Abstract
Diatom data have been collected in large-scale biological assessments in the United States, such as the U.S. Environmental Protection Agency’s National Rivers and Streams Assessment (NRSA). However, the effectiveness of diatoms as indicators may suffer if inconsistent taxon identifications across different analysts obscure the relationships between assemblage composition and environmental variables. To reduce these inconsistencies, we harmonized the 2008–2009 NRSA data from nine analysts by updating names to current synonyms and by statistically identifying taxa with high analyst signal (taxa with more variation in relative abundance explained by the analyst factor, relative to environmental variables). We then screened a subset of samples with QA/QC data and combined taxa with mismatching identifications by the primary and secondary analysts. When these combined “slash groups” did not reduce analyst signal, we elevated taxa to the genus level or omitted taxa in difficult species complexes. We examined the variation explained by analyst in the original and revised datasets. Further, we examined how revising the datasets to reduce analyst signal can reduce inconsistency, thereby uncovering the variation in assemblage composition explained by total phosphorus (TP), an environmental variable of high priority for water managers. To produce a revised dataset with the greatest taxonomic consistency, we ultimately made 124 slash groups, omitted 7 taxa in the small naviculoid (e.g., Sellaphora atomoides) species complex, and elevated Nitzschia, Diploneis, and Tryblionella taxa to the genus level. Relative to the original dataset, the revised dataset had more overlap among samples grouped by analyst in ordination space, less variation explained by the analyst factor, and more than double the variation in assemblage composition explained by TP. Elevating all taxa to the genus level did not eliminate analyst signal completely, and analyst remained the most important predictor for the genera Sellaphora, Mayamaea, and Psammodictyon, indicating that these taxa present the greatest obstacle to consistent identification in this dataset. Although our process did not completely remove analyst signal, this work provides a method to minimize analyst signal and improve detection of diatom association with TP in large datasets involving multiple analysts. Examination of variation in assemblage data explained by analyst and taxonomic harmonization may be necessary steps for improving data quality and the utility of diatoms as indicators of environmental variables.
Keywords: Bioassessment, Diatoms, Random forest, Taxonomic inconsistency, Total phosphorus, Intercalibration, Data harmonization
1. Introduction
Diatoms are single-celled algae with siliceous cell walls (valves) that are important basal components of aquatic food webs and contributors to aquatic ecosystem function. Diatoms are also useful indicators of environmental conditions, because they are ubiquitous in aquatic environments, have taxon-specific environmental preferences, and have relatively rapid generation times (Potapova and Charles, 2002; Smol and Stoermer, 2010; Smucker et al., 2013). Diatom assemblage composition, along with other algal metrics such as biomass and non-diatom algal composition, can be used in the development of nutrient and biological criteria (Paul et al., 2017). As part of efforts to understand and monitor the condition of the nation’s freshwater resources, diatom data have been collected in large-scale assessments, including the U.S. Environmental Protection Agency’s (EPA’s) National Aquatic Resource Surveys (NARS) and the U.S. Geological Survey’s (USGS’s) National Water-Quality Assessment (NAWQA) program. These data are valuable resources for assessing biological condition, but the effectiveness of diatoms as indicators may suffer if inconsistent taxon identifications across different analysts obscure the relationships between assemblage composition and environmental variables.
Biological assessment aims to detect impairment in the structure and function of biological assemblages in response to environmental stressors. Nutrient chemical stressors continue to be widespread causes of impairment in rivers and streams. Nutrient pollution can contribute to large economic losses from negative effects on real estate, recreation, and human wellbeing (Dodds et al., 2009). Nearly half of river and stream length in the U.S. has high levels of surface water total phosphorus (TP) compared to least-disturbed regional reference sites (USEPA, 2016), and increases in TP over a decade appear ubiquitous across developed and undeveloped catchments (Stoddard et al., 2016). Indeed, TP is a high priority environmental variable for water managers. The strong relationship between algal assemblage structure with TP makes algae-based bioassessment useful for water managers. For example, Taylor et al. (2014) identified taxa with synchronous declines in abundance above threshold concentrations of TP, and many researchers have shown that a variety of metrics with strong correlations with TP can be developed from algal assemblage information (Porter et al., 2008; Smucker et al., 2013; Stevenson et al., 2013; Munn et al., 2018). Taxonomic inconsistency can obscure relationships between biota and environmental conditions. Without accurate and consistent taxonomic information, the practical use of diatoms in bioassessment can be limited.
Here, we address the challenge of reducing analyst bias in an EPA National Rivers and Streams Assessment (NRSA) dataset to both support indicator development based on past assessments and improve diatom data consistency in future assessments. In some datasets, efforts have been undertaken to resolve taxonomic issues (e.g., by “lumping,” or combining taxa that are often misidentified). However, the process used to resolve ambiguous names or reconcile outdated names has rarely been completely and transparently documented, which has hindered the replication of data processing and statistical analyses necessary for indicator development. Moreover, it is unclear whether genus-level or a mixed hierarchy approach (i.e., a combination of fine- and coarse-scale taxonomic resolution) is sufficient for bioassessment.
The purpose of this work was to harmonize taxonomy across analysts for the 2008–2009 NRSA dataset to reduce uncertainty and bias associated with taxonomic inconsistency. We then evaluated the effectiveness of harmonization and different taxonomic adjustments (i.e., genus-level and mixed hierarchy) by quantifying the variability explained by analysts in the original and revised datasets. Further, we examined how revising the dataset to reduce analyst signal can increase the statistical power of diatom data by reducing errors from inconsistent taxonomic identifications, and thereby reveal greater variation in assemblage composition explained by TP. Diatom data processed to maximize taxonomic consistency can help increase the quality of data that managers can use as biological indicators of aquatic ecosystems.
We hypothesized that a diatom dataset with greater taxonomic consistency would have:
decreased variation in assemblage composition explained by analyst, and
increased variation in assemblage composition explained by TP.
2. Materials and methods
2.1. Initial taxonomic revisions
We used a mixed hierarchy approach to harmonize and revise diatom names in the 2008–2009 NRSA diatom dataset (Fig. 1, Table 1). We first harmonized the taxa list with BioData version 13.2 (USGS, 2017). The USGS BioData program documents nomenclature for fish, invertebrates, and algae (USGS, 2017). Diatom taxonomy used in the NAWQA program was refined by experts through a series of workshops (ANSP, 1999–2007) with ongoing, versioned updates in BioData (USGS, 2017). The R function biodata_check() translates diatom species names to reflect current BioData taxonomy (Bishop, 2017; refer to A.1 for output of taxonomic conversion notes from biodata_check). For names that were not included in BioData, we used published taxonomic literature and the “Diatoms of the United States” website (Spaulding et al., 2010; website content now transferred to “Diatoms of North America” at diatoms.org) to update operational taxonomic units (OTUs) to a consistent nomenclatural system and to create “slash groups” of taxa documented as difficult to distinguish. We define a slash group as taxa that were combined while preserving the names of the individual taxa. For example, the varieties of Cocconeis placentula are difficult to distinguish when the raphe and rapheless valves are not intact (Potapova and Spaulding, 2013). The slash group Cocconeis placentula/Cocconeis placentula var. euglypta/Cocconeis placentula var. lineata represents this OTU. The resulting dataset (mixed1; Fig. 1, Table 1) represents harmonization effort prior to analyzing the data to assess variation explained by analyst, but includes updating names to current synonyms and using the literature to minimize potential taxonomic problems (refer to A.2 for the complete list of revisions and justifications). Because mixed1 OTUs were assigned to the most updated genus designations, the genus-level dataset (genus) was produced by elevating all OTUs in the mixed1 dataset to genus. All data manipulations and analyses in R were conducted using R version 3.4.1 (R Core Team 2017)
Fig. 1.
