Abstract
Purpose
Ocular surface microbiome changes can affect meibomian gland dysfunction (MGD) development. This study aimed to delineate differences among the microbiome of eyelid skin, conjunctiva, and meibum in healthy controls (HCs) and patients afflicted with MGD.
Methods
Shotgun metagenomic analysis was used to determine if there are differences between the microbial communities in ocular sites surrounding the meibomian gland in healthy individuals and patients afflicted with MGD.
Results
The meibum bacterial content of these microbiomes was dissimilar in these two different types of individuals. Almost all of the most significant taxonomic changes in the meibum microbiome of individuals with MGD were also present in their eyelid skin, but not in the conjunctiva. Such site-specific microbe pattern changes accompany increases in the gene expression levels controlling carbohydrate and lipid metabolism. Most of the microbiomes in patients with MGD possess a microbe population capable of metabolizing benzoate. Pathogens known to underlie ocular infection were evident in these individuals. MGD meibum contained an abundance of Campylobacter coli, Campylobacter jejuni, and Enterococcus faecium pathogens, which were almost absent from HCs. Functional annotation indicated that in the microbiomes of MGD meibum their capability to undergo chemotaxis, display immune evasive virulence, and mediate type IV secretion was different than that in the microbiomes of meibum isolated from HCs.
Conclusions
MGD meibum contains distinct microbiota whose immune evasive virulence is much stronger than that in the HCs. Profiling differences between the meibum microbiome makeup in HCs and patients with MGD characterizes changes of microbial communities associated with the disease status.
Keywords: meibomian gland dysfunction, microbiome, metagenomics, metabolism, pathogen, benzoate degradation
Meibomian gland dysfunction (MGD) is the most prevalent cause of evaporative dry eye disease, Rabensteiner et al. found that 70.3% of patients with dry eye exhibited signs of this disease.1 It is characterized by chronic, diffuse meibomian gland abnormality, usually resulting from obstruction of the terminal duct of the meibomian gland and/or abnormal changes in the quality and quantity of glandular secretion by this gland. Such dysfunction can result from glandular losses, opening abnormalities at the eyelid margin blocking meibomian gland release onto the ocular surface.2,3 Meibomian glandular lipid secretory abnormalities or lipid component changes are major manifestations of MGD. Until now, its underlying pathogenesis is poorly understood.
Meibomian gland lipid secretions are essential for preventing ocular surface desiccation because they reduce tear film evaporation by spreading over the aqueous layer covering the ocular surface. In addition, they are an important component of the ocular surface antibacterial system because they form a barrier, which protects the eyes from microbial infections. In patients with MGD, changes in the lipid component of the meibum secreted by the meibomian gland include increases in certain fatty acids, etc.4–7 Such changes can alter the physical properties of the tear film and also trigger receptor mediated cell signaling events that comprise pathological sequelae in this disease. The microbiome and ocular immune functional competence stems from adapting to the challenges that eyes encounter as a consequence of constant exposure to numerous environmental challenges. There is extensive interest in identifying the host-microbial interactions in this ecosystem because such insight can potentially lead to the design of novel agents that are more selective in therapeutic management of ocular diseases in a clinical setting.
Meibomian glandular lipid abnormalities are believed to be associated with changes in the makeup of ocular surface microbial populations.8 The accumulation of lipids resulting from the blockage of the meibomian gland duct limits its bactericidal capability, which, in turn, heightens the proliferation of ocular surface microorganisms.2 Bacterial culturing of the ocular surface of patients with MGD demonstrated that multiple strains exhibit significant potential pathogenic roles. They include coagulase-negative staphylococci (mainly Staphylococcus epidermidis), P. acnes, coryneform bacteria, and Staphylococcus aureus.9 It is particularly noteworthy that the cholesterol esterase and fatty wax esterase activities of Staphylococcus epidermidis are essential in promoting the pathogenesis of MGD.10,11 Pathogen identification is crucial for infection control. Nevertheless, extensive efforts are still needed to more extensively characterize the resident pathogens responsible for ocular diseases.
In recent years, identifying the changes in the microbial population on the human body surface have become areas of intense interest in numerous different research fields studying disease processes. Novel data analysis of methodology is accumulating along with more improved metagenomic research procedures.12–14 Considering the risks of ocular infection by any of the different aforementioned bacteria, there is a pressing need to gain additional insight into the content of ocular surface microbiomes. Diverse efforts have been used to characterize the microbes inhabiting this domain in a pathological condition.15–18 Nevertheless, most progress has been restricted to focusing on the composition of the microbial population using 16S rDNA/rRNA sequencing to characterize the microbiomes in ocular sites. Limited studies are adopting untargeted sequencing of all microbes present in meibum using shotgun metagenomics.
Here, we collected samples from meibum, eyelid skin, and conjunctiva of patients with MGD and healthy individuals. Shotgun metagenomic analysis was performed involving next generation sequencing to characterize pathogens within these sites, identify functional changes related to MGD development, and inter-relationships among these different microbial populations at these three different ocular sites. Site-specific differences were identified on the ocular surface between the microbial populations in the meibum of patients with MGD and healthy controls (HCs). Novel targets were uncovered that are possibly relevant in clarifying how to improve the management of this disease in a clinical setting.
Materials and Methods
Recruitment of Subjects
This study was conducted in accordance with the Declaration of Helsinki principles for medical research and was approved by the Institutional Review Board/Ethics Committee from the Eye Hospital of Wenzhou Medical University. All of the subjects were recruited at the Dry Eye Center in the Eye Hospital of Wenzhou Medical University and informed consent was obtained from each participant. In this study, 76 volunteers, including 61 patients diagnosed with MGD and 15 HCs were enrolled. For all participants, clinical ocular surface examination and symptomatic evaluation were preceded by ophthalmological evaluation at the Dry Eye Center in the Eye Hospital of Wenzhou Medical University, including completion of the McMonnies, the ocular surface disease index (OSDI) questionnaire, Schirmer's test for dry eye, tear meniscus height (TMH), tear break up time (TBUT), and degree of Meibomian gland absence (upper/ lower) were assessed. Grading of MGD is according to the guideline from the International Workshop on Meibomian Gland Dysfunction.19
Subject information was obtained, including gender, age, antibiotic usage within 6 months, and ocular and general health status. Detailed participant information is shown in Supplementary Table S1. The inclusion criteria for patients include: (1) patients with a complaint of one of the following symptoms: dryness, foreign body sensation, burning; (2) a diagnosis of MGD with two or more of the following signs in both eyes: redness or thickening of the lid margin, telangiectasia, reduced or no secretions, poor quality secretions, and gland obstruction; (3) did not have ocular or systemic diseases, ocular traumas, transplantations, or laser surgery; (4) treatment naive: did not have intervention therapy for MGD and dry eye (including medical and physical therapy), this is the first time to squeeze the gland for meibum collection, did not recently take any antibiotics, and/or receive steroid treatment within the previous 6 months; (5) did not have allergies to medications, pollen, etc.; and (6) no contact lenses were used within the past 6 months. For the HCs: (1) absence of any dry eye symptoms; (2) absence of any symptomology that are criteria for diagnosing MGD; and (3) corneal fluorescein staining was negative.
