Abstract
We have recently identified three molecules (tilorone, quinacrine and pyronaridine tetraphosphate) which all demonstrated efficacy in the mouse model of infection with mouse-adapted Ebola virus (EBOV) model of disease and had similar in vitro inhibition of an Ebola pseudovirus (VSV-EBOV-GP), suggesting they interfere with viral entry. Using a machine learning model to predict lysosomotropism these compounds were evaluated for their ability to possess a lysosomotropic mechanism in vitro. We now demonstrate in vitro that pyronaridine tetraphosphate is an inhibitor of Lysotracker accumulation in lysosomes (IC50 = 0.56 μM). Further, we evaluated antiviral synergy between pyronaridine and artesunate (Pyramax®), which are used in combination to treat malaria. Artesunate was not found to have lysosomotropic activity in vitro and the combination effect on EBOV inhibition was shown to be additive. Pyramax® may represent a unique example of the repurposing of a combination product for another disease.
Keywords: Antiviral, Ebola, Lysosomotropic, Machine learning
Highlights
-
•
We describe a machine learning model to predict lysosomotropic activity.
-
•
We demonstrate that pyronaridine, quinacrine and tilorone all inhibit lysotracker accumulation as predicted.
-
•
Artesunate does not appear to inhibit lysostracker accumulation as predicted.
-
•
We demonstrate additivity between pyronaridine and artesunate for their in vitro EBOV activity.
-
•
These findings suggest pyronaridine and artesunate may be combined and repurposed against EBOV.
1. Introduction
The outbreaks of Ebola virus (EBOV) disease (EVD) in Africa have come at great human and financial cost (Bornholdt et al., 2019; Ekins et al., 2015b). For example, the outbreak in 2014–2016 killed over 11,000 and it is estimated that it resulted in $53bn in economic damage (Jonas, 2019). The most recent outbreak in the Democratic Republic of the Congo, has killed more than 2200 people (Ilunga Kalenga et al., 2019). Even with approval of a vaccine for prevention of EVD (FDA, 2019) there is still an urgent need to advance development of filovirus-specific antiviral therapeutics. A clinical trial (NCT03719586) investigated ZMapp (a monoclonal antibody cocktail) (Qiu et al., 2014)), remdesivir (a small molecule), MAb114 (a monoclonal combination) (Corti et al., 2016)) and REGN-EB3 (monoclonal antibody combination) (Sivapalasingam et al., 2018). These results showed that the antibodies REGN-EB3 and mAb114 had overall statistically similar survival rates of 71% and 66%, respectively. Unfortunately, ZMapp and remdesivir were less effective with a 51% and 47% survival rates, respectively (Mulangu et al., 2019). The search for antiviral small molecule with improved efficacy in vivo for EBOV therefore continues.
In an effort to repurpose drugs for the treatment of EVD, we have developed a Bayesian machine learning (ML) approach with a set of 868 anti-EBOV molecules screened in vitro (Madrid et al., 2013, 2015b). The EBOV ML model enabled us to virtually screen several thousand compounds and identify three active compounds against EBOV: tilorone, quinacrine and pyronaridine tetraphosphate (Ekins et al., 2015a). The three molecules inhibited EBOV in HeLa cells and demonstrated significant in vivo activity in the mouse-adapted EBOV (ma-EBOV) efficacy model (Ekins et al., 2018, 2020; Lane et al., 2019a, 2019b) and pyronaridine was active in the guinea pig model of EBOV infection (Lane et al., 2020). The compounds also inhibited replication of multiple strains of EBOV and Marburg virus (MARV) (Lane et al., 2020). The trend for compounds to be active against both EBOV and MARV has been demonstrated previously revealing in vitro inhibition (IC50) correlation ((Madrid et al., 2013; Madrid et al., 2015a), Figure S1).
To date, neither we nor others have determined the mechanism of these antiviral compounds against EBOV. Previously we evaluated pyronaridine, tilorone and quinacrine in vitro for its anti-EBOV activity (Zaire strain) in the type I IFN-deficient Vero 76 cell line (Desmyter et al., 1968; Emeny and Morgan, 1979) and no antiviral activity was observed at any concentration below the 50% cytotoxicity concentration. In HeLa cells all three drugs demonstrated selectivity (Ekins et al., 2015a; Lane et al., 2019b). These observations support the hypothesis that their antiviral activity could be partially acting through or on the type I IFN-related innate immunity pathway (Ekins et al., 2018). We also tested a combination of pyronaridine with tilorone in HeLa cells and evaluated the data with the BRAID model which suggested they are likely synergistic (Twarog et al., 2016). Based on published data for tilorone and quinacrine, which are well known to be lysosomotropic agents, it was suspected that this may also be important and worthy of further study. In addition, pyronaridine is used as an antimalarial in combination with artesunate (Pyramax®). We had previously determined that artesunate also has micromolar in vitro inhibitory activity against EBOV (Anantpadma et al., 2019a). We now assess whether pyronaridine, artesunate, tilorone and quinacrine accumulate in lysosomes. We also assess the effect of combining pyronaridine with artesunate or its active metabolite dihydroartemisinin against EBOV in vitro.
2. Materials and methods
Chemicals and reagents. Pyronaridine tetraphosphate [4-[(7-Chloro-2-methoxybenzo[b][1,5]naphthyridin-10-yl)amino]-2,6-bis(1-pyrrolidinylmethyl)phenol phosphate (1:4)] (Ekins et al., 2015a) was purchased from BOC Sciences (Shirley NY). Tilorone was purchased from BOC Sciences. Quinacrine and Chloroquine were purchased from Cayman Chemicals (Ann Arbor, Michigan) and Sigma-Aldrich (St. Louis, MO), respectively. Artesunate was purchased from TRC Canada (North York, ON, Canada) and dihydroartemisinin (DHA) was purchased from Sigma-Aldrich (#D7439).
NIAID antiviral screening. Pyronaridine tetraphosphate, tilorone and quinacrine were also tested (using the NIAID DMID services) against representatives of several viruses using human cells. The general methods have been described previously (Ekins et al., 2020).
