Abstract
Coronavirus Disease 2019 (COVID-19) is a dangerous global threat that has no clinically approved treatment yet. Bioinformatics represent an outstanding approach to reveal key immunogenic regions in viral proteins. Here, five severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural proteins (NSPs) (NSP7, NSP8, NSP9, NSP12, and NSP13) were screened to identify potential human leukocyte antigen (HLA) binding peptides. These peptides showed robust viral antigenicity, immunogenicity, and a marked interaction with HLA alleles. Interestingly, several peptides showed affinity by HLA class I (HLA-I) alleles that commonly activates to natural killer (NK) cells. Notably, HLA biding peptides are conserved among SARS-CoV-2, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle Eastern respiratory syndrome coronavirus (MERS-CoV). Interestingly, HLA-I and HLA class II (HLA-II) binding peptides induced humoral and cell-mediated responses after in silico vaccination. These results may open further in vitro and in vivo investigations to develop novel therapeutic strategies against coronaviral infections.
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle Eastern respiratory syndrome coronavirus; S, spike glycoprotein; E, envelope protein; M, membrane protein; N, nucleocapsid protein; ORF1ab, open reading frame 1ab; ORF3a, open reading frame 3a; ORF6, open reading frame 6; ORF7a, open reading frame 7; ORF8, open reading frame 8; ORF10, open reading frame 10; NSP, non-structural proteins; HLA, human leukocyte antigen; HLA-I, human leukocyte antigen class I; HLA-II, human leukocyte antigen class II; NSP7, non-structural protein 7; NSP8, non-structural protein 8; NSP9, non-structural protein 9; NSP12, non-structural protein 12; NSP13, non-structural protein 13; IFN-g, interferon-gamma; IEDB-AR, immune epitope database and analysis resource; 3D, three-dimensional; PDB, protein data bank; P, peptide; Ig, immunoglobulin; CTC, cytotoxic T cell; THC, T helper cell; IL-2, interleukin 2; SARS, severe acute respiratory syndrome; MERS, Middle Eastern respiratory syndrome; aKIR, activating killer immunoglobulin-like receptors; NK, natural killer cell; CTRL+, control positive; NCBI, National Center for Biotechnology Information
Keywords: Non-structural proteins, COVID-19, SARS, MERS, Epitopes, In silico
1. Introduction
SARS-CoV-2 is the causative agent of Coronavirus Disease 2019 (COVID-19) [1] and is classified in the family Coronaviridae and the genus Betacoronavirus. This genus includes other pathogenic coronaviruses for human that share a marked sequence identity to SARS-CoV-2 such as severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle Eastern respiratory syndrome coronavirus (MERS-CoV), whose explosive outbreak in recent years has also led to global public health crisis—severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), respectively [2], [3].
The SARS-CoV-2 single-stranded positive sense RNA genome encodes for both structural and non-structural proteins [2]. The former comprises the spike glycoprotein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) [3]. The latter, on the other hand, includes the open reading frame 1ab (ORF1ab), ORF3a, ORF6, ORF7a, ORF8, and ORF10. In turn, the ORF1ab expresses a polyprotein whose proteolytical cleaved forms 16 non-structural proteins (NSPs, numbered 1 to 16) [3]. The expression and activity of several NSPs constitute a fundamental aspect for SARS-CoV-2 pathogenesis [2]. For instance, the protein complex formed by NSP7, NSP8, and NSP12 give rise to the RNA polymerase, which is necessary for genome replication [4], whereas NSP13 represents the virus helicase, which unwinds duplex RNA [4]. On the other hand, it is thought that NSP9 binds to RNA, thereby improving viral replication [5].
Several works have shown that NSPs from other RNA viruses are fundamental factors to elicit effective immune responses [6], [7]. For instance, the presentation of conserved helicase peptides from several flaviviruses in the context of human leukocyte antigen class I (HLA-I) is sufficient to inhibit viral replication [6]. Therefore, peptides derived from critical SARS-CoV-2 NSPs such as those involved in viral replication and pathogenesis (e.g., NSP7, NSP8, NSP9, NSP12, and NSP13) represent optimal targets to develop future prophylactic and therapeutics approaches against COVID-19. Recent studies have reported promising viral peptides from other SARS-CoV-2 NSPs, including ORF3a [8], ORF6, ORF7, and ORF8 [9]. Likewise, SARS-CoV-2 structural proteins (S, E, M, and N) haven been extensively explored for potential immunogenic peptides [8], [9], [10], [11], [12], [13]. In fact, increasing data suggest that peptides from these structural proteins can be efficiently used in multi-epitope vaccine models [12], [13]. Regardless of these remarkable contributions, our knowledge on the existence of similar peptides in SARS-CoV-2 NSP7, NSP8, NSP9, NSP12, and NSP13 is still poor. Therefore, this immunoinformatics work aimed to identify HLA-I and HLA class II (HLA-II) binding peptides from these key SARS-CoV-2 NSPs, thereby providing more immunogenic targets with strong potential to fight COVID-19.