Sampling
All samples were collected from April to September 2017. For each subject, a random eye was chosen for sampling. First, the lower eyelid skin was gently wiped 2 to 3 times using one Specimen Collection Flocked Swabs (Huachenyang [Shenzhen] Technology Co., Ltd., Shenzhen, Guangdong, China); the lower bulbar conjunctiva sac of some subjects also served as an internal control, as described by Zhang et al.20 For meibomian gland sampling, first, proparacaine hydrochloride (Tianlong Pharmaceutical [Suzhou] Co., Ltd., Suzhou, Jiangsu, China) was applied as a topical anesthetic, then the meibomian gland was massaged and pressed using sterile tweezers to release its secretion, which was collected with Flocked Swabs. The swabs were placed into 1.5 mL tubes (Axygen Biotechnology [Hangzhou] Co., Ltd., Hangzhou, Zhejiang, China) containing 200 µl DNase-Free double distilled water (ddH2O; Ambion, Thermo Fisher Scientific Inc., Cleveland, OH, USA). The samples were then quickly stored at −80 deg Celsius (°C) until use.
Sequencing Experiments
DNA extraction and whole genome amplification followed the protocols described in our previous study.20 For each sample, 200 ng DNA was used in paired-end sequencing (2 × 150 base pair, bp) on an HiSeq sequencer (Illumina, Inc., San Diego, CA, USA) and 10G raw data were obtained.
Quality Control of Sequencing Data
The low-quality bases of the raw reads were trimmed based on the quality information. Trimmomatic (version 0.36) was used to trim and discard the adaptor sequences.21 The bases at the beginning and end of each reads were discarded. The Trimmomatic slides from the 5' end in windows with the length at 4 bases; when the average quality in the window is lower than the setting threshold at 15, the read will be cut. Moreover, truncated reads, which were shorter than 36 bp, were discarded. The sequencing statistics for each sample are summarized in Supplementary Table S2.
Ribosomal RNA Gene Depletion for Datasets
The SortmeRNA (version 2.1b) was used to filter the rRNA gene sequence after quality control (QC) with default parameters and full database (silva-arc-16s-db, silva-arc-23s-db, silva-bac-16s-db, silva-bac-23s-db, silva-euk-18s-db, silva-euk-18s-db, silva-euk-28s-db, rfam-5.8s-db, and rfam-5s-db).22
Taxonomic Assignment of Sequencing Reads, Quantification of Taxonomic Categories, and Pathogen Identification
The Centrifuge (version 1.0.4b) uses the Burrows-Wheeler transform (BWT) and Ferragina-Manzini (FM) index as the indexing scheme for the National Center for Biotechnology Information (NCBI) NT database (nt_2018_3_3) to classify sequencing reads, and allows each read assigned by multiple taxonomic categories.23 Then, we assigned the read to a single taxonomic category using the lowest common ancestor of all matching hits with parameters “–min- hitlen 22 -k 1.” We subsequently removed sequencing reads with hit length < 60 base pairs and assigned them to eukaryotic kingdom clade (taxID: 2759) before taxonomic composition analysis at any taxonomic rank (phylum, class, order, family, genus, and species). According to the assigned taxonomic results at the species level, we quantified pathogens in the Karius Pathogen List (https://kariusdx.com/pathogenlist/3.4).
Functional Profiling of Shotgun Sequencing Reads
The UProc (version 2.0.0-rc1) was used to calculate the functional classification and relative abundance of sequencing reads.24 UProc translated DNA into protein for all the six frames, then compared the reads with oligopeptides at the protein level, and used Mosaic Matching calculation and Mosaic Matching Score to identity the most matching protein family. The SUPER-FOCUS (version 0.31) was used to annotate with the SUPER-FOCUS functional classification system.25 During annotation, DIAMOND blasted the protein sequence data and fetched the seed classification information. The antibiotic genes were annotated with ResistoMap with the Comprehensive Antibiotic Research Database (CARD) database for sequence alignment.
De Novo Assembly
Trinity (version 2.8.4) was used for de novo contig assembly without reference genomes from sequencing reads with parameters “–max_memory 300G –min_contig_length 200 –CPU 40 –bflyCPU 40 –inchworm_cpu 40 –full_cleanup –no_normalize_reads.”26
Gene Function Annotation for Assembled Contigs
The coding sequences in each assembled Trinity contigs were predicted with MetaProdigal (version 2.6.3).27 The Clusters of Orthologous Groups (COG) of proteins and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations were performed with evolutionary genealogy of genes (eggNOG: Non-supervised Orthologous Groups)-mapper (version 1.0.3) with an eggNOG database (version 4.5).28 The gene abundance (transcript per million reads, here, is gene per million reads) was estimated with Salmon (version 0.11.3),29 and the derivations of each functional gene were predicted with Centrifuge, and the antibiotic resistance genes were identified using Resistance Gene Identifier (RGI, version 4.0.2) with the CARD database (version 2.0.0).30 The proteins with homologous sequences in the ARDB database is performed using USEARCH with parameters “-b 60 -i 30 -e 1e-10.”
Differential Abundance and Statistical Analyses
To identify annotations with differential abundance between the two different groups, the abundance level for each annotation in units of Reads Per Million was calculated using Salmon (version 0.11.3). DESeq2 (version 1.10.1) was used for differential analysis.31 The significance difference level between the two groups was selected using the criterion: P values should be < 0.05. Significant differences in the relative abundance of different taxa present in both groups were found after application of the Metastats statistical method with a false discovery rate (FDR) correction, adjusted following the Benjamini-Hochberg method.
Other statistical analysis was performed with R (version R-3.6).32 Welch's t-test was applied for group to group comparisons of pathogen positive rate, and the Bonferroni test for P value corrections. Chi-square test was applied to compare the amount of eggNOG entries with decreased abundances or increased abundances between the MGD and HC groups. Those differences with a P value < 0.05 were considered statistically significant.
Data Visualization
The Principal Components Analysis (PCA) and bar-plot figures were generated using R (version R-3.6).32 The heatmap figures were generated using pheatmap and ComplexHeatmap packages.33,34
Results
Decreased Microbial Abundance in MGD Meibum and Eyelid Skin
In all, we enrolled 61 Chinese Han patients with MGD (treatment naive, the criteria see Method) and 15 volunteers as HCs (Fig. 1, see Supplementary Table S1). They provided DNA samples from swabs of meibum, the eyelid skin, and conjunctiva following the procedure shown in Figure 1, Method. After performing QC and removing host sequences, 117 metagenome datasets were obtained. Each dataset contained over 100 K sequencing reads for metagenome assembly and subsequent annotations (see Supplementary Table S2). In taxonomic profiling, taxa were determined based on nucleic acid annotation and marker gene presence, respectively (Supplementary Fig. S1). In de novo assembly, each dataset had 32,692 contigs on average, with their mean length ranging from 225 bp to 528 bp (Supplementary Table S3).
Figure 1.
