens | NA | NA | NA | NA | NA |
| Brainstem | NA | NA | NA | NA | NA |
| Cortical thickness | NA | NA | NA | NA | NA |
| Brain lesion number | NA | NA | NA | NA | NA |
| Brain lesion volume | NA | NA | NA | NA | NA |
| MUCCA | NA | NA | NA | NA | NA |
| pRNFL | NA | NA | NA | NA | NA |
| GCIP | NA | NA | NA | NA | NA |
| INL | NA | NA | NA | NA | NA |
| EDSS | NA | NA | NA | NA | NA |
| No. of ON | 10 | 37.639 | 7.996 | 0 | 55.635 |
| No. of myelitis | 8 | 39.863 | 8.468 | 0 | 56.331 |
| No. of other attacks | 4 | 9.118 | 1.937 | 0 | 15.055 |
| No. of combined attacks | 7 | 45.390 | 9.642 | 0 | 62.032 |
| No. of total attacks | 5 | 17.284 | 3.672 | 0 | 25.955 |
| Disease duration | 8 | 48.417 | 10.285 | 0 | 66.702 |
Directed multimodal network analysis from types of attacks and disease duration to MRI, OCT and EDSS scores. Bolded sums indicate the most connected nodes in the network.
EDSS = expanded disability status scale; GCIP = ganglion cell inner plexiform; INL = inner nuclear layer; MUCCA = mean upper cervical cord area; NBV = normalized brain volume; ON = optic neuritis; pRNFL = peripapillary retinal nerve fibre layer.
Multimodal network analysis with 95% probability of observed correlation
Next, we increased the probability threshold to 95% to limit false positives. When looking at significant connections between attacks and disease duration with the other MRI, OCT and clinical nodes, we found that the number of combined attacks and disease duration remained the most connected to changes in other measures, shown in Table 4. Meanwhile, disease-related attacks were not found to be linked to brain lesion counts or volume, nor thalamic or hippocampal volumes. The number of other attacks (area postrema or brainstem syndrome) also did not show a significant influence on MRI, OCT or clinical disability measures in this cohort. Nodes that were removed from the network due to non-statistical significance were the inner nuclear layer, hippocampus, thalamus, putamen, amygdala, brainstem, brain lesion count and volume and the number of other attacks (Fig. 1C).
Table 4.
Network analysis measures with a 95% probability of observed correlation
| Node | Degree | Output | Closeness | Betweenness | Sum |
|---|---|---|---|---|---|
| NBV | NA | NA | NA | NA | NA |
| Thalamus | NA | NA | NA | NA | NA |
| Caudate nucleus | NA | NA | NA | NA | NA |
| Putamen | NA | NA | NA | NA | NA |
| Pallidum | NA | NA | NA | NA | NA |
| Hippocampus | NA | NA | NA | NA | NA |
| Amygdala | NA | NA | NA | NA | NA |
| Nucleus accumbens | NA | NA | NA | NA | NA |
| Brainstem | NA | NA | NA | NA | NA |
| Cortical thickness | NA | NA | NA | NA | NA |
| Brain lesion count | NA | NA | NA | NA | NA |
| Brain lesion volume | NA | NA | NA | NA | NA |
| MUCCA | NA | NA | NA | NA | NA |
| pRNFL | NA | NA | NA | NA | NA |
| GCIP | NA | NA | NA | NA | NA |
| INL | NA | NA | NA | NA | NA |
| EDSS | NA | NA | NA | NA | NA |
| No. of ON | 1 | 13.217 | 1.628 | 0 | 15.845 |
| No. of myelitis attacks | 2 | 23.933 | 2.948 | 0 | 28.881 |
| No. of other attacks | 0 | 0 | 0 | 0 | 0 |
| No. of combined attacks | 5 | 39.634 | 4.882 | 0 | 49.516 |
| No. of total attacks | 2 | 9.023 | 1.111 | 0 | 12.134 |
| Disease duration | 6 | 44.089 | 5.431 | 0 | 55.520 |
Directed multimodal network analysis from types of attacks and disease duration to MRI, OCT and EDSS scores. Bolded sums indicate the most connected nodes in the network.