2. Materials and methods
2.1. Data collection and availability
Amino acid sequences from NSPs of SARS-CoV-2, SARS-CoV, and MERS-CoV were downloaded from the National Center for Biotechnology Information (NCBI) using the following accession numbers: YP_009725303.1 (NSP7), YP_009725304.1 (NSP8), YP_009725305.1 (NSP9), YP_009725307.1 (NSP12), and YP_009725308.1 (NSP13) for SARS-CoV-2; NP_828865.1 (NSP7), NP_828866.1 (NSP8), NP_828867.1 (NSP9), NP_828869.1 (NSP12), and NP_828870.1 (NSP13) for SARS-CoV; YP_009047235.1 (NSP7), YP_009047236.1 (NSP8), YP_009047237.1 (NSP9), YP_009047223.1 (NSP12), YP_009047224.1 (NSP13) for MERS-CoV.
2.2. Prediction of HLA binding peptides from SARS-CoV-2 NSPs
A recent bioinformatics SARS-CoV-2 research [8] with minor modifications was used to identify HLA binding peptides. Briefly, HLA-I binding peptides (9 amino acids of length) were predicted using the NetMHCpan EL 4.0 algorithm [14]. The HLA-I alleles used for this analysis represent some of the most common alleles in human population: HLA-A*01:01, HLA-A*02:01, HLA-A*02:05, HLA-A*03:01, HLA-A*03:02, HLA-A*11:01, HLA-A*24:02, HLA-B*07:02, HLA-B*07:05, HLA-B*08:01, HLA-B*13:02, HLA-B*14:02, HLA-B*35:01, HLA-B*40:01, HLA-B*44:02, HLA-B*44:03, HLA-C*01:02, HLA-C*02:02, HLA-C*03:03, HLA-C*03:04, HLA-C*04:01, HLA-C*06:02, and HLA-C*12:02. HLA-II epitopes (15 amino acids of length) were predicted using the Consensus method [15]. The HLA-II alleles considered in the study were HLA-DRB1*01:01, HLA-DRB1*01:02, HLA-DRB1*03:01, HLA-DRB1*03:06, HLA-DRB1*03:07, HLA-DRB1*04:01, HLA-DRB1*04:02, HLA-DRB1*04:04, and HLA-DRB1*04:05, HLA-DRB1*07:01. In general, HLA-I and HLA-II binding peptides were selected with a percentile rank of prediction cut-off < 20. Peptides with lower percentile values have a higher affinity by HLA molecules [16].
2.3. General assessment of predicted HLA binding peptides
Viral antigenicity (cut-off ≥ 0.5), allergenicity and toxicity predictions were conducted on Vaxijen (www.ddg-pharmfac.net/vaxijen/) [17], AlergenFP (http://ddg-pharmfac.net/AllergenFP/index.html) [18], and ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) [19], respectively. Moreover, HLA-II binding peptides were also selected by their potential capability to induce interferon-gamma (IFN-g), which was evaluated on IFNepitope web server (http://crdd.osdd.net/raghava/ifnepitope/) [20].
2.4. Conservancy analysis
Conservation of HLA binding peptides within NSPs (NSP7, NSP8, NSP9, NSP12, and NSP13) across SARS-CoV-2, SARS-CoV and MERS-CoV was studied by an epitope conservancy analysis on the Immune Epitope Database and Analysis Resource (IEDB-AR) (http://tools.immuneepitope.org/main/) [21]. Conservancy is described as the fraction of protein sequences that have the peptide whereas the identity is referred to the degree of correspondence (similarity) between several sequences [21]. Results were represented as a barplot using Rstudio software (Version 3.5.3).
2.5. Interaction between HLA alleles and viral peptides: molecular docking
HLA-I and HLA-II binding peptides were docked with representative HLA-I and HLA-II alleles. The molecular docking simulation study was performed as follows. First, the three-dimensional (3D) structures of 9-mer and 15-mer peptides were obtained from PEPFOLD server (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) [22]. Second, the receptor‐ligand docking simulations were conducted on ClusPro (version 2.0) server (https://cluspro.bu.edu/) [23]. For this purpose, the Protein Data Bank (PDB) accession numbers of HLA-A*02:01 (1DUZ) [24] and DRB1*01:01 (1AQD) [25] along with the PDB format file of each peptide were used as input on ClusPro server. Third, the receptor-ligand docking complexes were visualized on VMD software (Version 1.9.3) [26]. The best allele-peptide complexes were selected based upon visual inspection and the ClusPro criteria, as well as they were compared with the co-crystal ligands of the HLA allele, which are considered as control peptides in this study. Finally, the Gibbs free energy (ΔG) of the each HLA-viral peptide complex was predicted on the PRODIGY server (https://bianca.science.uu.nl//prodigy/) [27]. Free energy values were represented as barplots using Rstudio software (Version 3.5.3).
2.6. Immune response simulations
To evaluate the potential use of HLA binding peptides on future vaccine trials, a vaccine amino acid sequence was develop with the predicted HLA-I and HLA-II binding peptides, which were linked using AAY and GPGPG linkers, respectively, as previously reported [28]. This vaccine construct was subjected to immune response simulations on the C-ImmSim server (http://150.146.2.1/C-IMMSIM/index.php) [29]. Three injections were applied four weeks apart as described by Nain et al. 2020 [28]. For immune interpretation purpose, the Simpson index D was used.