Lysosomotropic machine learning model. The Assay Central™ software has been previously described (Anantpadma et al., 2019b; Dalecki et al., 2019; Ekins et al., 2019a, 2019b; Hernandez et al., 2018; Lane et al., 2018; Russo et al., 2018; Sandoval et al., 2018; Wang et al., 2019; Zorn et al., 2019) which uses the source code management system Git to gather and store structure-activity datasets collated in Molecular Notebook (Molecular Materials Informatics, Inc. in Montreal, Canada). The output is a high-quality dataset and a Bayesian model using extended-connectivity fingerprints of maximum diameter 6 (ECFP6) descriptors. Each model includes several metrics to evaluate and compare predictive performance as previously described in a relevant publication (Russo et al., 2018), including Receiver Operator Characteristic, Recall, Precision, F1 Score, Cohen's Kappa (Carletta, 1996; Cohen, 1960), and Matthews Correlation Coefficient (Matthews, 1975). Applicability is representative of the overlap between the training and the test set. It is the quotient of the total number of ECFP6 fingerprints of the test molecule represented in the model divided by the total number of ECFP6 fingerprints of that test molecule. Generation and interpretation of prediction scores has been previously described (Clark et al., 2015; Clark and Ekins, 2015). The model consisted of curated data from a key paper from Nadanaciva et al. (2011), where their quantitative approach to measuring lysosomotropic properties allowed for a direct activity threshold cut-off and was defined as an IC50 (decrease in LysoTracker Red staining) of ≥70 μM. A negative series of drugs that lack lysosomotropic properties from Kazmi et al. was also curated and added as inactive compounds (Kazmi et al., 2013) to the model.
Lysosomotropic method. A previous published lysosomotropic assay by Nadanaciva et al. was used as the basis for the following work (Nadanaciva et al., 2011).
2.1. MCF7 cell culture conditions
The human metastatic mammary gland cell line MCF7 was obtained from American Type Culture Collection (ATCC# HTB-22). Cells were grown in Eagle's minimum essential medium (Corning) supplemented with 10% fetal bovine serum (Gibco), 100 unit/ml penicillin and 100 μg/ml streptomycin (Corning) in a humidified incubator at 37 °C and 5% CO2.
2.2. Lysosomotropic assay
MCF7 cells were seeded into black walled clear bottom 96-well plates at 15,000 cells/well in 100 μl growth media and incubated for 48 h (h). Cells were treated with drugs at 2-fold dilutions, with an initial testing concentration of 50 μM and an additional series of 9 tested dilutions (final 0.098 μM). Based on solubility restrictions, compounds for stocks were either solubilized in DMSO (tilorone, quinacrine, artesunate) or water (pyronaridine, chloroquine). Control wells included cells treated with DMSO or water. To start the assay, 0.5 μl of appropriate compound stock or control was added using Biomek NXp (Beckman Coulter) and incubated at 37 °C, 5% CO2 for 3 h. LysoTracker Red (75 nM) (ThermoFisher) was then added and incubated for 30 min followed by a wash in phosphate buffered saline (PBS). The cells were immediately fixed with 10% formalin at room temperature for 15 min. Cells were then stained with Hoechst (5 μg/ml) (Sigma-Aldrich) in PBS for 10 min at room temperature. Following cell staining they were washed in PBS. Each experimental run tested a series of compounds in triplicate and was repeated on two different days (n = 6 for each compound series) with multiple DMSO (n = 12) and water (n = 24) controls per plate.
Imaging was done using a CellInsight CX5 High Content Screening Platform (Thermo Scientific) with 10X objective. Fluorescence was measured with Hoechst (nuclei) and LysoTracker Red (lysosomes) in channel 1 and 2, respectively. A total of 3–4 fields were captured for all wells. For analysis, nuclei were identified, and a circular mask was extended out 5 pixels to represent the cell. Total intensity of the fluorescent signal from Lysotracker Red within the mask area was then used to represent the lysosomal staining in the cells. Data was normalized to controls and then analyzed with GraphPad Prism version 8.00. Error bars of dose-response curves represent the SEM of the replicates.
2.3. Cell viability for lysosomotropic assay
MCF7 cells were seeded in white walled clear bottom 96-well plates at 15,000 cells/well in 100 μl growth media and incubated for 48 h. To start the assay, 0.5 μl of compound stock or control was added using Biomek NXp and incubated at 37 °C, 5% CO2 for 3.5 h. Following compound incubation, 80 μl of CellTiter-Glo (Promega) was added. The plates were shaken on an orbital shaker at 300 rpm for 20 min and then read on an Envision 2104 Multilabel reader (PerkinElmer). Experiments were repeated in triplicate and data was analyzed with Graphpad Prism version 8.00. Error bars of dose-response curves represent the SEM of the replicates.
In vitro combination studies methods. The in vitro infection inhibition of EBOV/Mak (Makona, IRF0165, 1.98E7 PFU/mL) was performed in HeLa and Huh 7 cells. HeLa cells were seeded at 3 × 104 cells/well in 96-well plates. After 24 h the drugs were added to cells in a 6x6 matrix with 2-fold serial dilutions with a starting concentration of 30 μM. The experiment was run on 3–4 replicate plates. The experiment was run on 2 different days. Cells were infected with virus 1 h after the addition of the drugs in BSL4-containment at a multiplicity of infection (MOI) of 0.21 (Huh 7) or 0.5 (HeLa). After 48 h, plates were fixed and virus was detected with a mouse antibody specific for EBOV VP40 protein (#B-MD04-BD07-AE11, made by US Army Medical Research Institute of Infectious Diseases, Frederick MD under Centers for Disease Control and Prevention contract) (Cong et al., 2016) followed by staining with anti-mouse IgG-peroxidase labeled antibody (KPL, Gaithersburg, MD, #074–1802). Luminescence was read on a Spark 20M plate reader (Tecan US, Morrisville, NC). The signal of treated, infected wells was normalized to uninfected control wells and measured (in percent) relative to untreated infected wells. Non-linear regression analysis was performed, and the 50% inhibitory concentrations (EC50s) were calculated from fitted curves (log [agonist] versus response [variable slope] with constraints to remain above 0% and not exceed 100%) (GraphPad Software version 8.0, La Jolla, CA). The EBOV drug screen assay was performed with three replicates for each drug concentration. Error bars of dose-response curves represent the SEM of the replicates.
2.4. Cell viability for combination studies
For quantitation of drug toxicity, HeLa cells were mock infected (no virus) and treated with drug dilutions under the same conditions as the infected cells. After 48 h, cell viability was measured using the CellTiter Glo Luminescent Cell Viability Assay kit according to manufacturer's protocol (Promega, Madison, WI).