EDSS = expanded disability status scale; GCIP = ganglion cell inner plexiform; INL = inner nuclear layer; MUCCA = mean upper cervical cord area; NBV = normalized brain volume; ON = optic neuritis; pRNFL = peripapillary retinal nerve fibre layer.
Multivariable linear analysis
We then used weighted path distances to select the closest connections between imaging and clinical markers (Fig. 2A and B). All connections in the 95% probability network with a path length <2 were tested in multivariable linear regression models, corrected for by age, to derive effect sizes of the associations. Path lengths <2 in a binary shortest distance calculation indicate a direct connection. While for instance, if two nodes are not connected, but share a connection with another node, the shortest distance (path length) between these two nodes would be two (Opsahl et al., 2010). Thus, we specifically chose directly connected nodes for further investigation. Here, NBV, pallidum volume, cortical thickness and EDSS score were only significantly associated with patient age, but not with any clinical measures (Table 5).
Figure 2.
Nodal connections and their distances calculated from multimodal network analysis. (A) Distances calculated from the 80% probability multimodal network analysis of each attack node (x-axis) and their connection to different MRI, OCT and clinical disability measures (y-axis); (B) Distances calculated from the 95% probability multimodal network analysis of each node connection to different MRI, OCT and clinical disability measures. Magenta squares = <1 unit away, grey squares = >1 unit away. Distances (shortest path lengths) have no measurement units, as the connection lengths are based on the weight of each hyperedge (F-statistic value). EDSS = expanded disability status scale; GCIP = ganglion cell inner plexiform; INL = inner nuclear layer; MUCCA = mean upper cervical cord area; ON = optic neuritis; pRNFL = peripapillary retinal nerve fibre layer.
Table 5.
Multivariable linear models for changes in AQP4-IgG+ NMOSD MRI, OCT and clinical measures
| ON attacks | Myelitis attacks | Other attacks | Combined attacks | Total number of attacks | Disease Duration (DD) | Interactions | Age | F-statistic | Adjusted R2 | Model P-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NBV | NA | NA | NA |
|
NA |
|
|
|
9.922 | 0.5352 | 4.5e−5 |
| Caudate nucleus | NA | NA | NA |
|
NA |
|
|
|
8.277 | 0.4843 | 1.7e−4 |
| Pallidum |
|
|
NA | NA | NA | NA |
|
|
3.310 | 0.1956 | 0.0215 |
| Nucleus accumbens | NA | NA | NA |
|
NA |
|
|
|
3.579 | 0.2497 | 0.0182 |
| Cortical thickness | NA | NA | NA |
|
NA | NA |
|
13.88 | 0.4041 | 3.4e−5 | |
| MUCCA | NA |
|
NA |
|
|
|
|
|
2.935 | 0.4479 | 0.0180 |
| pRNFL | NA | NA | NA | NA | NA |
|
NA |
|
1.748 | 0.06365 | 0.1997 |
| GCIP | NA | NA | NA |
|
NA | NA | NA |
|
2.055 | 0.08755 | 0.1542 |
| EDSS | NA | NA | NA | NA | NA |
|
NA |
|
7.252 | 0.3013 | 0.0030 |
Multivariable linear regression models of associations between attack types and disease duration with MRI, OCT and EDSS score, with adjustment for age. Bolded test statistics indicate statistical significance.
DD = disease duration; EDSS = expanded disability status scale; GCIP = ganglion cell inner plexiform; MUCCA = mean upper cervical cord volume; NBV = normalized brain volume; ON = optic neuritis; pRNFL = peripapillary retinal nerve fibre layer; β = standardized coefficients in multivariable linear regressions.