3. Results
3.1. Prediction of HLA binding peptides
31 HLA-I binding peptides were identified as the best (numbered, P1 to P31) (Table 1 ). These viral peptides showed robust viral antigenicity (≥0.5) and absence of either allergenic or toxic residues (Table 1). The higher number of peptides was observed in NSP12 and NSP13 (10 and 11 peptides, respectively). Interestingly, overlapping residues were observed among P7, P8, and P9, and among P22, P23, P24, P25, and P26 (Table 1). Regarding the HLA-I interacting alleles, most of the viral peptides showed promiscuity by several HLA-I molecules including A*02:01, B*08:01, C*02:02, and C*12:02 (Table 1).
Table 1.
Potential HLA-I binding peptides identified in five non-structural proteins of SARS-CoV-2.
| Protein | Peptide ID | Peptide sequence | Position (star-end) | Percentile Rank | Viral antigenicity | Toxicity and Allergenicity | HLA-I interacting allele |
|---|---|---|---|---|---|---|---|
| NSP7 | P1 | LRVESSSKL | 20–28 | 1.60 | 0.60 | Negative | C*06:02 |
| P2 | LSMQGAVDI | 60–68 | 1.50 | 0.78 | Negative | C*01:02, C*02:02, C*04:01, C*12:02 | |
| P3 | EMLDNRATL | 74–82 | 0.17 | 0.63 | Negative | B*08:01, C*01:02, C*02:02, C*06:02, C*12:02, | |
| NSP8 | P4 | AAFATAQEA | 13–21 | 1.10 | 0.53 | Negative | C*12:02, C*02:02, |
| P5 | CVPLNIIPL | 114–122 | 0.87 | 1.37 | Negative | C*01:02 | |
| P6 | LAWPLIVTA | 180–188 | 0.38 | 1.02 | Negative | A*02:01, C*02:02, C*04:01, C*12:02, B*35:01 | |
| NSP9 | P7 | GLNNLNRGM | 93–102 | 1.80 | 1.01 | Negative | A*02:01, C*02:02, C*02:09, C*12:02 |
| P8 | NNLNRGMVL | 95–103 | 0.09 | 0.52 | Negative | B*08:01, C*06:02 | |
| P9 | GMVLGSLAA | 100–108 | 1.20 | 0.60 | Negative | A*02:01 | |
| P10 | VLGSLAATV | 102–100 | 0.20 | 0.66 | Negative | A*02:01, A*02:05, C*02:09, C*02:02 | |
| NSP12 | P11 | WEPEFYEAM | 116–124 | 0.21 | 0.53 | Negative | A*02:01 |
| P12 | MRNAGIVGV | 196–204 | 0.02 | 1.63 | Negative | C*06:02 | |
| P13 | LVYAADPAM | 372–380 | 0.02 | 0.52 | Negative | C*03:02, C*12:02, B*35:01, C*02:09, C*02:02, C*08:01 | |
| P14 | TVKPGNFNK | 409–417 | 0.02 | 1.38 | Negative | A*11:01 | |
| P15 | NAAISDYDY | 447–455 | 0.07 | 1.29 | Negative | B*35:01, A*01:01, C*03:03 | |
| P16 | FAYTKRNVI | 538–546 | 0.06 | 1.03 | Negative | B*08:01, C*03:02, C*12:02, C*08:01, C*02:02, C*02:09, C*01:02, C*06:02 | |
| P17 | IAATRGATV | 579–587 | 0.16 | 0.89 | Negative | B*08:01, C*01:02, C*02:02, C*03:02, C*02:09, C*08:01, C*12:02, | |
| P18 | ATVVIGTSK | 585–593 | 0.02 | 0.76 | Negative | A*11:01 | |
| P19 | RLANECAQV | 654–662 | 0.16 | 0.93 | Negative | A*02:01 | |
| P20 | MLDMYSVML | 899–907 | 0.08 | 0.56 | Negative | A*02:01, A*01:01, C*01:02, C*08:01, | |
| NSP13 | P21 | HVISTSHKL | 33–41 | 0.10 | 0.69 | Negative | C*01:02, C*02:02, C*03:02, C*12:02, B*35:01, |
| P22 | TEETFKLSY | 141–149 | 0.65 | 0.65 | Negative | A*01:01 | |
| P23 | YGIATVREV | 149–157 | 0.37 | 1.43 | Negative | C*02:02, C*03:02, C*12:02, | |
| P24 | PTLVPQEHY | 238–246 | 0.43 | 0.78 | Negative | A*01:01 | |
| P25 | TLVPQEHYV | 239–247 | 0.04 | 0.54 | Negative | A*02:01, C*02:02 | |
| P26 | EHYVRITGL | 244–252 | 0.33 | 0.57 | Negative | B*08:01 | |
| P27 | SHFAIGLAL | 289–297 | 0.35 | 1.39 | Negative | C*02:02, C*03:02, C*12:02, B*40:01, | |
| P28 | VVFDEISMA | 371–379 | 0.11 | 0.74 | Negative | A*02:01, A*02:05, C*02:02, C*12:02 | |
| P29 | VVNARLRAK | 386–394 | 0.08 | 1.93 | Negative | A*03:01, A*11:01 | |
| P30 | EIVDTVSAL | 447–455 | 0.26 | 0.66 | Negative | C*02:02, C*03:02, C*04:01, C*12:02, B*35:01 | |
| P31 | ITRAKVGIL | 565–573 | 0.25 | 0.91 | Negative | B*08:01 | |
46 HLA-II binding peptides were predicted (numbered, P32 to P76). In this regard, 1 peptide was identified for NSP7, 13 peptides for NSP8, 9 peptides for NSP9, 17 peptides for NSP12, and 6 peptides for NSP13 (Table 2 ). Similar to HLA-I binding peptides, strong viral antigenicity and lack of allergenicity and toxicity were observed in these 15-mer viral peptides. Of note, each HLA-II binding peptides was classified as a potential inductor of IFN-g (Table 2). Likewise, viral peptides from NSP8, NSP9, NSP12, and NSP13 showed the presence of overlapping residues. On the other hand, DRB1*01:01, DRB1*03:01, and DRB1*04:01were identified as the most common HLA-II interacting alleles (Table 2).