Combination Analysis using BRAID and SynergyFinder. The BRAID analysis (Twarog et al., 2016) service calculates synergy by fitting data to a seven-variable function. The variable κ represents a quantitative synergy value where κ < 0 implies antagonism, κ = 0 implies additivity, and κ > 0 implies synergy. As an additional reference, “strong synergy” corresponds to κ = 2.5, “mild synergy” corresponds to κ = 1, “mild antagonism” corresponds to κ = −0.66, and “strong antagonism” corresponds to κ = −1. To assess if the combined inhibitory effect of pyronaridine and or artesunate/dihydroartemisinin on EBOV was synergistic, additive, or antagonistic we analyzed a 6x6 checkboard assay with these pairs of drugs at various combined concentrations in HeLa and Huh 7 cells. It is noted that inhibition data under toxic concentrations (consistently >50% cell death) were removed from the analysis. Inclusively, this consisted of only individual and combined experiments with concentrations of pyronaridine that exceeded its CC50 (i.e. 5 μM concentrations in HeLa cells only). All toxicity data was retained for BRAID analysis.
The SynergyFinder analysis service (Ianevski et al., 2020) similarly calculates the degree of combination, synergy or antagonism by comparing the observed drug combination response against the expected response, while assuming there is no interaction between the two drugs. These scores were calculated using the Loewe reference additivity mode (Loewe, 1953). The threshold to define a good synergy score is variable, but the program developers suggest that synergy scores near 0 gives limited confidence on synergy or antagonism and a score < −10 or >10 are expected to be antagonist or synergistic, respectively.
3. Results
NIAID antiviral screening. Pyronaridine, quinacrine and tilorone were previously demonstrated to be active against EBOV in HeLa cell but not Vero cells (Lane et al., 2019b). We have now tested these three compounds against Adenovirus 5, Human papillomavirus 11, Chikungunya virus, Dengue virus 2, Powassan virus, Rift valley virus, Yellow Fever virus and human cytomegalovirus in additional human cells through the use of NIAID screening resources. None however showed selectivity in the cell lines and concentrations tested, but this may be due to high cytotoxicity and or the insufficient range of concentrations tested in these cell lines (Table S1A-C).
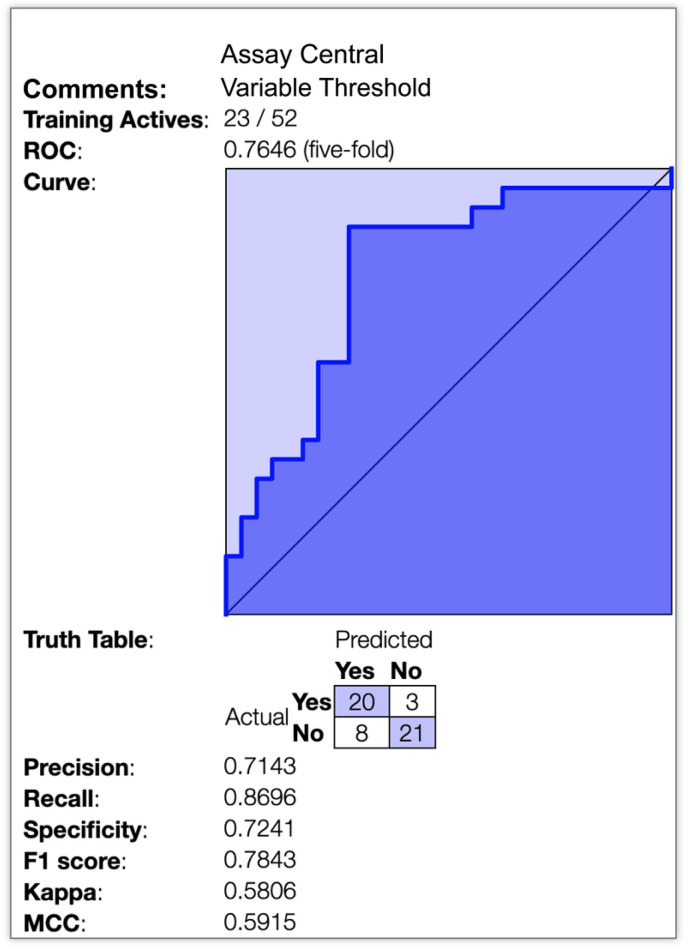
Lysosomotropic machine learning model predictions. A Bayesian machine learning model with 52 compounds (23 were classed as lysosomotropic) was generated from published data using Assay Central™ with 5-fold cross validation ROC = 0.765 (Fig. 1 ). Additional model statistics suggest that the model is potentially useful for scoring compounds to predict lysosomal accumulation (Table 1 ). Tilorone, pyronaridine, quinacrine and artesunate were used as a prospective test set as these were not in the model training set.
Fig. 1.
Lysosomotropic machine learning model. 5-fold cross validation receiver operator curve as well as multiple metrics depicting the internal validation of this Bayesian model (ECFP6).
Table 1.
Physicochemical properties and Assay Central lysosomotropic machine learning predictions for compounds tested in vitro. Larger prediction scores have a higher probability of activity. An applicability score of 1 indicates that all the fragments are in the model and may indicate the molecule is in the training set (chloroquine is in the training set) (Calculated with ACD/Labs PhysChem Batch program$ (Ploemen et al., 2004)). Predicted pka's (negative log of the acid dissociation constant) were obtained from DrugBank, which were initially calculated using Chemaxon. AlogP (predicted log octanol-water partition coefficient was calculated via Discovery Studio).
