Abstract
Purpose
To quantify organelles impacting imaging in the cell body and intact apical processes of human retinal pigment epithelium (RPE), including melanosomes, lipofuscin–melanolipofuscin (LM), mitochondria, and nuclei.
Methods
A normal perifovea of a 21-year-old white male was preserved after rapid organ recovery. An aligned image stack was generated using serial block-face scanning electron microscopy and was annotated by expert readers (TrakEM, ImageJ). Acquired measures included cell body and nuclear volume (n = 17); organelle count in apical processes (n = 17) and cell bodies (n = 8); distance of cell body organelles along a normalized apical–basal axis (n = 8); and dimensions of organelle-bounding boxes in apical processes in selected subsamples of cell bodies and apical processes.
Results
In 2661 sections through 17 cells, apical processes contained 65 ± 24 melanosomes in mononucleate (n = 15) and 131 ± 28 in binucleate cells (n = 2). Cell bodies contained 681 ± 153 LM and 734 ± 170 mitochondria. LM was excluded from the basal quartile, and mitochondria from the apical quartile. Lengths of melanosomes, LM, and mitochondria, respectively were 2305 ± 528, 1320 ± 574, and 1195 ± 294 nm. The ratio of cell body to nucleus volume was 4.6 ± 0.4. LM and mitochondria covered 75% and 63%, respectively, of the retinal imaging plane.
Conclusions
Among RPE signal sources for optical coherence tomography, LM and mitochondria are the most numerous reflective cell body organelles. These and our published data show that most melanosomes are in apical processes. Overlapping LM and previously mitochondria cushions may support multiple reflective bands in cell bodies. This atlas of subcellular reflectivity sources can inform development of advanced optical coherence tomography technologies.
Keywords: retinal pigment epithelium, human, electron microscopy, lipofuscin, melanosomes, melanolipofuscin, mitochondria, optical coherence tomography, color fundus photography
The retinal pigment epithelium (RPE) is a cuboidal epithelium facing the neurosensory retina to its apical side and Bruch’s membrane and choroid to the basal side; it consists of a cell body and apical processes. It serves essential functions for the physiological maintenance of photoreceptors and choroidal vasculature,1,2 including phagocytosis of outer segments,3,4 transport of nutrients and retinoids from the choriocapillaris to the neurosensory retina,1,3,5 and serving as a metabolic gatekeeper for photoreceptors and Müller glia.6,7 The RPE is central in the pathogenesis of numerous retinal diseases, among which age-related macular degeneration (AMD) is the most prevalent8 and a leading cause of vision impairment and legal blindness worldwide.9–11 Earliest clinical detection of pathologic processes in the RPE is key to preventing vision-threatening damage. Identifying a progression timeline is a research priority.12
The RPE is visualized clinically by its organelles, including melanosomes and melanolipofuscin for color fundus photography, lipofuscin and melanolipofuscin for blue autofluorescence, and all of these plus mitochondria for optical coherence tomography (OCT). For pigmentation, the choroid is the major source of between-individual variability.13,14 For both autofluorescence15 and OCT,16 signals from the RPE are intense, and for OCT they involve high reflectivity, shadowing of posterior structures, and reduction and loss of shadowing due to degeneration and atrophy.17–19 Advances in OCT technology have allowed unprecedented visualization of RPE details, including size, morphology, and intraretinal migration.16 Although not reflective or autofluorescent itself, the RPE nucleus is also relevant to imaging, because the space it occupies within the cell body is not available to other organelles.20–22 Incorporating these considerations into clinical image interpretation allows us to quantify and map RPE organelles.
Transmission electron microscopy and quantitative light microscopy laid a foundation for human RPE cell biology.14,23,24 Data from these venerable studies are fragmentary with respect to questions raised by modern retinal imaging. Super-resolution microscopy25,26 has shown hundreds of organelles in RPE cell bodies. Further, advanced OCT imaging of living human eyes reveals multiple reflectivity bands between photoreceptors and choriocapillaris,27–31 suggesting discrete zones of reflectors stacked across the apical-to-basal extent of individual RPE cells. Older microscopy studies described different cells at various or unspecified retinal locations, with some samples lacking apical processes, scale bars, or photodocumentation. The total organelle census in any one cell is thus unclear. New data are especially welcome for the delicate RPE apical processes, which contact photoreceptor outer segments, contain dozens of melanosomes, and likely contribute to the interdigitation zone (IZ) of OCT. In post-mortem specimens subject to artifact it is difficult to observe their full vertical extent (∼15 µm).23
A comprehensive and precise z-axis mapping of human RPE organelles that could inform retinal imaging has recently become feasible by the introduction of three-dimensional serial block-face scanning electron microscopy. This technology involves robotic sectioning of an epoxy resin block mounted in a field emission scanning electron microscope. Aligned volumetric image stacks are generated by alternately imaging the block face with backscattered electrons and then removing a slice 65 to 125 nm thick.32–35 We obtained a rapidly preserved eye of a human organ donor with the retina attached to the RPE. We aimed to illustrate and quantify the total number, dimensions, and spatial distribution of organelles relevant to imaging in perifoveal RPE cells of this specimen. We observed that melanosomes are outnumbered by other cell body organelles and preferentially localize to apical processes. New information about subcellular reflectivity sources will aid development of advanced OCT technologies.
Methods
Tissue Recovery, Characterization, and Preparation
A globe from a 21-year-old white male victim of a motorcycle accident was recovered within minutes of the termination of life support during organ recovery, using preservation methods adapted from those used for retinal neurophysiology studies.36,37 In brief, the enucleated eye was transected at the limbus, drained of vitreous, placed in warm oxygenated culture medium (Ames’ Medium; Sigma-Aldrich, St. Louis, MO, USA), and maintained in this medium at 37°C. The retina with RPE–choroid intact was dissected from the sclera and isolated from the globe by a cut at the optic nerve head. The isolated retinal RPE was then immersion fixed in 4% glutaraldehyde in 0.1-M phosphate buffer after fixation cuts were made in the retina so it could be laid flat in the form of a typical wholemount. Under a dissecting microscope, the optic disc, foveal depression, foveal center, arcuate bundles, and major meridians were visually localized. The foveal center was carefully dissected for other purposes (central 1-mm diameter), and then small perifoveal pieces (∼1–2 mm in length) were cut around this region (1–2 mm eccentricity) and preserved for further processing. Perifoveal tissue used in this study was rinsed in cacodylate buffer (0.1 M, pH 7.4) and incubated for 1 hour in a 1.5% potassium ferrocyanide and 2% osmium tetroxide (OsO4) solution in 0.1-M cacodylate buffer. After washing, the tissue was placed in a freshly made thiocarbohydrazide solution (0.1 g in 10 ml double-distilled H2O heated to 600°C for 1 hour) for 20 minutes at room temperature (RT). After another rinse at RT, the tissue was incubated in 2% OsO4 for 30 minutes at RT. The samples were rinsed again and stained en bloc in 1% uranyl acetate overnight at 40°C; they were then washed and stained with Walton's lead aspartate for 30 minutes. After a final wash, retinal pieces were dehydrated in a graded alcohol series, placed in propylene oxide at RT for 10 minutes, and then embedded in Durcupan resin (Sigma-Aldrich). Semi-thin vertical sections (0.5–1 µm thick) were cut and stained with toluidine blue and examined to define a region of interest to be imaged during serial block-face sectioning.