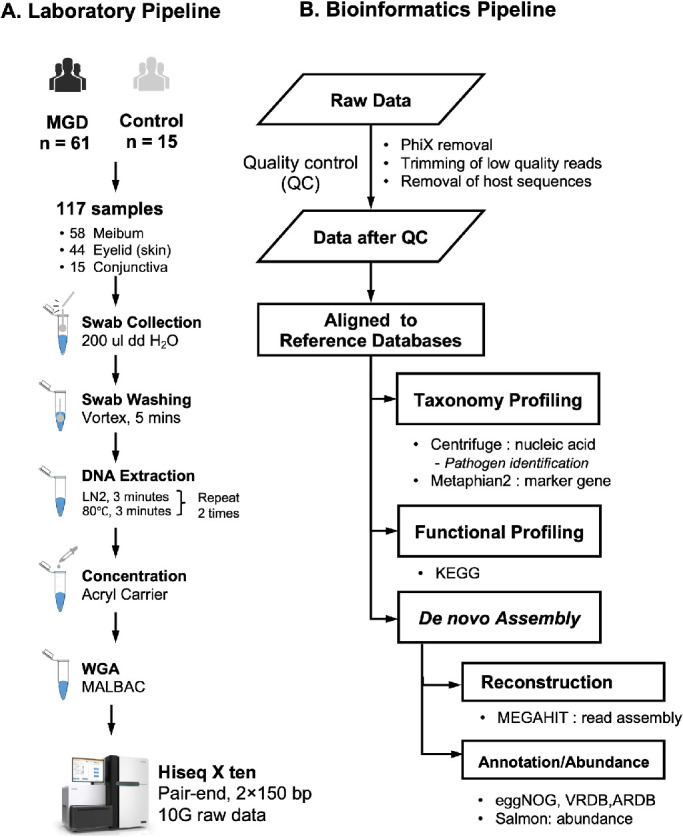
Study design. (A) Laboratory Pipeline. Swabs were collected from 76 subjects, including 61 MGD patients and 15 HC volunteers. Totally, 117 samples had qualified sequencing results (Method), including 58 were from meibum, 44 from skin of eyelid, and 15 from the conjunctiva. Liquid nitrogen (LN) was used for DNA extraction, and Acryl carrier was used to enrich the DNA fragment and decrease the volumes of DNA solution. Whole-genome amplification (WGA) was applied to obtain an adequate amount of DNA for sequencing library construction. (B) Bioinformatic Pipeline. Quality control process low quality reads, PhiX, and host sequences were removed in the quality control process. Multiple tools and public databases were applied to annotate sequencing fragments and de novo assembly.
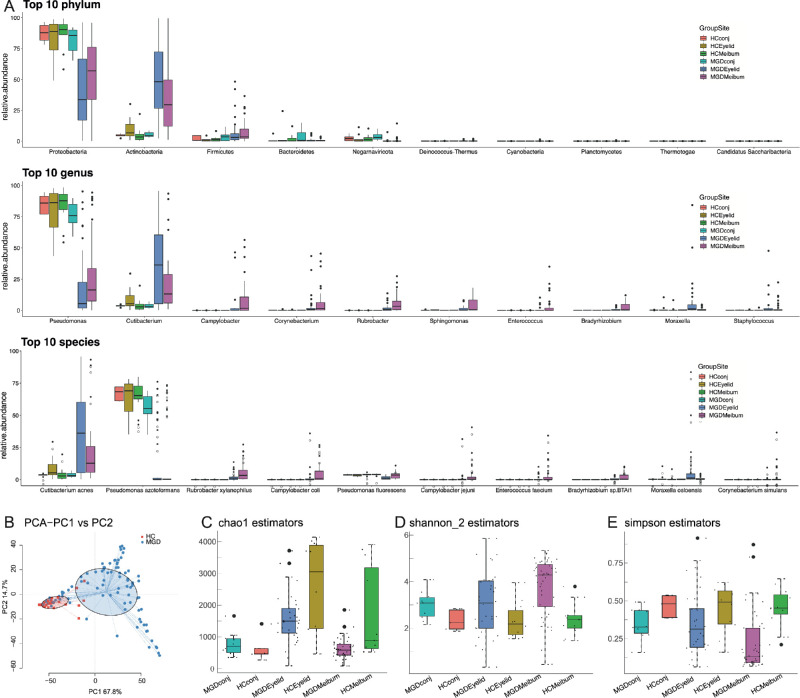
In MGD meibum, microbial populations having the same phylum were preponderant at the three ocular sites: Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and Negarnaviricota (Fig. 2A). At the genus level, the predominant genera at the three ocular sites are Pseudomonas, Cutibacterium, Campylobacter, Corynebacterium, and Rubrobacter. At the species level, Cutibacterium acnes, Pseudomonas azotoformans, Rubrobacter xylanophilus, Campylobacter coli, and Pseudomonas fluorescens were preponderant (see Fig. 2A). Samples were clustered into distinct groups according to different levels of taxonomic classification and their disease status (Fig. 2B, see Supplementary Fig. S1). As expected, the eyelid skin microbiome had the highest community richness, whereas conjunctival flora was sparse. In patients with MGD, their meibum microbiome community population was much smaller than in the HC group (chao1, Fig. 2C), whereas their community diversities were similar (Shannon, Fig. 2D; Simpson, Fig. 2E). This population decline was attributable to decreases in the abundance of the more preponderant microbes in these patients. Interestingly, alpha diversity was similar in the meibum and eyelid skin microbiomes (see Figs. 2C–2E).
Figure 2.
Overall comparison of taxonomic classification in the microbiome of all samples. (A) Microbial diversity in meibum, eyelid skin and conjunctiva. (B) Principle coordinate analysis (PCoA) plot of microbial communities at the genus level (centrifuge). (C-E) Alpha diversity measured by KO abundance for MGD patients and HC individuals.
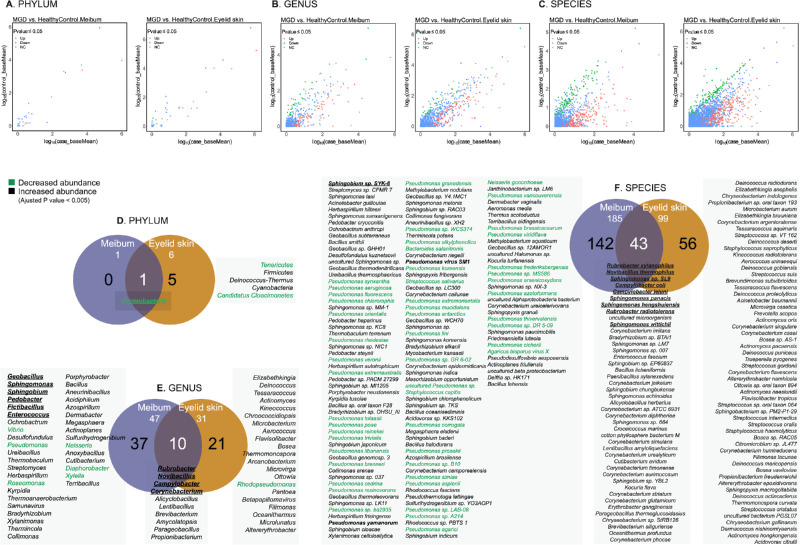
Changes in the microbial community were observed in the microbiomes of MGD meibum and eyelid skin at different taxonomic classification levels (Figs. 3A–3C). Compared to HCs, the most significant microbial change at the phylum level in the MGD meibum was the significantly decreasing abundance of Proteobacteria (log2 Fold Change = -4, adjusted P value = 2.8 × 10−15, Supplementary Table S4), which was also seen in the MGD eyelid skin microbiome (Fig. 3D, Supplementary Table S5). A comparison of the MGD and HC groups shows that the top 10 genera whose abundance was not the same were present at higher levels in MGD meibum. These genera included Rubrobacter, Novibacillus, Campylobacter, Geobacillus, Sphingomonas, Corynebacterium, Sphingobium, Pedobacter, Fictibacillus, and Enterococcus (log2 Fold Change = approximately 3–14, adjusted P value < 1 × 10-6, Fig. 3E, see Supplementary Table S4). Among these genera, the same changes were observed in the top three and Corynebacterium in the eyelid skin microbiome, whereas a greater abundance of Geobacillus, Sphingomonas, Sphingobium, Pedobacter, Fictibacillus, and Enterococcus was specific to the microbiome of the MGD meibum. At the species level, the top 10 were Rubrobacter xylanophilus, Novibacillus thermophilus, Sphingomonas sp. SL9, Campylobacter coli, Campylobacter jejuni, Sphingomonas panacis, Sphingomonas hengshuiensis, Rubrobacter radiotolerans, Sphingobium sp. SYK-6, and Sphingomonas wittichii, all of them were more abundant in MGD samples (log2 Fold Change = approximately 5–14, adjusted P value < 1 × 10-9, see Supplementary Table S3). Among them, only Sphingobium sp. SYK-6 was more abundant in the MGD meibum, whereas the other nine species were also more preponderant in the microbiomes of the MGD eyelid skin (Fig. 3F).