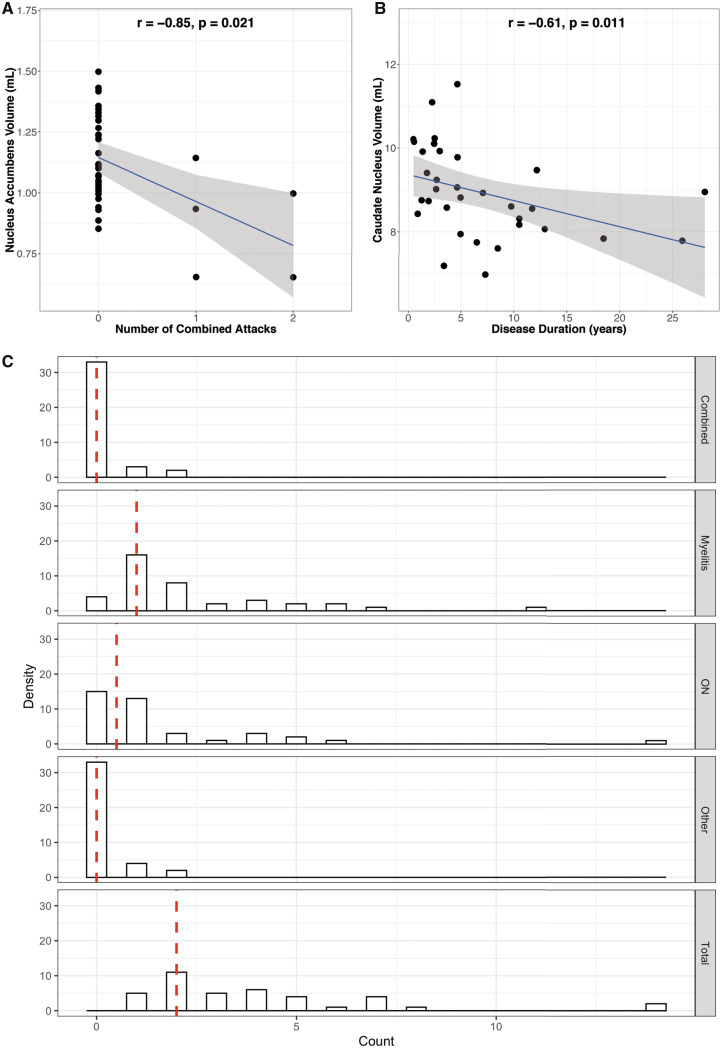
Meanwhile, the number of combined attacks (i.e. simultaneous myelitis and ON or myelitis and brainstem syndrome) was significantly associated with a decrease in the nucleus accumbens volume (β = −0.85, P = 0.021, Fig. 3A). The number of myelitis attacks was associated with decreased MUCCA (β = −4.1, P = 0.023), while the number of other attacks and total number of attacks did not affect any other node, independently. Finally, disease duration was associated with a decrease in caudate nucleus volume (β = −0.61, P = 0.011, Fig. 3B). Using leave-one-out-cross-validation (Sammut and Webb, 2010), it was found over 6-folds that there were prediction errors of 0.0465 (mean square error = 4.2% of the mean) and 1.37 (mean squared error = 15.3% of the mean) for the nucleus accumbens and caudate nucleus multivariable linear models, respectively. Attack type count distributions for this patient cohort are shown in Fig. 3C.
Figure 3.
Associations and distributions of disease-related attacks. Multivariable linear regression models of (A) the nucleus accumbens volume versus the number of combined attacks and (B) the caudate nucleus volume versus the disease duration in AQP4-IgG+ NMOSD patients, corrected for age. (C) Distribution of attack type counts in this cohort. Abbreviations: ON = optic neuritis.
Discussion
Our study identified the following imaging biomarker candidates in AQP4-IgG+ NMOSD patients, where damage seemed to occur indirectly from the attack site:
decreases in the nucleus accumbens volume is associated with increased number of combined attacks, and
decreases in the caudate nucleus volume associates with increased disease duration.
Regarding attack-site related damage, our study confirmed previously reported associations of myelitis and MUCCA (Chien et al., 2018b) in a 95% probability network, as well as ON and GCIP and peripapillary retinal nerve fibre layer (Oertel et al., 2017) when looking broader in the 80% probability network. Age was the most important factor in measures of the NBV, pallidum volume, cortical thickness and EDSS score.