Serial Block-Face Scanning Electron Microscopy
We identified a region of interest approximately 2 mm from the foveal center for block-face imaging in a scanning electron microscope (SEM). The block was trimmed, gold-coated by standard methods, and mounted in a Gatan 3View SEM (Carl Zeiss Meditec, Jena, Germany). The block face was imaged in the axial direction (across retinal layers) in 40 µm × 40 µm tiles (10% overlap between tiles) that included Bruch's membrane, RPE, and outer segments. Two block faces with a total of 2661 sections were imaged after removal of slices at 75 nm (RPE cells 1–3) or 65 nm (RPE cells 4–17) thickness in the axial cross-sectional plane. Scanning was performed with a 5-nm x-y resolution and a dwell time of 1 µs. The resulting TIFF images were contrast normalized, stitched into cross-sections, and aligned into a volume using the align multilayer mosaic option available with TrakEM2 software38 (plug-in for ImageJ FIJI; National Institutes of Health, Bethesda, MD, USA). Images from block-face scanning produce little or no distortion due to sectioning, because the undisturbed block face is imaged and sections are not cut and mounted on a grid as in standard transmission EM serial reconstruction. For this reason, we utilized the TrakEM2 “Rigid” alignment option both within a layer and across layers. This method permitted translation and rotation of image tiles within a layer and layers relative to each other; however, size scaling and shear (affine adjustments) were not allowed. No additional regularization was performed.
Data Analysis Via ImageJ 3D Viewer
All reconstructions were manually guided, semi-automated, and computer assisted. We mapped features positions within two-dimensional (2D) cross-sections in the axial plane with the ImageJ TrakEM plug-in,39,40 using the main window to display cross-sections while annotating. Features followed through cross-sections were traced using the pen tool by generating area lists. The 2D segmentations were then stitched together across sections to generate three-dimensional (3D) objects, as follows.
To determine total number and distribution of organelles within individual RPE cells, the basolateral cell membranes of 17 fully imaged cell bodies were reconstructed in 300 ± 57 2D cross-sections per cell. We selected cells for analysis based on well-defined borders in cross-sections. Then, melanosomes, lipofuscin, and melanolipofuscin granules in cell bodies and apical processes ensheathing cone/rod photoreceptors and nuclei and mitochondria in cell bodies were traced. Organelles were classified using established morphologic criteria as described in our previous publication26 and expanded in the Results section. Tasks were performed by a primary reader (MN) and a supervising editor (AP). To ensure accurate and reproducible assessment of granule number and types, results from three randomly chosen individual cells were compared. Granule annotation in each cell varied by ≤2% between readers. The decisions were simplified in this study compared to our previous publication,26 because we did not differentiate among osmophilic cell body organelles in this study.
Cell reconstructions were performed using TrakEM to first create skeletons of individual organelles. The grader marked centers of each organelle in cross-sections while scrolling through the z-stack of axial cross-sections, starting by setting the root node at one end of the structure. The course of each structure was followed in the z-stack by setting branch nodes, and the termination was marked by setting the end node. The x and y coordinates establishing the position of each organelle within a tissue space were determined by the location of the geometric center within the image. The z coordinate was derived from the sequence of the image in the z-stack. Previously marked features were displayed to provide context and prevent duplicate assignments. The tracing and classification of granules and organelles could be proofread and edited if necessary.
In addition, selected organelles were colored using the brush tool of the area-tree to allow full 3D reconstruction and volumetric measurements. Similar to tracing, the various features were colored in each 2D cross-section displaying a node marking the respective feature. When all slices of the z-stack containing specific organelles had been fully processed, 3D reconstruction was performed using the 3D viewer of the TrakEM software with a resample setting of 1. To display the exact relation of specific photoreceptors to their contacting apical processes and granule contents, the outer segments of two rods and one cone were fully reconstructed as described above.
Visualization and Calculation of Coverage Area
Cell bodies, nuclei, and organelles were represented by 3D polyhedra traced in planar slices and reconstructed using TrakEM. A custom Java program read .obj TrakEM output files and created projections of each object type (cell body, nucleus, organelle) for figures displaying the relative apical–basal depths and coverage of the retinal imaging plane by specific organelle classes. Coverage is expressed as a ratio of the area of organelles projected onto Bruch's membrane to the area projected by the cell body.
To calculate dimensions of individual organelles, approximate bounding boxes were fit to each object, based on the reconstructed polyhedron produced by TrakEM. The length, width, and height of each bounding box described the gross spatial extent of each polyhedron. We experimented with several methods for fitting bounding boxes and selected one based on principle components analysis (PCA). Given a collection of 3D points (i.e., vertices of the polyhedron), we found three eigenvectors of a covariance matrix, which formed a three-axis reference frame. All points were then projected onto each of these axes to find the minimum and maximum projected value and thus the lengths of the projections. These were then sorted to report a length, width, and height for each reconstructed organelle. An alternative procedure attempted to fit minimum volume bounding boxes. An exact solution to this problem would have been prohibitively time consuming on our data, so an approximation was based on a 3D extension of methods used to find 2D bounding boxes.41 Performances on this dataset were very similar. Because the PCA method was 1000 times faster than the minimum-fit method, we report PCA-generated results. Counts and dimensions are provided as mean ± SD.
Results
Results are based on manually guided, semi-automated 3D organelle mapping in 17 perifoveal RPE cells: cell body and nuclear volume (n = 17 cells); organelle count in apical processes (n = 17) and cell bodies (n = 8, all mononucleate); distance of organelles along a normalized apical–basal axis (n = 8); and dimensions of organelle-bounding boxes in selected subsets of cell bodies and apical processes (total cell body organelles, n = 1; 10% random sample of all cell body organelles, n = 15; apical process organelles, n = 2). Organelle counts are reported as mean ± SD and percent coefficient of variation (standard deviation/mean).
Rapid tissue recovery coupled with immediate oxygenation yielded excellent retinal fine structure42 (Supplementary Video). As seen in Figure 1C, the neurosensory retina was completely attached to the RPE. Detailed ultrastructure could be observed for cone and rod photoreceptor outer segments, apical processes of RPE surrounding photoreceptors, and RPE cell body borders and organelles. An artifact common in human retina due to proportionately thin outer segments (relative to laboratory rodent retina) is referred to as compaction (i.e., closing up of the subretinal space). Compaction can bend outer segments, either singly or multiply, like pleats. Outer segment cross-sections thus appear as circular or circular plus longitudinal, rather than strictly vertical and longitudinal as in the absence of compaction. An ∼4-µm-thick zone of myriad tiny cross-sections at the apical surface of the RPE cell body represents laying-over apical processes, also due to compaction. Even in this state, however, individual processes could be traced back to the cell body through serial sections. Boundaries of individual cells were visible by actin cytoskeleton (red lines in Fig. 1C). Cell nuclei were spherical and located in the basal cell body (turquoise in Fig. 1C). Two of the analyzed cells were binucleate.
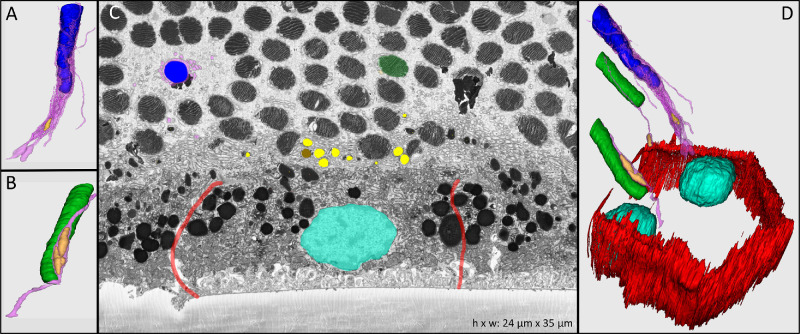
Figure 1.