Figure 3.
Comparison of significant changes at the different taxonomic levels in meibum and eyelid microbiomes of patients with MGD. Dot plots show the mean abundance of each taxa in different classifications (A: phylum, B: genus, C: species) in patients with MGD (x-axis) and healthy control (HC) individuals (y-axis). The colors of dots indicate if the taxon has a significantly higher (red)/lower (green) abundance in patients with MGD (adjusted P value < 0.05, Benjamini-Hochberg), or their abundance is at similar levels in MGD and HC groups (blue). Venn plots show the common and specific taxa (D: phylum, E: genus, F: species) with most significant abundance changes (adjusted P value < 0.005, Benjamini-Hochberg) in microbiomes from the meibum and eyelid skin of MGD patients. The taxa shown in green have a significantly lower abundance in MGD meibum. Bold italics denote top 10 taxa with differential abundance in the MGD meibum microbiome. The taxa ranking is based on their significance level.
At the genus and species levels, other changes were more highly significant (adjusted P value < 0.005, Benjamini-Hochberg, see Figs. 3E, 3F) in the MGD meibum (genus = 47; species =185) than in the eyelid skin (genus = 31; species = 99). Specifically, all of the taxa that were common to the two aforementioned sites were more abundant in patients with MGD than HC volunteers. However, in some strains of the Pseudomonas genus, there were species with altered abundance that were specific to MGD meibum. These strains account for 88% (44/50, green, see Fig. 3F) of the species with decreased abundance in the MGD meibum at this significance level. Nevertheless, two other species of Pseudomonas (P.), P. yamanorum, and P. virus SM1 (bold italic, see Fig. 3F), are exceptions, whose abundance in meibum was greater in patients with MGD than in HCs.
Pathogen Preponderance in Meibum and Disease Status
In HC samples, 968 different strains of pathogens were detected (Method), whereas MGD samples contained 2400 pathogens (pathogen; see Supplementary Table S1). However, on the average, the population of each pathogen in the MGD group at the two sites (conjunctiva and eyelid skin) was smaller than in the HC group (mean, MGD group = 26 vs. HC group = 37, P = 0.02, Welch t-test). Interestingly, the MGD meibum also had far fewer pathogens than that in the HC group (mean, MGD group = 13 vs. HC group = 36, P = 0.0014, Welch t-test, Supplementary Fig. S2A). Besides, in the patients with MGD, the magnitude of decreases in the number of different types of pathogens was slightly correlated with increases in disease severity (P = 0.063, Welch t-test; Supplementary Fig. S2B). Additionally, there was either no gender or age difference between the numbers of pathogens in the meibum and those at two ocular sites (conjunctiva and eyelid skin) between the MGD and HC groups (Supplementary Fig. S2C and Supplementary Fig. S3).
Twenty pathogens were identified whose positive rate was different in patients with MGD from that in the HCs (P < 0.05, Welch t-test; Table 1, Supplementary Fig.S2D). Furthermore, 28 pathogens were detected whose positive rate was > 10% in the meibum samples (Table 2, Supplementary Fig. S2E). The most preponderant pathogens in all meibum samples were Pseudomonas fluorescens (P. fluorescens), in over 90% of the samples (see Table 2, see Supplementary Fig. S2E), Cupriavidus metallidurans (76%), and Pseudomonas putida (52%). Nevertheless, only Pseudomonas fluorescens was present in 90% of the MGD and 100% of the HC samples (P = 0.044; see Table 2). Pathogens whose positive rate was much higher in MGD than in HC meibum were Campylobacter coli (P = 4.4 × 10−10, Welch t-test; see Table 1), Campylobacter jejuni (P = 1.2 × 10−8, Welch t-test), and Enterococcus faecium (P = 8.3 × 10−8, Welch t-test). Furthermore, their abundance was > 16-fold higher in the MGD than in the HC meibum (adjusted P value < 2 × 10−8; see Supplementary Table S4). Additionally, Pseudomonas aeruginosa (P = 4.8 × 10−6, Welch t-test), Pseudomonas mosselii (P = 1.4 × 10−4, Welch t-test), Escherichia coli (P = 0.001, Welch t-test), Stenotrophomonas maltophilia (P = 0.008, Welch t-test), and Neisseria sicca (P = 0.016, Welch t-test) had a lower positive rate in the MGD meibum and samples from the eyelid and conjunctiva.
Table 1.
Pathogens With Differential Positive Rate in Meibum Between Patients With MGD and HCs
| Pathogen | MGD (n = 47*) | HC (n = 11*) | P Value |
|---|---|---|---|
| Pseudomonas fluorescens (G−) | 42 (89%) | 11 (100%) | 0.024 |
| Campylobacter coli (G−) | 27 (57%) | 0 (0%) | 4.4E-10 |
| Campylobacter jejuni (G−) | 24 (51%) | 0 (0%) | 1.2E-08 |
| Enterococcus faecium (G+) | 22 (47%) | 0 (0%) | 8.3E-08 |
| Malassezia globosa | 22 (47%) | 10 (91%) | 0.001 |
| Pseudomonas protegens (G−) | 18 (38%) | 8 (73%) | 0.045 |
| Escherichia coli (G−) | 12 (26%) | 9 (82%) | 0.001 |
| Ralstonia pickettii (G−) | 12 (26%) | 8 (73%) | 0.008 |
| Pseudomonas aeruginosa (G−) | 11 (23%) | 10 (91%) | 4.8E-06 |
| Pseudomonas mosselii (G−) | 6 (13%) | 9 (82%) | 1.4E-04 |
| Bacillus licheniformis (G+) | 6 (13%) | 0 (0%) | 0.013 |
| Yarrowia lipolytica | 5 (11%) | 0 (0%) | 0.024 |
| Enterobacter cloacae complex (G−) | 3 (6%) | 5 (45%) | 0.034 |
| Comamonas testosteroni (G−) | 2 (4%) | 5 (45%) | 0.026 |
| Serratia marcescens (G-) | 2 (4%) | 5 (45%) | 0.026 |
| Stenotrophomonas maltophilia (G−) | 1 (2%) | 6 (55%) | 0.008 |
| Neisseria sicca (G−) | 0 (0%) | 5 (45%) | 0.016 |
| Neisseria meningitidis (G−) | 0 (0%) | 4 (36%) | 0.038 |
| Roseomonas gilardii (G−) | 0 (0%) | 4 (36%) | 0.038 |
| Staphylococcus capitis (G+) | 0 (0%) | 4 (36%) | 0.038 |
*MGD patients or HCs with meibum samples available.