| Name | pKa (predicted) | Pka (Experimental) | AlogP | Lysosomotropic Prediction Score | Lysosomotropic Applicability Score |
|---|---|---|---|---|---|
| Chloroquine | 10.32 (Strongest Base) | 4.0, 8.4 and 10.2 (Schroeder and Gerber, 2014) | 4.34 | 1.09 | 1 |
| Artesunate | 3.77 (Strongest Acid), −4.2 (Strongest Base) | 4.6 (Augustijns et al., 1996) | 1.84 | 0.31 | 0.21 |
| Quinacrine | 10.33 (Strongest Base) | N/A | 5.67 | 1.00 | 0.68 |
| Tilorone | ~8.6$ | N/A | 4.56 | 0.75 | 0.69 |
| Pyronaridine | 7.96 (Strongest Acid), 10.08 (Strongest Base) | 7.08, 7.39, 9.88 and 10.30 (Adegoke et al., 2006) | 6.19 | 0.68 | 0.51 |
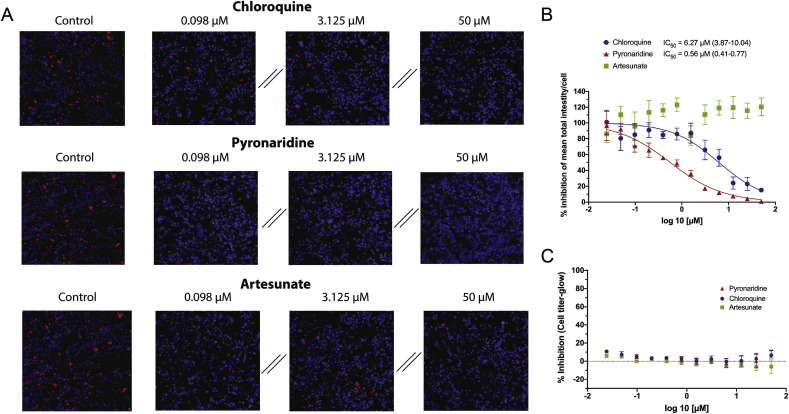
In vitro inhibition of lysosomal accumulation of Lysotracker. Pyronaridine tetraphosphate was found to be a potent inhibitor of Lysotracker accumulation in MCF7 lysosomes in vitro (IC50 = 0.56 μM). In contrast, artesunate showed no appreciable inhibition of Lysotracker (Fig. 2 ). Tilorone (IC50 = 3.09 μM) (Ekins et al., 2020), chloroquine (IC50 = 6.21 μM) (Ekins et al., 2020) and finally quinacrine (IC50 = 7.31 μM) were less potent inhibitors of Lysotracker accumulation in MCF7 lysosomes in vitro (Figure S2). All compounds were correctly predicted, with tilorone, quinacrine and pyronaridine predicted to be lysosomotropic (prediction score > 0.5 = lysosomotropic), while artesunate was predicted to not be lysosomotropic (prediction score < 0.5 = non-lysosomotropic) (Table 1). Chloroquine was already in the training set and so was excluded from this analysis.
Fig. 2.
Inhibition analysis of total fluorescent intensity/cell of lysotracker red by chloroquine, pyronaridine and artesunate in MCF7 Cells. Lysotracker accumulation in lysosomes is pH dependent, therefore a reduction in signal from the lysotracker suggests a pH increase in these organelles. This is proposed to be caused by accumulation of the charged base of the lysosomotropic compound in the lysosome, which in a lower pH environment becomes neutralized and trapped in the organelle. A) Representative images showing Lysotracker lysosomal accumulation inhibition at various concentrations. B) Graphical representation and quantification (Parentheses represent 95% CI) of the dose-dependent effect of on Lysotracker accumulation in lysosomes (Error bars represent SEM). Outliers were identified using the ROUT method (Q = 10%) and consequentially removed. C) Measure of cellular toxicity at concentrations and times mimicking the inhibition assays.
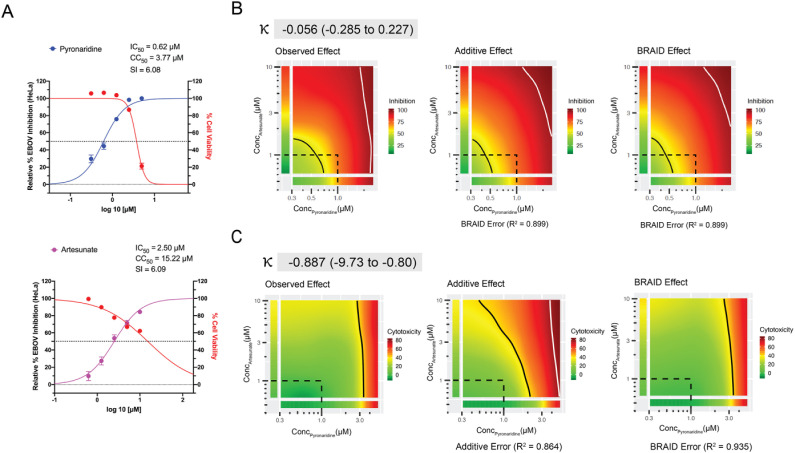
Combination Analysis. The BRAID analysis of pyronaridine and artesunate in vitro inhibition data from the checkerboard assay indicates additivity of these molecules in HeLa cells (Fig. 3 ). Artesunate ameliorates the toxicity of pyronaridine in the checkboard assay and therefore indirectly potentiates pyronaridine. The non-linear regression, 4-parameter curve fit (Hill equation) for the artesunate control in Huh 7 cells suggested a plateau at ~60% inhibition (Figure S3), therefore the combination data for this cell-line and pyronaridine/artesunate pair was not included in the analysis. Pyronaridine and DHA similarly shows an additive effect in both HeLa and Huh 7 cells, both with a parallel reduction in toxicity based on the BRAID analysis (Figure S4). A secondary analysis using Synergyfinder (Figures S5-S6) also suggests that this combination indirectly potentiates pyronaridine in HeLa cells, but in Huh 7 cells these interpretations are ambiguous (Figure S7).
Fig. 3.
Combination data for the pyronaridine and artesunate checkerboard assay in HeLa cells. A) Inhibition/cytotoxicity plots for the pyronaridine and artesunate controls (compound tested in the absence of the other compound). Controls were run in triplicate at 5 concentrations per plate, so the total number from replicates for each compound varied (Pyronaridine, n = 27; Artesunate, n = 18). Error bars represent the SEM at each concentration tested. B) Graphical representations (from left to right) of the inhibition plots of the smoothed raw data, predicted additive inhibition and predicted inhibition using the 7-parameter BRAID analysis. It is noted that inhibition data under toxic concentrations (>50% cell death) were removed from the analysis. The “Additive” or “BRAID” error represents the corresponding accuracy of fit with the “Observed Effect”. κ represents the combinatory effect where κ = 0 implies additivity, and κ > 0 implies synergy, “strong synergy” corresponds to κ = 2.5, “mild synergy” corresponds to κ = 1, “mild antagonism” corresponds to κ = −0.66, and “strong antagonism” corresponds to κ = −1. C) Representation of the cytotoxicity (toxicity is representative of % cell death from control) arranged in the same manner as inhibition.
4. Discussion
Within the last 5 years we have seen two major EVD outbreaks in Africa. These led to renewed efforts to develop treatments for this virus. The active and inactive compounds of several in vitro high throughput drug screens (Johansen et al., 2013; Madrid et al., 2013, 2015a) have been used to develop computational models for predicting anti-EBOV activity of compounds. More recently, combinations of approved drugs found in these and other studies have suggested synergistic combinations (Bekerman et al., 2017; Dyall et al., 2018b; McCarthy et al., 2016; Sun et al., 2017). To date, none of these many efforts for EBOV have resulted in a clinical antiviral candidate. Several small molecule antivirals were felled at the hurdle of animal models, specifically the transition from mouse to the guinea pig model. Compounds that have failed to show in vivo efficacy against EBOV following this well-trodden route include chloroquine (Dowall et al., 2015; Madrid et al., 2015a), azithromycin (Madrid et al., 2015a), amiodarone (Dyall et al., 2018a), BGB324 (Dowall et al., 2016), NCK8 (Dowall et al., 2016) and 17-DMAG (Dowall et al., 2016). As discussed previously (Lane et al., 2020), this may be due to differences in drug metabolism making the guinea pig model inappropriate for EBOV.