Human RPE in cross-section and in manually reconstructed 3D view. In the eye of a 21-year-old organ donor preserved minutes after withdrawal of life support, attachment of the neurosensory retina to the RPE was preserved, and the relevant ultrastructure was differentiated in the perifoveal area. Although the photoreceptors are attached to the RPE, the outer segments are bent and appear as cross-sections. Granules in the cell body are highly electron dense. Individual subtypes are not distinguishable. Colors: red, borders of cell body; turquoise, cell nucleus; yellow, melanosomes in apical processes; brown, lipofuscin granules in apical processes; green, rod photoreceptor outer segments; blue, cone photoreceptor outer segments; pink, apical processes. (A) Cone outer segment with surrounding RPE apical processes containing a melanosome. (B) Rod outer segment with RPE apical processes containing two melanosomes. (C) Single cross-sectional transmission electron micrograph showing RPE attached to outer segments; Bruch's membrane is not present. (D) Volumetric manual reconstruction of a binucleate (turquoise) RPE cell body including cone (blue) and rod (green) photoreceptor outer segments and melanosome granules (yellow) located in apical processes (pink).
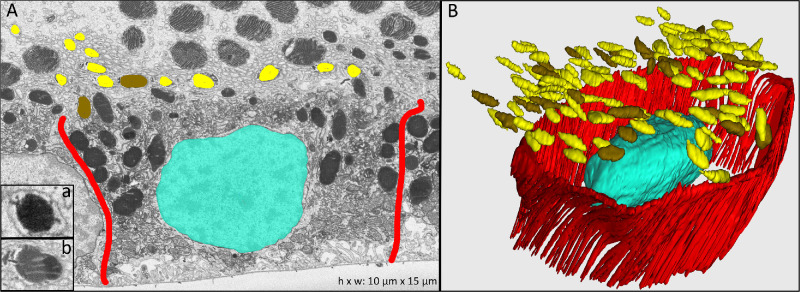
Semi-automated reconstruction of cellular ultrastructure in each cross-sectional image allowed the 3D representation seen in Figure 1. Bundled apical processes (Fig. 1, pink) departed the RPE cell body (Fig. 1A, limited by red lines) and ensheathed single photoreceptor outer segments (green, rods; blue, cones). Melanosomes (yellow in Fig. 1) were located within apical processes, appearing circular in cross-section (Fig. 2A) or spindle-shaped in longitudinal section (Figs. 1A, 1B, 1D) with homogeneously high internal electron density. Lipofuscin granules could also be identified in the apical processes and were characterized by a spherical shape, electron-dense interior, and electron-dense rim or condensations around the perimeter (resembling earmuffs) (Fig. 2B). The plasma membrane of the apical process surrounding each single melanosome and lipofuscin granule was manually tracked to its respective RPE cell body. Figure 2 shows melanosome (yellow) and lipofuscin (brown) granules in apical processes of one representative RPE cell (single section and complete reconstruction shown in Figs. 2A and 2B, respectively). The vast majority of granules in the apical processes were melanosomes, with a mean number of 65 ± 24 (36.9%) in mononucleate and 131 ± 28 (21.4%) in binucleate RPE cells. The mean number of lipofuscin granules was 12 ± 6 in mononucleate and 26 ± 1 in binucleate RPE cells. The length, width, and height of melanosomes within apical processes were 2305 ± 528 nm, 937 ± 137 nm, and 738 ± 97 nm, respectively. Lipofuscin granules measured 2355 ± 574 nm in length, 994 ± 149 nm in width, and 788 ± 109 nm in height.
Figure 2.
Melanosomes and lipofuscin granules in RPE apical processes. (A) Electron-microscopy cross-section with inset a showing a representative melanosome and inset b a lipofuscin granule. Cell borders of one RPE cell are highlighted in red. (B) Manually reconstructed 3D view of one RPE cell with associated complete reconstructions of melanosome and lipofuscin granules located in the apical processes (not shown). Colors: red, RPE cell borders; turquoise, cell nucleus; yellow, melanosomes; brown, lipofuscin.
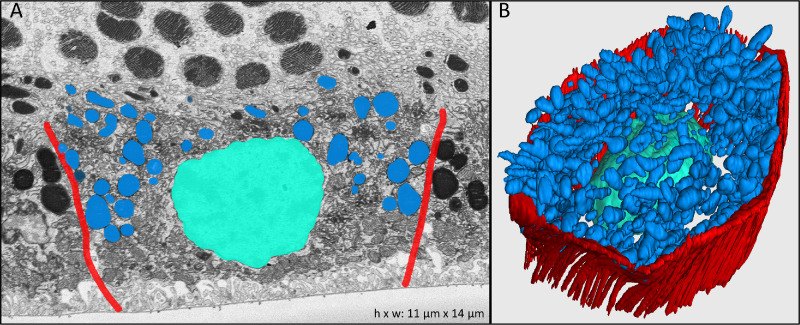
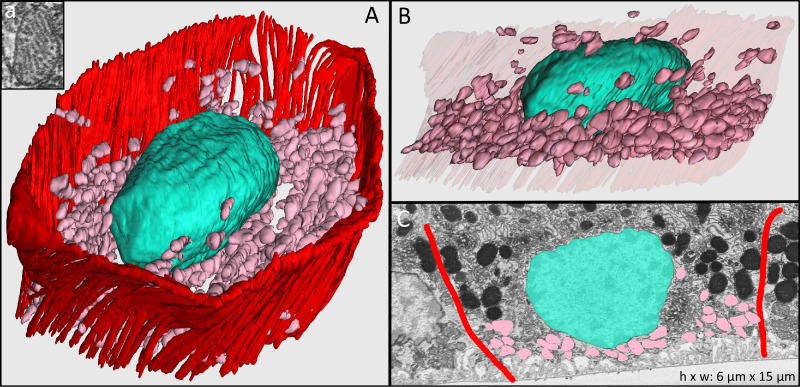
The RPE cell body contains distinct cushions of organelles within it. Within the cell body, melanosomes, lipofuscin, and melanolipofuscin were uniformly electron dense and therefore could not be reliably distinguished from each other, unlike the apical processes in these cells and unlike our recent experience with cell bodies when we used a different post-fixation method.26 Reasons for the different granule appearances in the previous and current study may relate to the mordanting capability of the prior post-fixation method,43 short time before post-processing of this specimen, differences in photographic imaging contrast, real differences between the composition of organelles, or a combination of factors. Herein, we refer to the combined organelle population as granules, a term used in high-resolution autofluorescence investigations.25,44 The combined population of granules (mean per cell, 681 ± 153; 22.5%) form one thick cushion in the apical cell body (Figs. 3A, 3B, blue). The mean length, width, and height of these organelles were 1320 ± 574 nm, 920 ± 340 nm, and 756 ± 278 nm, respectively. The basal cell body is lined by mitochondria, as demonstrated in Figure 4 (pink). Mitochondria appear as ovoid structures with an electron-dense internal membrane forming shelf-like cristae (Fig. 4, insert). We found a mean of 734 ± 170 (23.2%) mitochondria per RPE cell body, with mean length, width, and height of 1195 ± 294 nm, 870 ± 164 nm, and 628 ± 130 nm, respectively.