Table 2.
Pathogens With Positive Rate Over 10% Across All Meibum Samples
| Pathogen | Total Meibum Samples (n = 58) | MGD (n = 47) | HC (n = 11) | P Value Welch t-test |
|---|---|---|---|---|
| Pseudomonas fluorescens (G−) | 53 (91%) | 42 (89%) | 11 (100%) | 0.02 |
| Cupriavidus metallidurans (G−) | 44 (76%) | 36 (77%) | 8 (73%) | 0.81 |
| Malassezia globosa | 32 (55%) | 22 (47%) | 10 (91%) | 8.9E-04 |
| Pseudomonas putida (G−) | 30 (52%) | 22 (47%) | 8 (73%) | 0.12 |
| Campylobacter coli (G−) | 27 (47%) | 27 (57%) | 0 (0%) | 4.4E-10 |
| Pseudomonas protegens (G−) | 26 (45%) | 18 (38%) | 8 (73%) | 0.05 |
| Campylobacter jejuni (G−) | 24 (41%) | 24 (51%) | 0 (0%) | 1.2E-08 |
| Enterococcus faecium (G+) | 22 (38%) | 22 (47%) | 0 (0%) | 8.3E-08 |
| Corynebacterium jeikeium (G+) | 22 (38%) | 20 (43%) | 2 (18%) | 0.10 |
| Alternaria alternata | 22 (38%) | 16 (34%) | 6 (55%) | 0.25 |
| Escherichia coli (G−) | 21 (36%) | 12 (26%) | 9 (82%) | 8.6E-04 |
| Pseudomonas aeruginosa (G−) | 21 (36%) | 11 (23%) | 10 (91%) | 4.8E-06 |
| Citrobacter freundii (G−) | 20 (34%) | 14 (30%) | 6 (55%) | 0.17 |
| Ralstonia pickettii (G−) | 20 (34%) | 12 (26%) | 8 (73%) | 0.01 |
| Staphylococcus epidermidis(G+) | 19 (33%) | 13 (28%) | 6 (55%) | 0.14 |
| Corynebacterium simulans (G+) | 18 (31%) | 15 (32%) | 3 (27%) | 0.77 |
| Corynebacterium striatum (G+) | 18 (31%) | 15 (32%) | 3 (27%) | 0.77 |
| Corynebacterium aurimucosum (G+) | 16 (28%) | 13 (28%) | 3 (27%) | 0.98 |
| Klebsiella pneumoniae (G−) | 15 (26%) | 10 (21%) | 5 (45%) | 0.18 |
| Pseudomonas mosselii (G−) | 15 (26%) | 6 (13%) | 9 (82%) | 1.4E-04 |
| Corynebacterium diphtheriae (G+) | 14 (24%) | 11 (23%) | 3 (27%) | 0.81 |
| Corynebacterium ureicelerivorans (G+) | 11 (19%) | 7 (15%) | 4 (36%) | 0.21 |
| Toxoplasma gondii | 10 (17%) | 7 (15%) | 3 (27%) | 0.43 |
| Bacteroides vulgatus (G−) | 10 (17%) | 5 (11%) | 5 (45%) | 0.06 |
| Aeromonas hydrophila (G−) | 9 (16%) | 6 (13%) | 3 (27%) | 0.35 |
| Corynebacterium riegelii (G+) | 7 (12%) | 5 (11%) | 2 (18%) | 0.57 |
| Corynebacterium urealyticum (G+) | 7 (12%) | 5 (11%) | 2 (18%) | 0.57 |
| Bacillus licheniformis (G+) | 6 (10%) | 6 (13%) | 0 (0%) | 0.01 |
G+, Gram-positive; G, Gram-negative.
Metabolic Features of Microbiome of MGD Meibum and Eyelid Skin
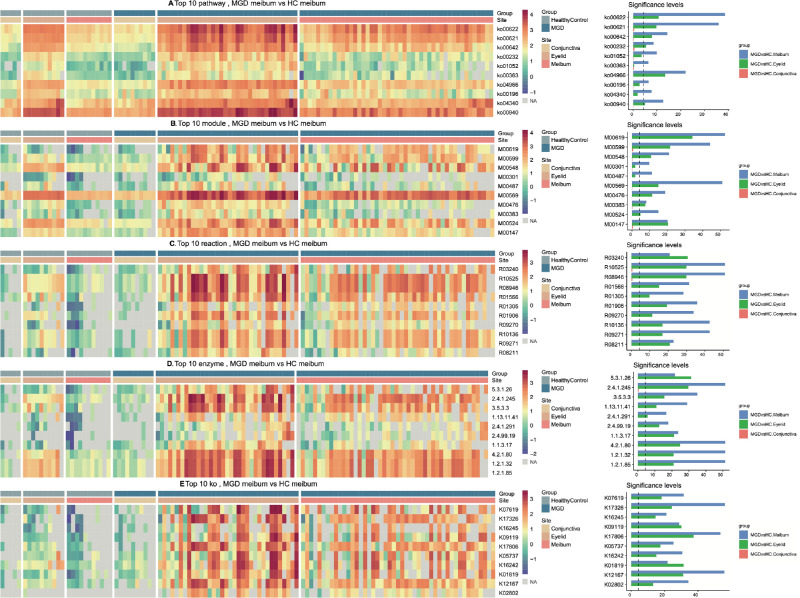
The assignation results for function annotation in the KEGG list includes the over-represented gene sets at different classification levels in the MGD meibum and eyelid skin. On the other hand, no differential entry was found for conjunctiva (adjusted P value > 0.05; Supplementary Tables S6–S7). Similar to taxonomy profiling, the top 10 gene sets with the largest fold abundance change in the MGD meibum microbiome in each annotation category (Figs. 4A–4E) also had significant alterations in the MGD eyelid samples, but not in the MGD conjunctival microbiome. The most notable microbiome changes in both the MGD meibum and eyelid skin were those pathways mediating microbial metabolism. Among the top 10 KEGG pathways, 4 were involved in the degradation process, including xylene (ko00622), dioxin (ko00621), ethylbenzene (ko00642), and bisphenol (ko00363).
Figure 4.
Over-representation of functions in microbiomes from patients with MGD and HCs. Annotations according to KEGG database at different layers: (A) Pathway; (B) Module; (C) Reaction; (D) Enzyme; (E) KEGG Orthology, KO. Full annotations for each entry are listed in Supplementary Table S11. The rank of the top 10 entries is listed in a descending order according to fold change of abundance in MGD meibum. Right panel: x-axis, -log2 (adjusted P value), dashed vertical line: adjusted P value = 0.05.