Table 2.
Potential HLA-II binding peptides identified in five non-structural proteins of SARS-CoV-2.
| Protein | Peptide ID | Peptide sequence | Position (star-end) | Percentile Rank | Viral antigenicity | Toxicity and Allergenicity | IFN-g induction | HLA-I interacting allele |
|---|---|---|---|---|---|---|---|---|
| NSP7 | P32 | VVLLSVLQQLRVESS | 11–25 | 0.60 | 0.50 | Negative | Positive | DRB1*01:01, DRB1*01:02, DRB1*03:01, DRB1*03:07, DRB1*04:01, DRB1*04:02, DRB1*04:04, |
| NSP8 | P33 | IASEFSSLPSYAAFA | 2–16 | 0.13 | 0.54 | Negative | Positive | DRB1*01:01, DRB1*04:01, DRB1*04:04 |
| P34 | SEFSSLPSYAAFATA | 4–18 | 0.15 | 0.54 | Negative | Positive | DRB1*01:01, DRB1*04:01, DRB1*04:04 | |
| P35 | FSSLPSYAAFATAQE | 6–20 | 7.20 | 0.51 | Negative | Positive | DRB1*01:01, DRB1*04:01, DRB1*04:04 | |
| P36 | GCVPLNIIPLTTAAK | 113–127 | 3.00 | 1.12 | Negative | Positive | DRB1*03:06, DRB1*03:07, DRB1*04:04, DRB1*04:01 | |
| P37 | PLNIIPLTTAAKLMV | 116–130 | 6.10 | 0.82 | Negative | Positive | DRB1*03:06, DRB1*03:07, DRB1*04:04, DRB1*04:01 | |
| P38 | WEIQQVVDADSKIVQ | 154–168 | 7.60 | 0.53 | Negative | Positive | DRB1*03:01, DRB1*03:06, DRB1*03:07 | |
| P39 | IQQVVDADSKIVQLS | 156–170 | 7.20 | 0.59 | Negative | Positive | DRB1*03:01, DRB1*03:06, DRB1*03:07 | |
| P40 | PNLAWPLIVTALRAN | 178–192 | 8.50 | 0.92 | Negative | Positive | DRB1*01:01 | |
| P41 | NLAWPLIVTALRANS | 179–193 | 3.50 | 0.99 | Negative | Positive | DRB1*03:06, DRB1*03:07, DRB1*04:04 | |
| P42 | LAWPLIVTALRANSA | 180–194 | 3.10 | 0.80 | Negative | Positive | DRB1*01:01, DRB1*03:06, DRB1*03:07, DRB1*04:04 | |
| P43 | AWPLIVTALRANSAV | 181–195 | 3.30 | 0.72 | Negative | Positive | DRB1*01:01, DRB1*03:06, DRB1*03:07, DRB1*04:04 | |
| P44 | WPLIVTALRANSAVK | 182–196 | 2.60 | 0.74 | Negative | Positive | DRB1*01:01, DRB1*03:06, DRB1*03:07, DRB1*04:04 | |
| P45 | PLIVTALRANSAVKL | 183–197 | 0.45 | 0.66 | Negative | Positive | DRB1*01:01, DRB1*01:02, DRB1*03:01, DRB1*03:06, DRB1*03:07, DRB1*04:01, DRB1*04:04 | |
| NSP9 | P46 | GPKVKYLYFIKGLNN | 82–96 | 7.20 | 0.70 | Negative | Positive | DRB1*04:04 |
| P47 | PKVKYLYFIKGLNNL | 83–97 | 6.10 | 0.62 | Negative | Positive | DRB1*01:01, DRB1*04:04 | |
| P48 | KVKYLYFIKGLNNLN | 84–98 | 4.40 | 0.91 | Negative | Positive | DRB1*04:01, DRB1*01:01, DRB1*04:04 | |
| P49 | KYLYFIKGLNNLNRG | 86–100 | 0.38 | 0.64 | Negative | Positive | DRB1*01:01, DRB1*04:01, DRB1*04:04 | |
| P50 | YLYFIKGLNNLNRGM | 87–101 | 0.31 | 0.52 | Negative | Positive | DRB1*01:01, DRB1*04:01, DRB1*04:04 | |
| P51 | LYFIKGLNNLNRGMV | 88–102 | 0.31 | 0.52 | Negative | Positive | DRB1*01:01, DRB1*04:01, DRB1*04:04 | |
| P52 | NNLNRGMVLGSLAAT | 95–109 | 1.20 | 0.82 | Negative | Positive | DRB1*03:06, DRB1*03:07, DRB1*04:02 | |
| P53 | NLNRGMVLGSLAATV | 96–110 | 1.20 | 0.83 | Negative | Positive | DRB1*03:06, DRB1*03:07, DRB1*04:02 | |
| P54 | LNRGMVLGSLAATVR | 97–111 | 1.20 | 0.92 | Negative | Positive | DRB1*04:04 | |