The most intriguing finding of our study is the potential evidence of neurodegeneration in non-attack localized regions in NMOSD, the nucleus accumbens and the caudate nucleus. Previous studies found possible covert damage in the optic radiation and retina, but no occult damage in lesion-free brain regions (Pache et al., 2016; Oertel et al., 2017; Pasquier et al., 2019). Interestingly, an earlier study looked at markers for cognitive impairment in NMOSD patients and found that nucleus accumbens volume was associated with overall cognition and cognitive impairment in these patients. Moreover, cognitively impaired patients with NMOSD showed atrophy in the caudate nucleus (Kim et al., 2017). Although we did not specifically consider cognitive impairment in this study, Kim et al.’s findings alongside ours, serve as independent verification that the nucleus accumbens and caudate nucleus are affected in AQP4-IgG+ NMOSD patients. The nucleus accumbens has been implicated in neuropathic pain and neuropsychiatric conditions and is suggested to be modulated in an aquaporin-4-dependent manner (Wu et al., 2018). Previously, we also found moderate to severely depressed NMOSD patients are more susceptible to insufficiently mediated neuropathic pain and fatigue (Chavarro et al., 2016; Asseyer et al., 2018). Together, these data suggest that nucleus accumbens and caudate nucleus affection in NMOSD may be a true positive finding, warranting further confirmation, although the underlying mechanism remains elusive. Due to these interesting findings, we are encouraged to investigate the potential changes in cognitive, executive and mood functions of NMOSD patients and their associations with DGM volumes calculated from MRI scans.
The main suggested mechanism of damage by aquaporin-4 antibodies in NMOSD is immune-mediated attacks against astrocytes, subsequent metabolic breakdown at the attack site and resulting axonal transsection throughout structural pathways, such as the anterior visual pathway (Kuchling et al., 2018). In line with attack-related damage, associations of decreased MUCCA with an increased number of clinical myelitis attacks were previously found (Chien et al., 2018b). Regarding retinal measures, previous studies have established peripapillary retinal nerve fibre layer and GCIP as potential measures of NMOSD attack-related damage (Oertel et al., 2018). Our results are in accordance with previous findings, thus strengthening the likelihood that this multimodal network analysis technique is yielding true positive results.
Age is a well-known factor in brain atrophy measures (Opfer et al., 2018), which we also observed. In fact, age has been found to affect the majority of grey matter atrophy measures typically associated with disease, e.g. in multiple sclerosis (Barkhof et al., 2009). Our study indicates that there may also be disease-related atrophy in the pallidum, cortical thickness and NBV in larger cohorts, when adjusted for age. Despite earlier studies finding no group differences with HC (Finke et al., 2016; Pasquier et al., 2019), these studies did not look at the complex interactions of MRI or OCT measures in relation to clinical attacks and disability measures, which multimodal network analysis allows for. Therefore, this study serves as an exemplary showcase for directed graph theory-based multimodal network analysis.
The cross-sectional exploratory nature and small sample size, including the low number of other attacks (brainstem or area postrema syndrome), are important limitations. To account for both physiological fluctuations and noisy data, we performed the multimodal network analysis using F-statistics calculated from an ANOVA with Type III sum of squares, including HC and patients. This method allows for accurate testing of main effects and interactions, as well as accounting for between-group and within-group variances (Kim, 2014). Inclusion of HC data and interaction effects in the models to minimize physiological and disease-unrelated effects is a clear strength of our approach. This method could be applied to studies where patients were treated with different attack preventing therapies to allow for evaluation of treatment–imaging relationships. We were not able to investigate this in our study due to the cross-sectional and retrospective nature, as well as subgrouping the heterogeneous attack preventing therapies would not have led to sufficient group sizes for any meaningful analysis. It is thus, a limitation that we cannot fully differentiate if there are central nervous system volume effects based on attack preventing therapy, neurodegeneration, astrocytopathy or inflammation. However, one may find it feasible to use therapies as interaction effects in future larger and longitudinal studies. We were also limited by the low number of attacks in our patient cohort, where very few patients had combined attacks; however, we performed a leave-one-out-cross-validation analysis of the multivariable linear regression models used for the nucleus accumbens and caudate nucleus volumes. This validation analysis gave low mean squared error values, thus, indicating that despite a skewness towards lower attack counts, the complex regression models fit quite well and can aid in the prediction of volumetric changes based on real-world combined attack and disease duration data.
In summary, using graph theory-based multimodal network analysis, we identified two DGM imaging biomarker candidates in AQP4-IgG+ NMOSD. Rather than studying the localized region of disease-related attacks, multimodal network analysis gives insights into connectivity and interactions between multiple regions in the central nervous system that can lead to knowledge gain of damage as part of a complex, dynamic system. The network approach provides an avenue for future investigations of potential biomarkers, or associated changes to monitor for clinical trials to treat many neurologically complex diseases in large and heterogeneous datasets.