We further explored the molecular processes underlying 509 enzymes, which were mapped into KEGG pathways. The enzymes (see lists in Supplementary Table S7) in the MGD meibum microbiome whose abundance increased more than twofold from their levels in the HC meibum (adjusted P value < 0.05). Over half of them are involved in KEGG Metabolic pathways (63%, 319/509), 100 of them in the biosynthesis of secondary metabolites, 95 others in KEGG pathways of microbial metabolism in diverse environments, and 49 in the biosynthesis of antibiotic reference pathways (Supplementary Table S8). Specifically, all of the 15 pathways involved in carbohydrate metabolism had at least 4 enzymes with increased abundance. The second significant KEGG pathway category is involved with lipid metabolism. Ten of the 17 pathways contained at least 2 enzymes that were mapped into pathways mediating fatty acid elongation, biosynthesis and degradation, glycerolipid metabolism, glycerophospholipid metabolism, ether lipid metabolism, sphingolipid metabolism, biosynthesis of unsaturated fatty acids, carbon fixation pathways in prokaryotes, and nitrogen metabolism (see Supplementary Table S8).
For xenobiotic biodegradation and metabolism, 30 enzymes with increased sequencing read abundance mapped into this pathway category. Interestingly, the crucial one was likely to mediate benzoate degradation. First, 13 of these enzymes (43%, 13/30) directly mapped into this pathway (Supplementary Fig. S4A). Second, the subsequent eight enzymes with increased abundance catalyze the reactions that precede by one or two steps entrance into the benzoate degradation pathway, including E1.14.13.7 (phenol 2-monooxygenase, NADPH) and E1.14.13.82 (vanillate monooxygenase) in aminobenzoate degradation (Supplementary Fig. S4B), E3.7.1.8 (2,6-dioxo-6-phenylhexa-3-enoate hydrolase) in dioxin degradation (Supplementary Fig. S4C), E2.3.1.16 (acetyl-CoA C-acyltransferase) in ethylbenzene degradation (Supplementary Fig. S4D), E1.13.11.2 (catechol 2,3-dioxygenase) in styrene degradation (Supplementary Fig. S4E), and E1.14.13.1 (salicylate 1-monooxygenase) in dioxin degradation (see Supplementary Fig. S4C), naphthalene degradation (Supplementary Fig. S4F), and polycyclic aromatic hydrocarbon degradation (Supplementary Fig. S4G). In addition, four enzymes involved in the initial step in this process are in a branch leading to benzoate degradation, including E1.1.1.2 (alcohol dehydrogenase, NADP+) and E3.1.1.17 (gluconolactonase) in caprolactam degradation (Supplementary Fig. S4H), E1.14.12.10 (benzoate 1, 2-dioxygenase) in fluorobenzoate degradation pathway (Supplementary Fig. S4I), and E4.1.99.11 (benzylsuccinate synthase) in the toluene degradation pathway (Supplementary Fig. S4J).
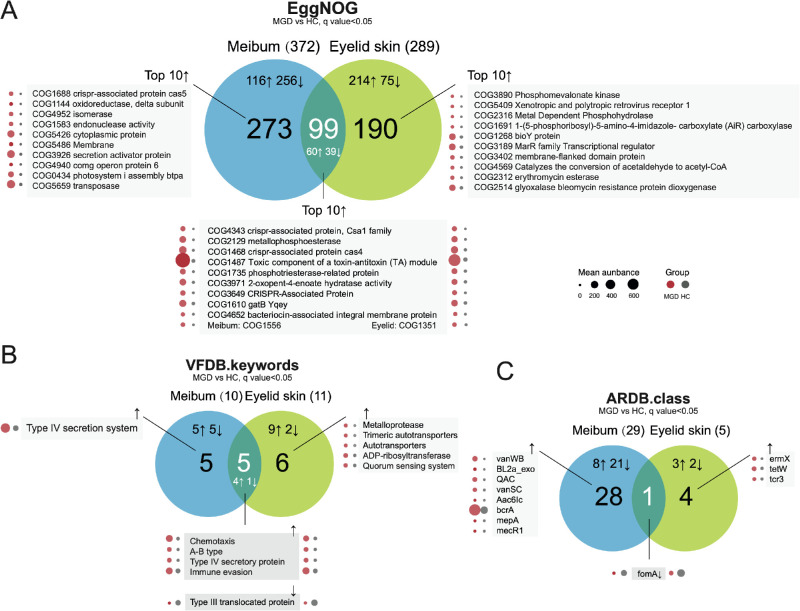
Functional Features Identified in EggNOG Database, Virulence Factors Database, and Antibiotic Resistance Genes Database
EggNOG annotation for contigs obtained in de novo assembly demonstrated significantly more functional entries with decreased abundance in the microbiomes of MGD meibum than those in the MGD eyelid skins (P < 0.0001, χ2 test, Fig. 5A; details in Supplementary Tables S9, S10), which is similar with the observation in taxonomy profiling. The featured virulence factor for MGD meibum microbiome was the type IV secretion system (T4SS), achieving about a fivefold change in abundance (q = 0.017, FDR; Fig. 5B, Supplementary Table S11). For antibiotic resistance genes, both sites had significantly reduced representation of the FomA gene (Fig. 5C), involved in Fosfomycin resistance.
Figure 5.
Over-representation of functional entries in (A) EggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database, (B) virulence Factors Database (VFDB), and (C) Antibiotic Resistance Genes Database (ARDB) in microbiomes of MGD meibum and eyelid skin. ↑ increase; ↓ decrease.
Discussion
As a result of contact with the external environment, many studies suggest that microbial populations are stable on the ocular surface and in the meibomian gland. Our objective here was to determine the makeup of the microbiome at these sites and also to delineate more extensively their microbiota functional features. Such insight was expected to improve our understanding of their microbial metabolic activity and virulence. This could ultimately foster efforts to identify novel effective antibiotic treatment options. To reach this goal, microbial population makeup was compared in meibum samples with those at the conjunctiva and in the eyelid skin obtained from individuals with MGD and in HCs. The results show that there is a resemblance between the bacterial populations in the meibum and eyelid microbiome. This finding is supportive of speculation indicating that these two sites are connected. Nevertheless, the most significant feature of the MGD meibum microbiome characterization stems from the results of microbial community diversity analysis. It was surprisingly revealed that in the MGD samples there was less pathogen diversity and community richness than in the HCs. Finally, our more in-depth analysis of the MGD and HC ocular surface microbiomes shows that there is a marked disparity between them. Our results are different from others in that the number of bacterial types was significantly higher in the severe MGD group than that in the HC based on 16S rRNA sequencing.35 This disagreement may be attributable to variance in methods and sample size. Additional studies are warranted work to clarify the origin of this discrepancy.
Even using whole genome amplification for these sparse microbiomes in ocular sites, few patients in our study could provide adequate microbiome data of the meibum and two ocular sites. To reduce DNA losses incurred in the extraction process, we did not treat the samples with RNAse before whole genome amplification, which may instead lead to RNA contamination. This difference may explain why the phylum Negarnaviricota was present, which includes all negative-sense single-stranded RNA viruses in the results (see Fig. 2A). However, their read abundance was too small for reliable virus identification. Future ocular virobiome studies may provide novel insights regarding their involvement in eye diseases.