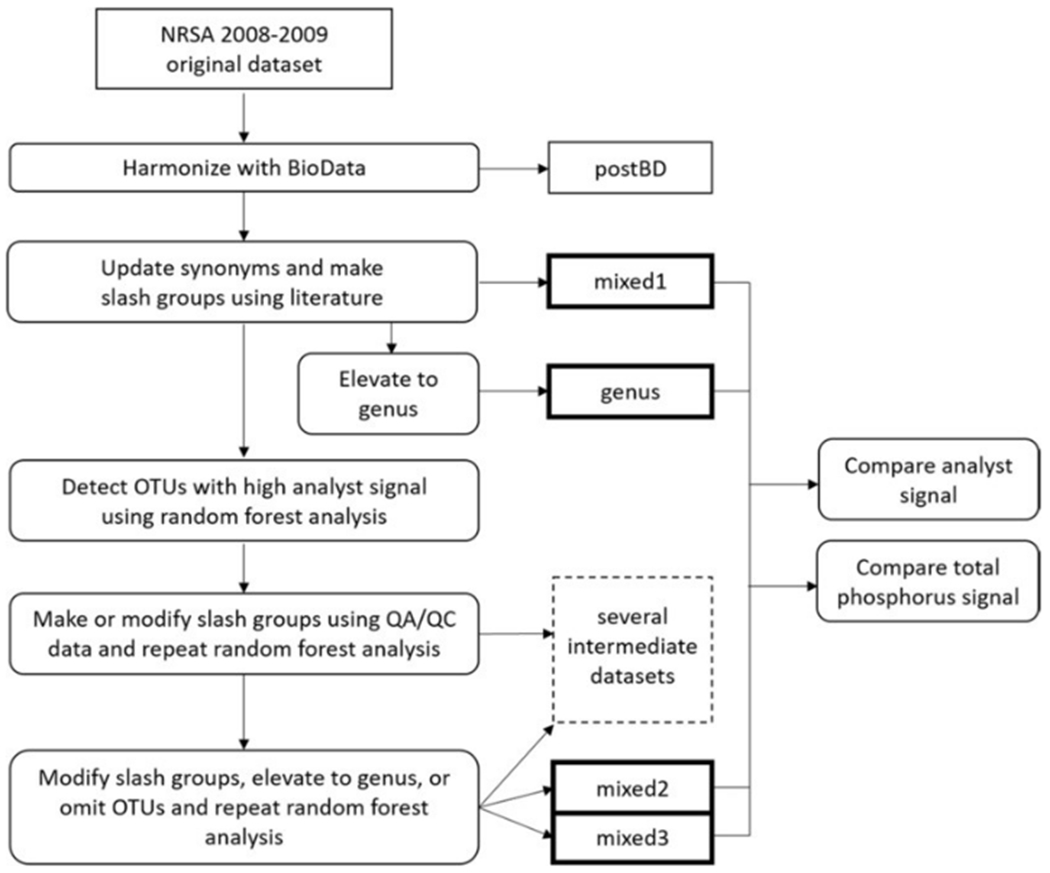
Process of harmonizing the 2008–2009 NRSA diatom dataset using a mixed hierarchy approach. Rectangles represent datasets and bold rectangles are revised datasets compared in final analyses. NRSA = National Rivers and Streams Assessment; OTU = Operational Taxonomic Unit; QA/QC = Quality Assurance/Quality Control. Refer to Table 1 for more details on actions taken to produce revised datasets.
Table 1.
Modification of the original 2008–2009 NRSA diatom data into revised datasets; for each dataset, information is provided on number of samples, number of OTUs, whether revisions were based on results of random forest, number of slash groups, number of OTUs elevated to genus, number of OTUs omitted, and an explanation of changes made to the dataset to produce the next revised dataset. Datasets in bold compared in subsequent analyses.
| Dataset | Samples | OTUs | Random forest | Slash groups | OTUs elevated to genus | OTUs omitted | Explanation of actions taken to produce revised datasets |
|---|---|---|---|---|---|---|---|
| original | 2292 | 1526a | - | - | - | - | Harmonized with BioData taxa list to produce postBD. |
| postBD | 1722 | 409 | - | - | - | - | Updated 56 OTUs with synonyms and made 65 slash groups based on references to produce mixed1. |
| genus | 1396 | 90 | - | - | 351 | - | Elevated all OTUs in mixed1 to genus. |
| mixed1 | 1714 | 351 | - | 65 | - | - | Added or modified 28 slash groups in mixed1 to produce inter1. |
| inter1b | 1714 | 306 | Y | 87 | - | - | Added or modified 8 slash groups in inter1 to produce inter2. |
| inter2 | 1714 | 297 | Y | 86 | - | - | Modified 1 slash group, elevated 14 Diploneis and 156 Nitzschia OTUs to genus, and omitted 7 small naviculoid OTUs in inter2 to produce inter3. |
| inter2 | 1714 | 297 | Y | 86 | - | - | Modified 4 slash groups, elevated 11 Diploneis and 130 Nitzschia OTUs to genus, and omitted 7 small naviculoid OTUs in inter2 to produce mixed2 (fewer OTUs elevated to genus compared to mixed3). |
| mixed2 | 1685 | 262 | Y | 74 | 143 | 7 | - |
| inter3 | 1683 | 259 | Y | 75 | 172 | 7 | Modified 1 slash group, elevated 21 Tryblionella OTUs to genus in inter3 to produce inter4. |
| inter4 | 1683 | 251 | Y | 67 | 193 | 7 | Modified 2 slash groups in inter4 to produce mixed3. |
| mixed3 | 1683 | 249 | Y | 68 | 193 | 7 | - |
Y indicates yes;
dash (−) indicates not applicable;
number of OTUs includes taxa with <50 occurrences;
“inter” indicates “intermediate dataset”.
2.2. Random forest analyses
We used random forest analysis (Breiman, 2001) to detect OTUs that may have been applied differently across analysts. Random forest provides a flexible tool for modeling relationships between many predictor variables and a single response. It is robust to different variable distributions and can effectively identify the relative importance of predictors in the presence of correlated variables and interactive effects. The relative abundance of an OTU may vary among sites due to differences in environment or due to analyst inconsistency. Random forest models can estimate the relative importance of analyst versus environmental variables in predicting the relative abundance of each OTU.