Efforts to improve the efficiency and cost effectiveness of EBOV drug discovery have involved our attempts to identify several molecules using ML which have progressed through in vitro and in vivo testing (Ekins et al., 2015a, 2018; Ekins and Madrid, 2020; Lane et al., 2019a, 2019b, 2020). We have also previously used these ML models to predict in vitro efficacy for drugs that were then tested against EBOV (Anantpadma et al., 2019a; Ekins et al., 2015a). The mechanism for tilorone, quinacrine and pyronaridine against EBOV are still unknown. Others have demonstrated that compounds with physicochemical properties such as a basic pKa (>6.5) and cLogP of >2 tend to be lysosomotropic (Nadanaciva et al., 2011) and they accumulate in the lysosomes. We have now taken a ML approach to predict potential for a lysosomotropic mechanism using published in vitro data (Kazmi et al., 2013; Nadanaciva et al., 2011) along with ECFP6 molecular fingerprints and a Bayesian algorithm. We have also performed several in vitro studies to validate the predictive ability of this model as well as infer the potential mechanism of these EBOV drugs. Based on this new data pyronaridine is clearly a potent lysosomotropic agent, more so than all the other molecules tested. There is a strong correlation between published anti-EBOV activity and the lysosomotropic property (Table S2) for a large number of drugs. All of the compounds that were considered actives in our model were researched to identify whether they had been previously tested against EBOV and or MARV either with a psuedovirus/VLP and or a competent virus inhibition assay. 21 of 23 of these compounds had been tested previously and all inhibited these viruses, with IC50's in the nM to low μM range. This is certainly not a comprehensive list of every lysosomotropic compound, but this strongly supports the notion that the lysosomotropic characteristic is directly related to the antiviral activity of the compounds within this model.
The lysosomotropic characteristic has previously been shown to be relevant for EBOV in many other studies (Madrid et al., 2013, 2015a). Molecules with the cationic amphiphilic feature concentrate in lysosomes but the mechanism of how they inhibit Ebola virus entry has not been fully resolved as this could be via increased cholesterol accumulation in endosomes and lysosomes, lysosomal membrane stability or inhibition of acid sphingomyelinase (Salata et al., 2017; Xiao et al., 2018). Alternatively, cationic amphiphilic containing molecules may also bind to a hydrophobic pocket of the EBOV-GP and destabilize the molecule in vitro. Zimmer et al., showed 1-Benzyl-3-cetyl-2-methylimidazolium Iodide (NH125), a lysosomotropic drug has a broad-spectrum of inhibition against viral entry (Moeschler et al., 2018) and Selakovic et al., described cationic amphiphilic diazachrysene analogs which accumulate in lysosomes and had potent in vitro and in vivo activity against EBOV (Selakovic et al., 2019).
Artesunate which is also similarly active against EBOV in HeLa cells in vitro (EC50 8.21 μM, CC50 > 50) (Anantpadma et al., 2019a) was predicted and found not to share this physiological characteristic and was subsequently not lysosomotropic when tested in vitro. The initial combination of these two drugs to form Pyramax® was to avoid drug resistance of Plasmodium parasites, the causative agents of malaria (Croft et al., 2012). The combinations of these two molecules are now described as additive in inhibiting EBOV replication in vitro but with a reduced cytotoxicity as compared to the individual treatments (Fig. 2, Fig. 3). Previous work has suggested that it is possible to identify pairs of drugs that block EBOV infection in vitro via the same methodology as used here (Dyall et al., 2018b) and these prior data have been used with additional software to suggest a variation in prioritizing drug pairs based on selective efficacy (Ianevski et al., 2020b), which considers both synergy and toxicity. This software independently confirms our observations with the BRAID analysis performed here (Figure S5-7).
This current study has implications outside of EBOV, with the increased interest in finding new antivirals and in particular the current focus on chloroquine and hydroxychloroquine (Jeon et al., 2020; Liu et al., 2020; Wang et al., 2020; Weston et al., 2020). Pyronaridine has recently also been shown to have some limited activity against SAR-CoV-2 in Vero cells (Jeon et al., 2020), but from our experience Vero cells may not be as appropriate as human cells to test compounds such as pyronaridine (low in vitro selectivity index, but high in vivo antiviral activity). Based on our previous findings, this leaves the distinct possibility that there has been an underestimate of pyronaridine's antiviral inhibition potential. Our current data also suggests that we should be testing artesunate versus additional viruses as well as in combination with pyronaridine.
Frequently, single drugs are repurposed for new uses (Baker et al., 2018), to our knowledge there is no precedent for a two drug combination being repurposed for the same indication. We have previously estimated that the dose used for treating malaria patients may have a beneficial effect in EBOV patients (Lane et al., 2019b), further indicating the potential for direct repurposing without the need to change dose, route or formulation. From our experience and insights gained during the discovery of the antiviral properties of pyronaridine, tilorone and quinacrine we propose our ML approach could be optimized by adding the additional ML model described here for the lysosomotropic mechanism. This would enable us to create a computational pipeline to identify new antivirals more rapidly that could have this lysosomotropic property and hence direct the antiviral mechanism of action investigations. Molecules with this mechanism may also have more utility as broad-spectrum antivirals. Future work may involve further mechanistic analysis to understand the pleiotropic effects of these compounds relating to the inhibition of entry, the interaction with the immune system and whether the additive effect of pyronaridine and artesunate can translate in vivo. Our next logical steps would therefore involve testing the combination of pyronaridine and artesunate in the mouse model of mouse adapted EBOV infection to assess whether the additivity seen in vitro is also observed (Lane et al., 2019b).
Funding
We kindly acknowledge NIH funding: R21TR001718 from NCATS. In addition, 1R43GM122196–01 and R44GM122196-02A1 “Centralized assay datasets for modelling support of small drug discovery organizations” from NIGMS and NIEHS for 1R43ES031038-01 “MegaTox for analyzing and visualizing data across different screening systems”. “Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R43ES031038. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. “This work was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases (NIAID); Integrated Research Facility (NIAID, Division of Clinical Research); Battelle Memorial Institute's prime contract with NIAID (Contract # HHSN272200700016I). H.Z., J.D., and M.R.H performed this work as employees of Battelle Memorial Institute (BMI). The findings and conclusions in this report do not necessarily reflect the views or policies of the US Department of Health and Human Services or of the institutions and companies affiliated with the authors.