| NSP12 | P55 | NAGIVGVLTLDNQDL | 198–212 | 1.70 | 1.30 | Negative | Positive | DRB1*04:05, DRB1*04:03 |
| P56 | RLSFKELLVYAADPA | 365–379 | 3.50 | 0.59 | Negative | Positive | DRB1*04:01, DRB1*04:03 DRB1*01:01 |
|
| P57 | LSFKELLVYAADPAM | 366–380 | 3.00 | 0.65 | Negative | Positive | DRB1*04:01 DRB1*04:03, DRB1*01:01 DRB1*04:05 |
|
| P58 | AASGNLLLDKRTTCF | 382–396 | 2.40 | 0.89 | Negative | Positive | DRB1*03:01 | |
| P59 | ASGNLLLDKRTTCFS | 383–397 | 1.70 | 0.94 | Negative | Positive | DRB1*03:01 | |
| P60 | SGNLLLDKRTTCFSV | 384–398 | 1.40 | 1.04 | Negative | Positive | DRB1*03:01 | |
| P61 | GNLLLDKRTTCFSVA | 385–399 | 1.90 | 1.20 | Negative | Positive | DRB1*03:01 | |
| P62 | NLLLDKRTTCFSVAA | 386–400 | 3.80 | 1.20 | Negative | Positive | DRB1*03:01 | |
| P63 | KHFFFAQDGNAAISD | 438–452 | 1.10 | 0.54 | Negative | Positive | DRB1*04:01, DRB1*01:03 DRB1*04:03, DRB1*01:01 |
|
| P64 | KRNVIPTITQMNLKY | 532–546 | 0.96 | 1.25 | Negative | Positive | DRB1*04:03 | |
| P65 | ITQMNLKYAISAKNR | 539–553 | 1.20 | 1.51 | Negative | Positive | DRB1*04:03, DRB1*01:03 DRB1*04:01 |
|
| P66 | QMNLKYAISAKNRAR | 541–555 | 0.32 | 1.50 | Negative | Positive | DRB1*04:03, DRB1*01:03, DRB1*04:01, DRB1*04:05 | |
| P67 | MNLKYAISAKNRART | 542–556 | 0.44 | 1.44 | Negative | Positive | DRB1*04:03, DRB1*04:01 DRB1*01:03 |
|
| P68 | NLKYAISAKNRARTV | 543–557 | 0.96 | 1.34 | Negative | Positive | DRB1*04:03, DRB1*01:03 DRB1*04:01 |
|
| P69 | MLRIMASLVLARKHT | 629–643 | 0.43 | 0.53 | Negative | Positive | DRB1*04:03, DRB1*03:01 DRB1*01:01, DRB1*01:03 DRB1*04:01, DRB1*04:05 |
|
| P70 | LRIMASLVLARKHTT | 630–644 | 0.53 | 0.67 | Negative | Positive | DRB1*03:01, DRB1*04:03 DRB1*01:01, DRB1*01:03, DRB1*04:01 |
|
| P71 | ASQGLVASIKNFKSV | 771–785 | 0.85 | 0.58 | Negative | Positive | DRB1*04:03, DRB1*04:01 DRB1*03:01 |
|
| NSP13 | P72 | EVLSDRELHLSWEVG | 156–170 | 2.90 | 1.54 | Negative | Positive | DRB1*03:01 |
| P73 | TTYKLNVGDYFVLTS | 215–229 | 2.50 | 0.60 | Negative | Positive | DRB1*03:02 | |
| P74 | LNVGDYFVLTSHTVM | 219–233 | 3.00 | 0.61 | Negative | Positive | DRB1*01:01, DRB1*01:03 | |
| P75 | DYFVLTSHTVMPLSA | 223–237 | 0.96 | 0.56 | Negative | Positive | DRB1*01:01, DRB1*01:03 | |
| P76 | RFNVAITRAKVGILC | 560–574 | 3.60 | 1.32 | Negative | Positive | DRB1*04:02, DRB1*01:03 | |
| P77 | KVGILCIMSDRDLYD | 569–583 | 0.98 | 1.51 | Negative | Positive | DRB1*03:01, DRB1*03:02 | |
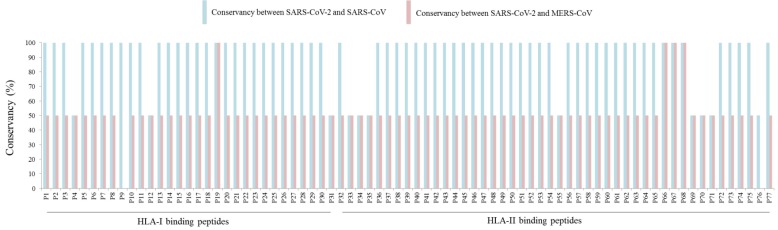
Notably, HLA-I and HLA-II binding peptides are 100% conserved between SARS-CoV and SARS-CoV-2 with exception of P4, P12, P31, P33, P34, P35, P55, P69, P70, P71, and P76, that showed 50% of conservancy (Fig. 1 ). In contrast, most of the SARS-CoV-2 peptides common to MERS-CoV reached 50% of conservancy, with exception of P9 and P76 (0% of conservancy each) and P19, P66, P67, and P68 (100% of conservancy each) (Fig. 1).