The original NRSA 2008–2009 dataset had 2292 samples and 1526 OTUs (Table 1). The total number of unique sites was 2115; samples from repeat visits to the same site were omitted from the analysis. To prepare the data for random forest analysis, we excluded samples with fewer than 400 valves counted after OTUs were adjusted. While the target valve count for NRSA was 600 valves per slide, the target was not always reached because of low density of diatoms or high density of debris in the samples. We included samples with at least 400 valves to retain a relatively large sample size. Thus, the total number of samples included in the analyses varied with the number of OTUs in the modified and genus-level datasets (Table 1). We then selected OTUs occurring in at least 50 samples to ensure that sufficient data were available to fit the random forest model. Count data were converted to relative abundances. In addition to analyst, we included predictor variables characterizing water chemistry, streambed substrate, water flow, canopy cover, human disturbance, stream size, and geographic location of the samples (refer to A.3 for the full list of predictor variables). The final necessary processing steps for inclusion of predictor variables in random forest analysis included log10(x + 1) or square root transformations for variables with skewed distributions and standardization of all continuous variables to unit mean and zero variance.
We fit a separate random forest model for each OTU and estimated the importance of each predictor variable by comparing the mean square error of the fitted model to the mean square error of the model with the value of that predictor variable randomly permuted (Breiman, 2001). The OTUs for which a random permutation of analyst accounted for the greatest increase in mean square error were flagged as being potentially problematic. In other words, random forest detected OTUs for which analyst explained the most variation in relative abundance, compared to all other predictor variables.
The OTUs detected by random forest as having the strongest analyst signals were each examined using QA/QC data available for a subset of the samples (143 slides). These 143 slides were counted once by a primary analyst, and again by a secondary analyst. There were three primary analysts and two secondary analysts. The QA/QC data were sorted by taxa with the greatest differences in number of valves counted on the same slide by the primary and secondary analysts. When possible, data from at least 5 slides were examined for each taxon to compare the taxa list and enumerations by the two analysts. Data from slides with mismatches in taxa names (i.e., primary analyst counted many valves of the OTU of interest while secondary analyst counted zero valves of the same OTU because they used a different OTU name) were prioritized in this step to maximize detection of cases where the same taxon was reported as different names by different analysts. Because diatom QA/QC transects do not perfectly match the transect of the primary analyst, differences between analysts’ counts can have multiple sources of variation (Lavoie and Campeau, 2016).
Multiple instances of taxa mismatch or lumping vs. splitting taxa were used to justify the combining of two or more OTUs into slash groups. References (e.g., Krammer and Lange-Bertalot, 1986, 1988, 1991a, 1991b; Spaulding et al., 2010) and voucher images from NRSA and other assessments in the Academy of Natural Sciences database (ANSP, 1998–2017) were examined to verify morphological similarity between OTUs that were combined. We repeated the process of random forest modeling and creation of slash groups to produce datasets with mixed hierarchy (intermediate datasets; Fig. 1). To further resolve analyst differences, slash groups and OTUs consistently exhibiting strong analyst signals were elevated to higher levels within the taxonomic hierarchy; when elevation to genus did not resolve analyst differences, the slash groups contributing to analyst differences were omitted (intermediate datasets; Fig. 1). We repeated random forest modeling and modification of slash groups until there was no further improvement in analyst signal (mixed2 and mixed3). The mixed2 dataset resulted from efforts to retain more taxonomic resolution by elevating fewer OTUs to genus compared to mixed3, while still resolving OTUs with high analyst signal. Refer to B.1–B.5 for the R scripts, C.1 for the full table of revisions in each of the datasets produced by the above process, and C.2 for the QA/QC data.
2.3. Comparison of analyst signals
To compare the strength of analyst signals in the harmonized and genus-level datasets, we used non-metric multidimensional scaling (NMDS) ordination plots to visually assess the effect of harmonization and taxonomic resolution on diatom assemblage data, as well as analysis of similarity (ANOSIM) to assess the magnitude of Bray-Curtis dissimilarities between samples associated with different analysts relative to within analyst group dissimilarities. We used the “anosim” function in the vegan package to perform ANOSIM (Oksanen et al., 2017). Because ANOSIM p-values are highly dependent on sample size, we interpreted only the R values to compare relative magnitudes of dissimilarity among sample groups. R values close to zero indicate little dissimilarity among groups and values close to unity indicate large dissimilarity among groups; R values near 0.2 indicate overlap in many OTUs among groups, but separable differences in relative abundance of the OTUs (Clarke and Warwick, 2001). We also assessed the goodness of fit of analyst as a factor in the NMDS plots using the “envfit” function in the vegan package, which averages ordination scores (i.e., obtains centroids) for factor levels and performs permutations to calculate a squared correlation coefficient (Oksanen et al., 2017).
2.4. Comparison of total phosphorus signals
We expected harmonization to reduce noise associated with errors resulting from taxonomic inconsistency and thereby increase variation in assemblage composition explained by environmental drivers. To examine whether harmonization reduced noise and improved diatom-inferred environmental signals, and to limit the effects of other factors affecting assemblage composition (Reavie et al., 2014), we selected OTUs from the harmonized and genus datasets for which TP was among the top 5 most important predictors of relative abundance from random forest analysis. With this data subset, we quantified the variation in assemblage composition explained by TP using permutational multivariate ANOVA (PERMANOVA) using the “adonis” function in the vegan package (Oksanen et al., 2017). We produced species response curves across the TP gradient for the selected OTUs using generalized additive modeling using the “goeveg” package (Goral and Schellenberg, 2017). Supplementary data and R scripts are available in an open access data repository at doi: https://doi.org/10.23719/1503373.
3. Results
3.1. Mixed1 and genus datasets
The original NRSA 2008–2009 dataset had 2292 samples and 1526 OTUs (Table 1). The total number of unique sites was 2115; samples from repeat visits to the same site were dropped. Because of some missing data, merging with environmental data brought the number of samples down to 1828. Of these, 106 samples with ≤400 valves were dropped, which brought the number of samples down to 1722. After harmonization with BioData, the number of OTUs decreased to 1474. Of these OTUs, 409 had at least 50 occurrences (i.e., at least 1 valve counted in ≥50 samples) in the dataset required to model OTU relative abundances with random forest (Table 1). We manually reviewed the 409 OTUs and updated 56 OTUs with synonyms (e.g., Psammothidium reversum/Achnanthes reversa) and made 65 slash groups to combine taxa with known issues based on information from references (e.g., Amphora inariensis/Amphora pediculus; Stepanek and Kociolek, 2011). With these changes, the number of OTUs decreased to 351 in the mixed1 dataset. Elevating the OTUs in the mixed1 dataset to genus level resulted in 90 genera.