Acknowledgements
We would like to thank all participants who volunteered for these observational studies and the study nurses, MRI technicians (S. Pikol and C. Kraut), OCT technicians and residents who aided in collecting data analyzed in this study.
Funding
Funding for this work was provided from the Deutsche Forschungsgemeinschaft DFG (EXC 257) that was awarded to F.P. and A.U.B. We also acknowledge support with publication fees from the DFG and the Open Access Publication Funds of Charité - Universitätsmedizin Berlin.
Competing interests
F.C.O. was employed by Nocturne, unrelated to this project. J.K. received conference registration fees from Biogen and financial research support from Krankheitsbezogenes Kompetenznetzwerk Multiple Sklerose (KKNMS), not related to this work. M.S. reports no competing interests. K.R. received research support from Novartis, Merck Serono and German Ministry of Education and Research as well as speaking fees and travel grants from Bayer Healthcare, Biogen Idec, Merck Serono, sanofi-aventis/Genzyme, Teva Pharmaceuticals, Roche, Novartis and Guthy Jackson Charitable Foundation. J.B-S. received speaking fees and travel grants from Bayer Healthcare, Sanofi-Aventis/Genzyme, Biogen and Teva Pharmaceuticals. F.P. is on the study steering committee of Novartis OCTIMS study; received speaker honoraria and travel funding from Bayer, Novartis, Biogen, Teva, Sanofi-Aventis/Genzyme, Merck Serono, Alexion, Chugai Pharmaceutical, MedImmune and Shire; is an associate editor of Neurology® Neuroimmunology & Neuroinflammation and an academic editor of PLoS One; consulted for Sanofi Genzyme, Biogen, MedImmune, Shire and Alexion; and received research support from Bayer, Novartis, Biogen, Teva, Sanofi-Aventis/Genzyme, Alexion, Merck Serono, the German Research Council, Werth Stiftung of the City of Cologne, the German Ministry of Education and Research, Arthur Arnstein Stiftung Berlin, EU FP7 Framework Program, the Guthy Jackson Charitable Foundation, the Arthur Arnstein Foundation and the National Multiple Sclerosis Society. A.U.B. is cofounder and shareholder of medical technology start-ups Nocturne GmbH and Motognosis GmbH. He is named as inventor on several patent applications describing multiple sclerosis serum biomarkers, retinal image analysis and visual perceptive computing for measuring motor dysfunction. The remaining authors declare no conflicts of interest.
Glossary
- AQP4-IgG+
aquaporin-4-immunoglobulin G seropositive
- DGM
deep grey matter
- EDSS
expanded disability status scale
- GCIP
ganglion cell inner plexiform
- HC
healthy control
- INL
inner nuclear layer
- MRI
magnetic resonance imaging
- MUCCA
mean upper cervical cord area
- NBV
normalized brain volume
- NMOSD
neuromyelitis optica spectrum disorders
- OCT
optical coherence tomography
- ON
optic neuritis
- pRNFL
peripapillary retinal nerve fibre layer
References
- Asseyer S, Schmidt F, Chien C, Scheel M, Ruprecht K, Bellmann-Strobl J.. Pain in AQP4-IgG-positive and MOG-IgG-positive neuromyelitis optica spectrum disorders. Mult Scler J Exp Transl Clin 2018; 4: 1–12. doi: 10.1177/2055217318796684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhof F, Calabresi PA, Miller DH, Reingold SC.. Imaging outcomes for neuroprotection and repair in multiple sclerosis trials. Nat Rev Neurol 2009; 5: 256–66. [DOI] [PubMed] [Google Scholar]
- Chavarro VS, Mealy MA, Simpson A, Lacheta A, Pache F, Ruprecht K, et al. Insufficient treatment of severe depression in neuromyelitis optica spectrum disorder. Neurol Neuroimmunol Neuroinflamm 2016; 3: e286.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C, Brandt AU, Schmidt F, Bellmann-Strobl J, Ruprecht K, Paul F, et al. MRI-based methods for spinal cord atrophy evaluation: a comparison of cervical cord cross-sectional area, cervical cord volume, and full spinal cord volume in patients with aquaporin-4 antibody seropositive neuromyelitis optica spectrum disorders. Am J Neuroradiol 2018. 39: 1362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C, Scheel M, Schmitz-Hübsch T, Borisow N, Ruprecht K, Bellmann-Strobl J, et al. Spinal cord lesions and atrophy in NMOSD with AQP4-IgG and MOG-IgG associated autoimmunity. Mult Scler J 2018; 1–11. doi: 10.1177/1352458518815596. [DOI] [PubMed] [Google Scholar]