Previous studies reported that the microbial community composition changes with age. This finding is consistent with ours because the skin microbiome constituents were dissimilar between the group aged under 40 years old and the one over 60 years old.36 To reduce such variance, we enrolled instead young HC volunteers and 42 patients with MGD who were under 40 years old. Specifically, the majority of them were not clustered into an HC group for PCA analysis (see Fig. 2B). Moreover, the number of pathogens identified in conjunctival and eyelid samples was not age-related (see Supplementary Fig. S3). Pathogens Campylobacter coli, Campylobacter jejuni, and Enterococcus faecium, which were prevalent in our MGD meibum samples are known to cause eye infections.37–39
Although there seems to be a negative correlation between the number of pathogens and ocular disease status, this does not mean that patients with MGD were at a reduced infection risk. Pseudomonas fluorescens was less abundant in MGD meibum (see Fig. 3F), but it had the highest positive rate across all meibum samples (91%; see Table 1, see Supplementary Fig. S2E). Nevertheless, infections caused by P. fluorescens seem to be rare. They were only detected in a few cases diagnosed with endophthalmitis,40,41 bacterial keratitis,42 or infectious crystalline keratopathy.43 Recently, a study suggested that the routine culture temperature may be too high for P. fluorescens to grow, which reduces the likelihood of evaluating its pathogenic impact.44 For Pseudomonas putida abundance, its decline was at a moderate significance level (log2 Fold Change = -1.2, adjusted P value = 0.024) in the MGD meibum compared to its HC counterpart. Whereas, its positive rate was over 50% in meibum samples irrespective of disease status (MGD versus HC, P = 0.12, Welch t-test; see Supplementary Table S6). Cupriavidus metallidurans had a positive rate of 73% in all meibum samples, but it only causes infection under extreme conditions.45 Pseudomonas aeruginosa, a well-known pathogen, which is responsible for 6% to 39% of bacterial keratitis cases in the United States,46 its positive rate was 40% in our meibum samples; Staphylococcus epidermidis, had a positive rate of 48.6% in MGD samples, which was based on the results of a traditional culture method.17 It was present in 33% of the meibum samples irrespective of whether or not they were obtained either from MGD or HC samples (MGD versus HC, P = 0.17, Welch t-test; see Supplementary Table S6). However, this equivalence may be erroneous due to sample heterogeneity or nonuniform culture methodology among these studies.
The changes in the meibum composition in patients with MGD are believed to affect their microbial population makeup. However, almost all significant functional annotation changes identified in MGD meibum were also seen in MGD eyelid skin, indicating that the underlying factors may be shared by the meibomian gland and sebaceous gland. Increased needs for more diverse metabolic pathways in MGD microbiomes may account for the extensive changes in the microenvironment. Fulfillment of bioenergetic requirements of all organisms has top priority, which helps explain why genes promoting carbohydrate related biological processes, carbon fixation, and nitrogen metabolism are all over-represented (see Supplementary Table S8). The most significant unique metabolic trait of the MGD microbiome is its capacity to catabolize benzoate, which belongs to Xenobiotics biodegradation and metabolism in the KEGG database. Most other changes in this pathway category are in agreement with their contribution to switch downstream the reaction direction toward benzoate degradation. Bacterial degradation of benzoate is believed to be part of the biological strategy that underlies the ability of organisms to adapt and survive despite swings in environmental oxygen concentration.47 This adaptability to survive despite changes in oxygen levels may stem from variations in the composition of MGD meibum.
Microbe profiling alterations may account for how immune cells get activated to increase in abundance to elicit an inflammatory response in tears and glandular tissue. This cascade of events affects the differentiation of glandular cells and their ability to synthesize and secrete lipids.3 These inflammatory responses by immune cells in the glandular environment are believed to be a key link in inducing pathological changes. Our study did not identify which microbes exclusively flourish in the MGD meibum. Nevertheless, the microbial community in the MGD samples seems to express more genes that induce chemotaxis and immune evasion (see Fig. 5C). Meanwhile, the changes in the meibum composition may also impede the responses by the immune activated cells to eliminate the pathogens in the meibum, which continuously release virulence factors at a stable elevated level. Therefore, MGD meibum harboring a microbial community containing more T4SS acts as a sustained intractable stimulant. This notion may partly explain why current efforts in alleviating symptoms are mainly effective if they include procedures that change the meibum composition. This can be accomplished through heating or improving the ability of meibomian glandular cells to produce and release more meibum out of the gland. Moreover, in future studies, our observations should be validated using larger diverse populations from diverse geographic locations around the world. Future efforts ought to consider individual uniqueness or microbiome population specificity.
Availability of Data and Material
The raw sequence data were deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers PRJCA002217, that are publicly accessible at http://bigd.big.ac.cn/gsa.
Supplementary Material
Acknowledgments
Supported by Zhejiang Provincial Natural Science Foundation of China (LY18H120005), National Natural Science Foundation of China (8157088 and 81970770), and Innovation Promotion Association CAS (2016098). They had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author Contributions: F.X.Z. and D.K.Z. interpreted results and drafted the manuscript. F.X.Z., D.K.Z., and L.Z. performed analyses, developed analysis methods, and power calculations. F.X.Z. and C.X.G. enrolled patients and collected all the clinical information. C.C.T. conducted sequencing experiments. F.X.Z. and C.C.T. collected and prepared samples for sequencing analysis, Z.L.Z, C.C.Z., and W.J.F. performed clinical examination and collected results of clinical assays. P.S.R. and C.Q.Z. reviewed and edited the manuscript. X.J.T. interpreted results and provided clinical and bioinformatic expertise. W.C. designed the study, supervised all experiments and analysis, reviewed, and edited the manuscript. All authors approved the final version of the manuscript.