3.2. Mixed2 and mixed3 datasets
The percent of taxa with analyst as the most important predictor of relative abundance decreased with dataset modification (Fig. 2). Random forest detected 39 OTUs in the mixed1 dataset with analyst as the most important predictor. To resolve these 39 OTUs, we made or modified 28 slash groups to produce the first intermediate (inter1) dataset. We repeated the random forest analysis, then made or modified 8 slash groups to produce inter2. Changes to inter2 to produce mixed2 were done while attempting to retain more taxonomic resolution by elevating a more conservative number of OTUs to genus (only the OTUs in inter2 with analyst as the predictor explaining the most variation in relative abundance were elevated). Several OTUs in the genus Nitzschia could not be resolved without creating increasingly large species complexes, so we elevated 130 Nitzschia OTUs in inter2 to genus. We retained several Nitzschia OTUs that may be consistently identified (e.g., Nitzschia dissipata/Nitzschia dissipata var. media, Nitzschia kurzeana/Nitzschia obtusa, Nitzschia sigmoidea/Nitzschia vermicularis, and a slash group containing several taxa with some morphological similarities to Nitzschia fonticola, Nitzschia frustulum, and Nitzschia perminuta. Additionally, analyst signal did not improve even when Sellaphora was elevated to the genus level, so 7 “small naviculoid” OTUs were omitted from inter2, including: Adlafia minuscula, Craticula molestiformis, Craticula subminuscula, Eolimna minima, Eolimna tantula, Mayamaea agrestis, and Mayamaea permitis. Further, attempts to resolve inconsistency of the Diploneis elliptica OTU by making slash groups based on QA/QC data were unsuccessful because of the low number of occurrences (63 occurrences in the full NRSA dataset). Thus, we elevated 11 Diploneis OTUs to the genus level in inter2.
Fig. 2.
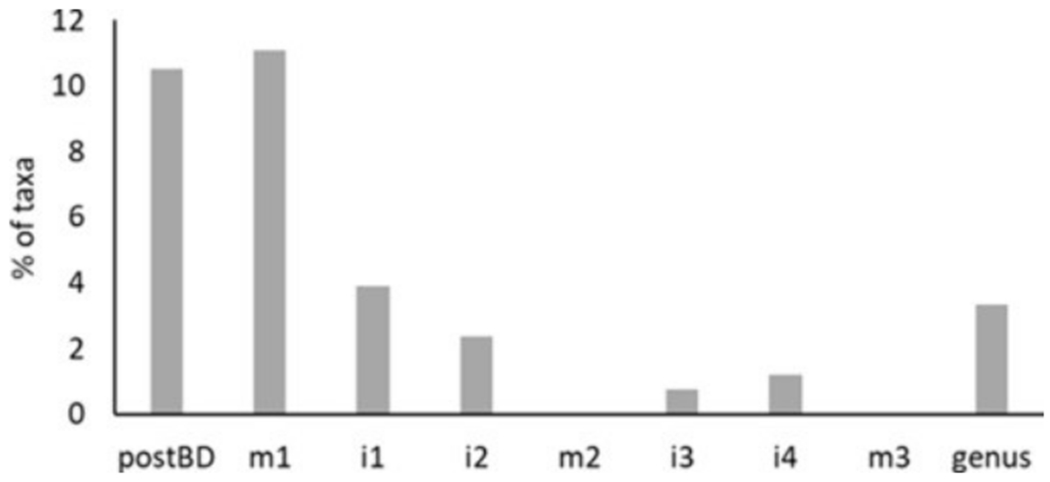
Percent of taxa with analyst as the most important predictor variable explaining variation in relative abundance, as determined by random forest analysis, in all dataset versions. The abbreviations “postBD,” “m,” and “i” indicate “post-BioData,” “mixed,” and “intermediate,” respectively.
We did not use a conservative approach for mixed3. Instead, we prioritized minimizing analyst signal by elevating all 157 OTUs in Nitzschia and all 15 OTUs in Diploneis to their respective genera and omitting the 7 “small naviculoid” OTUs to produce inter3. We repeated random forest analysis 2 more times. Tryblionella gracilis was another OTU that could not be resolved with slash groups and thus, all 21 Tryblionella OTUs were elevated to the genus level to produce inter4. After modifying 2 slash groups in inter4 to produced mixed3, application of random forest models to mixed3 showed that analyst was no longer the most important predictor of any OTU (Fig. 2). In mixed3, analyst was the second most important predictor of relative abundance for 6 out of 249 OTUs. For these 6 OTUs, the most important predictors included longitude, latitude, sulfate, or TN. For more detailed random forest output, see D.1.
3.3. Comparison of predictors
The predictor most frequently identified by random forest as being important in determining diatom relative abundance was conductivity (Table 2). In mixed1, the next most frequent predictor was analyst (39 OTUs), while longitude was the second most frequent predictor in mixed3 and genus. In genus, models for three genera, Sellaphora, Mayamaea, and Psammodictyon, identified analyst as the most important predictor. Models for Adlafia, Geissleria, and Cocconeis identified analyst as the second most important predictor.
Table 2.
Most frequent variables identified by random forest analysis as the most important predictors explaining variation in OTU relative abundance. Variables with the same number of OTUs are listed at the same rank. Analyst variable is shown in bold (= 0 OTUs in mixed2 and mixed3). For more details on variables, see Appendix A.3.
| Dataset (number of OTUs) | ||||
|---|---|---|---|---|
| Rank | mixed1 (351) | genus (90) | mixed2 (262) | mixed3 (249) |
| 1 | Conductivity (72) | Conductivity (16) | Conductivity (60) | Conductivity (51) |
| 2 | Analyst (39) | Longitude (11) | Longitude (33) | Longitude (36) |
| 3 | Longitude (38) | TN, Watershed area (7) | Potassium, pH, TN (15) | TN (16) |
| 4 | pH, Sulfate (21) | LOWFLOW, pH (6) | Sulfate (13) | pH (15) |
| 5 | Potassium (19) | TP, Sulfate (5) | BANKFULLFLOW (12) | Potassium (13) |
| 6 | TN (18) | Potassium (4) | TP, Nitrate, Watershed area (11) | LDCBF_G08, Sulfate (12) |
| 7 | LDCBF_G08, Watershed area (14) | Analyst, Ammonia, LDCBF_G08 (3) | Latitude, LOWFLOW (7) | TP, LOWFLOW (11) |
| 8 | TP (11) | Eco9CPL, Latitude, PCT_SAFN, Shade (2) | LDCBF_G08, PCT_SAFN (6) | Watershed area (10) |
| 9 | BANKFULLFLOW (10) | BANKFULLFLOW, DOC, MAVFLOWV, Nitrate, SiO2, Turbidity (1) | Ammonia, Turbidity (5) | BANKFULLFLOW (9) |
| 10 | LOWFLOW (8) | - | Dissolved organic carbon, Eco9UMW, MAVFLOWV (4) | Nitrate (6) |
LDCBF_G08 is log10(streambed critical diameter at bank-full flow).
LOWFLOW and BANKFULLFLOW are bed shear stress at low flow and bank-full flow, respectively.
MAVFLOWV is mean annual flow computed by Vogel method.
TP and TN are total phosphorus and total nitrogen, respectively.