Figure 3.
Lipofuscin, melanolipofuscin, and melanosome granules in the RPE cell body. (A) Electron-microscopy cross-section of one RPE cell highlighting the combined population of lipofuscin, melanolipofuscin, and melanosome granules. Due to tissue post-processing, individual granule types could not be differentiated. (B) Manually reconstructed 3D view of the RPE cell shown in A. Colors: red, RPE cell borders; turquoise, cell nucleus; blue, combined lipofuscin, melanolipofuscin, and melanosome granules in the cell body.
Figure 4.
Mitochondria in RPE cell body. (A) Hundreds of manually reconstructed mitochondria (pink) fill the basal part of a cell body in the 21-year-old male donor eye. Inset a shows the cross-section of a single mitochondrion in an electron microscopy image. (B) Cross-sectional manual reconstruction highlights the spatial distribution of mitochondria within the cell body. (C) Electron microscopy image of one representative cross-section showing borders of the RPE cell body (red), nucleus (turquoise), and mitochondria (pink).
Mitochondria and granules in eight comprehensively reconstructed cell bodies are shown together in an en face projection view in Supplementary Figure S1. To facilitate comparison of organelle content with OCT technologies showing multiple RPE-related reflective bands, we plotted in Figure 5 and Supplementary Figure S2 the frequency of granules and mitochondria as a function of percent distance from the apical-most to the basal-most organelle found in each cell body (excluding apical processes). For the eight reconstructed cells, the mean distance between these landmarks was 15.8 ± 1.0 µm (6.6% of the mean); distances for individual cells are provided in the figure captions. Figure 5 shows in a representative cell that granules and mitochondria have overlapping distributions in the apical–basal axis, creating three zones of distinct organelle content. Mitochondria are excluded from the apical-most quartile and granules are excluded from the basal-most quartile, but both organelle classes are present in the middle half. The third quartile of the mitochondria distribution has fewer organelles, because the nucleus occupies space at that level. Supplementary Figure S2 shows that results from all eight cells analyzed in this manner are consistent.
Figure 5.
Apical-to-basal distribution of organelles in RPE cell bodies. Shown are one RPE cell (#06, left and middle panels) and a mean of eight cells (right panel; all eight cells are shown in Supplementary Figs. S1 and S2). All granules (lipofuscin, melanolipofuscin, melanosomes) in the cell body (excluding apical processes) and mitochondria are displayed in an axial projection (middle panel; see en face projection views in Supplementary Fig. S1). The projection view is scaled to match the normalized distance axis in the graph. Organelles are shown as 1-µm-diameter circles. The graphs plot the number of granules and mitochondria in ∼3-µm-wide bins as a function of percent distance from the apical-most to the basal-most organelle (17.4 µm for cell #06). Four quartiles of distance represented by the grid lines delimit three zones of differing organelle content: mitochondria are excluded from the apical-most quartile and granules are excluded from the basal-most quartile, but both organelle classes are present in the middle half. The third quartile of the mitochondria distribution has fewer organelles (represented by a curved indentation), because the nucleus occupies space at that level.
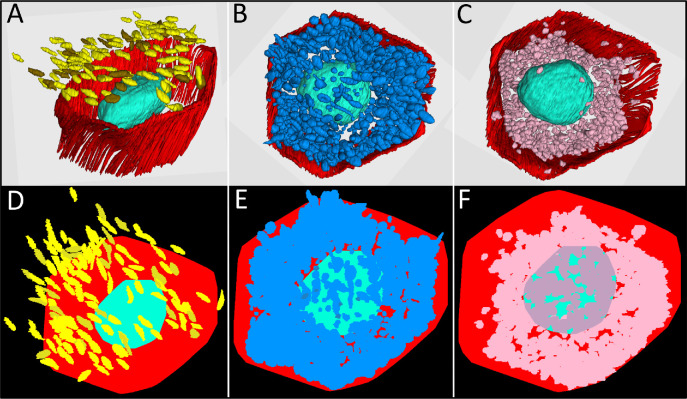
We next determined that the mean volumes of cell bodies (including nuclei) and nuclei alone in 17 RPE cells were 2416 ± 263 µm3 and 510 ± 174 µm3, respectively; thus, the ratio between cell volume and nucleus volume was 4.6 ± 0.4. Figures 6A to 6C show three organelle classes at the same magnification and perspective. Figures 6D to 6F demonstrate the percentage of the imaging plane covered by reflective granules and mitochondria. Considering this mean volume, mitochondria alone comprised 8% of the total volume. We found that lipofuscin and melanolipofuscin covered 75% of the retinal imaging plane within a single RPE cell (Fig. 6E), whereas mitochondria accounted for coverage of 63% (Fig. 6F). Coverage was not computed for melanosomes in the apical microvilli, which extended outside the cell body borders in this specimen due to compaction (Fig. 6D).
Figure 6.
RPE cell coverage area of reflective granules and mitochondria. The top row shows 3D reconstructions of melanosome (yellow) and lipofuscin (brown) granules in the apical processes of RPE cells (A), combined population of lipofuscin, melanolipofuscin, and melanosome granules (blue) in the RPE cell body (B), and mitochondria (pink) in the RPE cell body (C). RPE cell borders are indicated in red and cell nucleus in turquoise. The bottom row (D–F) indicates the modeled coverage area of the respective granules and mitochondria (see color code for A–C) visualized from an apical view. (D) Coverage area of melanosomes and lipofuscin granules in RPE apical processes with some granules outside the cell borders due to bending of apical processes occurring during processing of tissue. (E) Reflective granules (lipofuscin, melanolipofuscin, melanosome) and (F) mitochondria in the cell body with dark pink areas indicating mitochondria located on the basal side of the nucleus.
Discussion
We herein document the number, dimensions, and spatial distribution of imaging-relevant organelles in RPE apical processes and cell bodies, using manually guided, semi-automated annotation of volume electron microscopy. Our previous study26 demonstrated more lipofuscin and melanolipofuscin than melanosomes in foveal RPE cell bodies of four donors aged 16 to 84 years (lipofuscin + melanolipofuscin vs. melanosomes: 478 vs. 29; 311 vs. 24; 92 vs. 12; 517 vs. 7). In the current study, we found dozens of melanosomes in the apical processes in the perifoveal RPE of a single 21-year-old donor. Taken together, our published26 and current data imply that apical processes contain the majority of human RPE melanosomes. Further, these organelles that produce and store melanin45 and are known light scatterers46 are vastly outnumbered in cell bodies by lipofuscin, melanolipofuscin, and mitochondria. In cell bodies, these three organelles form two overlapping apical and basolateral cushions. Enumerating organelles in this manner is an important first step in building quantitative models of reflectivity, one that will also require information about scattering and absorption properties. Our data are relevant to in vivo RPE visualization via OCT and fundus autofluorescence and are overall timely due to new concepts of RPE organelle dynamism25,26 and motility.30
Our ultrastructural exploration was guided by an understanding that OCT images are generated by interference of light backscattered from tissue and from a comparative reference mirror.47 Light scattering, defined as redirection of light from an original direction, occurs at larger refractive index gradients introduced, for example, by water–lipid interfaces such as at membrane-bounded organelles. As RPE granule dimensions are similar to imaging wavelengths, this phenomenon is known in optical physics as Mie scattering.46,48–50 Both the intense OCT signal and strong shadowing of posterior structures from the RPE layer are most likely generated by the combination of melanosomes, lipofuscin, melanolipofuscin, and mitochondria, whereas nuclei backscatter light only weakly. Conversely organelle loss or redistribution and cellular thinning, migration, or death in disease16,25,51,52 can contribute to hypertransmission of imaging light into the choroid. Optical models based on Mie scattering require information about the distribution, shape, and size of the scattering particles,53–55 which we provide here.