- Cook LJ, Rose JW, Alvey JS, Jolley AM, Kuhn R, Marron B, et al. Collaborative International Research in Clinical and Longitudinal Experience Study in NMOSD. Neurol Neuroimmunol Neuroinflamm 2019; 6: e583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Herranz A, Balk LJ, Oberwahrenbrock T, Saidha S, Martinez-Lapiscina EH, Lagreze WA, et al. The APOSTEL recommendations for reporting quantitative optical coherence tomography studies. Neurology 2016; 86: 2303–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahnke R, Yotter RA, Gaser C.. Cortical thickness and central surface estimation. NeuroImage 2013; 65: 336–48. [DOI] [PubMed] [Google Scholar]
- Finke C, Heine J, Pache F, Lacheta A, Borisow N, Kuchling J, et al. Normal volumes and microstructural integrity of deep gray matter structures in AQP4+ NMOSD. Neurol Neuroimmunol Neuroinflamm 2016; 3: e229.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer V, Radetz A, Ciolac D, Muthuraman M, Gonzalez-Escamilla G, Zipp F, et al. Graph theoretical framework of brain networks in multiple sclerosis: a review of concepts. Neuroscience 2017; 403: 35–53. [DOI] [PubMed] [Google Scholar]
- Heath LS, Sioson AA.. Multimodal networks: structure and operations. IEEE/ACM Trans Comput Biol and Bioinf 2009; 6: 321–32. [DOI] [PubMed] [Google Scholar]
- Heath LS, Sioson AA.. Semantics of multimodal network models. IEEE/ACM Trans Comput Biol and Bioinf 2009; 6: 271–80. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Paul F, Lana-Peixoto MA, Tenembaum S, Asgari N, Palace J, et al. MRI characteristics of neuromyelitis optica spectrum disorder: an international update. Neurology 2015; 84: 1165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-Y. Analysis of variance (ANOVA) comparing means of more than two groups. Restor Dent Endod 2014; 39: 74–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Kwak K, Hyun J-W, Jeong IH, Jo H-J, Joung A, et al. Widespread cortical thinning in patients with neuromyelitis optica spectrum disorder. Eur J Neurol 2016; 23: 1165–73. [DOI] [PubMed] [Google Scholar]
- Kim S-H, Park EY, Park B, Hyun J-W, Park NY, Joung A, et al. Multimodal magnetic resonance imaging in relation to cognitive impairment in neuromyelitis optica spectrum disorder. Sci Rep 2017; 7: 9180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer S, Renard F, Achard S, Lana-Peixoto MA, Palace J, Asgari N, et al. Use of advanced magnetic resonance imaging techniques in neuromyelitis optica spectrum disorder. JAMA Neurol 2015; 72: 815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchling J, Backner Y, Oertel FC, Raz N, Bellmann-Strobl J, Ruprecht K, et al. Comparison of probabilistic tractography and tract-based spatial statistics for assessing optic radiation damage in patients with autoimmune inflammatory disorders of the central nervous system. NeuroImage Clin 2018; 19: 538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambiotte R, Rosvall M, Scholtes I.. From networks to optimal higher-order models of complex systems. Nat Phys 2019; 15: 313.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leave-one-out cross-validation In: Sammut C, Webb GI, editors. Encyclopedia of machine learning. Boston, MA: Springer US; 2010. p. 600–1. [Google Scholar]
- Mealy MA, Mossburg SE, Kim S-H, Messina S, Borisow N, Lopez-Gonzalez R, et al. Long-term disability in neuromyelitis optica spectrum disorder with a history of myelitis is associated with age at onset, delay in diagnosis/preventive treatment, MRI lesion length and presence of symptomatic brain lesions. Mult Scler Relat Disord 2019; 28: 64–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Kuwabara S, Paul F.. Worldwide prevalence of neuromyelitis optica spectrum disorders. J Neurol Neurosurg Psychiatry 2018; 89: 555–6. [DOI] [PubMed] [Google Scholar]