Eco9CPL and Eco9UMW are coastal plains and upper Midwest ecoregions, respectively.
PCT_SAFN is percent of streambed surface consisting of sand and fine sediments.
Dash indicates no additional OTUs remaining.
3.4. Comparison of analyst signal
The best solutions for three-dimensional NMDS produced ordinations with stress = 0.19, 0.18, 0.17, and 0.16 for mixed1, genus, mixed2, and mixed3, respectively (Fig. 3). Mixed3 produced a NMDS plot with more overlap of the ellipsoid hull boundaries among samples grouped by analyst in ordination space compared to mixed1, mixed2, or genus (Fig. 3, see E.1 for ordination plots of original dataset and postBD). The analyst factor goodness of fit in the NMDS plots was lowest in mixed3 (r2 = 0.25, 0.16, 0.15, and 0.14 for mixed1, genus, mixed2, and mixed3, respectively). Mixed3 also had the lowest ANOSIM R, indicating the most similarity among samples grouped by analyst, relative to the other datasets (R = 0.17, 0.11, 0.08, and 0.06 for mixed1, genus, mixed2, and mixed3, respectively).
Fig. 3.
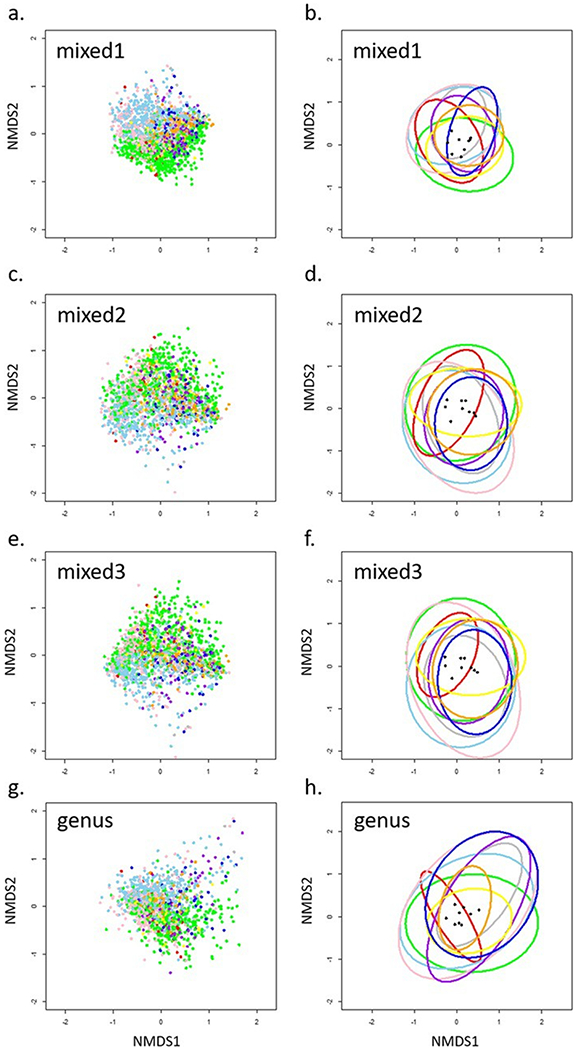
Nonmetric multidimensional scaling ordination plots of sites and ellipsoid hulls (ellipses that enclose all points of a group) around sites of NRSA 2008–2009 data: a) mixed1 sites, b) mixed1 hulls, c) mixed2 sites, d) mixed2 hulls, e) mixed3 sites, f) mixed3 hulls, g) genus sites, h) genus hulls. Colors indicate sites associated with 9 different analysts. Black points are centroids of the ellipsoid hulls. See E.1 for ordination plots of original data.
3.5. Comparison of total phosphorus signal
Mixed1, mixed2, and mixed3 had 54, 38, and 35 OTUs, respectively, for which TP was among the top 5 predictors of relative abundance. Of these, models for 11 to 13 OTUs identified TP as the most important predictor in mixed1, mixed2, and mixed3 (Table 3). Harmonization more than doubled the magnitude of variation explained by TP (PERMANOVA r2 = 0.08 and 0.20 for mixed1 and mixed3, respectively), indicating that the harmonization process helped to elucidate the strength of the relationship between assemblage composition and TP. Mixed2 had no improvement in variation explained by TP (PERMANOVA r2 = 0.08). Genus had 16 genera for which TP was among the top 5 predictors and PERMANOVA r2 = 0.05. Models for 5 of these 16 genera identified TP as the most important predictor (Table 3). Among the top 5 predictors, TP was the fourth and second predictor associated with Sellaphora and Mayamaea, respectively, but as reported above, analyst was the most important predictor of these two genera. See E.2 for species response curves.
Table 3.
Operational taxonomic units (OTUs) identified by random forest as having TP as the most important predictor of relative abundance in mixed1 (revised but not corrected for taxonomic inconsistency), mixed2 (harmonized with fewer OTUs elevated to genus), mixed3 (harmonized), and genus.
| Dataset | Operational Taxonomic Units | O | P |
|---|---|---|---|
| mixed1 | Achnanthidium minutissimum (Kütz.) Czarn. 1994 | 1466 | 10.4 |
| Ulnaria ulna (Nitzsch) Compère 2001 | 890 | 0.60 | |
| Gyrosigma acuminatum (Kütz.) Rabenh. 1853 | 318 | 0.11 | |
| Rhopalodia gibba (Ehrenb.) O.Müll. 1895 | 280 | 0.23 | |
| Caloneis silicula (Ehrenb.) Cleve 1894 | 235 | 0.10 | |
| Diadesmis confervacea Kütz. 1844 | 167 | 0.26 | |
| Navicula viridulacalcis Lange-Bert. in Rumrich, Lange-Bert. and Rumrich 2000 | 154 | 0.06 | |
| Aulacoseira italica (Ehrenb.) Simonsen 1979 | 120 | 0.09 | |
| Cymbella cistula (Ehrenb.) O.Kirchner 1878 | 108 | 0.09 | |
| Sellaphora bacillum (Ehrenb.) D.G.Mann 1989 | 80 | 0.02 | |
| Sellaphora stroemii (Hustedt) H.Kobayasi 2002 | 63 | 0.02 | |
| mixed2 | Achnanthidium minutissimum/Achnanthidium jackii/Achnanthidium reimeri/Achnanthidium deflexum/Achnanthidium rivulare | 1511 | 14.4 |
| Gyrosigma acuminatum (Kütz.) Rabenh. 1853 | 318 | 0.12 | |
| Encyonema microcephala | 251 | 0.30 | |
| Diadesmis confervacea Kütz. 1844 | 167 | 0.28 | |
| Navicula viridulacalcis Lange-Bert. in Rumrich, Lange-Bert. and Rumrich 2000 | 154 | 0.06 | |
| Diatoma tenuis C.Agardh 1812 | 112 | 0.07 | |
| Cymbella cistula (Ehrenb.) O.Kirchner 1878 | 108 | 0.09 | |
| Encyonopsis subminuta Krammer and E.Reichardt 1997 | 84 | 0.08 | |
| Gyrosigma strigilis (W.Smith) J.W.Griffin & Henfrey 1856 | 80 | 0.04 | |
| Sellaphora bacillum (Ehrenb.) D.G.Mann 1989 | 80 | 0.02 | |
| Sellaphora stroemii (Hustedt) H.Kobayasi 2002 | 63 | 0.02 | |
| mixed3 | Achnanthidium minutissimum/Achnanthidium jackii/Achnanthidium reimeri/Achnanthidium deflexum/Achnanthidium rivulare | 1511 | 14.4 |
| Fragilaria sepes/Ulnaria acus | 470 | 0.34 | |
| Gyrosigma acuminatum (Kütz.) Rabenh. 1853 | 318 | 0.12 | |
| Diadesmis confervacea Kütz. 1844 | 167 | 0.28 | |
| Navicula viridulacalcis Lange-Bert. in Rumrich, Lange-Bert. and Rumrich 2000 | 154 | 0.06 | |
| Aulacoseira italica (Ehrenb.) Simonsen 1979 | 120 | 0.09 | |
| Diatoma tenuis C.Agardh 1812 | 112 | 0.07 | |
| Encyonopsis subminuta Krammer and E.Reichardt 1997 | 84 | 0.08 | |
| Gyrosigma strigilis (W.Smith) J.W.Griffin & Henfrey 1856 | 80 | 0.04 | |
| Sellaphora bacillum (Ehrenb.) D.G.Mann 1989 | 80 | 0.02 | |
| Sellaphora stroemii (Hustedt) H.Kobayasi 2002 | 63 | 0.02 | |
| genus | Ulnaria (Kütz.) Compère 2001 | 1169 | 1.41 |
| Cymbella Agardh 1830 | 851 | 0.89 | |
| Gomphoneis Cleve 1894 | 502 | 0.57 | |
| Encyonopsis Krammer 1997 | 369 | 0.45 | |
| Diadesmis Kütz. 1844 | 191 | 0.33 |
O is number of occurrences;
P is mean percent abundance.