A key interface in vision is the ensheathing of cone and rod photoreceptor outer segment tips with fine processes protruding from the apical RPE cell body. The end tips of rod outer segments terminate close to the RPE cell body in most mammalian retinas, but cones typically stop short, resulting in apical processes of two different lengths. Thus, these two interfaces can be clearly separated with ultra-high-resolution OCT or adaptive optics OCT (AO-OCT).56,57 Apical processes participate in outer segment phagocytosis and contribute to structural stability of and molecular, metabolic, and electrolytic exchange with photoreceptors, including retinoid transfer. Apical processes are damaged when photoreceptors detach. Even if attached and intact, they and outer segments can bend, as in our case, challenging attempts to characterize normal organelle content. Our knowledge of human apical processes owes much to Steinberg and Wood, who analyzed enucleated eyes of four patients with tumors (4, 38, 45, and 60 years old) by single-section transmission electron microscopy in 1977.23 OCT validation is best served by a combination of subcellular resolution to reveal organelles over a large area and a comprehensive visualization of all cellular components. This goal is facilitated by the “connectomics” revolution of volume electron microscopy in neuroscience,58 including the retina.59–62 In contrast, selective stains for specific tissue elements are subject to selection bias and cannot reveal non-stained elements.63 A recent confocal microscopy study described a zone of outer segment phagosomes inside apical RPE cell bodies.63 With the benefit of higher magnification and resolution, we interpret these findings to be artifactually bent outer segment tips surrounded by compacted apical processes—that is, outside the cell body as seen in Figure 2.
Like Steinberg and Wood who observed numerous melanosomes in human apical processes, we found that processes attached to mononucleate and binucleate RPE cells contained 65 and 131 melanosomes, respectively, along with 12 and 16 lipofuscin granules, respectively, seen in this location for the first time. In some species, virtually all RPE melanosomes localize to apical processes.46,64,65 In non-human primates, high-resolution and magnification light microscopic images of RPE with attached retina66 (Fig. 1C; see also Fig. 3E of Rudolf et al.67) show vertically oriented and abundant spindle-shaped melanosomes paralleling outer segments, a thin zone lacking melanosomes, and a few more in the apical cell body. Unlike other human ocular tissues that contain spherical melanosomes, RPE melanosomes are spindle shaped,68 perhaps an evolutionary adaptation to the slender apical processes. Melanosomes have higher refractive indices (1.7 for melanin) than cytoplasm (1.37) and therefore represent potential scatterers of imaging light69,70 and a source of OCT signal.46,71,72 In some species, melanosomes translocate toward apical process tips during dark adaptation and back toward the cell body during light adaptation.73–75 Experimental studies in frog and ground squirrel showed that translocated melanosomes are accompanied by a corresponding change in the vertical extent of reflectivity,46,65 directly confirming this signal source. Interestingly, albino mice have fourth bands that are thinner than in pigmented animals.76,77
Our data pertain to the clinical consensus nomenclature for commercial SD-OCT,78 which recognizes in normal eyes four reflective and two hyporeflective bands starting at the external limiting membrane and moving outward. The third reflective band (IZ78) was the last revealed of the major bands and was either variably named or not named at all in early publications. Determining the structural basis of the IZ has been elusive,79,80 in part because the delicacy of apical processes impedes their exploration in laboratory studies. High magnification, vertically expanded images of intact eyes acquired with commercial and prototype OCT technologies27–31 suggest the presence of additional hyporeflective bands external to the IZ. The ultrastructural correlates of these bands are beyond our current scope. Organelles may be absent from specific levels, such as at the base of the apical processes or in the cell body just below them, which we cannot assess in our material due to compaction. Nevertheless, Figure 5 shows an apical-to-basal distribution of cell body organelles at a finer scale than available with low-resolution or single-section microscopy. We conclude that RPE organelles are stacked in partially overlapping cushions with proportions that vary along the apical–basal axis. Two organelle classes can combine to create three cushions of distinct composition (Fig. 5). The apical-most cushion of granules might be further subdivided in appropriately osmicated specimens. Adding melanosomes of the apical process (Figs. 2, 6) allows at least four distinct organelle cushions. Thus OCT technology with better than 5-µm axial resolution may provide subcellular biologic resolution. This logic, previously applied to photoreceptors,81,82 may also be applied to the vertically compartmentalized RPE, which is less than half the height of the inner and outer segments, even with apical processes. It is also possible that differing compositions of membranes bounding these organelles may contribute to variation in refractive index gradients (and thus reflectivity).
We showed a remarkable number of mitochondria, >700 per RPE cell, and these were ovoid in shape, unlike the long, branching organelles sometimes seen in cultured cells.83 The apical extent of the mitochondrial cushion was unexpected based on publications showing mitochondria alone at the basolateral aspect.84,85 Mitochondria fulfill key functions for RPE maintenance and survival by serving as the main producers of cellular energy required by these multipurpose cells and contributing to oxidative phosphorylation, beta-oxidation of fatty acids, calcium homeostasis, and regulation of excitability.86–89 Mitochondrial morphology is dynamically controlled to respond to energy requirements caused by different environmental stimuli, with fission occurring in apoptotic cells.90–92 Light scattering by mitochondria is influenced by apoptosis or oxidative stress.48,93 In cell culture experiments employing organelle-specific deletion, mitochondria and lysosomes have been shown to be strong light scatterers conforming to Mie theory.48,49,94 Commercial SD-OCT95 indicates that mitochondria are major light scatterers in human cone inner segments, but that finding has not been confirmed by AO-OCT.96
The nucleus occupies space otherwise taken up by organelles generating strong signals, and the small nuclear volume relative to the cell body found in our study has implications for autofluorescence imaging. In adaptive optics-assisted 488-nm and 787-nm autofluorescence of human RPE, a central hypoautofluorescent area occupies most of the en face projection view and is ringed by a narrow band of punctate autofluorescence in both excitation wavelengths.21,22,72 This central area has been attributed to the RPE nucleus. Our current and past data (see Fig. 2 of Pollreisz et al.26 and Fig. 2 of Starnes et al.97) collectively suggest that the nucleus is too small to account for this appearance. Rather, we suggest that the hypoautofluorescent area includes a tuft of upright apical processes containing melanosomes.
Strengths of this study include an innovative connectomics approach and a detailed 3D dataset with positions and dimensions of individual organelles in entire RPE cells, including apical processes. Limitations include the restriction of analysis to one perifoveal region of one donor eye, thus restricting generalizability, and the inability to differentiate among osmophilic granules within the RPE cell body. Despite these specimen-specific limitations, this initial atlas of RPE organelles is relevant to state-of-the-art clinical imaging, complementary efforts to quantify autofluorescent organelles,98 benchmarking of cells destined for replacement therapies, and mechanistic in vivo studies of retinal diseases. The organelle types quantified here have all been considered to have either helpful or harmful properties for AMD, amenable to pharmaceutical modulation99,100 (ClinicalTrials.gov number NCT03891875) and monitoring through imaging. Our data thus also have implications for therapeutic strategies. By introducing artificial intelligence methods to speed data capture,101 sample size can be increased for future studies of cone- and rod-rich areas in human eyes, eyes of different racial groups,102 eyes with AMD, and relevant animal models.