- Oberwahrenbrock T, Traber GL, Lukas S, Gabilondo I, Nolan R, Songster C, et al. Multicenter reliability of semiautomatic retinal layer segmentation using OCT. Neurol Neuroimmunol Neuroinflamm 2018; 5: e449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel FC, Kuchling J, Zimmermann H, Chien C, Schmidt F, Knier B, et al. Microstructural visual system changes in AQP4-antibody-seropositive NMOSD. Neurol Neuroimmunol Neuroinflamm 2017; 4: e334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel FC, Schließeit J, Brandt AU, Paul F.. Impairment in neuromyelitis optica spectrum disorders: a review of clinical and neuroradiological features. Front Neurol 2019; 10: 608.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel FC, Zimmermann H, Paul F, Brandt AU.. Optical coherence tomography in neuromyelitis optica spectrum disorders: potential advantages for individualized monitoring of progression and therapy. EPMA J 2018; 9: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opfer R, Ostwaldt A-C, Sormani MP, Gocke C, Walker-Egger C, Manogaran P, et al. Estimates of age-dependent cutoffs for pathological brain volume loss using SIENA/FSL—a longitudinal brain volumetry study in healthy adults. Neurobiol Aging 2018; 65: 1–6. [DOI] [PubMed] [Google Scholar]
- Opsahl T, Agneessens F, Skvoretz J.. Node centrality in weighted networks: generalizing degree and shortest paths. Soc Netw 2010; 32: 245–51. [Google Scholar]
- Pache F, Zimmermann H, Finke C, Lacheta A, Papazoglou S, Kuchling J, et al. Brain parenchymal damage in neuromyelitis optica spectrum disorder—a multimodal MRI study. Eur Radiol 2016; 26: 4413–22. [DOI] [PubMed] [Google Scholar]
- Pasquier B, Borisow N, Rasche L, Bellmann-Strobl J, Ruprecht K, Niendorf T, et al. Quantitative 7T MRI does not detect occult brain damage in neuromyelitis optica. Neurol Neuroimmunol Neuroinflamm 2019; 6: e541.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M.. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 2011; 56: 907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O.. Complex network measures of brain connectivity: uses and interpretations. NeuroImage 2010; 52: 1059–69. [DOI] [PubMed] [Google Scholar]
- Schippling S, Balk LJ, Costello F, Albrecht P, Balcer L, Calabresi PA, et al. Quality control for retinal OCT in multiple sclerosis: validation of the OSCAR-IB criteria. Mult Scler 2015; 21: 163–70. [DOI] [PubMed] [Google Scholar]
- Schmidt F, Zimmermann H, Mikolajczak J, Oertel FC, Pache F, Weinhold M, et al. Severe structural and functional visual system damage leads to profound loss of vision-related quality of life in patients with neuromyelitis optica spectrum disorders. Mult Scler Relat Disord 2017; 11: 45–50. [DOI] [PubMed] [Google Scholar]
- Schmidt P, Gaser C, Arsic M, Buck D, Förschler A, Berthele A, et al. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage 2012; 59: 3774–83. [DOI] [PubMed] [Google Scholar]
- Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, et al. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. NeuroImage 2002; 17: 479–89. [DOI] [PubMed] [Google Scholar]
- Wingerchuk DM, Banwell B, Bennett JL, Cabre P, Carroll W, Chitnis T, et al. International consensus diagnostic criteria forneuromyelitis optica spectrum disorders. Neurology 2015; 85: 177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X-B, Jing P-B, Zhang Z-J, Cao D-L, Gao M-H, Jiang B-C, et al. Chemokine receptor CCR2 contributes to neuropathic pain and the associated depression via increasing NR2B-mediated currents in both D1 and D2 dopamine receptor-containing medium spiny neurons in the nucleus accumbens shell. Neuropsychopharmacol 2018; 43: 2320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from this study can be shared at the request of other investigators for purposes of replicating procedures and results.