Abstract
Human Coronaviruses (HCoV), periodically emerging across the world, are potential threat to humans such as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) – diseases termed as COVID-19. Current SARS-CoV-2 outbreak have fueled ongoing efforts to exploit various viral target proteins for therapy, but strategies aimed at blocking the viral proteins as in drug and vaccine development have largely failed. In fact, evidence has now shown that coronaviruses undergoes rapid recombination to generate new strains of altered virulence; additionally, escaped the host antiviral defense system and target humoral immune system which further results in severe deterioration of the body such as by cytokine storm. This demands the understanding of phenotypic and genotypic classification, and pathogenesis of SARS-CoV-2 for the production of potential therapy. In lack of clear clinical evidences for the pathogenesis of COVID-19, comparative analysis of previous pandemic HCoVs associated immunological responses can provide insights into COVID-19 pathogenesis. In this review, we summarize the possible origin and transmission mode of CoVs and the current understanding on the viral genome integrity of known pandemic virus against SARS-CoV-2. We also consider the host immune response and viral evasion based on available clinical evidences which would be helpful to remodel COVID-19 pathogenesis; and hence, development of therapeutics against broad spectrum of coronaviruses.
Keywords: Human coronaviruses, COVID-19, Angiotensin-converting enzyme 2, Interleukin, Cytokine storm, Interferons
Graphical abstract
1. Introduction
Human coronaviruses (HCoVs) are the members of coronaviruses (CoVs) responsible for multiple respiratory diseases of varying severity such as common cold, bronchiolitis, and pneumonia (Pene et al., 2003). Today, HCoVs are well known for rapid evolution due to high nucleotide substitution and recombination rate (Vijgen et al., 2005). HCoVs have been appeared periodically in different places around the world and linked with major outbreaks of human fatal pneumonia since the beginning of the 21st century (Wu et al., 2020). First CoV outbreak as severe acute respiratory syndrome coronavirus (SARS-CoV) initiated in November 2002 at Foshan, China (Ge et al., 2015) and later turned into global infection in 2003 with a lethal rate of 10% worldwide (Lee et al., 2003). Following one decade, second HCoV pandemic was caused by Middle East respiratory syndrome coronavirus (MERS-CoV), originated in June 2012 at Jeddah, Saudi Arabia (Ge et al., 2015), with a global fatality rate of 35% (de Groot et al., 2013). Recent third major HCoV explosion, occurred in December 2019 at Wuhan province of China, caused by highly homologous novel strain of SARS-CoV, classified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); designated the infection as COVID-19 (Coronavirus Disease 2019) pandemic (Zhu et al., 2020). These HCoVs outbreaks are classified as continuous threat to humans and world economy because of their unpredicted emergence, rapid and easy proliferation that lead to catastrophic consequences.
The virus and its host shared a complex relationship, including numerous viral and host factors, for initiation of viral infection; subsequently in potential pathogenesis (Lim et al., 2016). As intracellular obligate parasites, viruses have also advanced various strategies to hijack host-cell machineries (Lim et al., 2016). For HCoVs, there is no potential therapeutic agents such as drug or vaccine; thus, strict implementation of high vigilance such as for SARS-CoV-2 has been recommended for prevention and to check the infection (Cheng et al., 2020). Researchers and Authorities (WHO, CDC Atlanta and others) across the globe are working to combat the current ongoing SARS-CoV-2 outbreaks, and identifying the possible origin of this novel virus to develop effective therapeutics (Cohen, 2020; Cyranoski, 2020; Lu, 2020). Therefore, identifying the route of origin and pathogenesis of major pathogenic HCoVs may provide cognizance in the development of potential therapeutics. In this context, this review provides recent insights on origin of three pandemic HCoVs, i.e. SARS-CoV, MERS-CoV, and SARS-CoV-2, followed by comparative pathogenesis and attempts to summarizes the essential viral factors that manipulate the host cells to advance its infection and new progeny formation. Additionally, we have also given understandings on the possible multiple host responses reported against HCoVs and performance of viral factors to escape virus in their combat against one another.
2. Taxonomy and phylogeny of pandemic HCoV
HCoVs are studied as enveloped virus which contain non-segmented, single-stranded, positive-sense RNA genome and primarily host vertebrates (Masters, 2006). These viruses have been confronted on several occasions with prerequisite to classify a newly emerged virus associated to a severe or even fatal disease in humans under existing genera or a new species (Gorbalenya et al., 2020). In this context, current classification distributed 39 species of CoVs in 27 subgenera, 5 genera, and 2 subfamilies that categorized under family Coronaviridae, suborder Cornidovirineae, order Nidovirales, and realm Riboviria (Gorbalenya et al., 2020; Siddell et al., 2019). Herein, HCoVs are categorized under subfamily Coronavirinae of family Coronaviridae, are genotypically and serologically alienated into four major genera; AlphaCoV, BetaCoV, GammaCoV, and DeltaCoV, by International Committee for Taxonomy of Viruses (Wu et al., 2020).
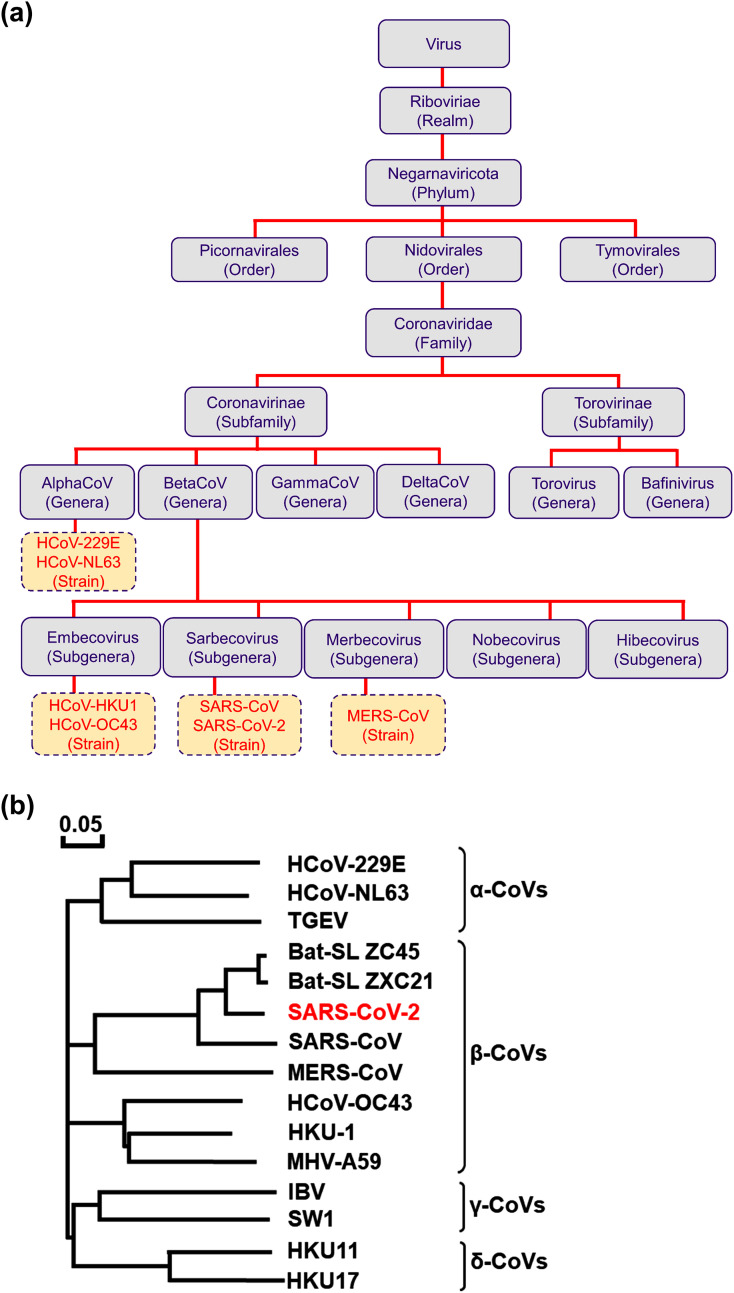
Albeit, history of CoVs began in 1940’s (Cheever et al., 1949), first human CoVs were identified in 1960’s as infectious agents for mild respiratory diseases named as HCoV-229E and HCoV-OC43 (Hamre and Procknow, 1966; McIntosh et al., 1967). The outbreak of SARS-CoV in 2002 led to an active research on novel CoVs and identified HCoV-NL63 and HCoV-HKU1 in 2004 and 2005, respectively (van der Hoek et al., 2004; Woo et al., 2005). Besides, other HCoVs discovered till date are MERS-CoV, SARS-CoV and SARS-CoV-2. The first four CoVs, i.e. HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU, are commonly dispersed and subsidize about one-third of communal cold in humans (Kanwar et al., 2017). In severe cases, however, they can cause bronchiolitis and life-threatening pneumonia in immunocompromised individuals and children (Pene et al., 2003; Walsh et al., 2013). Additionally, these CoVs were found associated with enteric and neurological diseases (Arbour et al., 2000; Arbour et al., 1999; Jacomy et al., 2006; Smuts, 2008; Vabret et al., 2003). However, SARS-CoV, MERS-CoV, and SARS-CoV-2 are well known as pandemic HCoVs. In taxonomy, HCoV-229E and HCoV-NL63 are placed under AlphaCoV whilst HCoV-HKU1, HCoV-OC43, SARS-CoV, and MERS-CoV are classified as BetaCoV; both groups mainly infect mammals while GammaCoV and DeltaCoV are specific to birds, but occasionally can also infect mammals (Woo et al., 2012). Previously, the Coronaviridae Study Group (CSG) identified SARS-CoV and MERS-CoV strains into a new species under the new informal subgroup of BetaCoV genus (van Boheemen et al., 2012). However, recent introduction of subgenus rank in virus taxonomy have established the two informal subgroups of SARS-CoV and MERS-CoV as subgenera Sarbecovirus and Merbecovirus (de Groot et al., 2013; Gorbalenya et al., 2004), respectively. Remarkably, newly emerged SARS-CoV-2 differs from previously reported zoonotic pandemic viruses, viz. SARS-CoV and MERS-CoV; and hence, taxonomic position of SARS-CoV-2 under subgenera Sarbecovirus may be tentative to change based on further evidences (Gorbalenya et al., 2020) ( Fig. 1 ).
Fig. 1.
The taxonomic (a) classification and positions for the known seven HCoVs, and (b) phylogenetic tree analysis of CoVs constructed based on S gene using Molecular Evolutionary Genetics Analysis 6 software under neighbor-joining method and 1000 bootstrap values (Biswas et al., 2020).