Supplementary Material
Acknowledgments
Supported by NIH Grant R01EY028282 (Dacey), a grant from the Macula Society Research and by institutional support to the Department of Ophthalmology and Visual Sciences at the University of Alabama at Birmingham from Research to Prevent Blindness, Inc., and EyeSight Foundation of Alabama.
Disclosure: A. Pollreisz, None; M. Neschi, None; K.R. Sloan, MacRegen (I); M. Pircher, None; T. Mittermueller, None; D.M. Dacey, None; U. Schmidt-Erfurth, None; C.A. Curcio, Heidelberg Engineering (F), Genentech/Hoffman-LaRoche (F), MacRegen (I)
References
- 1.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005; 85: 845–881. [DOI] [PubMed] [Google Scholar]
- 2. Bok D. The retinal pigment epithelium: a versatile partner in vision. J Cell Sci. 1993; 17(suppl): 189–195. [DOI] [PubMed] [Google Scholar]
- 3.Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda). 2010; 25: 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bok D. Processing and transport of retinoids by the retinal pigment epithelium. Eye. 1990; 4: 326–332. [DOI] [PubMed] [Google Scholar]
- 5. Kanow MA, Giarmarco MM, Jankowski CS, et al.. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017; 6: e28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adijanto J, Du J, Moffat C, Seifert E, Hurley JB, Philp NJ. The retinal pigment epithelium utilizes fatty acids for ketonegenesis: implications for metabolic coupling with the outer retina. J Biol Chem. 2014; 289: 20570–20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ait-Ali N, Fridlich R, Millet-Puel G, et al.. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell. 2015; 161: 817–832. [DOI] [PubMed] [Google Scholar]
- 8.Bhutto I, Lutty G. Understanding age-related macular degeneration (AMD): relationships between the photoreceptor/retinal pigment epithelium/Bruch's membrane/choriocapillaris complex. Mol Aspects Med. 2012; 33: 295–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panthier C, Querques G, Puche N, et al.. Evaluation of semiautomated measurement of geographic atrophy in age-related macular degeneration by fundus autofluorescence in clinical setting. Retina. 2013; 34: 576–582. [DOI] [PubMed] [Google Scholar]
- 10. Congdon N, O'Colmain B, Klaver CC, et al.. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004; 122: 477–485. [DOI] [PubMed] [Google Scholar]
- 11. Nangia V, Jonas JB, George R, et al.. Prevalence and causes of blindness and vision impairment: magnitude, temporal trends and projections in South and Central Asia. Br J Ophthalmol. 2018; 103: 871–877. [DOI] [PubMed] [Google Scholar]
- 12. Handa JT, Bowes Rickman C, Dick AD, et al.. A systems biology approach towards understanding and treating non-neovascular age-related macular degeneration. Nat Commun. 2019; 10: 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt SY, Peisch RD. Melanin concentration in normal human retinal pigment epithelium. Regional variation and age-related reduction. Invest Ophthalmol Vis Sci. 1986; 27: 1063–1067. [PubMed] [Google Scholar]
- 14. Weiter JJ, Delori FC, Wing GL, Fitch KA. Retinal pigment epithelial lipofuscin and melanin and choroidal melanin in human eyes. Invest Ophthalmol Vis Sci. 1986; 27: 145–152. [PubMed] [Google Scholar]
- 15. Rudolf M, Vogt SD, Curcio CA, et al.. Histologic basis of variations in retinal pigment epithelium autofluorescence in eyes with geographic atrophy. Ophthalmology. 2013; 120: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Curcio CA, Zanzottera EC, Ach T, Balaratnasingam C, Freund KB. Activated retinal pigment epithelium, an optical coherence tomography biomarker for progression in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2017; 58: BIO211–BIO226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho J, Witkin AJ, Liu J, et al.. Documentation of intraretinal retinal pigment epithelium migration via high-speed ultrahigh-resolution optical coherence tomography. Ophthalmology. 2011; 118: 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Q, de Sisternes L, Leng T, Zheng L, Kutzscher L, Rubin DL. Semi-automatic geographic atrophy segmentation for SD-OCT images. Biomed Opt Express. 2013; 4: 2729–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sayegh RG, Kiss CG, Simader C, et al.. A systematic correlation of morphology and function using spectral domain optical coherence tomography and microperimetry in patients with geographic atrophy. Br J Ophthalmol. 2014; 98: 1050–1055. [DOI] [PubMed] [Google Scholar]
- 20. Morgan JI, Dubra A, Wolfe R, Merigan WH, Williams DR. In vivo autofluorescence imaging of the human and macaque retinal pigment epithelial cell mosaic. Invest Ophthalmol Vis Sci. 2009; 50: 1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu T, Jung H, Liu J, Droettboom M, Tam J. Noninvasive near infrared autofluorescence imaging of retinal pigment epithelial cells in the human retina using adaptive optics. Biomed Opt Express. 2017; 8: 4348–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Granger CE, Yang Q, Song H, et al.. Human retinal pigment epithelium: in vivo cell morphometry, multispectral autofluorescence, and relationship to cone mosaic. Invest Ophthalmol Vis Sci. 2018; 59: 5705–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steinberg RH, Wood I, Hogan MJ. Pigment epithelial ensheathment and phagocytosis of extrafoveal cones in human retina. Philos Trans R Soc Lond B Biol Sci. 1977; 277: 459–474. [DOI] [PubMed] [Google Scholar]
- 24. Feeney L. Lipofuscin and melanin of human retinal pigment epithelium. Fluorescence, enzyme cytochemical and ultrastructural studies. Invest Ophthalmol Vis Sci. 1978; 17: 583–600. [PubMed] [Google Scholar]
- 25. Ach T, Tolstik E, Messinger JD, Zarubina AV, Heintzmann R, Curcio CA. Lipofuscin re-distribution and loss accompanied by cytoskeletal stress in retinal pigment epithelium of eyes with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015; 56: 3242–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pollreisz A, Messinger JD, Sloan KR, et al.. Visualizing melanosomes, lipofuscin, and melanolipofuscin in human retinal pigment epithelium using serial block face scanning electron microscopy. Exp Eye Res. 2018; 166: 131–139. [DOI] [PubMed] [Google Scholar]
- 27. Wilk MA, McAllister JT, Cooper RF, et al.. Relationship between foveal cone specialization and pit morphology in albinism. Invest Ophthalmol Vis Sci. 2014; 55: 4186–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chong SP, Zhang T, Kho A, Bernucci MT, Dubra A, Srinivasan VJ. Ultrahigh resolution retinal imaging by visible light OCT with longitudinal achromatization. Biomed Opt Express. 2018; 9: 1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang T, Kho AM, Srinivasan VJ. Improving visible light OCT of the human retina with rapid spectral shaping and axial tracking. Biomed Opt Express. 2019; 10: 2918–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Z, Kurokawa K, Hammer DX, Miller DT. In vivo measurement of organelle motility in human retinal pigment epithelial cells. Biomed Opt Express. 2019; 10: 4142–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Azimipour M, Migacz JV, Zawadzki RJ, Werner JS, Jonnal RS. Functional retinal imaging using adaptive optics swept-source OCT at 1.6 MHz. Optica. 