Disclosure: F. Zhao, None; D. Zhang, None; C. Ge, None; L. Zhang, None; P.S. Reinach, None; X. Tian, None; C. Tao, None; Z. Zhao, None; C. Zhao, None; W. Fu, None; C. Zeng, None; W. Chen, None
References
- 1. Rabensteiner DF, Aminfar H, Boldin I, Schwantzer G, Horwath-Winter J. The prevalence of meibomian gland dysfunction, tear film and ocular surface parameters in an Austrian dry eye clinic population. Acta Ophthalmol. 2018; 96: e707–e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knop E, Knop N, Millar T, Obata H, Sullivan DA. The international workshop on meibomian gland dysfunction: report of the subcommittee on anatomy, physiology, and pathophysiology of the meibomian gland. Invest Ophthalmol Vis Sci. 2011; 52: 1938–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nichols KK, Foulks GN, Bron AJ, et al.. The international workshop on meibomian gland dysfunction: executive summary. Invest Ophthalmol Vis Sci. 2011; 52: 1922–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arita R, Mori N, Shirakawa R, et al.. Meibum color and free fatty acid composition in patients with meibomian gland dysfunction. Invest Ophthalmol Vis Sci. 2015; 56: 4403–4412. [DOI] [PubMed] [Google Scholar]
- 5. Pucker AD, Haworth KM. The presence and significance of polar meibum and tear lipids. Ocul Surf. 2015; 13: 26–42. [DOI] [PubMed] [Google Scholar]
- 6. Arita R, Mori N, Shirakawa R, et al.. Linoleic acid content of human meibum is associated with telangiectasia and plugging of gland orifices in meibomian gland dysfunction. Exp Eye Res. 2016; 145: 359–362. [DOI] [PubMed] [Google Scholar]
- 7. Paranjpe V, Tan J, Nguyen J, et al.. Clinical signs of meibomian gland dysfunction (MGD) are associated with changes in meibum sphingolipid composition. Ocul Surf. 2019; 17: 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Green-Church KB, Butovich I, Willcox M, et al.. The international workshop on meibomian gland dysfunction: report of the subcommittee on tear film lipids and lipid-protein interactions in health and disease. Invest Ophthalmol Vis Sci. 2011; 52: 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huber-Spitzy V, Baumgartner I, Bohler-Sommeregger K, Grabner G. Blepharitis–a diagnostic and therapeutic challenge. A report on 407 consecutive cases. Graefes Arch Clin Exp Ophthalmol. 1991; 229: 224–227. [DOI] [PubMed] [Google Scholar]
- 10. Dougherty JM, Osgood JK, McCulley JP. The role of wax and sterol ester fatty acids in chronic blepharitis. Invest Ophthalmol Vis Sci. 1991; 32: 1932–1937. [PubMed] [Google Scholar]
- 11. Shine WE, McCulley JP. Role of wax ester fatty alcohols in chronic blepharitis. Invest Ophthalmol Vis Sci. 1993; 34: 3515–3521. [PubMed] [Google Scholar]
- 12. Aguiar-Pulido V, Huang W, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol Bioinform Online. 2016; 12: 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turaev D, Rattei T. High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved. Curr Opin Biotechnol. 2016; 39: 174–181. [DOI] [PubMed] [Google Scholar]
- 14. Sedlar K, Kupkova K, Provaznik I. Bioinformatics strategies for taxonomy independent binning and visualization of sequences in shotgun metagenomics. Comput Struct Biotechnol J. 2017; 15: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Itahashi M, Higaki S, Fukuda M, Shimomura Y. Detection and quantification of pathogenic bacteria and fungi using real-time polymerase chain reaction by cycling probe in patients with corneal ulcer. Arch Ophthalmol. 2010; 128: 535–540. [DOI] [PubMed] [Google Scholar]
- 16. Schabereiter-Gurtner C, Maca S, Rolleke S, et al.. 16S rDNA-based identification of bacteria from conjunctival swabs by PCR and DGGE fingerprinting. Invest Ophthalmol Vis Sci. 2001; 42: 1164–1171. [PubMed] [Google Scholar]
- 17. Zhang SD, He JN, Niu TT, et al.. Bacteriological profile of ocular surface flora in meibomian gland dysfunction. Ocul Surf. 2017; 15: 242–247. [DOI] [PubMed] [Google Scholar]
- 18. Ozkan J, Nielsen S, Diez-Vives C, Coroneo M, Thomas T, Willcox M. Temporal stability and composition of the ocular surface microbiome. Sci Rep. 2017; 7: 9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geerling G, Tauber J, Baudouin C, et al.. The international workshop on meibomian gland dysfunction: report of the subcommittee on management and treatment of meibomian gland dysfunction. Invest Ophthalmol Vis Sci. 2011; 52: 2050–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H, Zhao F, Hutchinson DS, et al.. Conjunctival microbiome changes associated with soft contact lens and orthokeratology lens wearing. Invest Ophthalmol Vis Sci. 2017; 58: 128–136. [DOI] [PubMed] [Google Scholar]
- 21. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012; 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- 23. Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016; 26: 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meinicke P. UProC: tools for ultra-fast protein domain classification. Bioinformatics. 2015; 31: 1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silva GG, Green KT, Dutilh BE, Edwards RA. SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics. 2016; 32: 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grabherr MG, Haas BJ, Yassour M, et al.. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011; 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2012; 28: 2223–2230. [DOI] [PubMed] [Google Scholar]
- 28. Huerta-Cepas J, Forslund K, Coelho LP, et al.. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 2017; 34: 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017; 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alcock BP, Raphenya AR, Lau TTY, et al.. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020; 48: D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Team RC. R: A language and environment for statistical computing. 2013.
- 33. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32: 2847–2849. [DOI] [PubMed] [Google Scholar]
- 34. Kolde R, Kolde MR. Package ‘pheatmap’. R Package 2015. https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf.
- 35. Jiang X, Deng A, Yang J, et al.. Pathogens in the meibomian gland and conjunctival sac: microbiome of normal subjects and patients with meibomian gland dysfunction. Infect Drug Resist. 2018; 11: 1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shibagaki N, Suda W, Clavaud C, et al.. Aging-related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci Rep. 2017; 7: 10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuriyan AE, Sridhar J, Flynn HW Jr., et al.. Endophthalmitis caused by Enterococcus faecalis: clinical features, antibiotic sensitivities, and outcomes. Am J Ophthalmol. 2014; 158: 1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Janssen R, Krogfelt KA, Cawthraw SA, van Pelt W, Wagenaar JA, Owen RJ. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin Microbiol Rev. 2008; 21: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keat A, Rowe I. Reiter's syndrome and associated arthritides. Rheum Dis Clin North Am. 1991; 17: 25–42. [PubMed] [Google Scholar]
- 40. Robert PY, Chainier D, Garnier F, et al.. Alcaligenes xylosoxidans endophthalmitis following phacoemulsification and intraocular lens implantation. Ophthalmic Surg Lasers Imaging. 2008; 39: 500–504. [DOI] [PubMed] [Google Scholar]
- 41. Essex RW, Charles PG, Allen PJ. Three cases of post-traumatic endophthalmitis caused by unusual bacteria. Clin Exp Ophthalmol. 2004; 32: 445–447. [DOI] [PubMed] [Google Scholar]
- 42. Kitzmann AS, Goins KM, Syed NA, Wagoner MD. Bilateral herpes simplex keratitis with unilateral secondary bacterial keratitis and corneal perforation in a patient with pityriasis rubra pilaris. Cornea. 2008; 27: 1212–1214. [DOI] [PubMed] [Google Scholar]
- 43. Huerva V, Sanchez MC. Infectious crystalline keratopathy caused by Pseudomonas fluorescens. Eye Contact Lens. 2015; 41: e9–e10. [DOI] [PubMed] [Google Scholar]
- 44. Mitra S, Rath S, Das S, Basu S. Ocular infection by a psychrophile: Pseudomonas fluorescens. Indian J Med Microbiol. 2019; 37: 289–291. [DOI] [PubMed] [Google Scholar]
- 45. Langevin S, Vincelette J, Bekal S, Gaudreau C. First case of invasive human infection caused by Cupriavidus metallidurans. J Clin Microbiol. 2011; 49: 744–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sy A, Srinivasan M, Mascarenhas J, et al.. Pseudomonas aeruginosa keratitis: outcomes and response to corticosteroid treatment. Invest Ophthalmol Vis Sci. 2012; 53: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valderrama JA, Durante-Rodriguez G, Blazquez B, Garcia JL, Carmona M, Diaz E. Bacterial degradation of benzoate: cross-regulation between aerobic and anaerobic pathways. J Biol Chem. 2012; 287: 10494–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data were deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers PRJCA002217, that are publicly accessible at http://bigd.big.ac.cn/gsa.