4. Discussion
Overall, harmonization efforts used to produce the mixed3 dataset resulted in improved taxonomic consistency. Analyst groups overlapped more in ordination space, with relatively lower ANOSIM R and lower goodness of fit. The genus dataset reduced, but did not eliminate, analyst signal. Even after harmonization, models for three genera identified analyst as the most important predictor. Analyst signal in Psammodictyon could be reduced by combination with a morphologically similar genus, Nitzschia, but this strategy did not work with small naviculoid taxa within Sellaphora and Mayamaea (data not shown). The coarse taxonomic resolution also resulted in fewer sensitive indicators of TP. The use of genus-level data may best be limited to applications that consider gradients related to genus-level characteristics of diatom adaptations, such as motility and pH tolerance (Hill et al., 2001), rather than as a method to resolve taxonomic inconsistencies.
Our results show that a mixed hierarchy approach that uses different harmonization strategies for different taxa can resolve inconsistent assemblage data while retaining information that may be lost using coarse taxonomic resolution (e.g., genus-level data). Mixed hierarchy approaches have been used successfully in other diatom-based assessments in Canada and Europe (Lavoie et al., 2009, Kelly and Ector 2012). Problematic diatom species have been identified and are treated as species complexes in Europe (Kahlert et al., 2016). Bioassessments using other indicators, such as macroinvertebrates, also accept multiple levels of taxonomic resolution depending on resource and analyst expertise levels. These programs usually aim for the lowest practical taxonomic resolution, and can include a combination of macroinvertebrate species, genera, and families (e.g., Barbour et al., 1999, KDOW, 2015). Like the slash groups used here, macroinvertebrate bioassessments also use slash groups if clear distinctions cannot be made between morphologically similar taxa (e.g., “Cricotopus/Orthocladius” taxa that are nearly indistinguishable at the larval stage) (USEPA, 2012).
Autecological information should inform harmonization strategies. A drastic harmonization strategy used in this study was omitting 7 OTUs of small naviculoid taxa from the dataset. Creating slash groups or elevating to genera did not improve taxonomic consistency in this group. Omitting these small naviculoids from the dataset helped minimize variation in assemblage data explained by analyst and reveal variation explained by TP. However, omitting these taxa may result in misclassification of sites on the basis of other environmental variables, such as organic pollution or general human disturbance (Kahlert et al., 2009). Small naviculoid taxa with morphology similar to Sellaphora nigri (De Not.) Wetzel and Ector, 2015 (commonly reported as Eolimna minima) often dominate freshwater benthic assemblages with organic pollution or human disturbance (van Dam et al., 1994, Wetzel et al., 2015). Wetzel et al. (2015) found a high diversity of morphology and wide variety of names applied by researchers to taxa in this group and Kahlert et al. (2016) noted several European countries where these taxa were a problematic group in intercalibration exercises. These small-celled taxa have sometimes been overlooked during enumeration when analysts did not use high quality microscopes (Kahlert et al., 2009). Analysts contributing to the NRSA dataset in the current study used high quality microscopes and were unlikely to have overlooked small-celled taxa. However, training and sharing of knowledge among analysts on how to distinguish these difficult taxa during the early stages of enumeration would have improved taxonomic consistency (Kahlert et al., 2009, 2016).
Harmonization improved taxonomic consistency and increased the variation explained by TP. The variation in assemblage composition explained by TP increased from 8% to about 20%, improving the detection and precision of the relationship between diatom assemblages and TP. The proportion of variation unexplained by TP may seem high, given that we selected a subset of OTUs for which random forest analysis identified TP as an important predictor. However, our result was greater or similar to that in other studies quantifying variation in diatom assemblages explained by TP. Reavie et al. (2014) improved the performance of a diatom-based transfer function for the Laurentian Great Lakes by selecting taxa with directional responses along the TP gradient that were minimally confounded by other environmental variables. They found TP explained 6.8% of the total variation (Reavie et al., 2014). The variation in diatom assemblage in a national-scale dataset that was explained by all included environmental variables (including TP) using partial canonical correspondence analysis was 11% (Potapova and Charles, 2002). In the same study, limiting the analysis to sites from a smaller geographic area (Northern Forests) increased the variation explained by environmental variables to 36.5%, but the variation explained by TP was not reported (Potapova and Charles, 2002).
A single environmental variable is unlikely to explain all the variation in observational assemblage data, but reducing noise related to taxonomic identification errors was important for uncovering a more precise relationship between diatom assemblages and TP. Reasons for the observed proportion of unexplained variation may include the many interacting factors that influence the statistical relationships between in-stream water chemistry and algal responses to nutrients, such as spatial and temporal variability in environmental variables (Wold and Hershey, 1999), such as light regime, land use, human disturbance, and climate (Beck et al., 2017).