2019; 6: 300–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mills EA, Davis CH, Bushong EA, et al.. Astrocytes phagocytose focal dystrophies from shortening myelin segments in the optic nerve of Xenopus laevis at metamorphosis. Proc Natl Acad Sci U S A. 2015; 112: 10509–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mustafi D, Kevany BM, Genoud C, et al.. Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 2011; 25: 3157–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohno N, Kidd GJ, Mahad D, et al.. Myelination and axonal electrical activity modulate the distribution and motility of mitochondria at CNS nodes of Ranvier. J Neurosci. 2011; 31: 7249–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tait SW, Oberst A, Quarato G, et al.. Widespread mitochondrial depletion via mitophagy does not compromise necroptosis. Cell Rep. 2013; 5: 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dacey DM, Petersen MR. Dendritic field size and morphology of midget and parasol ganglion cells of the human retina. Proc Natl Acad Sci U S A. 1992; 89: 9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dacey DM. The mosaic of midget ganglion cells in the human retina. J Neurosci. 1993; 13: 5334–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cardona A, Saalfeld S, Schindelin J, et al.. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012; 7: e38011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schindelin J, Arganda-Carreras I, Frise E, et al.. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012; 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sloan KR. Analysis of “dot product space” shape descriptions. IEEE Trans Pattern Anal Mach Intell. 1982; 4: 87–90. [DOI] [PubMed] [Google Scholar]
- 42. Calkins DJ, Schein SJ, Tsukamoto Y, Sterling P. M and L cones in macaque fovea connect to midget ganglion cells by different numbers of excitatory synapses. Nature. 1994; 371: 70–72. [DOI] [PubMed] [Google Scholar]
- 43. Guyton JR, Klemp KF. Ultrastructural discrimination of lipid droplets and vesicles in atherosclerosis: value of osmium-thiocarbohydrazide-osmium and tannic acid-paraphenylenediamine techniques. J Histochem Cytochem. 1988; 36: 1319–1328. [DOI] [PubMed] [Google Scholar]
- 44. Ach T, Best G, Rossberger S, Heintzmann R, Cremer C, Dithmar S. Autofluorescence imaging of human RPE cell granules using structured illumination microscopy. Br J Ophthalmol. 2012; 96: 1141–1144. [DOI] [PubMed] [Google Scholar]
- 45. Barral DC, Seabra MC. The melanosome as a model to study organelle motility in mammals. Pigment Cell Res. 2004; 17: 111–118. [DOI] [PubMed] [Google Scholar]
- 46. Zhang QX, Lu RW, Messinger JD, Curcio CA, Guarcello V, Yao XC. In vivo optical coherence tomography of light-driven melanosome translocation in retinal pigment epithelium. Sci Rep. 2013; 3: 2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adhi M, Duker JS. Optical coherence tomography - current and future applications. Curr Opin Ophthalmol. 2013; 24: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson JD, Cottrell WJ, Foster TH. Index-of-refraction-dependent subcellular light scattering observed with organelle-specific dyes. J Biomed Opt. 2007; 12: 014010. [DOI] [PubMed] [Google Scholar]
- 49. Wilson JD, Foster TH. Characterization of lysosomal contribution to whole-cell light scattering by organelle ablation. J Biomed Opt. 2007; 12: 030503. [DOI] [PubMed] [Google Scholar]
- 50. Baumann B, Baumann SO, Konegger T, et al.. Polarization sensitive optical coherence tomography of melanin provides intrinsic contrast based on depolarization. Biomed Opt Express. 2012; 3: 1670–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schütze C, Ritter M, Blum R, et al.. Retinal pigment epithelium findings in patients with albinism using wide-field polarization-sensitive optical coherence tomography. Retina. 2014; 34: 2208–2217. [DOI] [PubMed] [Google Scholar]
- 52. Xu X, Liu X, Wang X, et al.. Retinal pigment epithelium degeneration associated with subretinal drusenoid deposits in age-related macular degeneration. Am J Ophthalmol. 2017; 175: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheong WF, Prahl SA, Welch AJ. A review of the optical properties of biological tissues. IEEE J Quantum Electron. 1990; 26: 2166–2185. [Google Scholar]
- 54. Schmitt JM, Kumar G. Optical scattering properties of soft tissue: a discrete particle model. Appl Opt. 1998; 37: 2788–2797. [DOI] [PubMed] [Google Scholar]
- 55. Jacques SL. Optical properties of biological tissues: a review. Phys Med Biol. 2013; 58: R37–R61. [DOI] [PubMed] [Google Scholar]
- 56. Srinivasan VJ, Monson BK, Wojtkowski M, et al.. Characterization of outer retinal morphology with high-speed, ultrahigh-resolution optical coherence tomography. Invest Ophthalmol Vis Sci. 2008; 49: 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Felberer F, Kroisamer JS, Baumann B, et al.. Adaptive optics SLO/OCT for 3D imaging of human photoreceptors in vivo. Biomed Opt Express. 2014; 5: 439–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Helmstaedter M, Briggman KL, Denk W. High-accuracy neurite reconstruction for high-throughput neuroanatomy. Nat Neurosci. 2011; 14: 1081–1088. [DOI] [PubMed] [Google Scholar]
- 59. Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013; 500: 168–174. [DOI] [PubMed] [Google Scholar]
- 60. Della Santina L, Kuo SP, Yoshimatsu T, et al.. Glutamatergic monopolar interneurons provide a novel pathway of excitation in the mouse retina. Curr Biol. 2016; 26: 2070–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim JS, Greene MJ, Zlateski A, et al.. Space-time wiring specificity supports direction selectivity in the retina. Nature. 2014; 509: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011; 471: 183–188. [DOI] [PubMed] [Google Scholar]
- 63. Cuenca N, Ortuno-Lizaran I, Pinilla I. Cellular characterization of OCT and outer retinal bands using specific immunohistochemistry markers and clinical implications. Ophthalmology. 2018; 125: 407–422. [DOI] [PubMed] [Google Scholar]
- 64. Knabe W, Skatchkov S, Kuhn HJ. “Lens mitochondria” in the retinal cones of the tree-shrew Tupaia belangeri. Vision Res. 1997; 37: 267–271. [DOI] [PubMed] [Google Scholar]
- 65. Sajdak BS, Bell BA, Lewis TR, et al.. Assessment of outer retinal remodeling in the hibernating 13-lined ground squirrel. Invest Ophthalmol Vis Sci. 2018; 59: 2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yiu G, Wang Z, Munevar C, et al.. Comparison of chorioretinal layers in rhesus macaques using spectral-domain optical coherence tomography and high-resolution histological sections. Exp Eye Res. 2018; 168: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rudolf M, Curcio CA, Schlötzer-Schrehardt U, et al.. Apolipoprotein A-I mimetic peptide L-4F removes Bruch's membrane lipids in aged non-human primates. Invest Ophthalmol Vis Sci. 2019; 60: 461–472. [DOI] [PubMed] [Google Scholar]
- 68. Durairaj C, Chastain JE, Kompella UB. Intraocular distribution of melanin in human, monkey, rabbit, minipig and dog eyes. Exp Eye Res. 2012; 98: 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dunn AK, Smithpeter CL, Welch AJ, Richards-Kortum RR. Finite-difference time-domain simulation of light scattering from single cells. J Biomed Opt. 1997; 2: 262–266. [DOI] [PubMed] [Google Scholar]
- 70. Barton JK, Gossage KW, Xu W, et al.. Investigating sun-damaged skin and actinic keratosis with optical coherence tomography: a pilot study. Technol Cancer Res Treat. 2003; 2: 525–535. [DOI] [PubMed] [Google Scholar]