Before the emergence of SARS-CoV, construction of phylogenetic tree for CoVs was based on Pol (Polymerase) or Nucleocapsid (N) gene as standard practice. Initially, this method proposed SARS-CoV as a member of GammaCoV (Rota et al., 2003). However, further evidences on amino-terminal domain of SARS-CoV spike protein discovered that 19 out of 20 cysteine residues were structurally conserved with in BetaCoV group while only 5 conserved residues were matched within AlphaCoV and GammaCoV group (Rota et al., 2003). Subsequently, whole genome-based phylogenetic analysis indicated SARS-CoV as member of BetaCoV lineage B. Likewise, MERS-CoV was also identified as novel BetaCoV lineage C with high pathogenicity (Memish et al., 2013). Remarkably, RNA-dependent RNA polymerase (RdRP) and spike (S) gene sequence based phylogenetic analysis also differentiated SARS-CoV as member of BetaCoV subgroup (Eickmann et al., 2003). The sequence analysis of SARS-CoV-2 established 88% identity to sequence of two bat-derived SARS-like CoVs:bat-SL-CoVZC45 and bat-SL-CoVZXC21, and distant similarity of about 79 and 50% to SARS-CoV and MERS-CoV, respectively (Fig. 1) (Lu et al., 2020; Zhou et al., 2020b).
3. Origin and transmission of CoVs to humans
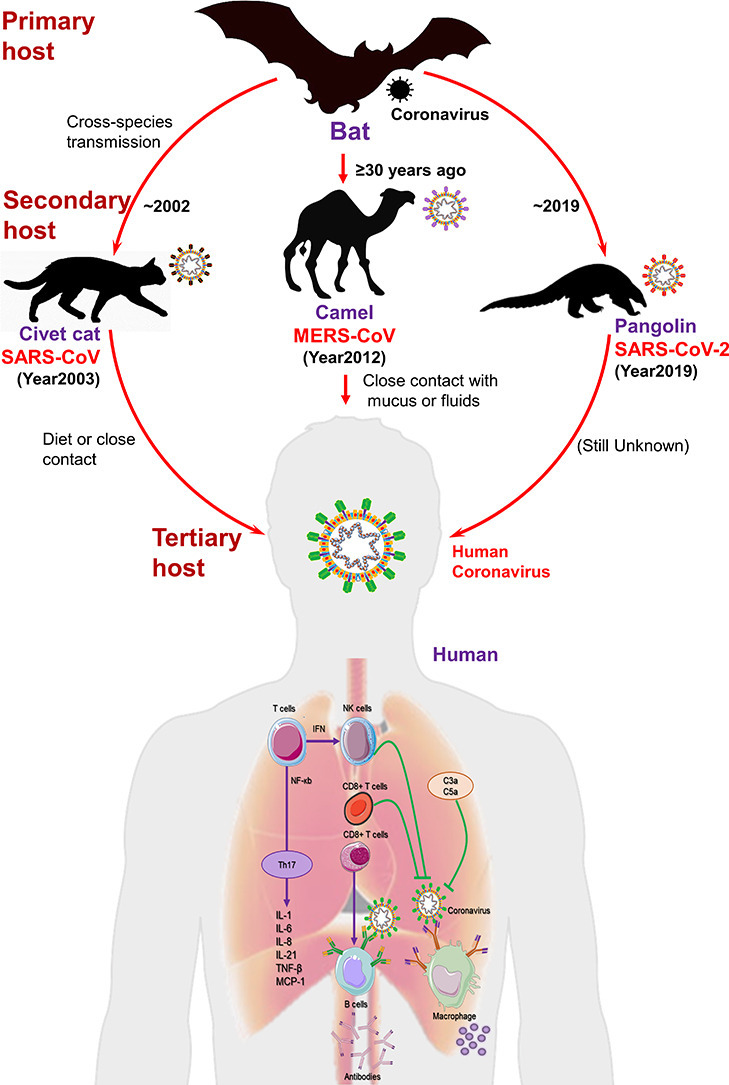
Animal CoVs have been known since the late 1930s like transmissible gastroenteritis virus of swine (TGEV), bovine coronavirus (BCoV), feline infectious peritonitis virus (FIPV), and infectious bronchitis virus (IBV) (Saif, 2004). Recent studies associated HCoVs evolution with expedited urbanization and poultry farming; these practices permitted frequent interchange of species and simplified crossing of species barrier, and genomic recombination in these viruses (Jones et al., 2013). Bats harbored a great diversity of CoVs; viral sampling conducted by the EcoHealth Alliance in China alone identified about 400 new strains of CoVs (Olival et al., 2015). Besides, study of CoVs diversity harbored by bats in eastern Thailand revealed forty-seven CoVs (Wacharapluesadee et al., 2015). Moreover, bats were documented with unique immune systems that allowed them to cope with a variety of viruses by comparison to other terrestrial mammals (Xie et al., 2018). Also, a high diversity of zoonotic AlphaCoVs and BetaCoVs were detected in the circulating bats of Western Europe (Ar Gouilh et al., 2018). Generally, bats harbored RNA viruses but can be infected by DNA viruses (Xie et al., 2018). Although, clear transmission of CoVs from bats to humans is not yet fully discovered but transmission by intermediary host to humans via direct contact has been widely suggested as one of the probable modes of transmission.
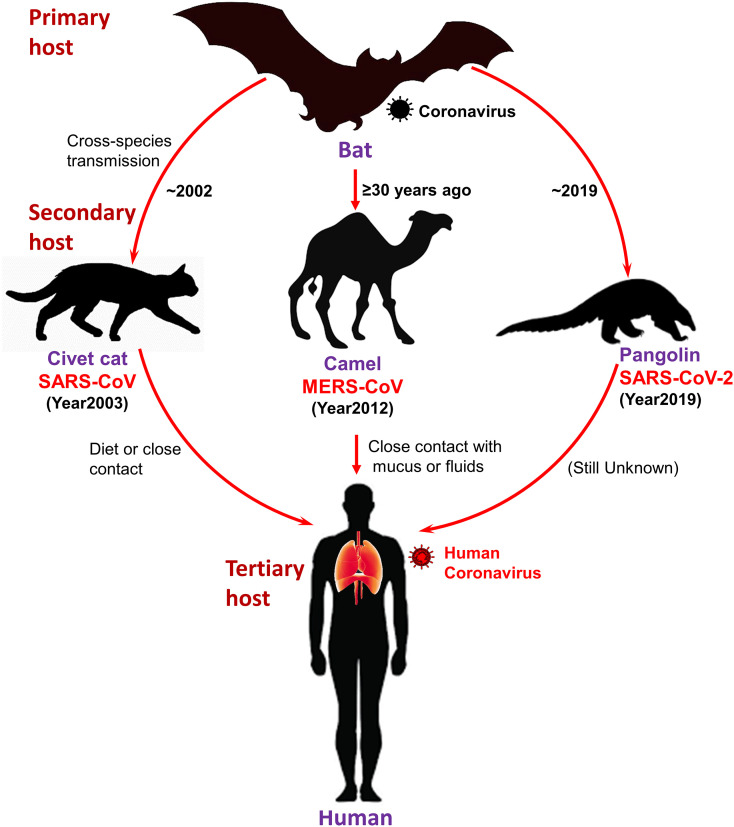
The first SARS-CoV case was documented in November 2002 in Foshan, China (Ge et al., 2015). It became an epidemic, affected 28 countries around the world with 8,096 cases and 774 deaths (Ge et al., 2015). During the SARS epidemic, the first indication for the source of SARS-CoV viral strain was detected in masked palm civets (Paguma larvata) and raccoon dog (Nyctereutes procyonoides). Later discovery of antibodies for virus in Chinese ferret badgers (Melogale moschata) in a live-animal market in Shenzhen, China were suspected as source of human infection (Drexler et al., 2014; Guan et al., 2003; Song et al., 2005). However, later findings predicted these animals merely as incidental hosts in absence of concrete results to support the motion of SARS-CoV-like viruses in palm civets among the wild or in the breeding facilities (Fig. 2 ) (Wang et al., 2006). Later, genetically differentiated CoVs linked to SARS-CoV were also found in Chinese rhinolophid bats (Rhinolophus spp.), indicated these animals as reservoir to this novel HCoV (Lau et al., 2005; Li et al., 2005). These findings along with concomitant explanation of Ebola virus in African flying foxes (Pteropus spp.) (Leroy et al., 2005), triggered the research into bats as hosts of emerging viral pathogens. Among the various factors endorsing bats as animal reservoirs of mammalian viruses were also defined in terms of their densely packed colonies, longevity, close social interaction, and ability to fly (Calisher et al., 2006; Luis et al., 2013).
Fig 2.
Schematic representation of three HCoVs, viz. SARS-CoV, MERS-CoV, and SARS-CoV-2, transmission to human from bat through intermediate hosts.
Likewise, first human case of MERS-CoV was reported in June 2012 at Jeddah, Saudi Arabia (Ge et al., 2015). As of November 2019, 2,494 cases of MERS-CoV have been reported in 27 countries, resulting in 858 fatalities (WHO, 2020). Initially, search for MERS-CoV reservoir was focused on bats; however, serological survey in dromedary camels (Camelus dromedarius) from Oman and the Canary Islands exhibited a significant prevalence of MERS-CoV-neutralizing antibodies (Reusken et al., 2013). Additionally, dromedary camels were detected with MERS-CoV RNA at a farm in Qatar related to two human cases of MERS; virus particles were also detected from dromedary camels in Qatar and Saudi Arabia (Hemida et al., 2014; Raj et al., 2014). Serological evidences supported that dromedary camels have been harboring MERS-CoV-like virus in Eastern Africa, Northern Africa, and Middle East, dating back as far as 1983 (Muller et al., 2014). Additionally, in Saudi Arabia, dromedary camels were detected with various viral genetic lineages (Sabir et al., 2016), including which crossed the species barrier and caused the outbreaks in humans (Omrani et al., 2015). Collectively, these evidences strongly suggested the contribution of dromedary camels as considerable reservoir to MERS-CoV; the ancestral virus were predicted to cross the species barrier into dromedary camels from bats more than ≥30 years ago as supported by serological evidences (Muller et al., 2014) (Fig. 2).