Declaration of competing interest
SE is CEO of Collaborations Pharmaceuticals, Inc. TRL and DHF are employees at Collaborations Pharmaceuticals, Inc. Collaborations Pharmaceuticals, Inc. has obtained FDA orphan drug designations for pyronaridine, tilorone and quinacrine for use against Ebola.
Acknowledgments
Dr. Mindy Davis is gratefully acknowledged for assistance with the NIAID virus screening capabilities, Task Order number B22. We acknowledge numerous discussions with Dr. Jason E. Comer, Dr. Shane Massey, Dr. Manu Anantpadma, Dr. Robert A. Davey and Dr. Joel S. Freundlich.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104908.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adegoke O.A., Babalola C.P., Oshitade O.S., Famuyiwa A.A. Determination of the physicochemical properties of pyronaridine - a new antimalarial drug. Pak. J. Pharm. Sci. 2006;19:1–6. [PubMed] [Google Scholar]
- Anantpadma M., Lane T., Zorn K.M., Lingerfelt M.A., Clark A.M., Freundlich J.S., Davey R.A., Madrid P., Ekins S. Ebola virus bayesian machine learning models enable new in vitro leads. ACS Omega. 2019;4:2353–2361. doi: 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantpadma M., Lane T., Zorn K.M., Lingerfelt M.A., Clark A.M., Freundlich J.S., Davey R.A., Madrid P.B., Ekins S. Ebola virus bayesian machine learning models enable new in vitro leads. ACS Omega. 2019;4:2353–2361. doi: 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustijns P., D'Hulst A., Van Daele J., Kinget R. Transport of artemisinin and sodium artesunate in Caco-2 intestinal epithelial cells. J Pharm Sci. 1996;85:577–579. doi: 10.1021/js960001i. [DOI] [PubMed] [Google Scholar]
- Baker N.C., Ekins S., Williams A.J., Tropsha A. A bibliometric review of drug repurposing. Drug Discov. Today. 2018;23:661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Neveu G., Shulla A., Brannan J., Pu S.Y., Wang S., Xiao F., Barouch-Bentov R., Bakken R.R., Mateo R., Govero J., Nagamine C.M., Diamond M.S., De Jonghe S., Herdewijn P., Dye J.M., Randall G., Einav S. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 2017;127:1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt Z.A., Herbert A.S., Mire C.E., He S., Cross R.W., Wec A.Z., Abelson D.M., Geisbert J.B., James R.M., Rahim M.N., Zhu W., Borisevich V., Banadyga L., Gunn B.M., Agans K.N., Wirchnianski A.S., Goodwin E., Tierney K., Shestowsky W.S., Bohorov O., Bohorova N., Velasco J., Ailor E., Kim D., Pauly M.H., Whaley K.J., Alter G., Walker L.M., Chandran K., Zeitlin L., Qiu X., Geisbert T.W., Dye J.M. A two-antibody pan-ebolavirus cocktail confers broad therapeutic protection in ferrets and nonhuman primates. Cell Host Microbe. 2019;25:49–58. doi: 10.1016/j.chom.2018.12.005. e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletta J. Assessing agreement on classification tasks: the kappa statistic. Comput. Ling. 1996;22:249–254. [Google Scholar]
- Clark A.M., Dole K., Coulon-Spektor A., McNutt A., Grass G., Freundlich J.S., Reynolds R.C., Ekins S. Open source bayesian models. 1. Application to ADME/tox and drug discovery datasets. J. Chem. Inf. Model. 2015;55:1231–1245. doi: 10.1021/acs.jcim.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.M., Ekins S. Open source bayesian models. 2. Mining a "big dataset" to create and validate models with ChEMBL. J. Chem. Inf. Model. 2015;55:1246–1260. doi: 10.1021/acs.jcim.5b00144. [DOI] [PubMed] [Google Scholar]
- Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. [Google Scholar]
- Cong Y., Dyall J., Hart B.J., DeWald L.E., Johnson J.C., Postnikova E., Zhou H., Gross R., Rojas O., Alexander I., Josleyn N., Zhang T., Michelotti J., Janosko K., Glass P.J., Flint M., McMullan L.K., Spiropoulou C.F., Mierzwa T., Guha R., Shinn P., Michael S., Klumpp-Thomas C., McKnight C., Thomas C., Eakin A.E., O'Loughlin K.G., Green C.E., Catz P., Mirsalis J.C., Honko A.N., Olinger G.G., Jr., Bennett R.S., Holbrook M.R., Hensley L.E., Jahrling P.B. Evaluation of the activity of lamivudine and zidovudine against Ebola virus. PloS One. 2016;11 doi: 10.1371/journal.pone.0166318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Misasi J., Mulangu S., Stanley D.A., Kanekiyo M., Wollen S., Ploquin A., Doria-Rose N.A., Staupe R.P., Bailey M., Shi W., Choe M., Marcus H., Thompson E.A., Cagigi A., Silacci C., Fernandez-Rodriguez B., Perez L., Sallusto F., Vanzetta F., Agatic G., Cameroni E., Kisalu N., Gordon I., Ledgerwood J.E., Mascola J.R., Graham B.S., Muyembe-Tamfun J.J., Trefry J.C., Lanzavecchia A., Sullivan N.J. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- Croft S.L., Duparc S., Arbe-Barnes S.J., Craft J.C., Shin C.S., Fleckenstein L., Borghini-Fuhrer I., Rim H.J. Review of pyronaridine anti-malarial properties and product characteristics. Malar. J. 2012;11:270. doi: 10.1186/1475-2875-11-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalecki A.G., Zorn K.M., Clark A.M., Ekins S., Narmore W.T., Tower N., Rasmussen L., Bostwick R., Kutsch O., Wolschendorf F. High-throughput screening and Bayesian machine learning for copper-dependent inhibitors of Staphylococcus aureus. Metall. 2019;11:696–706. doi: 10.1039/c8mt00342d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmyter J., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J. Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall S.D., Bewley K., Watson R.J., Vasan S.S., Ghosh C., Konai M.M., Gausdal G., Lorens J.B., Long J., Barclay W., Garcia-Dorival I., Hiscox J., Bosworth A., Taylor I., Easterbrook L., Pitman J., Summers S., Chan-Pensley J., Funnell S., Vipond J., Charlton S., Haldar J., Hewson R., Carroll M.W. 2016. Antiviral Screening of Multiple Compounds against Ebola Virus. Viruses 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall S.D., Bosworth A., Watson R., Bewley K., Taylor I., Rayner E., Hunter L., Pearson G., Easterbrook L., Pitman J., Hewson R., Carroll M.W. Chloroquine inhibited Ebola virus replication in vitro but failed to protect against infection and disease in the in vivo Guinea pig model. J. Gen. Virol. 