Efforts to standardize diatom identification should begin early in bioassessment programs, rather than after sample enumeration. While research to better understand poorly distinguished groups of diatom taxa continues, it is important to thoroughly document OTUs with voucher images (e.g., Bishop et al., 2017). During enumeration, these voucher images represent a common resource for analysts to use and contribute to the project flora. Voucher flora should adequately represent the variation in morphology that occurs with size diminution of diatom valves to help analysts make more consistent identifications. Even those taxa that have not yet been described can be documented as OTUs and included in statistical analyses with confidence. After enumeration, voucher images offer permanent documentation of taxonomic decisions made for individual projects, providing a valuable reference for harmonization of datasets over time and across projects that expand the spatial scale of the study system. Coordinated communication and harmonization workshops before and during enumeration can also be very helpful for minimizing inconsistency. Both Kahlert et al. (2016) and Lavoie and Campeau (2016) observed the importance of communication between analysts and regional monitoring groups to reduce inconsistencies in diatom datasets. Moreover, the shared knowledge gained from intercalibration exercises among analysts was more important than analysts’ experience level with diatoms for producing more consistent datasets (Kahlert et al., 2009).
Researchers working with datasets created without standardized identification methods or combining data from multiple locations or monitoring programs to increase spatial coverage can adapt the method described here to assess and minimize taxonomic inconsistency. The R scripts and functions help to automate several steps of the harmonization process (https://doi.org/10.23719/1503373). Documentation of taxonomic updates and justifications for making slash groups are available in Appendices A through E, to help researchers improve taxonomic consistency in their data. While the specific actions to harmonize taxonomy in this study uncovered a more precise relationship between diatoms and TP, different actions (e.g., making slash groups, elevating to genus, omitting OTUs) may be appropriate for variables other than TP. Quantitative assessment of uncertainty and potential bias in the data is important to ensure data quality and appropriate application of large diatom datasets to development of water quality indicators.
5. Conclusions
Harmonization of diatom data through a series of defined steps can re-establish confidence in data quality, which has been uncertain in the past because of inconsistent identification across different analysts. Although the influence of analyst bias may not be completely removed from datasets, this work reveals the extent of the problem and provides a method to minimize analyst signal. Reducing analyst signal helped to minimize the confounding effect of taxonomic inconsistency across analysts to better detect and elucidate the magnitude of association between diatoms and TP. Examination of variation in assemblage composition explained by analyst and taxonomic harmonization may improve the quality of large datasets. Quantitative assessment of variation in assemblage composition explained by analyst provides a transparent indication of potential bias in the dataset. Taxonomic harmonization reduces potential bias in the dataset and can improve interpretation of how assemblage data associate with environmental variables. Taxonomic harmonization may influence conclusions about diatom responses to TP reached by diatom-based monitoring and assessment efforts using large datasets involving multiple analysts.
Supplementary Material
Highlights.
R tools automated several steps of harmonization.
Harmonization resolved all taxa with high analyst signal.
Analyst signal was still detectable, but minimized.
Revised data revealed a stronger association between diatoms and total phosphorus.
Acknowledgments
We thank Marina Potapova for providing QA/QC data and feedback on methods. We also thank several colleagues and reviewers who provided feedback and support, including: two anonymous reviewers, Nathan Smucker, Micah Bennett, Sue Norton, Kate Schofield, Britta Bierwagen, Scot Hagerthey, Phil Kaufmann, Daren Carlisle, Yong Cao, Amina Pollard, Susan Jackson, and Janice Alers-Garcia.
Footnotes
Publisher's Disclaimer: Disclaimer
The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ecolind.2019.01.061.
R scripts are available in EPA’s ScienceHub open access data repository at https://doi.org/10.23719/1503373.
References
- Academy of Natural Sciences of Drexel University in Philadelphia (ANSP). 1998–2017. 23 ANSP Full Taxa List. Phycology Section, accessed [19 April 2018], at URL http://diatom.ansp.org/taxaservice/ShowTaxonomy.ashx?taxonomy_id=23.
- Academy of Natural Sciences of Drexel University in Philadelphia (ANSP). 1999–2007. NAWQA Workshops, Patrick Center for Environmental Research, accessed [21 March 2018], at URL http://diatom.ansp.org/nawqa/Workshops.aspx. [Google Scholar]
- Barbour MT, Gerritsen J, Snyder BD and Stribling JB, Rapid Bioassessment Protocols for use in Streams and Wadeable Rivers: Periphyton, Benthic Macroinvertebrates and Fish, second ed., 1999, U.S. Environmental Protection Agency, Office of Water; Washington, D.C., EPA 841-B-99-002. [Google Scholar]
- Beck WS, Rugenski AT and Poff NL, Influence of experimental, environmental, and geographic factors on nutrient-diffusing substrate experiments in running waters, Freshw. Biol 62, 2017, 1667–1680. [Google Scholar]
- Bishop IW, 2017. biodata_check. Github repository, accessed [31 October 2017], at URL https://github.com/bishopia/biodata_check. [Google Scholar]
- Bishop IW, Esposito RM, Tyree M and Spaulding SA, A diatom voucher flora from selected southeast rivers (USA), Phytotaxa 332, 2017, 101–140. [Google Scholar]
- Breiman L, Random forests, Mach. Learn 45, 2001, 5–32. [Google Scholar]
- Clarke KR and Warwick RM, Change in marine communities: an approach to statistical analysis and interpretation, 2nd edition Primer-E Ltd, Plymouth. [Google Scholar]
- Dodds WK, Bouska WW, Eitzmann JL, Pilger TJ, Pitts KL, Riley AJ, Schloesser JT and Thornbrugh DJ, Eutrophication of U.S. freshwaters: analysis of potential economic damages, Environ. Sci. Technol 43, 2009, 12–19. [DOI] [PubMed] [Google Scholar]
- Goral F and Schellenberg J (2017). goeveg: Functions for Community Data and Ordinations. R package version 0.3.3. https://CRAN.R-project.org/package=goeveg. [Google Scholar]
- Hill BH, Stevenson RJ, Pan Y, Herlihy AT, Kaufmann PR and Johnson CB, Comparison of correlations between environmental characteristics and stream diatom assemblages characterized at genus and species levels, J North Am. Benthol. Soc 20, 2001, 299–310. [Google Scholar]