Recent SARS-CoV-2 outbreak highlights the hidden wild zoonotic reservoir of deadly viruses and possible threat of spillover zoonoses (Malik et al., 2020). In 2019, a food market that sold live animals in Wuhan, China was linked to the outbreak of SARS-CoV-2. Through genetic analysis, bats were again suggested as native host of SARS-CoV-2 as it shared 96% genome with two SARS-like CoVs, viz. bat-SL-CoVZX45 and bat-SL-CoVZX2, from bats (Xu et al., 2020; Zhou et al., 2020b). However, intermediate host and route which assisted viral transmission to humans is not yet fully understood. Initially, Ji, et al., proposed snakes as an intermediate host of SARS-CoV-2 transmission from bats to humans which involved homologous recombination within the viral spike (S) protein (Ji et al., 2020). Later, pangolins (Manis spp.) were suggested as potential intermediate host for SARS-CoV-2 as it shared 99% genetic homology with CoVs in pangolins (Fig. 2) (Yi et al., 2020). However, 1% variance all over the two-genome sequence was considered as significant difference; and thus, conclusive and concrete evidences are still under investigation (Yi et al., 2020). Moreover, scientists also showed concerned about its validation genetics technique (codon usage bias) (Callaway and Cyranoski, 2020). Hence, diversity of CoVs in bat population needs further investigation in detail along with critical surveillance and monitoring of bats to prevent future outbreaks in animals and public.
After infection in human, HCoVs are transmitted among the human population by close person-to-person contact. For example, close contact with infected person under in 3 feet distance has been concluded as major factor for CoV transmission (Peiris et al., 2003). The pandemic viruses, SARS-CoV, MERS-CoV and SARS-CoV-2, transmitted between humans mostly by nosocomial transmission; supported by evidences that health care workers and patients had higher frequency of infection against their relatives (de Wit et al., 2016). The collected data predicted that only 22-39% of SARS and 13-21% of MERS transmission occurred between family members. Also, SARS-CoV transmission from infected patients to health care workers was very frequent (33–42%) and MERS-CoV transmission between patients was recorded as the most common course of infection (62-79% of cases) (Chowell et al., 2015). Such a predominance of nosocomial transmission was predicted due to significant virus shedding that occurred following the onset of infection when most patients were already under medical care (Cowling et al., 2015; Peiris et al., 2003). A study on hospital surfaces following treatment of MERS-CoV infected patients were detected with viral RNA for several days even after patients no longer detected for viral infection (Bin et al., 2016). Moreover, many patients with SARS-CoV or MERS-CoV were infected through super spreaders (Chowell et al., 2015; Kucharski and Althaus, 2015). In addition, SARS-CoV was also suspected for transmission through air (airborne spread) or some other unkown transmission routes (Peiris et al., 2003).
4. HCoVs structure
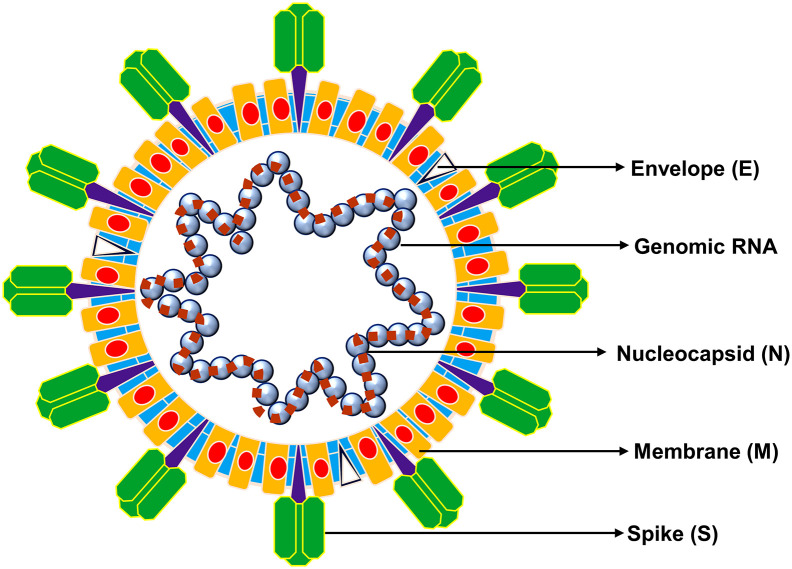
HCoVs contained phosphorylated nucleocapsid (N) protein with genomic RNA as core enveloped by phospholipid bilayers to form spherical or pleomorphic particles of 80-120 nm size and characterized by outer surface projected spike (S) proteins (Barcena et al., 2009; Neuman et al., 2006). Generally, HCoVs are composed of proteins, viz. spike (S), membrane (M), envelope (E), nucleocapsid (N) proteins, and hemagglutinin (HA), where S, M, and E proteins are embedded in viral envelop while N protein protects viral RNA genome located as core of virus (Fig. 3 ) (Zhou et al., 2020b). S protein is characterized as heavily glycosylated protein and contained the receptor binding domain (RBD), which is the most variable structure in CoVs (Zhou et al., 2020b); mediates viral entry into host cells (Bosch et al., 2003). The two different types of spike proteins have been discovered in CoVs: spike glycoprotein trimmer (S) that can be found in all CoVs, and hemagglutinin-esterase (HE) that found in specific CoVs (Belouzard et al., 2012; Cui et al., 2019). Some CoVs, including SARS-CoV and SARS-CoV-2, contain polybasic cleavage site (RRAR/S) at the junction of two subunits (S1 and S2) of S protein. This allowed digestion by host furin-like protease during viral replication (Bosch et al., 2003) and contributed in determination of viral infectivity towards host range (Nao et al., 2017). Although function of polybasic cleavage site in SARS-CoV-2 is not yet discovered; experimental studies with SARS-CoV and MERS-CoV have determined that insertion of a furin cleavage site (FCS) at S1-S2 junction enhanced the cell-cell fusion without affecting viral entry (Follis et al., 2006) and efficient digestion enabled MERS-CoV like CoVs from bats to infect human cells (Menachery et al., 2020). Furthermore, M protein exists in higher quantities against E protein in virion structure (Nal et al., 2005) and critically required in orchestrating viral shape, assembly, and in generation of mature viral envelopes (Siu et al., 2008). Moreover, it also functions in intracellular virion formation without the requirement of S protein. In some occasions, such as in presence of tunicamycin, CoVs were reported to generate noninfectious virions without spike but contained M protein in envelop (de Haan et al., 1998). Likewise, structural protein, E protein assisted in mature virions secretion from host cells, in addition to other functions like ion channel activity, inhibits the host cell stress response, and implicates pathogenesis in host cell through virus (Ruch and Machamer, 2012). N protein essentially assisted in genomic RNA packing during viral particle assembly (Chang et al., 2006; Hurst et al., 2009). Whereas, HE, which is a hemagglutinin similar to influenza virus hemagglutinin, conducts the acetyl-esterase activity (Klausegger et al., 1999). Recently, role of HE was also logged to catalyst both viral invasion across host cell membrane and in pathogenesis (Ashour et al., 2020).
Fig. 3.
A typical structure of CoV (80-120 nm) exhibiting various structural proteins, i.e. S, M, E, and genomic RNA packed inside the particle by N protein.
5. Genome of HCoVs
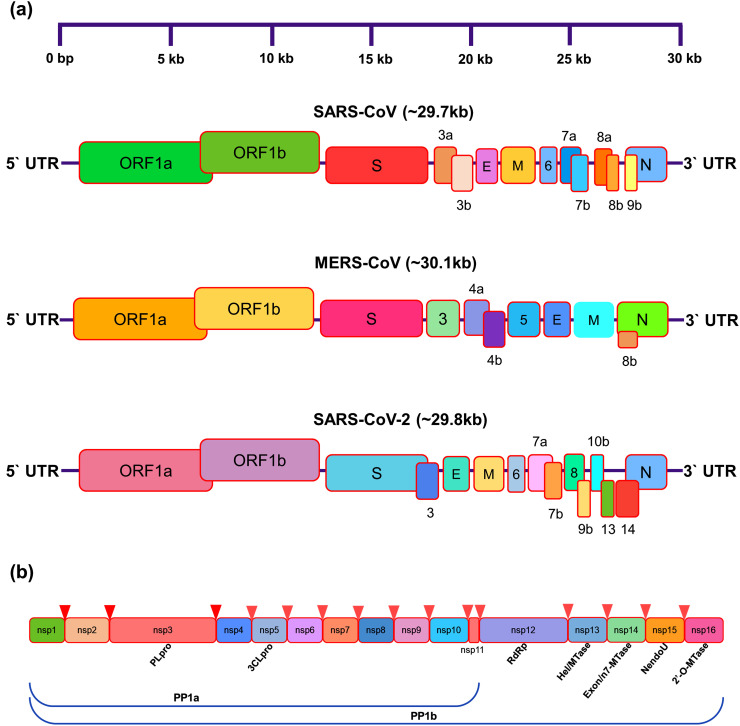
HCoVs, including SARS-CoV, MERS-CoV, and SARS-CoV-2, contained largest non-segmented positive-sense single-stranded RNA genome of size between 26.2 and 31.7 kb with varied G+C contents of 32% to 43% (Mousavizadeh and Ghasemi, 2020; Yang et al., 2006). The genome organization for a CoV is 5′-leader-UTR-replicase/transcriptase-spike (S)-envelope (E)-membrane (M)-nucleocapsid (N)-3′UTR-poly (A) tail (Fig. 4 ). The 5′UTR (untranslated region) and 3′UTR are involved in inter- and intramolecular interactions, essentially required in RNA–RNA interactions and for binding of viral and cellular proteins (Yang and Leibowitz, 2015). The ORF1a and ORF1b occupied first two-thirds of RNA genome and produced replicase/transcriptase polyprotein which undergoes autoproteolytic activity with the aid of Papain-like proteinase (PLpro) and 3C-like main protease (3CLpro or Mpro) to form 16 non-structural proteins (nsps) (Fehr and Perlman, 2015). PLpro conducts cleavage at N-terminus of replicase polyprotein to produce nsp1, nsp2, and nsp3 required for viral replication (Harcourt et al., 2004). However, Mpro is first protein which autoproteolytically separates polyprotein replicase 1ab (~790 kDa, recognition sequence located at Leu-Gln↓Ser,Ala,Gly; where ↓marks stand for cleavage site) to produced mature viral enzymes and further cleaved downstream nsps at 11 sites to release nsp4-nsp16 (Fig. 4) (Yang et al., 2005; Zhang et al., 2020). These events indicated the direct role of Mpro to mediate maturation of nsps, which are basically required for virus life cycle in host cells. Hence, PLpro and Mpro have been suggested as potential targets in drug development against HCoVs. The later reading frames on the genome encoded the four major structural proteins; S, E, M, and N, which are common proteins among all CoVs (Snijder et al., 2003). All the viral structural and accessory proteins are decoded by subgenomic (sg) RNAs of CoVs where accessory proteins are interspersed between ORFs strands but their number and functional activities are restricted to CoV strains (Fehr et al., 2015; Fehr and Perlman, 2015). Specifically, SARS-CoV genome translated 8 accessory gene (3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b), MERS-CoV encoded 5 accessory genes (3, 4a, 4b, 5, and 8b), and though exact number of functional proteins remains to be established in SARS-CoV-2 but based on previous study on SARS-CoV, SARS-CoV-2 has been predicted with translation of 4 structural proteins (Yoshimoto, 2020) and at least 6 or 9 accessory proteins (3, 6, 7a, 7b, 8, 9b, 10b, 13, 14) (Liu et al., 2014; Wu et al., 2020c; Zhou et al., 2020b). Also, ORF1ab position in SARS-CoV-2 genome (251-21541 nt) was noticed with change at starting codon position by comparison to SARS-CoV (265-21486 nt) and MERS-CoV (279-21514 nt). Interestingly, conserved amino acid substitution at positions 439, 501, 493, 485, and 486 were also logged in RBD domain of SARS-CoV-2 S protein, which was considered as important in SARS-CoV (Lu et al., 2020). It is important to add that genomic material of HCoVs is highly susceptible to frequent recombination process which have been directly associated with establishment of altered virulence in new HCoV strains (Hilgenfeld, 2014).