2015;96:3484–3492. doi: 10.1099/jgv.0.000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Johnson J.C., Hart B.J., Postnikova E., Cong Y., Zhou H., Gerhardt D.M., Michelotti J., Honko A.N., Kern S., DeWald L.E., O'Loughlin K.G., Green C.E., Mirsalis J.C., Bennett R.S., Olinger G.G., Jr., Jahrling P.B., Hensley L.E. In vitro and in vivo activity of amiodarone against Ebola virus. J. Infect. Dis. 2018;218:S592–S596. doi: 10.1093/infdis/jiy345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Nelson E.A., DeWald L.E., Guha R., Hart B.J., Zhou H., Postnikova E., Logue J., Vargas W.M., Gross R., Michelotti J., Deiuliis N., Bennett R.S., Crozier I., Holbrook M.R., Morris P.J., Klumpp-Thomas C., McKnight C., Mierzwa T., Shinn P., Glass P.J., Johansen L.M., Jahrling P.B., Hensley L.E., Olinger G.G., Jr., Thomas C., White J.M. Identification of combinations of approved drugs with synergistic activity against Ebola virus in cell cultures. J. Infect. Dis. 2018;218:S672–S678. doi: 10.1093/infdis/jiy304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Freundlich J., Clark A., Anantpadma M., Davey R., Madrid P. Machine learning models identify molecules active against Ebola virus in vitro. F1000Res. 2015;4:1091. doi: 10.12688/f1000research.7217.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Gerlach J., Zorn K.M., Antonio B.M., Lin Z., Gerlach A. Repurposing approved drugs as inhibitors of Kv7.1 and Nav1.8 to treat pitt hopkins syndrome. Pharm. Res. (N. Y.) 2019;36:137. doi: 10.1007/s11095-019-2671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Lane T.R., Madrid P.B. Tilorone: a broad-spectrum antiviral invented in the USA and commercialized in Russia and beyond. Pharm. Res. (N. Y.) 2020;37:71. doi: 10.1007/s11095-020-02799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Lingerfelt M.A., Comer J.E., Freiberg A.N., Mirsalis J.C., O'Loughlin K., Harutyunyan A., McFarlane C., Green C.E., Madrid P.B. Efficacy of tilorone dihydrochloride against Ebola virus infection. AAC (Antimicrob. Agents Chemother.) 2018;62 doi: 10.1128/AAC.01711-17. e01711-01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Madrid P.B. Tilorone: a broad-spectrum antiviral for emerging viruses. AAC (Antimicrob. Agents Chemother.) 2020;64 doi: 10.1128/AAC.00440-20. e00440-00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Puhl A.C., Zorn K.M., Lane T.R., Russo D.P., Klein J.J., Hickey A.J., Clark A.M. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 2019;18:435–441. doi: 10.1038/s41563-019-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Southan C., Coffee M. Finding small molecules for the 'next Ebola. F1000Res. 2015;4:58. doi: 10.12688/f1000research.6181.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny J.M., Morgan M.J. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979;43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- FDA . 2019. First FDA-Approved Vaccine for the Prevention of Ebola Virus Disease, Marking a Critical Milestone in Public Health Preparedness and Response. [Google Scholar]
- Hernandez H.W., Soeung M., Zorn K.M., Ashoura N., Mottin M., Andrade C.H., Caffrey C.R., de Siqueira-Neto J.L., Ekins S. High throughput and computational repurposing for neglected diseases. Pharm. Res. (N. Y.) 2018;36:27. doi: 10.1007/s11095-018-2558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020 Jul 2;48(W1):W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A., Timonen S., Kononov A., Aittokallio T., Giri A.K. SynToxProfiler: an interactive analysis of drug combination synergy, toxicity and efficacy. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1007604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilunga Kalenga O., Moeti M., Sparrow A., Nguyen V.K., Lucey D., Ghebreyesus T.A. The ongoing Ebola epidemic in the democratic republic of Congo, 2018-2019. N. Engl. J. Med. 2019 Jul 25;381(4):373–383. doi: 10.1056/NEJMsr1904253. [DOI] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. 2020. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FD-Approved Drugs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.M., Brannan J.M., Delos S.E., Shoemaker C.J., Stossel A., Lear C., Hoffstrom B.G., DeWald L.E., Schornberg K.L., Scully C., Lehár J., Hensley L.E., White J.M., Olinger G.G. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005471. 190ra179-190ra179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas O. Pandemic bonds: designed to fail in Ebola. Nature. 2019;572:285. doi: 10.1038/d41586-019-02415-9. [DOI] [PubMed] [Google Scholar]
- Kazmi F., Hensley T., Pope C., Funk R.S., Loewen G.J., Buckley D.B., Parkinson A. Lysosomal sequestration (trapping) of lipophilic amine (cationic amphiphilic) drugs in immortalized human hepatocytes (Fa2N-4 cells) Drug Metab. Dispos. 2013;41:897–905. doi: 10.1124/dmd.112.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T., Russo D.P., Zorn K.M., Clark A.M., Korotcov A., Tkachenko V., Reynolds R.C., Perryman A.L., Freundlich J.S., Ekins S. Comparing and validating machine learning models for Mycobacterium tuberculosis drug discovery. Mol. Pharm. 2018;15:4346–4360. doi: 10.1021/acs.molpharmaceut.8b00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.R., Comer J.E., Freiberg A.N., Madrid P.B., Ekins S. Repurposing quinacrine against Ebola virus infection in vivo. AAC (Antimicrob. Agents Chemother.) 2019;63 doi: 10.1128/AAC.01142-19. e01142-01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.R., Massey C., Comer J.E., Anantpadma M., Freundlich J.S., Davey R.A., Madrid P.B., Ekins S. Repurposing the antimalarial pyronaridine tetraphosphate to protect against Ebola virus. Infection PLoS Negl Trop Dis. 2019;13 doi: 10.1371/journal.pntd.0007890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.R., Massey C., Comer J.E., Freiberg A.N., Zhou H., Dyall J., Holbrook M.R., Anantpadma M., Davey R.A., Madrid P.B., Ekins S. Antiviral Res In press; 2020. Pyronaridine Tetraphosphate Efficacy against Ebola Virus Infection in Guinea Pig. 2020.2003.2020.001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–290. [PubMed] [Google Scholar]