Fig. 1.
Conservancy analysis of HLA binding peptides.
3.2. HLA allele-viral peptide interaction
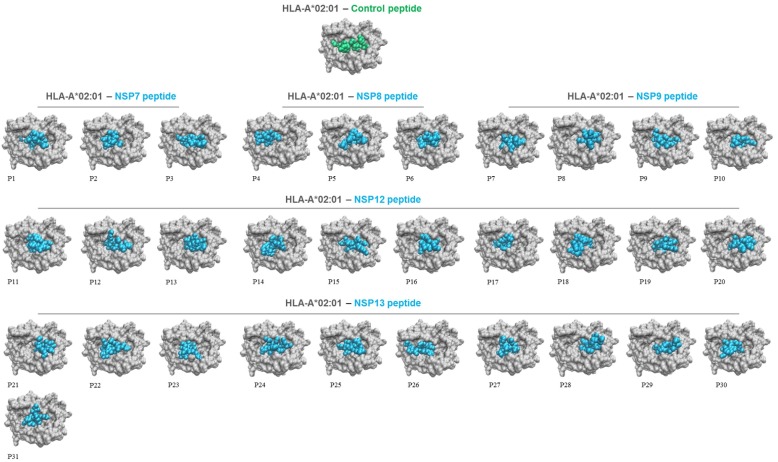
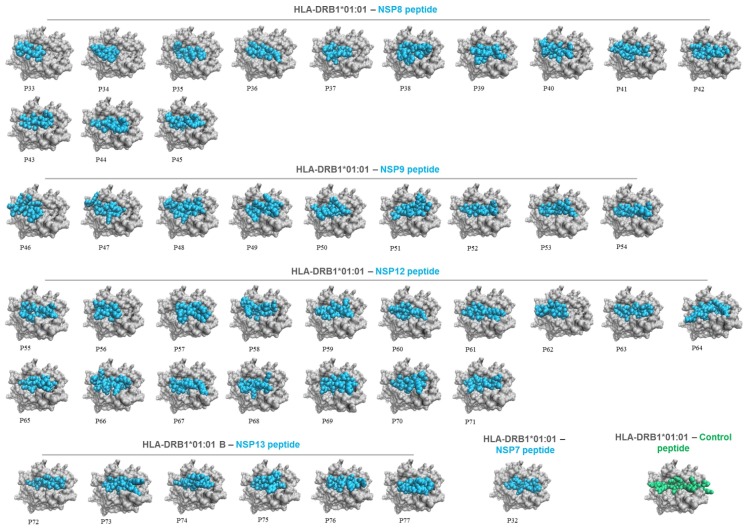
Molecular docking simulations were conducted to evaluate the presentation of HLA binding peptides in the context of HLA molecules. To achieve this aim, A*02:01 and DRB1*01:01 were selected as representative HLA-I and HLA-II alleles, respectively. The analysis revealed that both 9-mer (Fig. 2 ) and 15-mer (Fig. 3 ) peptides rightly interact with the groove of their respective HLA molecule in a similar way to control peptides, as well as they showed different HLA binding patterns (Figs. 2 and 3).
Fig. 2.
Top view of HLA-A*02:01 presenting 9-mer viral peptides (numbered, P1 to P31) from selected SARS-CoV-2 NSPs. HLA alleles, viral peptides, and control peptides are shown in grey, cyan, and green, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Top view of HLA-DRB1*01:01 presenting 15-mer viral peptides (numbered, P32 to P77) from selected SARS-CoV-2 NSPs. HLA alleles, viral peptides, and control peptides are shown in grey, cyan, and green, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
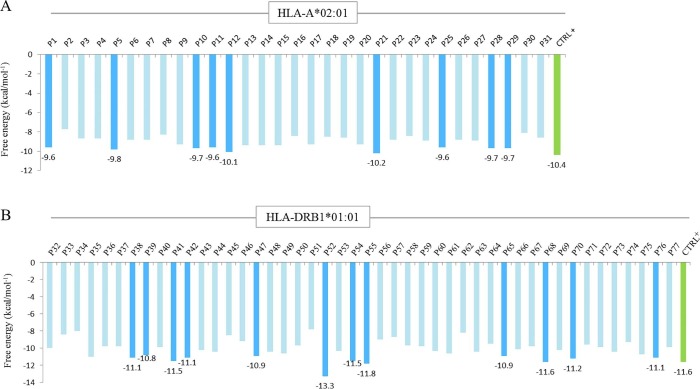
Remarkably, these HLA-viral peptide complexes showed strong potential interactions (free energy values <−6 kcal/mol−1) comparable to control peptides (Fig. 4 A,B). In this regard, P1, P5, P10, P11, P12, P21, P25, P28, P29, P38, P39, P41, P42, P47, P52, P54, P55, P65, P68, P70, and P76 showed the lowest affinity values (Fig. 4A,B). Interestingly, P52 obtained the lower free energy value (-13.3 kcal/mol−1) compared to other HLA-II binding peptides (Fig. 4B).