Fig. 4.
Schematic representation for (a) genome organization and (b) functional domains in the genome of SARS-CoV, MERS-CoV, and SARS-CoV-2.
6. Life cycle of corona virus
All the SARS-like CoVs exhibits typical streadgy for replication and translation following infection in host cell. This section briefly summarizes SARS-CoV, MERS-CoV and SARS-CoV-2 with respect to their comparative lifecycle in host cell, which begans on attachment of virus with host cell receptor and finished with release of newly generated progeny from infected cells.
6.1. Host-cells attachment and entry
The binding of HCoV to host-cell receptor is the initial step in viral infection which determined the severity of infection and pathogenesis. A better understanding of coupling of viral structural proteins with targeted receptor on host cells and other involved proteins, such as host protease, can help to predict the specific zoonotic CoVs infection in humans.
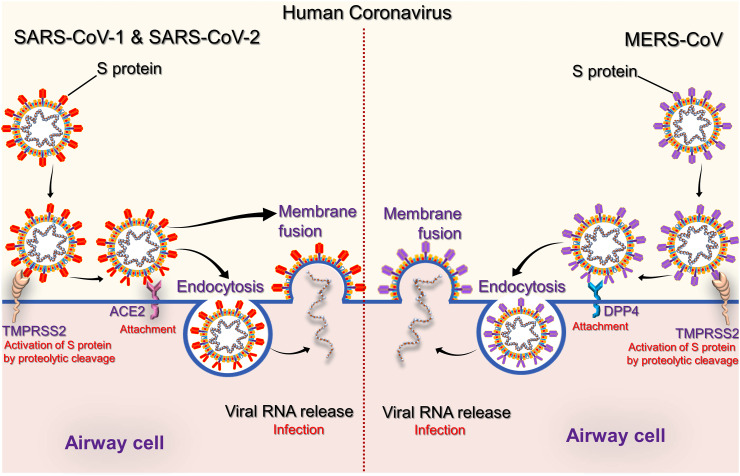
HCoVs infection begins with attachment of viral particle on host cell surface by densely glycosylated S protein, which is a trimeric class I fusion protein and consist of two major subunits; a receptor binding domain (S1; also known as RBD) and a second domain (S2) mediated viral fusion with host cell membrane. The fusion process of viral membrane with host cell membrane is triggered when S1 domain binds with host-cell receptor; for example, angiotensin-converting enzyme 2 (ACE2) for SARS-CoV (Li et al., 2003) and SARS-CoV-2 (Wu et al., 2020b, Wu et al., 2020c); in addition an alternative receptor CD209L (a C-type lectin, also called L-SIGN) with low affinity for SARS-CoV (Jeffers et al., 2004), and dipeptidyl peptidase-4 (DPP4, also known as CD26) for MERS-CoV (Raj et al., 2013). Thus, cleavage of spike glycoprotein is performed by host cell proteases enables interaction of S2 domain for virus entry into the cell (de Wit et al., 2016).
In the respiratory tract, ACE2 receptor is widely distributed on the epithelial cells of trachea, bronchi, bronchial serous glands, and alveoli (Liu et al., 2011), as well as alveolar monocytes and macrophages (Kuba et al., 2005); SARS-CoV attacked these cells and mature virions are later released to infect other new target cells (Zhang et al., 2004). ACE2 is also diffusely expressed on endothelial cells of arteries and veins, cerebral neurons, immune cells, tubular epithelial cells of kidneys, mucosal cells of intestines, and epithelial cells of renal tubules, presented as a variety of susceptible targets to SARS-CoV infection (Gu and Korteweg, 2007; Guo et al., 2008). Whereas, MERS-CoV is known to target respiratory tract, liver small intestines, kidney, prostate, and activated leukocytes (Widagdo et al., 2016). Remarkably, unlike SARS-CoV in vitro studies, MERS-CoV was found to infect human dendritic cells (Chu et al., 2014) and macrophages (Zhou et al., 2014); therefore, virus disrupted the immune system. Besides, T (lymphocyte) cells are another potential target for MERS-CoV due to presence of high amounts of CD26 (Chu et al., 2016). Hence, this CoV was predicted to dysregulate antiviral T-cell responses because of stimulation of T-cell apoptosis (Chu et al., 2016; Yeung et al., 2016). Recent studies suggested that SARS-CoV-2 employs ACE2 as main receptor as in SARS-CoV infection with higher affinity, suggesting the likelihood of same group of host-cells being targeted and infected (Zhou et al., 2020b; Zou et al., 2020).
After attachment to host-cell surface, virus entry into cell has been deciphered by two different paths based on availability of host cell protease to activate receptor-attached spike protein (Fig. 5 ) (Simmons et al., 2013). In first path, CoVs invaded host cell as an endosome which is mediated by clathrin-dependent and-independent endocytosis (Fig. 5) (Kuba et al., 2010; Wang et al., 2008a). This phenomenon induced conformational changes in viral particle which subsequently fused viral envelope with endosomal wall (Simmons et al., 2013). Alternatively, in second pathway, direct invasion of virus particles into host cell are mediated via proteolytic cleavage of receptor-attached spike protein by host's transmembrane serine protease 2 (TMPRSS2) or transmembrane serine protease 11D (TMPRSS11D) on the cell surface (Heurich et al., 2014; Zumla et al., 2016). Herein, S2 domain of spike protein accomplished direct membrane fusion between virus and plasma membrane as initially observed in SARS-CoV (Simmons et al., 2013). Belouzard et al. discovered the critical proteolytic cleavage event of S protein of SARS-CoV at position (S20) which facilitated membrane fusion process and viral infectivity (Belouzard et al., 2009). Likewise, MERS-CoV was also studied for abnormal two-step furin activation to enable virus for fusion with host cell membrane (Millet and Whittaker, 2014).
Fig. 5.
Schematic representation for HCoVs attachment and entry into airway cells. The envelope spike glycoprotein binds to its cellular receptor ACE2 for SARS-CoV and DPP4 for MERS-CoV.
6.2. Genome translation
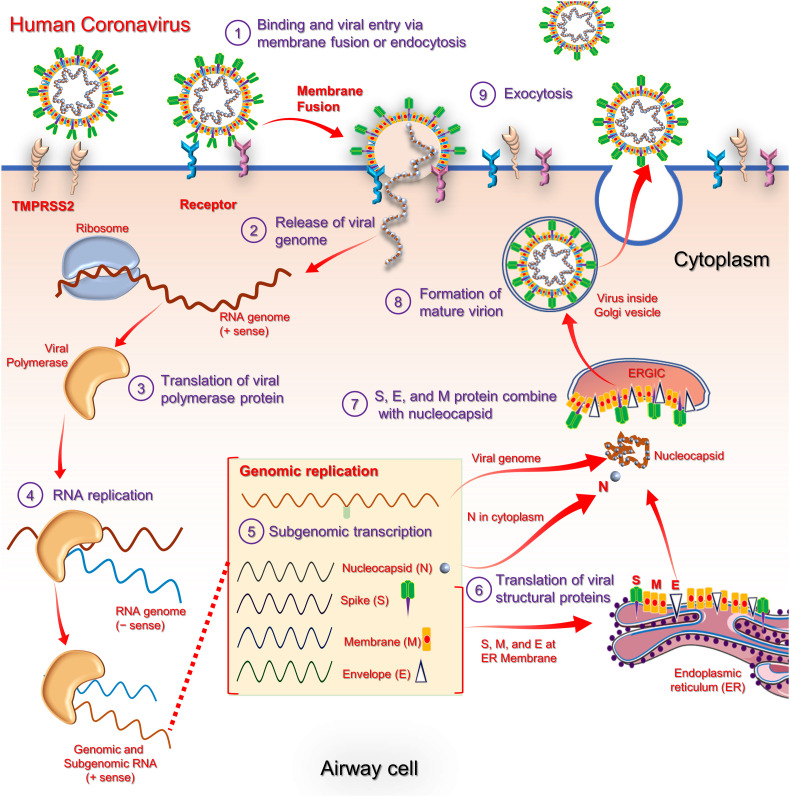
After virus and host cell membrane fusion event, virus released the nucleocapsid packed genomic RNA into cellular cytoplasm under the influence of induced structural conformation changes (Fehr and Perlman, 2015). Then, viral genome acted as a mRNA and cell's ribosome translates two-thirds of this RNA, crossponds to ORF1a and ORF1b into two large overlapping polyproteins (pp): pp1a and pp1ab. The larger polyprotein pp1ab translated from a -1 ribosomal frame shift triggered by slippery sequence (UUUAAAC) and downstream RNA pseudo knot at end of ORF1a (Masters, 2006). This ribosomal frameshift enabled continuous translation of ORF1a followed by ORF1b (Fehr and Perlman, 2015). The encoded polyproteins possess the proteases; PLpro and Mpro which assisted in generation of 16 nsps (nsp1-nsp16) from polyprotein pp1ab, including several replication proteins such as RdRp, RNA helicase, and exoribonuclease (ExoN) (Fehr and Perlman, 2015). Moreover, multifunction and enzyme activities for specific nsps in the previously reported HCoV, i.e. SARS-CoV and MERS-CoV, have been discovered over the past years and summarized elsewhere (Masters, 2006; Ziebuhr, 2005; Song et al., 2019).