- Kahlert M, Albert RL, Anttila EL, Bengtsson R, Bigler C, Eskola T, Gälman V, Gottschalk S, Herlitz E, Jarlman A and Kasperoviciene J, Harmonization is more important than experience - results of the first Nordic-Baltic diatom intercalibration exercise 2007 (stream monitoring), J. Appl. Phycol 21, 2009, 471–482. [Google Scholar]
- Kahlert M, Ács É, Almeida SF, Blanco S, Dreßler M, Ector L, Karjalainen SM, Liess A, Mertens A, van der Wal J and Vilbaste S, Quality assurance of diatom counts in Europe: towards harmonized datasets, Hydrobiologia 772, 2016, 1–4. [Google Scholar]
- Kelly MG and Ector L Effect of streamlining taxa lists on diatom-based indices: implications for intercalibrating ecological status, Hydrobiologia 695, 2012, 253–263. [Google Scholar]
- Kentucky Division of Water (KDOW), 2015. Laboratory procedures for macroinvertebrate sample processing and identification Revision 4.0. DOWSOP03005. Kentucky Department for Environmental Protection, Division of Water, Frankfort, Kentucky: 31 pp. Accessed [19 April 2018], at URL http://water.ky.gov/Documents/QA/Surface%20Water%20SOPs/BenthicMacroinvertebratesLabProcessingandIdentificationSOP.pdf. [Google Scholar]
- Krammer K and Lange-Bertalot H, Bacillariophyceae, In: Ettl J, Gerloff J, Heynig H and Mollenhauer D, (Eds.), 1 Teil: Naviculaceae. Süßwasserflora von Mitteleuropa 2/1, 1986, G. Fisher; Stuttgart, 876. [Google Scholar]
- Krammer K and Lange-Bertalot H, Bacillariophyceae, In: Ettl J, Gerloff J, Heynig H and Mollenhauer D, (Eds.), 2Teil: Bacillariaceae, Epithemiaceae, Surirellaceae. Süßwasserflora von Mitteleuropa 2/2, 1988, G. Fisher; Stuttgart, 596. [Google Scholar]
- Krammer K and Lange-Bertalot H, Bacillariophyceae, In: Ettl J, Gerloff J, Heynig H and Mollenhauer D, (Eds.), 3 Teil: Centrales, Fragilariaceae, Eunotiaceae. Süßwasserflora von Mitteleuropa 2/3, 1991a, G. Fisher; Stuttgart, 576. [Google Scholar]
- Krammer K and Lange-Bertalot H, Bacillariophyceae, In: Ettl J, Gerloff J, Heynig H and Mollenhauer D, (Eds.), 4 Teil: Achanthaceae. Süßwasserflora von Mitteleuropa 2/4, 1991b, G. Fisher; Stuttgart, 437. [Google Scholar]
- Lavoie I, Dillon PJ and Campeau S, The effect of excluding diatom taxa and reducing taxonomic resolution on multivariate analyses and stream bioassessment, Ecol. Ind 9, 2009, 213–225. [Google Scholar]
- Lavoie I and Campeau S, Assemblage diversity, cell density and within-slide variability: implications for quality assurance/quality control and uncertainty assessment in diatom-based monitoring, Ecol. Ind 69, 2016, 415–421. [Google Scholar]
- Munn MD, Waite I and Konrad CP, Assessing the influence of multiple stressors on stream diatom metrics in the upper Midwest, USA, Ecol. Ind 85, 2018, 1239–1248. [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H, 2017. vegan: Community Ecology Package. R package version 2.4-3. https://CRAN.R-project.org/package=vegan. [Google Scholar]
- Paul MJ, Walsh B, Oliver J, Thomas D, 2017. Algal indicators in streams: a review of their application in water quality management of nutrient pollution. U.S. Environmental Protection Agency; White Paper 822B17002, accessed [18 May 2018], at URL https://www.epa.gov/nutrient-policy-data/algal-indicators-streams-review-their-application-water-quality-management. [Google Scholar]
- Porter SD, Mueller DK, Spahr NE, Munn MD and Dubrovsky NM, Efficacy of algal metrics for assessing nutrient and organic enrichment in flowing waters, Freshw. Biol 53, 2008, 1036–1054. [Google Scholar]
- Potapova MG and Charles DF, Benthic diatoms in USA rivers: distributions along spatial and environmental gradients, J. Biogeogr 29, 2002, 167–187. [Google Scholar]
- Potapova MG, Spaulding S, 2013. Cocconeis placentula sensu lato. In: Diatoms of the United States. Retrieved April 12, 2018, from http://westerndiatoms.colorado.edu/taxa/species/cocconeis_placentula. [Google Scholar]
- Reavie ED, Heathcote AJ and Chraïbi VLS, Laurentian Great Lakes phytoplankton and their water quality characteristics, including a diatom-based model for paleoreconstruction of phosphorus, PLoS One 9, 2014, e104705 10.1371/journal.pone.0104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Accessed [31 October 2017], at URL, https://www.R-project.org/. [Google Scholar]
- Smol JP and Stoermer EF, The Diatoms: Applications for the Environmental and Earth Sciences, 2010, Cambridge University Press; Cambridge, 686. [Google Scholar]
- Smucker NJ, Becker M, Detenbeck NE and Morrison AC, Using algal metrics and biomass to evaluate multiple ways of defining concentration-based nutrient criteria in streams and their ecological relevance, Ecol. Ind 32, 2013, 51–61. [Google Scholar]
- Spaulding SA, Lubinski DJ, Potapova M, 2010. Diatoms of the United States. Accessed [21 March 2018], at URL http://westerndiatoms.colorado.edu. Currently, Diatoms of North America. Accessed [21 May 2018], at URL http://diatoms.org.
- Stevenson RJ, Zalack JT and Wolin J, A multimetric index of lake diatom condition based on surface-sediment assemblages, Freshwater Sci. 32, 2013, 1005–1025. [Google Scholar]
- Stoddard JL, Van Sickle J, Herlihy AT, Brahney J, Paulsen S, Peck DV, Mitchell R and Pollard AI, Continental-scale increase in lake and stream phosphorus: are oligotrophic systems disappearing in the United States? Environ. Sci. Technol 50, 2016, 3409–3415. [DOI] [PubMed] [Google Scholar]
- Taylor JM, King RS, Pease AA and Winemiller KO, Nonlinear response of stream ecosystem structure to low-level phosphorus enrichment, Freshw. Biol 59, 2014, 969–984. [Google Scholar]
- U.S. Environmental Protection Agency, National Rivers and Streams Assessment 2013-2014: Laboratory Operations Manual. Version 2.0. EPA-841-B-12-010, 2012, U.S. Environmental Protection Agency, Office of Water; Washington, DC, 225. [Google Scholar]
- U.S. Environmental Protection Agency, 2016. National Rivers and Streams Assessment 2008-2009: A Collaborative Survey (EPA/841/R-16/007) Office of Water and Office of Research and Development; Washington, DC: accessed [18 May 2018], at URL http://www.epa.gov/national‐aquatic‐resource‐surveys/nrsa. [Google Scholar]
- U.S. Geological Survey, 2017. BioData – Aquatic Bioassessment Data for the Nation available on the World Wide Web, accessed [31 October 2017], at URL https://my.usgs.gov/confluence/display/biodata/Complete+BioData+Taxonomy+Downloads. [Google Scholar]
- Van Dam H, Mertens A and Sinkeldam J, A coded checklist and ecological indicator values of freshwater diatoms from the Netherlands, Netherland J. Aquatic Ecol 28, 1994, 117–133. [Google Scholar]
- Wetzel CE, Ector L, Van de Vijver B, Compère P and Mann DG, Morphology, typification and critical analysis of some ecologically important small naviculoid species (Bacillariophyta), Fottea 15, 2015, 203–234. [Google Scholar]
- Wold AP and Hershey AE, Spatial and temporal variability of nutrient limitation in 6 North Shore tributaries to Lake Superior, J. North Am. Benthol. Soc 18, 1999, 2–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