- 71. Wilk MA, Huckenpahler AL, Collery RF, Link BA, Carroll J. The effect of retinal melanin on optical coherence tomography images. Transl Vis Sci Technol. 2017; 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu Z, Kocaoglu OP, Miller DT. 3D imaging of retinal pigment epithelial cells in the living human retina. Invest Ophthalmol Vis Sci. 2016; 57: OCT533–OCT543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mondragon R, Frixione E. Retinomotor movements in the frog retinal pigment epithelium: dependence of pigment migration on Na+ and Ca2+. Exp Eye Res. 1989; 48: 589–603. [DOI] [PubMed] [Google Scholar]
- 74. Burnside B, Adler R, O'Connor P. Retinomotor pigment migration in the teleost retinal pigment epithelium. I. Roles for actin and microtubules in pigment granule transport and cone movement. Invest Ophthalmol Vis Sci. 1983; 24: 1–15. [PubMed] [Google Scholar]
- 75. Lythgoe JN, Shand J. Endogenous circadian retinomotor movements in the neon tetra (Paracheirodon innesi). Invest Ophthalmol Vis Sci. 1983; 24: 1203–1210. [PubMed] [Google Scholar]
- 76. Berger A, Cavallero S, Dominguez E, et al.. Spectral-domain optical coherence tomography of the rodent eye: highlighting layers of the outer retina using signal averaging and comparison with histology. PLoS One. 2014; 9: e96494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Soukup P, Maloca P, Altmann B, Festag M, Atzpodien EA, Pot S. Interspecies variation of outer retina and choriocapillaris imaged with optical coherence tomography. Invest Ophthalmol Vis Sci. 2019; 60: 3332–3342. [DOI] [PubMed] [Google Scholar]
- 78. Staurenghi G, Sadda S, Chakravarthy U, Spaide RF. Proposed lexicon for anatomic landmarks in normal posterior segment spectral-domain optical coherence tomography: The IN*OCT Consensus. Ophthalmology. 2014; 121: 1572–1578. [DOI] [PubMed] [Google Scholar]
- 79. Zawadzki RJ, Jones SM, Olivier SS, et al.. Adaptive-optics optical coherence tomography for high-resolution and high-speed 3D retinal in vivo imaging. Opt Express. 2005; 13: 8532–8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Puche N, Querques G, Benhamou N, et al.. High-resolution spectral domain optical coherence tomography features in adult onset foveomacular vitelliform dystrophy. Br J Ophthalmol. 2010; 94: 1190–1196. [DOI] [PubMed] [Google Scholar]
- 81. Spaide R, Curcio CA. Anatomic correlates to the bands seen in the outer retina by optical coherence tomography: literature review and model. Retina. 2011; 31: 1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ross DH, Clark ME, Godara P, et al.. RefMob, a reflectivity feature model-based automated method for measuring four outer retinal hyperreflective bands in optical coherence tomography. Invest Ophthalmol Vis Sci. 2015; 56: 4166–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tan LX, Toops KA, Lakkaraju A. Protective responses to sublytic complement in the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2016; 113: 8789–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Woodell A, Coughlin B, Kunchithapautham K, et al.. Alternative complement pathway deficiency ameliorates chronic smoke-induced functional and morphological ocular injury. PLoS One. 2013; 8: e67894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gouras P, Ivert L, Neuringer M, Mattison JA. Topographic and age-related changes of the retinal epithelium and Bruch's membrane of rhesus monkeys. Graefes Arch Clin Exp Ophthalmol. 2010; 248: 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Eells JT, Wong-Riley MT, VerHoeve J, et al.. Mitochondrial signal transduction in accelerated wound and retinal healing by near-infrared light therapy. Mitochondrion. 2004; 4: 559–567. [DOI] [PubMed] [Google Scholar]
- 87. Yu DY, Cringle SJ. Retinal degeneration and local oxygen metabolism. Exp Eye Res. 2005; 80: 745–751. [DOI] [PubMed] [Google Scholar]
- 88. Barot M, Gokulgandhi MR, Mitra AK. Mitochondrial dysfunction in retinal diseases. Curr Eye Res. 2011; 36: 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang AL, Lukas TJ, Yuan M, Neufeld AH. Age-related increase in mitochondrial DNA damage and loss of DNA repair capacity in the neural retina. Neurobiol Aging. 2010; 31: 2002–2010. [DOI] [PubMed] [Google Scholar]
- 90. Mulvey CS, Sherwood CA, Bigio IJ. Wavelength-dependent backscattering measurements for quantitative real-time monitoring of apoptosis in living cells. J Biomed Opt. 2009; 14: 064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pasternack RM, Zheng JY, Boustany NN. Detection of mitochondrial fission with orientation-dependent optical Fourier filters. Cytometry A. 2011; 79: 137–148. [DOI] [PubMed] [Google Scholar]
- 92. Pasternack RM, Zheng JY, Boustany NN. Optical scatter changes at the onset of apoptosis are spatially associated with mitochondria. J Biomed Opt. 2010; 15: 040504. [DOI] [PubMed] [Google Scholar]
- 93. Beauvoit B, Evans SM, Jenkins TW, Miller EE, Chance B. Correlation between the light scattering and the mitochondrial content of normal tissues and transplantable rodent tumors. Anal Biochem. 1995; 226: 167–174. [DOI] [PubMed] [Google Scholar]
- 94. Wilson JD, Giesselman BR, Mitra S, Foster TH. Lysosome-damage-induced scattering changes coincide with release of cytochrome c. Opt Lett. 2007; 32: 2517–2519. [DOI] [PubMed] [Google Scholar]
- 95. Litts KM, Wang X, Clark ME, et al.. Exploring photoreceptor reflectivity via multimodal imaging of outer retinal tubulation in advanced age-related macular degeneration. Retina. 2017; 37: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jonnal RS, Kocaoglu OP, Zawadzki RJ, Lee SH, Werner JS, Miller DT. The cellular origins of the outer retinal bands in optical coherence tomography images. Invest Ophthalmol Vis Sci. 2014; 55: 7904–7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Starnes AC, Huisingh C, McGwin G, et al.. Multi-nucleate retinal pigment epithelium cells of the human macula exhibit a characteristic and highly specific distribution. Vis Neurosci. 2016; 33: E001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bermond K, Wobbe C, Tarau IS, Heintzmann R, Hillenkamp J, Curcio CA, Sloan KR, Ach T.. Autofluorescent granules of the human retinal pigment epithelium: age-related topographic and intracellular distribution. Invest Ophthalmol Vis Sci. 2020;5/23/20 online. PMID 32433758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Brilliant MH, Vaziri K, Connor TB Jr, et al.. Mining retrospective data for virtual prospective drug repurposing: l-DOPA and age-related macular degeneration. Am J Med. 2016; 129: 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rosenfeld PJ, Dugel PU, Holz FG, et al.. Emixustat hydrochloride for geographic atrophy secondary to age-related macular degeneration: a randomized clinical trial. Ophthalmology. 2018; 125: 1556–1567. [DOI] [PubMed] [Google Scholar]
- 101. Ai-Awami AK, Beyer J, Haehn D, et al.. NeuroBlocks–visual tracking of segmentation and proofreading for large connectomics projects. IEEE Trans Vis Comput Graph. 2016; 22: 738–746. [DOI] [PubMed] [Google Scholar]
- 102. Ko F, Foster PJ, Strouthidis NG, et al.. Associations with retinal pigment epithelium thickness measures in a large cohort: results from the UK Biobank. Ophthalmology. 2017; 124: 105–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.