6.3. Replication and transcription
Most of the newly translated nsps together with structural protein, i.e. N protein, formed the multi-protein replicase-transcriptase complex (RTC) which further conducted the viral genome replication and transcription (Fehr and Perlman, 2015). At the site of replicative organelles (Knoops et al., 2008), RTC complex contained RdRp as main replicase-transcriptase protein for the synthesis of negative-sense subgenomic (sg) RNA strands from viral RNA and transcription of negative-sense subgenomic RNA molecules from corresponding positive-sense mRNAs (Fehr and Perlman, 2015). The newly produced minus strands of genomic and sgRNAs are subsequently employed as templates for production of positive sense strands (mRNAs), specifically in generation of genomic RNA (genome replication) and sg mRNAs (transcription). These newly synthesized RNA strands acted as genome for generated new viral progeny. Also, several smaller mRNAs are produced from viral last third of genome which tracks reading frames ORF1a and ORF1b, interpreted into viral four structural proteins (S, E, M, and N) and along with accessory proteins (ORF3a to ORF9b) become part of viral progeny (Fehr and Perlman, 2015). Besides, other nsps in RTC complex also assist in viral replication and transcription process (Fehr and Perlman, 2015). For instance, nsp15 protein, a 3'-5' exoribonuclease, provides an extra fidelity to RTC complex via proofreading function in RNA replication, which lacked in RdRp. Likewise, nsp7 and nsp8 proteins generated a hexadecameric sliding clamp for RTC complex to catalyst the processivity of RdRp (Fehr and Perlman, 2015). These increased fidelity and processivity are essentially required in CoVs during RNA synthesis because of their relatively bulky RNA genome by comparison to other RNA viruses (Sexton et al., 2016).
6.4. Assembly and release
The viral RNA translation process occurred inside host cells’ endoplasmic reticulum lead to formation of structural proteins, viz. S, E and M, which moved along the secretory pathway into Golgi intermediate compartment. Herein, N protein packed the newly produced RNA genome and thereby shaped into a helical nucleocapsid. Following, virion assembly is triggered by M protein via multiple protein-protein interactions which assisted in incorporation of nucleocapsid, envelope, and spike proteins into virus particles (Fehr and Perlman, 2015). Later, viral progeny germinated into endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and released as secretory vesicles which fused with plasma membrane; finally secreted from host cell by exocytosis (Fig. 6 ) (Fehr and Perlman, 2015; Lim et al., 2016).
Fig. 6.
An overview for the life cycle of HCoVs in the host-cell (Song et al., 2019; Zumla et al., 2015)
7. Host response and CoVs immune evasion
Generally, human population lacks immunity against CoVs and hence, susceptible to novel CoV infections such as SARS-CoV-2. Since, immune response against SARS-CoV-2 is not fully deciphered, recent studies suggested to refer the previous reports on other HCoVs, especially SARS-CoV and MERS-CoV (Fig. 7 ) (Yi et al., 2020).
Fig. 7.
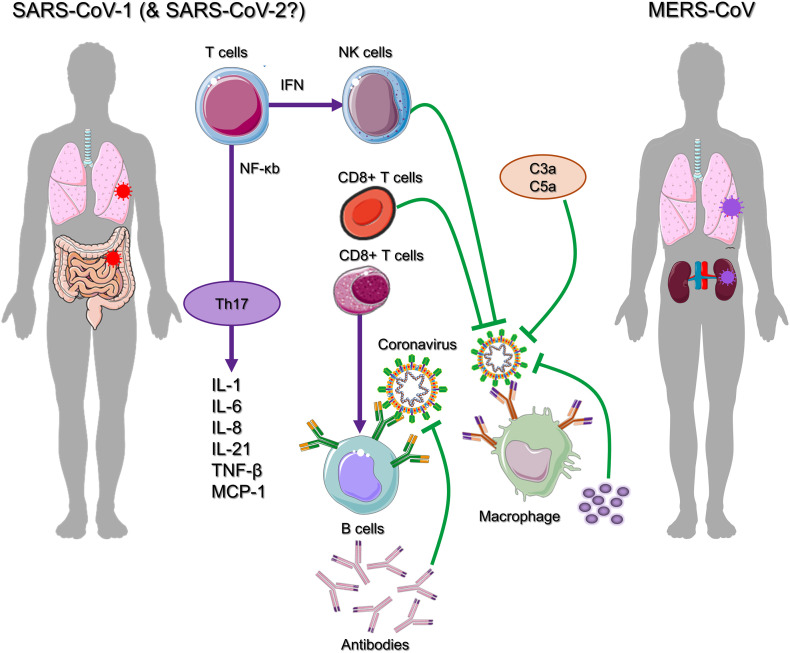
Cytokine pathogenesis of coronavirus disease (Yi et al., 2020).
In human body, interferon type I (IFN-I) or IFN-α/β production plays an antiviral role during early stages of viral infections (Channappanavar et al., 2019). As established earlier, pathogen-associated molecular patterns, often known as PAMPs are the molecular structures specific to virus infections such as uncapped mRNA or double-stranded RNA (dsRNA), are distinguished by pathogen recognition receptors (PRRs), including retinoic acid-inducible gene-I (RIG-I; also called as DDX58) like receptor (RLR), C-type lectin-like receptors, Toll-like receptor (TLR), or melanoma differentiation-associated protein 5 (MDA5; also called as IFIH1), PKR (protein kinase, RNA activated), and NOD-like receptor (NLR) (Ben Addi et al., 2008), of the host. These events in turn triggered the upregulation of IFN genes. Remarkably, TLRs are specially exhibited by immune cells, especially on myeloid dendritic cells (mDCs) and plasmacytoid cells (pDCs), while RLRs and PKR are considered to be in active state on all the nucleated cells. Thus, TLRs mainly mediated the detection of viral RNA in mDCs (such as TLRs3 and some TLR7) and pDCs by TLR7 (and TLR8 in human pDCs) (Schreibelt et al., 2010). Based on a specific PRR, various-partially cross-talking-signaling pathways resulted in promoters transactivation of antiviral genes (O'Neill et al., 2013). For instance, a prototypical PAMP relevant for CoVs is dsRNA, a by-product from genome replication and transcription (Weber et al., 2006; Zielecki et al., 2013). dsRNA can also be detected by TLR3 in the endosome while sensed by RNA helicases RIG-I, MDA5, and kinase PKR in the cytoplasm (Rasmussen et al., 2009; Yim and Williams, 2014; Yoneyama et al., 2016). Also, specific ssRNAs are presented as PAMPs, either if they displayed particular features or in the wrong location; for example, TLR7 detects GU-rich ssRNA in the endosome (Heil et al., 2004). Thus, numerous types of PRRs are regularly surveying the intracellular and extracellular space to sense virus infections in a sensitive and timely manner.
The detection of viral infection triggers complex signaling cascades such as MYD88 that induced manufacture of type I IFNs and stimulation of transcription factor nuclear factor-κB (NF-κB). In turn, active NF-κB induced transcription of pro-inflammatory cytokines (Fig. 7). Moreover, Type I IFNs signal by IFNα/β receptor (IFNAR) and following downstream signal activators and transducers of transcription (STAT) proteins triggered the manufacture of antiviral proteins that are programmed by interferon-stimulated genes (ISGs) like IFN-induced protein with tetratricopeptide repeats 1 (IFIT1). Altogether, this launches an antiviral immune response that restricts viral replication in infected and in neighbouring cells (de Wit et al., 2016). However, studies have showed that family of CoVs can significantly suppressed human immune responses against viral infection by evading immune detection machinery (Kikkert, 2020). For instance, papain-like protease (PLpro) domain of nsp3 of SARS-CoV interrelated with IFN regulatory factor 3 (IRF3) and inhibited its activation (Devaraj et al., 2007; Frieman et al., 2009). Besides, PLpro was reported to cause deubiquitination (or inhibit ubiquitination) of Tank-binding kinase 1 (TBK1), RIG-I, and IRF3 (Clementz et al., 2010; Devaraj et al., 2007; Frieman et al., 2009; Sun et al., 2012). The several functions and mechanisms adopted to counteract IFN induction, and strategies to develop resistance against IFN by previously pandemic HCoVs are summarized earlier (Kindler et al., 2016). Remarkably, SARS-CoV and MERS-CoV were reported to induced production of double-membrane vesicles that lack PRRs, replicated in these vesicles, and thereby escaped host detection system for viral dsRNA (de Wilde et al., 2013; Knoops et al., 2008). Moreover, CoVs induced very little IFN production in most of cell types (Kindler et al., 2016). Thus, high levels of dsRNA, which are generated during viral replication cycle (Weber et al., 2006; Zielecki et al., 2013), do not caused an adequate induction of IFN system.