Fig. 4.
Free energy for each HLA-viral peptide interaction. A: free energy values for the binding between HLA-A*02:01 and 9-mer viral peptides (numbered, P1 to P31). B: free energy values for the binding between HLA-DRB1*01:01 and 15-mer viral peptides (numbered, P32 to P377). Note that the lowest values are shown in cyan bars while values of control peptides (CTRL+) are shown in green bars. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Immune responses simulations
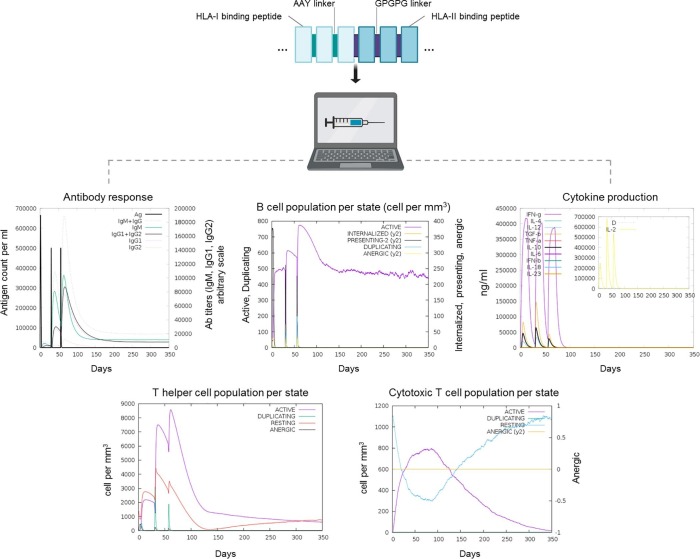
The immune responses simulations with the vaccine construct showed high levels of immunoglobulin M (IgM) as well as isotypes IgG1 and IgG2 along with reduced immunogen levels during the secondary humoral response. Moreover, active B cell population increased after each dose (Fig. 5 ). Likewise, a marked increase of cytotoxic T cell (CTC) and T helper cell (THC) populations were observed along with effector cell generation, which decreased after immunogen clearance (Fig. 5). Finally, higher levels of key cytokines such as IFN-g and interleukin 2 (IL-2) were documented after each dose (Fig. 5). Taken together, these results suggest that HLA-I and HLA-II binding peptides could elicit humoral and cellular-mediated immune responses to SARS-CoV-2.
Fig. 5.
Immune response simulations after in silico active immunization (3 doses) with the vaccine construct.
4. Discussion
The present study using an integrated in silico approach showed that five SARS-CoV-2 NSPs (NSP7, NSP8, NSP9, NSP12, and NSP13) harbour conserved viral peptides, whose properties—high viral antigenicity, absence of allergenic and toxic residues, potential IFN-g induction, interaction with HLA alleles, and suitable immune responses after in silico vaccination—make them potential targets for future prophylactic and/or therapeutics against the overwhelming COVID-19 pandemic. In this regard, HLA binding peptides from SARS-CoV-2 NSPs showed a robust viral antigenicity and immunogenicity similar to initial works based on peptide prediction from SARS-CoV-2 S, E, M, and N proteins [8], [10], [11]. Therefore, these peptides could successfully improve in vitro and in vivo immune responses against SARS-CoV-2. In fact, immune responses were observed after active immunization simulations, whose results (e.g., production of antibodies, T cell generation, and cytokines profile) are comparable to other vaccine candidates against infectious diseases [28], [30]. On the other hand, conservancy analysis on IEDB-AR revealed that most of the peptides are strongly conserved across highly pathogenic betacoronaviruses—SARS-CoV-2, SARS-CoV, and MERS-CoV. This result is in agreement, at least partially, with a former report that has demonstrated a robust amino acid sequence identity between SARS-CoV-2 NSPs and SARS-CoV NSPs [4]. Hence, HLA biding peptides here predicted could evoke—perhaps—simultaneous immune responses against other betacoronaviruses such as SARS-CoV, and MERS-CoV.
The clear and proper interaction between conserved viral peptides and HLA alleles suggest that they could elicit not only adaptive immune responses by CD8+ CTC and CD4+ THC but also an innate immune response by Natural Killer (NK) cells—one of the first lines of defence against viruses []. In fact, it has been shown a decisive role of activating killer immunoglobulin-like receptors (aKIR) in viral recognition by NK cells [31]. Moreover, it has been reported that the interaction between aKIRs and HLA-C molecules is conditioned by C1 and C2 epitopes, for instance, KIR2DS2 may recognize HLA-C allotypes carrying the C1 epitope (N80), including HLA-C*01:02, HLA-C*14:02, and HLA-C*16:01, whereas KIR2DS4 binds to HLA-C allotypes with C1 (N80) or C2 (K80) epitopes, including HLA-C*02:01, HLA-C*04:01, and HLA-C*05:01 [31]. Interestingly, Nayer and colleagues reported that KIR2DS2 directly recognizes conserved viral peptides in the context of HLA-C*01:02, thereby leading to the inhibition of the hepatitis C virus and dengue virus replication [6]. In the present study, several 9-mer viral peptides showed affinity by HLA-C*01:02 as well as by HLA-C*02:02, which activates to KIRD2DS4 [31]. However, further research is required to reveal peptide-specific recognition of SARS-CoV-2 and other betacoronaviruses by certain subsets within the NK cell compartment.