- Madrid P.B., Chopra S., Manger I.D., Gilfillan L., Keepers T.R., Shurtleff A.C., Green C.E., Iyer L.V., Dilks H.H., Davey R.A., Kolokoltsov A.A., Carrion R., Jr., Patterson J.L., Bavari S., Panchal R.G., Warren T.K., Wells J.B., Moos W.H., Burke R.L., Tanga M.J. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PloS One. 2013;8 doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2015;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A.A., Davey R.A., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2015;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- McCarthy S.D., Majchrzak-Kita B., Racine T., Kozlowski H.N., Baker D.P., Hoenen T., Kobinger G.P., Fish E.N., Branch D.R. A rapid screening assay identifies monotherapy with interferon-ss and combination therapies with nucleoside analogs as effective inhibitors of Ebola virus. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeschler S., Locher S., Zimmer G. 2018. 1-Benzyl-3-cetyl-2-methylimidazolium Iodide (NH125) Is a Broad-Spectrum Inhibitor of Virus Entry with Lysosomotropic Features. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., Ali R., Coulibaly S., Levine A.C., Grais R., Diaz J., Lane H.C., Muyembe-Tamfum J.J., Group P.W., Sivahera B., Camara M., Kojan R., Walker R., Dighero-Kemp B., Cao H., Mukumbayi P., Mbala-Kingebeni P., Ahuka S., Albert S., Bonnett T., Crozier I., Duvenhage M., Proffitt C., Teitelbaum M., Moench T., Aboulhab J., Barrett K., Cahill K., Cone K., Eckes R., Hensley L., Herpin B., Higgs E., Ledgerwood J., Pierson J., Smolskis M., Sow Y., Tierney J., Sivapalasingam S., Holman W., Gettinger N., Vallee D., Nordwall J., Team P.C.S. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaciva S., Lu S., Gebhard D.F., Jessen B.A., Pennie W.D., Will Y. A high content screening assay for identifying lysosomotropic compounds. Toxicol. Vitro. 2011;25:715–723. doi: 10.1016/j.tiv.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Ploemen J.P., Kelder J., Hafmans T., van de Sandt H., van Burgsteden J.A., Saleminki P.J., van Esch E. Use of physicochemical calculation of pKa and CLogP to predict phospholipidosis-inducing potential: a case study with structurally related piperazines. Exp. Toxicol. Pathol. 2004;55:347–355. doi: 10.1078/0940-2993-00338. [DOI] [PubMed] [Google Scholar]
- Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E., Johnson A., Morton J., Swope K., Bohorov O., Bohorova N., Goodman C., Kim D., Pauly M.H., Velasco J., Pettitt J., Olinger G.G., Whaley K., Xu B., Strong J.E., Zeitlin L., Kobinger G.P. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo D.P., Zorn K.M., Clark A.M., Zhu H., Ekins S. Comparing multiple machine learning algorithms and metrics for estrogen receptor binding prediction. Mol. Pharm. 2018;15:4361–4370. doi: 10.1021/acs.molpharmaceut.8b00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salata C., Calistri A., Parolin C., Baritussio A., Palu G. Antiviral activity of cationic amphiphilic drugs. Expert Rev. Anti Infect. Ther. 2017;15:483–492. doi: 10.1080/14787210.2017.1305888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval P.J., Zorn K.M., Clark A.M., Ekins S., Wright S.H. Assessment of substrate-dependent ligand interactions at the organic cation transporter OCT2 using six model substrates. Mol. Pharmacol. 2018;94:1057–1068. doi: 10.1124/mol.117.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R.L., Gerber J.P. Chloroquine and hydroxychloroquine binding to melanin: some possible consequences for pathologies. Toxicol Rep. 2014;1:963–968. doi: 10.1016/j.toxrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selakovic Z., Tran J.P., Kota K.P., Lazic M., Retterer C., Besch R., Panchal R.G., Soloveva V., Sean V.A., Jay W.B., Pavic A., Verbic T., Vasiljevic B., Kuehl K., Duplantier A.J., Bavari S., Mudhasani R., Solaja B.A. Second generation of diazachrysenes: protection of Ebola virus infected mice and mechanism of action. Eur. J. Med. Chem. 2019;162:32–50. doi: 10.1016/j.ejmech.2018.10.061. [DOI] [PubMed] [Google Scholar]
- Sivapalasingam S., Kamal M., Slim R., Hosain R., Shao W., Stoltz R., Yen J., Pologe L.G., Cao Y., Partridge M., Sumner G., Lipsich L. Safety, pharmacokinetics, and immunogenicity of a co-formulated cocktail of three human monoclonal antibodies targeting Ebola virus glycoprotein in healthy adults: a randomised, first-in-human phase 1 study. Lancet Infect. Dis. 2018;18:884–893. doi: 10.1016/S1473-3099(18)30397-9. [DOI] [PubMed] [Google Scholar]
- Sun W., He S., Martinez-Romero C., Kouznetsova J., Tawa G., Xu M., Shinn P., Fisher E., Long Y., Motabar O., Yang S., Sanderson P.E., Williamson P.R., Garcia-Sastre A., Qiu X., Zheng W. Synergistic drug combination effectively blocks Ebola virus infection. Antivir. Res. 2017;137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twarog N.R., Stewart E., Hammill C.V., A A.S. BRAID: a unifying paradigm for the analysis of combined drug action. Sci. Rep. 2016;6:25523. doi: 10.1038/srep25523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 Mar;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.F., Neiner A., Lane T.R., Zorn K.M., Ekins S., Kharasch E.D. Halogen substitution influences ketamine metabolism by cytochrome P450 2B6: in vitro and computational approaches. Mol. Pharm. 2019;16:898–906. doi: 10.1021/acs.molpharmaceut.8b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S., Haupt R., Logue J., Matthews K., Frieman M.B. FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 <em>in vitro</em>. bioRxiv. 2020;2020(2003):2025. 008482. [Google Scholar]
- Xiao J.H., Rijal P., Schimanski L., Tharkeshwar A.K., Wright E., Annaert W., Townsend A. Characterization of influenza virus pseudotyped with ebolavirus glycoprotein. J. Virol. 2018;92 doi: 10.1128/JVI.00941-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn K.M., Lane T.R., Russo D.P., Clark A.M., Makarov V., Ekins S. Multiple machine learning comparisons of HIV cell-based and reverse transcriptase data sets. Mol. Pharm. 2019;16:1620–1632. doi: 10.1021/acs.molpharmaceut.8b01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.