Moreover, antigen presentation subsequently stimulated body’s humoral and cellular immunity on viral invasion, which are mediated by virus-specific B and T cells. T lymphocytes, including Cluster of differentiation 4 (CD4+) and Cluster of differentiation 8 (CD8+) T cells, play a central role against viral infection, stimulated B cells to produce virus-specific antibodies while CD8+ T cells directly kills virus-infected cells. Also, humoral immunity, including complements such as C3a, C5a, and antibodies, are critical in fighting viral infection (Fig. 7) (Mathern and Heeger, 2015; Traggiai et al., 2004). Recent flow cytometric analysis of peripheral blood from SARS-CoV-2-infected patients showed substantially reduced CD4+ and CD8+ T cells number while their status was excessively activated as evident from high proportions of human leukocyte antigen – DR isotype (HLA-DR) (CD4 3.47%) and CD38 (CD8 39.4%) double positive fractions (Xu et al., 2020). Similarly, acute phase response in patients with SARS-CoV was associated with severe decrease of CD4+ T and CD8+ T cells. Even if there is no antigen, CD4+ and CD8+ memory T cells are known to persist for four years as observed in SARS-CoV recovered individuals and were able to perform T cell proliferation, delayed-type hypersensitivity (DTH) response and production of IFN-γ (Fan et al., 2009). Also, six years after SARS-CoV infection, specific T-cell memory response to SARS-CoV S peptide library was identified in 14 of 23 recovered SARS patients (Tang et al., 2011). The specific CD8+ T cells were documented to exhibit a similar effect on MERS-CoV clearance in mice (Zhao et al., 2014). These discoveries could be helpful to deduce a valuable information for the rational design of vaccines against SARS-CoV-2. For example, antibodies collected from a recovered patient were reported to neutralized MERS-CoV infection (Niu et al., 2018). Moreover, T helper cells also produced proinflammatory cytokines to assist the defending cells. However, CoV (SARS-CoV and MERS-CoV) were established to halt T cell function and induced their apoptosis (Chu et al., 2016; Mathern and Heeger, 2015; Yeung et al., 2016). Besides, SARS-CoV-2 assumed to attach with ACE2 as in SARS-CoV infection, target and infect the same set of host cells (Zhao et al., 2020; Zhou et al., 2020b; Zou et al., 2020). Additionally, ACE2-associated lung injury was documented in SARS-CoV infection (Imai et al., 2005; Kuba et al., 2005); S protein of SARS-CoV downregulates ACE2 (Glowacka et al., 2010; Wang et al., 2008b) and induce detachment of catalytically active ACE2 ectodomain (Haga et al., 2008; Jia et al., 2009). This results in non-functional pulmonary ACE2 receptor which has been advised to be linked with acute lung injury (Imai et al., 2008; Kuba et al., 2006); decreased activity of ACE2 further triggered renin-angiotensin system (RAS) dysfunction and caused enhance inflammation, and vascular permeability (Gheblawi et al., 2020). In animal studies with murine acute respiratory distress (ARD) model showed association of reduced ACE2 function with an increased lung edema, neutrophil accumulation, enhanced vascular permeability, and diminished lung function (Imai et al., 2005). Also, in human airway epithelia, constitutive shedding of ACE2 receptor by activity of a metalloprotease 17 (ADAM17), also called as tumour necrosis factor alpha (TNF-α) converting enzyme (TACE), and disintegrin secrete functionally active soluble ACE2 (sACE2) (Lambert et al., 2005). HCoVs infection and inflammatory cytokines like interleukin 1 beta (IL-1β) and TNF-α were related with enhance ACE2 shedding (Haga et al., 2008; Jia et al., 2009). Notably, in vitro studies presented close association of SARS-CoV S protein-induced ACE2 shedding with TNF-α production (Haga et al., 2008). Although, biological function of sACE2 is yet unknown, however, recent studies have suggested direct involvement of sACE2 in inflammatory responses against SARS-CoV, and possibly also connected with SARS-CoV-2 (Fu et al., 2020). Additionally, apoptosis of type 2 alveolar cells caused release of specific inflammatory mediators which further instigate macrophages to secrete three essential immune substances; interleukin-1 (IL-1), interleukin-6 (IL-6), and TNF-α; also called as cytokines. These immune substances in the bloodstream produced infection symptoms linked to HCoVs (Huang et al., 2020).
Recent in vitro cell experiments showed that delay release of cytokines and chemokines occurred in respiratory epithelial cells, dendritic cells (DCs), and macrophages at the early stage of SARS-CoV-2 infection (Choi and Joo, 2020). Like SARS-CoV, MERS-CoV also infected human airway epithelial cells, THP-1 cells (a monocyte cell line), human peripheral blood monocyte-derived macrophages and DCs, and induced late but elevated levels of proinflammatory cytokines and chemokines (Tynell et al., 2016; Zhou et al., 2014). Notably, SARS-CoV-2 infection is suspected to cause pyroptosis in lymphocytes and macrophages. It was observed that lymphocytopenia is often seen in severe SARS-CoV-2 patients and cytokine release syndrome (CRS) induced by SARS-CoV-2 was also considered to be facilitate by leukocytes other than T cells (Shi et al., 2020). A vast majority of patients (82.1%) infected with SARS-CoV-2 have been logged for peripheral blood lymphopenia (Guan et al., 2020), supported the pulmonary infiltration of lymphocytes and/or cell damage via pyroptosis or apoptosis (Huang et al., 2020). Previous pandemics triggered by SARS-CoV and MERS-CoV were also reported with massive production of chemokines and cytokines; SARS-CoV exhibited CCL2, CCL3, CCl5, CXCL8, CXCL9, CXCL10, etc.; and IL-12, IL-18, IL-6, IL-1beta, IL-33, IFN-alpha, IFN-gamma, TNF-α, and transforming growth factor (TGF)-beta. Likewise, MERS-CoV was reported for elevated expression of cytokines IFN-α, IL-6, and chemokine (CXCL-10, CCL-5, and CXCL-8) (Zheng et al., 2020). Hence, acute respiratory distress syndrome (ARDS) is caused by cytokine storm that triggers a destruction in host cells via immune system and subsequently results into multiple organs failure or death as stated in case of SARS-CoV-2 outbreak; similar observations were noted in case of SARS-CoV infection (Kumar et al., 2020). Albeit, there is no direct data is available to correlate the participation of pro-inflammatory cytokines and chemokines in lung pathology through SARS-CoV and MERS-CoV, correlative observations recorded from patients with severe disease suggested a role of hyper-inflammatory responses in HCoV pathogenesis (Fig. 7) (Channappanavar and Perlman, 2017). For instance, in MERS-CoV infection, plasmacytoid dendritic cells, but not mononuclear macrophages and DCs (Scheuplein et al., 2015), were induced to produce a large amount of IFNs (Choi and Joo, 2020).
Despite of similar virus titers in respiratory tract, SARS-CoV-infected old nonhuman primates are more likely to develop immune dysregulation than infected young primates, leading to more severe disease manifestations (Smits et al., 2010). It seems that excessive inflammatory response rather than virus titer is more relevant to the death of the old nonhuman primates (Smits et al., 2010). Similarly, in Bagg Albino/c (BALB/c) mice infected with SARS-CoV, disease severity in old mice was related to early and disproportionately strong upregulation of ARDS-related inflammatory gene signals (Rockx et al., 2009). The rapid replication of SARS-CoV in BALB/c mice induces delayed release of IFN-α/β, which was accompanied by influx of many pathogenic inflammatory mononuclear macrophages (Channappanavar et al., 2016). Depleting inflammatory monocyte-macrophages or neutralizing inflammatory cytokine TNF protected the mice from fatal SARS-CoV infection. Thus, inflammatory mediators contributes an vital role in pathogenesis of ARDS, which was estimated as an leading cause of death in patients infected with SARS-CoV or MERS-CoV (Drosten et al., 2013; Lew et al., 2003). It is now known that several proinflammatory cytokines consequences of cytokine storm contribute to the occurrence of ARDS (Channappanavar and Perlman, 2017; Reghunathan et al., 2005). These mechanisms also lead to activation of coagulation pathways. Massive inflammation can impaired all the three control mechanisms like antithrombin III, tissue factor pathway inhibitor, and protein C system (Jose et al., 2014); with reduced anticoagulant concentrations due to reduced production and increasing consumption. This defective procoagulant–anticoagulant balance predisposes in the development of microthrombosis, disseminated intravascular coagulation, and multiorgan failure; as evident from severe SARS-CoV-2 pneumonia with raised d-dimer concentrations being a poor prognostic feature and disseminated intravascular coagulation common in non-survivors (Tang et al., 2020; Zhou et al., 2020a). In this over all proposed mechanism, proteinase-activated receptor-1(PAR-1) acts as main thrombin receptor and mediates thrombin-induced platelet aggregation, and interplay between coagulation, inflammatory, and fibrotic responses; all of these factors are an important aspects of pathophysiology of fibroproliferative lung disease (Jose et al., 2014), such as seen in SARS-CoV-2. On the other hand, the proposed events of cytokine storms in case of SARS-CoV-2 was suggested to increased inflammation-related biomarkers, including C-reactive protein (CRP), ferroprotein, erythrocyte sedimentation rate (ESR), and IL-6 (Chen et al., 2020; Huang et al., 2020). In conclusion, based on previous HCoVs infection, severity and pathogenesis of SARSCoV-2 was suggested to divided into three stages; stage-1, an asymptomatic incubation time with or without detectable virus; stage-2, non-severe symptomatic period and detection of viral particles; stage-3, severe respiratory symptomatic stage and detection of high viral load (Wang et al., 2020).
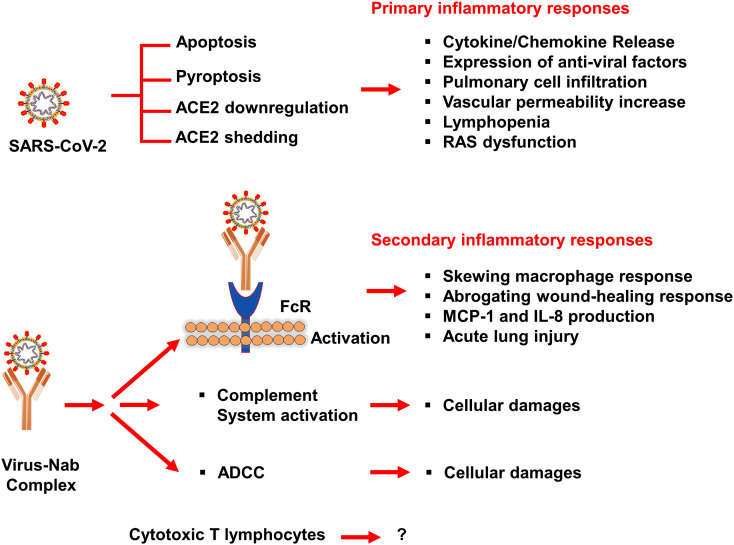
Furthermore, based on previous studies of SARS-CoV, the inflammatory responses in SARS-CoV-2 infection can be classified into primary and secondary responses (Shi et al., 2020). Initially, primary inflammatory responses are produced subsequently on viral infection, prior to arrival of neutralizing antibodies (NAb). The activation of these factors is primarily driven by viral-mediated ACE2 downregulation and shedding, vigorous viral replication, and host antiviral defense system. Secondary inflammatory responses are generated by the adaptive immunity and NAb. Moreover, virus-NAb complex was also suggested to triggered Fc receptor (FcR)-mediated inflammatory responses and acute lung injury that resulted in persistent viral replication and inflammatory responses from host macrophages (Fig. 8 ) (Fu et al., 2020). It was observed that patients could survive in primary inflammatory responses but secondary inflammatory responses could be fatal. Secondary inflammatory responses in which virus-Nab complex formed was considered as targeted to develop potential therapeutic strategy along with anti-inflammatory drugs (Fu et al., 2020).
Fig. 8.
Possible mechanisms of SARS-CoV-2-mediated inflammatory responses based on previous study of SARS-CoV (Fu et al., 2020).