This study was limited by 1) all data only reflect immunoinformatics analyses. Future in vitro and in vivo investigations are required to ensure safety and efficacy of these peptides prior to their use in human trials; 2) Future studies should address other SARS-CoV-2 NSPs derived from ORF1ab to determine their probable immunogenicity. However, NSP7, NSP8, NSP9, NSP12, and NSP13 used in this research represent optimal targets for anti-SARS-CoV-2 immune responses as exposed earlier; and 3) although the vaccine construct showed robust humoral and cell-mediated responses, further investigations are necessary to assess its physicochemical properties, 3D structure, and refinement.
In summary, these initial results describing the immunogenic potential of conserved peptides from SARS-CoV-2 NSPs are promising, and they could be strong candidates to evaluate functional NK cells, humoral, and T cell responses in experimental animal models. These results open a powerful window for future fundamental and translational studies focus on coronaviral diseases.
Declarations of interest
None.
Founding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgments
I wish to thank the support of the Venezuelan Institute for Scientific Research (IVIC) – Venezuela, and Laboratory of Cellular and Molecular Pathology-IVIC.
References
- 1.World Health Organization, Coronavirus disease (COVID-19) Pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed 17 June 2020).
- 2.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12:E372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimoto F. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein. J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020 doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naiyer M.M., Cassidy S.A., Magri A., Cowton V., Chen K., Mansour S., Kranidioti H., Mbiribindi B., Rettman P., Harris S., Fanning L.J., Mulder A., Claas F., Davidson A.D., Patel A.H., Purbhoo M.A., Khakoo S.I. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci. Immunol. 2017;2(15):eaal5296. doi: 10.1126/sciimmunol.aal5296. [DOI] [PubMed] [Google Scholar]
- 7.Stewart C.A., Laugier-Anfossi F., Vély F., Saulquin X., Riedmuller J., Tisserant A., Gauthier L., Romagné F., Ferracci G., Arosa F.A., Moretta A., Sun P.D., Ugolini S., Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. P. Natl. Acad. Sci. Usa. 2005;102(37):13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell. Host. Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiyotani A., Toyoshima Y., Nemoto K., Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020;65:569–575. doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92(5):495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilocca B., Soggiu A., Sanguinetti M., Babini G., De Maio F., Britti D., Zecconi A., Bonizzi L., Urbani A., Roncada P. Immunoinformatic analysis of SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19. Microb. Infect. 2020:182–187. doi: 10.1016/j.micinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalita P., Padhi A., Zhang K., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb. Pathog. 2020 doi: 10.1016/j.micpath.2020.104236. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., Lee S.S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J. Med. Virol. 2020 doi: 10.1002/jmv.25736. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul S., Lindestam Arlehamn C.S., Scriba T.J., Dillon M.B., Oseroff C., Hinz D., McKinney D.M., Carrasco Pro S., Sidney J., Peters B., Sette A. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J. Immunol. Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E., Nielsen M., Peters B. Immune epitope database analysis resource. Nucleic Acids. Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30:846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 19.S. Gupta, P. Kapoor, K. Chaudhary, A. Gautam, R. Kumar, Open Source Drug Discovery Consortium, G. P. Raghava, In silico approach for predicting toxicity of peptides and proteins, PloS one. 8 (2013) e73957. https://doi.org/10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed]
- 20.Dhanda S.K., Vir P., Raghava G.P. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 2013;8:30. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bui H.H., Sidney J., Li W., Fusseder N., Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007;8:361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maupetit J., Derreumaux P., Tuffery P. A fast and accurate method for large-scale de novo peptide structure prediction. J. Comput. Chem. 2010;31:726–738. doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- 23.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan A.R., Baker B.M., Ghosh P., Baddison W.E., Wiley D.C. The structure and stability of HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. immunol. 2000;164(12):6398–6405. doi: 10.4049/jimmunol.164.12.6389. [DOI] [PubMed] [Google Scholar]
- 25.Murthy V.L., Stern L.J. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: implications for the structural basis of specificity of pepetide binding structure. Structure. 1997;5(10):1385–1396. doi: 10.1016/s0969-2126(97)00288-8. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey W., Dalke A., Schulten K. VMD – Visual Molecular Dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: aweb server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 28.Nain Z., Karim M.M., Sen M.K., Adhikari U.K. Structural basis and designing of peptide vaccine using PE-PGRS family protein of Mycobacterium ulcerans-an integrated vaccinomics approach. Mol. Immunol. 2020;120:146–163. doi: 10.1016/j.molimm.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Rapin N., Lund O., Bernaschi M., Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PloS one. 2010;5 doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shey R.A., Ghogomu S.M., Esoh K.K., Nebangwa N.D., Shintouo C.M., Nongley N.F., Asa B.F., Ngale F.N., Vanhamme L., Souopgui J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci. Rep. 2019;9:4409. doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pende D., Falco M., Vitale M., Cantoni C., Vitale C., Munari E., Bertaina A., Moretta F., Del Zotto G., Pietra G., Mingari M.C., Locatelli F., Moretta L. Killer Ig-like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]