8. Conclusion
Accumulating data on HCoVs exhibits the importance of understanding the zoonotic viral transmission and pathogenesis in humans. Recent studies on virus have established the viral proteins as prime target in the drug designing and vaccine development; however, this review have highlighted the utmost importance of intermediate host for the viral transmission to human. Future approaches, therefore, should considered the strains from intermediate hosts in the therapeutic development pipelines to better understand the severity of virulence via genome recombination process in CoVs. We also concluded major damaged caused by the failure of host immune system against HCoV infection and further caused by insurgence of cytokines which lead to severe mutilation of the host body. Synergistic and/or multi-target strategies that check viral growth and malfunctioned immune system of the host cells simultaneously might also be successful in battle against HCoVs. Nevertheless, synergistic approaches must take into account crossponding to the complementary target pathways. When developing novel therapeutic strategies to check the immunoregulatory cytokines such as TNFβ and IL6, investigation should be considered on the viral strain and targeted organ specificity; for example, SARS-CoV-2 has more affinity to ACE2 which are scattering on different organs like lung and kidney while MERS-like CoV can even infect T-cells. In addition, we should be cognizant of the feedback responses that produce with any treatment and consider the possibility to leveraged in a combinatorial fashion, taking into consideration the outcomes of preceding standard care and/or target immunoregulatory molecules will affect the efficacy and incidence of virulence. Another utmost point to be consider is how to generalize the therapeutic decision under the different immunological responses to the viral infection; for instance, susceptibility of some persons and with asymptomatic symptoms or sever immune response to the infection. Given the relatively high emergence of HCoVs and potential threat to humans, future therapeutic strategies should involve deep immune profiling of the infected individuals and personalization of therapeutics when feasible to accelerate progress towards more effective treatment approaches. This goal is ever closer to becoming a reality as multiplexed imaging, immunophenotyping, and mutational analysis tools are increasing the high-throughput process. Although, many trials have failed before, but given the progress in our understanding on the interaction between host immune responses and viral escape plans, and the emerging sophisticated strategies, we have reasons to be hopeful for the future successful treatment against HCoVs.
Declaration of Competing Interest
Authors declares no conflict of interest.
References
- Ar Gouilh M., Puechmaille S.J., Diancourt L., Vandenbogaert M., Serra-Cobo J., Lopez Roig M., Brown P., Moutou F., Caro V., Vabret A., Manuguerra J.C. SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world. Virology. 2018;517:88–97. doi: 10.1016/j.virol.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour N., Ekande S., Cote G., Lachance C., Chagnon F., Tardieu M., Cashman N.R., Talbot P.J. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J. Virol. 1999;73:3326–3337. doi: 10.1128/jvi.73.4.3326-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour H.M., Elkhatib W.F., Rahman M.M., Elshabrawy H.A. Insights into the Recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9 doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena M., Oostergetel G.T., Bartelink W., Faas F.G.A., Verkleij A., Rottier P.J.M., Koster A.J., Bosch B.J. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc. Nat. Acad. Sci. U. S. A. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Nat. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses Basel. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Addi A., Lefort A., Hua X., Libert F., Communi D., Ledent C., Macours P., Tilley S.L., Boeynaems J.M., Robaye B. Modulation of murine dendritic cell function by adenine nucleotides and adenosine: involvement of the A(2B) receptor. Eur. J. Immunol. 2008;38:1610–1620. doi: 10.1002/eji.200737781. [DOI] [PubMed] [Google Scholar]
- Bin S.Y., Heo J.Y., Song M.S., Lee J., Kim E.H., Park S.J., Kwon H.I., Kim S.M., Kim Y.I., Si Y.J., Lee I.W., Baek Y.H., Choi W.S., Min J., Jeong H.W., Choi Y.K. Environmental contamination and Viral shedding in MERS patients during MERS-CoV outbreak in South Korea. Clin. Infect. Dis. Off. publication Infectious Diseases Society of America. 2016;62:755–760. doi: 10.1093/cid/civ1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A., Bhattacharjee U., Chakrabarti A.K., Tewari D.N., Banu H., Dutta S. Emergence of novel coronavirus and COVID-19: whether to stay or die out? Crit. Rev. Microbiol. 2020:1–12. doi: 10.1080/1040841X.2020.1739001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19 doi: 10.1128/CMR.00017-06. 531-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Cyranoski D.J.N. Vol. 577. 2020. Why snakes probably aren’t spreading the new China virus; p. 1. [DOI] [PubMed] [Google Scholar]
- Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lee S.J., Hsiao H.H., Wu W.J., Chang W.L., Lin C.H., Huang T.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever F.S., Daniels J.B., et al. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. J. Exp. Med. 1949;90:181–210. doi: 10.1084/jem.90.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C.C., Wong S.C., To K.K.W., Ho P.L., Yuen K.Y. Preparedness and proactive infection control measures against the emerging novel coronavirus in China. The Journal of Hospital Infection. 2020;104:254–255. doi: 10.1016/j.jhin.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.Y., Joo M. The pathogenesis and alternative treatment of SARS-CoV2. Integr Med Res. 2020;100421 doi: 10.1016/j.imr.2020.100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell G., Abdirizak F., Lee S., Lee J., Jung E., Nishiura H., Viboud C. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015;13:210. doi: 10.1186/s12916-015-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H.Y., Li C., Cheng Z.S., Lin X., Poon V.K.M., Sun T.H., Lau C.C.Y., Chan J.F.W., To K.K.W., Chan K.H., Lu L.W., Zheng B.J., Yuen K.Y. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014;454:197–205. doi: 10.1016/j.virol.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H.Y., Li C., Chan J.F.W., Cheng Z.S., Yang D., Wang D., Lee A.C.Y., Li C.G., Yeung M.L., Cai J.P., Chan I.H.Y., Ho W.K., To K.K.W., Zheng B.J., Yao Y.F., Qin C.A., Yuen K.Y. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Chen Z.B., Banach B.S., Wang Y.H., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K., Li K., Mesecar A.D., Baker S.C. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. New coronavirus threat galvanizes scientists. Science. 2020;367:492–493. doi: 10.1126/science.367.6477.492. [DOI] [PubMed] [Google Scholar]
- Cowling B.J., Park M., Fang V.J., Wu P., Leung G.M., Wu J.T. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Euro Surveillance Bulletin Europeen sur les Maladies Transmissibles European Communicable Disease Bulletin. 2015;20:7–13. doi: 10.2807/1560-7917.es2015.20.25.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. This scientist hopes to test coronavirus drugs on animals in locked-down Wuhan. Nature. 2020;577:607. doi: 10.1038/d41586-020-00190-6. [DOI] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A.M., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L.M., Snijder E.J., Stephens G.M., Woo P.C.Y., Zaki A.M., Zambon M., Ziebuhr J. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Kuo L., Masters P.S., Vennema H., Rottier P.J.M. Coronavirus particle assembly: Primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., van der Meer Y., Barcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T.K., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D., Junglen S., Muller M.A., Haas W., Guberina H., Rohnisch T., Schmid-Wendtner M., Aldabbagh S., Dittmer U., Gold H., Graf P., Bonin F., Rambaut A., Wendtner C.M. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H., Song H.C., Abrignani S., Covacci A., Rappuoli R. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- Fan Y.Y., Huang Z.T., Li L., Wu M.H., Yu T., Koup R.A., Bailer R.T., Wu C.Y. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods in Molecular Biology Clifton, N.J. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappa B signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol. Sin. 2020;35(3):266–271. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., Hu B., Shi Z.-L. Bat coronaviruses. Bats and Viruses. 2015:127–155. [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., Eichler J., Drosten C., Pohlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J.M. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for, C Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Korteweg C., McNutt M.A., Gu J. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 2008;133:4–12. doi: 10.1016/j.virusres.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K., Poon L.L., Perera R.A., Alhammadi M.A., Ng H.Y., Siu L.Y., Guan Y., Alnaeem A., Peiris M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83:7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Penninger J.M. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp. Physiol. 2008;93:543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy H., Fragoso G., Almazan G., Mushynski W.E., Talbot P.J. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology. 2006;349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Tan P., Shi L., Hickey M., Gakhar L., Chappell M.C., Wohlford-Lenane C., McCray P.B., Jr. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y., McKeever D., Mutua F., Young J., McDermott J., Pfeiffer D.U. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose R.J., Williams A.E., Chambers R.C. Proteinase-activated receptors in fibroproliferative lung disease. Thorax. 2014;69:190–192. doi: 10.1136/thoraxjnl-2013-204367. [DOI] [PubMed] [Google Scholar]
- Kanwar A., Selvaraju S., Esper F. Human coronavirus-HKU1 infection among adults in Cleveland. Ohio. Open Forum Infect Di. 2017;4 doi: 10.1093/ofid/ofx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M. Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V., Weber F. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv. Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausegger A., Strobl B., Regl G., Kaser A., Luytjes W., Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J. Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., van den Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Penninger J.M. Angiotensin-converting enzyme 2 in lung diseases. Curr. Opin. Pharmacol. 2006;6:271–276. doi: 10.1016/j.coph.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacology Therapeutics. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski A.J., Althaus C.L. The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Euro Surveillance Bulletin Europeen sur les Maladies Transmissibles European Communicable Disease Bulletin. 2015;20:14–18. doi: 10.2807/1560-7917.es2015.20.25.21167. [DOI] [PubMed] [Google Scholar]
- Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Coronavirus Disease 2019 (COVID-19) 2020. Host immune response and immunobiology of HUMAN SARS-CoV-2 infection; pp. 43–53. [Google Scholar]
- Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I., Hooper N.M., Turner A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J. Biol. Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To, K.F, Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Lew T.W., Kwek T.K., Tai D., Earnest A., Loo S., Singh K., Kwan K.M., Chan Y., Yim C.F., Bek S.L., Kor A.C., Yap W.S., Chelliah Y.R., Lai Y.C., Goh S.K. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. Jama. 2003;290:374–380. doi: 10.1001/jama.290.3.374. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.D., Shi Z.L., Yu M., Ren W.Z., Smith C., Epstein J.H., Wang H.Z., Crameri G., Hu Z.H., Zhang H.J., Zhang J.H., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S.Y., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4 doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q.A., Alvarez X., Wang H.B., Du Y.H., Zhu H., Jiang H., Zhou J.Y., Lam P., Zhang L.Q., Lackner A., Qin C.A., Chen Z.W. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of Rhesus Macaques. J. Virol. 2011;85:4025–4030. doi: 10.1128/JVI.02292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Bioscience Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Lu R.J., Zhao X., Li J., Niu P.H., Yang B., Wu H.L., Wang W.L., Song H., Huang B.Y., Zhu N., Bi Y.H., Ma X.J., Zhan F.X., Wang L., Hu T., Zhou H., Hu Z.H., Zhou W.M., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J.Y., Xie Z.H., Ma J.M., Liu W.J., Wang D.Y., Xu W.B., Holmes E.C., Gao G.F., Wu G.Z., Chen W.J., Shi W.F., Tan W.J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T.S., O'Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R.C., Mills J.N., Timonin M.E., Willis C.K.R., Cunningham A.A., Fooks A.R., Rupprecht C.E., Wood J.L.N., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? P Roy Soc B Biol Sci. 2013;280 doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.S., Sircar S., Bhat S., Sharun K., Dhama K., Dadar M., Tiwari R., Chaicumpa W. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. The Veterinary Quarterly. 2020;40:68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66 doi: 10.1016/S0065-3527(06)66005-3. 66, 193-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathern D.R., Heeger P.S. Molecules great and small: the complement system. Clinical Journal American Society Nephrology : CJASN. 2015;10:1636–1650. doi: 10.2215/CJN.06230614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H., Alhakeem R., Durosinloun A., Al Asmari M., Islam A., Kapoor A., Briese T., Daszak P., Al Rabeeah A.A., Lipkin W.I. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Dinnon K.H., Yount B.L., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L.S., Graham B., Randell S.H., Lipkin W.I., Baric R.S. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2020;94 doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S., Immunology Infection . 2020. Genotype and phenotype of COVID-19: Their roles in pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.J., Lattwein E., Hilali M., Musa B.E., Bornstein S., Drosten C. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C.M., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Nao N., Yamagishi J., Miyamoto H., Igarashi M., Manzoor R., Ohnuma A., Tsuda Y., Furuyama W., Shigeno A., Kajihara M., Kishida N., Yoshida R., Takada A. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. Mbio. 2017;8 doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Milligan R.A., Yeager M., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu P., Zhang S., Zhou P., Huang B., Deng Y., Qin K., Wang P., Wang W., Wang X., Zhou J., Zhang L., Tan W. Ultrapotent human neutralizing antibody repertoires against middle east respiratory syndrome coronavirus from a recovered patient. J. Infect. Dis. 2018;218:1249–1260. doi: 10.1093/infdis/jiy311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Weekley C.C., Daszak P. Are bats really 'special' as viral reservoirs? what we know and need to know. Bats and Viruses. 2015:281–294. [Google Scholar]
- Omrani A.S., Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction. Pathogens and Global Health. 2015;109:354–362. doi: 10.1080/20477724.2015.1122852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A.J., Golenbock D., Bowie A.G. The history of toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y., Group, H.U.S.S. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene F., Merlat A., Vabret A., Rozenberg F., Buzyn A., Dreyfus F., Cariou A., Freymuth F., Lebon P. Coronavirus 229E-related pneumonia in immunocompromised patients. Clinical Infectious Diseases Official Publication Infectious Diseases Society of America. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Farag E.A., Reusken C.B., Lamers M.M., Pas S.D., Voermans J., Smits S.L., Osterhaus A.D., Al-Mawlawi N., Al-Romaihi H.E., AlHajri M.M., El-Sayed A.M., Mohran K.A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., El-Maghraby M.M., Koopmans M.P., Haagmans B.L. Isolation of MERS coronavirus from a dromedary camel, Qatar, 2014. Emerg. Infect. Dis. 2014;20:1339–1342. doi: 10.3201/eid2008.140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.B., Reinert L.S., Paludan S.R. Innate recognition of intracellular pathogens: detection and activation of the first line of defense. Apmis. 2009;117:323–337. doi: 10.1111/j.1600-0463.2009.02456.x. [DOI] [PubMed] [Google Scholar]
- Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Smits-De Vries L., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Baas T., Zornetzer G.A., Haagmans B., Sheahan T., Frieman M., Dyer M.D., Teal T.H., Proll S., van den Brand J., Baric R., Katze M.G. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83:7062–7074. doi: 10.1128/JVI.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J.S., Lam T.T., Ahmed M.M., Li L., Shen Y., Abo-Aba S.E., Qureshi M.I., Abu-Zeid M., Zhang Y., Khiyami M.A., Alharbi N.S., Hajrah N.H., Sabir M.J., Mutwakil M.H., Kabli S.A., Alsulaimany F.A., Obaid A.Y., Zhou B., Smith D.K., Holmes E.C., Zhu H., Guan Y. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Revue Scientifique et Technique Office International des épizooties. 2004;23:643–660. doi: 10.20506/rst.23.2.1513. [DOI] [PubMed] [Google Scholar]
- Scheuplein V.A., Seifried J., Malczyk A.H., Miller L., Hocker L., Vergara-Alert J., Dolnik O., Zielecki F., Becker B., Spreitzer I., Konig R., Becker S., Waibler Z., Muhlebach M.D. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J. Virol. 2015;89:3859–3869. doi: 10.1128/JVI.03607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreibelt G., Tel J., Sliepen K.H.E.W.J., Benitez-Ribas D., Figdor C.G., Adema G.J., de Vries I.J.M. Toll-like receptor expression and function in human dendritic cell subsets: implications for dendritic cell-based anti-cancer immunotherapy. Cancer Immunol. Immunother. 2010;59:1573–1582. doi: 10.1007/s00262-010-0833-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton N.R., Smith E.C., Blanc H., Vignuzzi M., Peersen O.B., Denison M.R. Homology-based identification of a mutation in the coronavirus rna-dependent rna polymerase that confers resistance to multiple mutagens. J. Virol. 2016;90:7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G., Walker P.J., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018) Arch. Virol. 2019;164:943–946. doi: 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., de Lang A., van den Brand J.M., Leijten L.M., van I.W.F., Eijkemans M.J., van Amerongen G., Kuiken T., Andeweg A.C., Osterhaus A.D., Haagmans B.L. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smuts H. Human coronavirus NL63 infections in infants hospitalised with acute respiratory tract infections in South Africa. Influenza Other Respir. Viruses. 2008;2:135–138. doi: 10.1111/j.1750-2659.2008.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11 doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xing Y.L., Chen X.J., Zheng Y., Yang Y.D., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C., Chen Z.B. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012:7. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory b cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. Journal Thrombosis Haemostasis JTH. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynell J., Westenius V., Ronkko E., Munster V.J., Melen K., Osterlund P., Julkunen I. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte-derived macrophages and dendritic cells. J. Gen. Virol. 2016;97:344–355. doi: 10.1099/jgv.0.000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 2003;36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D.M.E., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. Mbio. 2012:3. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Maes P., Duson G., Van Ranst M. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 2005;43:5452–5456. doi: 10.1128/JCM.43.11.5452-5456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Duengkae P., Rodpan A., Kaewpom T., Maneeorn P., Kanchanasaka B., Yingsakmongkon S., Sittidetboripat N., Chareesaen C., Khlangsap N., Pidthong A., Leadprathom K., Ghai S., Epstein J.H., Daszak P., Olival K.J., Blair P.J., Callahan M.V., Hemachudha T. Diversity of coronavirus in bats from Eastern Thailand. Virol. J. 2015;12:57. doi: 10.1186/s12985-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Shin J.H., Falsey A.R. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J. Infect. Dis. 2013;208:1634–1642. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Guo F., Liu K., Wang H., Rao S., Yang P., Jiang C. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo W., Raj V.S., Schipper D., Kolijn K., van Leenders G.J.L.H., Bosch B.J., Bensaid A., Segales J., Baumgartner W., Osterhaus A.D.M.E., Koopmans M.P., van den Brand J.M.A., Haagmans B.L. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J. Virol. 2016;90:4838–4842. doi: 10.1128/JVI.02994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China (vol 579, pg 265, 2020) Nature. 2020;580:E7. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Li Y., Shen X., Goh G., Zhu Y., Cui J., Wang L.F., Shi Z.L., Zhou P. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23(297-301) doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3' and 5' ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.T., Xie W.Q., Xue X.Y., Yang K.L., Ma J., Liang W.X., Zhao Q., Zhou Z., Pei D.Q., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G.X., Chen S.J., Chen Z., Ma D.W., Bartlam M., Rao Z.H. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):1742–1752. doi: 10.1371/journal.pbio.0030324. [published correction appears in PLoS Biol. 2005 Nov;3(11):e428] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.T., Bartlam M., Rao Z.H. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr Pharm Design. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- Yeung M.L., Yao Y.F., Jia L.L., Chan J.F.W., Chan K.H., Cheung K.F., Chen H.L., Poon V.K.M., Tsang A.K.L., To K.K.W., Yiu M.K., Teng J.L.L., Chu H., Zhou J., Zhang Q., Deng W., Lau S.K.P., Lau J.Y.N., Woo P.C.Y., Chan T.M., Yung S.S., Zheng B.J., Jin D.Y., Mathieson P.W., Qin C.A., Yuen K.Y. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Lagniton P.N.P., Ye S., Li E., Xu R.H. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H.C.H., Williams B.R.G. Protein kinase R and the inflammasome. J. Interf. Cytokine Res. 2014;34:447–454. doi: 10.1089/jir.2014.0008. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Jogi M., Onomoto K. Regulation of antiviral innate immune signaling by stress-induced RNA granules. J. Biochem. 2016;159:279–286. doi: 10.1093/jb/mvv122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.F., Cui J.M., Huan X.J., Zheng H.Y., Huang J.H., Ling F., Li K.P., Zhang J.Q. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.L., Lin D.Z., Sun X.Y.Y., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368 doi: 10.1126/science.abb3405. 409-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.C., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J.X., Gale M.J., Baric R.S., Enjuanes L., Gallagher T., McCray P.B., Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA Expression Profiling of ACE2, The Receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020 doi: 10.1164/rccm.202001-0179LE. [published online ahead of print, 2020 Jul 14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H.Y., Cheng Z.S., Poon V.K.M., Sun T.H., Lau C.C.Y., Wong K.K.Y., Chan J.Y.W., Chan J.F.W., To K.K.W., Chan K.H., Zheng B.J., Yuen K.Y. Active replication of middle east respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. In: Coronavirus Replication and Reverse Genetics. Enjuanes L., editor. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2005. The coronavirus replicase; pp. 57–94. [Google Scholar]
- Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A.M., Matrosovich M., Becker S., Weber F. Human cell tropism and innate immune system interactions of human respiratory coronavirus emc compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 2013;87:5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J.W., Han P.Y., Hao J., Han Z.G. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med-Prc. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]