Abstract
Purpose
Prior studies have demonstrated that microglial activation is involved in the pathogenesis of primary open-angle glaucoma (POAG). Here we sought to identify genetic associations between POAG and variants in APOE and TREM2, genes associated with Alzheimer disease (AD) that critically regulate microglial neurodegeneration-associated molecular signature.
Methods
APOE genotypes were called using imputed data from the NEIGHBOR consortium (2120 POAG cases, 2262 controls) and a second cohort from the Massachusetts Eye and Ear Infirmary (MEEI; 486 cases, 344 controls). TREM2 coding variants were genotyped by means of the Illumina HumanExome BeadArray. The data set was analyzed for association with POAG overall, as well as the high-tension glaucoma (HTG) and normal-tension glaucoma (NTG) subgroups, using logistic regression adjusting for age and sex.
Results
In the combined NEIGHBOR-MEEI data set, significant association was observed for APOE ε4 in POAG overall (odds ratio [OR], 0.83; 95% confidence interval [CI], 0.74–0.94; P = 0.0022) and in both the HTG subgroup (OR, 0.81; 95% CI, 0.70–0.94; P = 0.0052) and NTG subgroup (OR, 0.71; 95% CI, 0.58–0.87; P = 0.0014). A rare TREM2 variant (A105V) was found only in HTG cases (3 of 2863 cases) and in none of the controls (P = 0.03). Three TREM2 rare variants associated with AD were not significantly associated with POAG (P > 0.05).
Conclusions
We have found that the APOE ε4 allele is associated with a reduced risk of POAG. Interestingly, the same allele is adversely associated with AD, suggesting a mechanistic difference between neurodegenerative diseases of the eye and the brain. TREM2 variants associated with AD did not significantly contribute to POAG risk.
Keywords: glaucoma, genetics, microglia, APOE, TREM2
Primary open-angle glaucoma (POAG) is a genetically and clinically heterogeneous neurodegenerative disease whose main hallmark is retinal ganglion cell (RGC) apoptosis. Glaucoma pathogenesis remains poorly understood, and there are currently no clinically approved therapies that directly promote RGC survival.1 Prior research has demonstrated that in terms of genetic risk, there exists overlap between glaucoma and neurodegenerative diseases of the brain. For example, some of the same genes that harbor known risk alleles for amyotrophic lateral sclerosis (ALS), such as optineurin (OPTN), TANK-binding kinase 1 (TBK1), and ataxin2 (ATXN2), have also been shown to confer genetic risk in glaucoma.2–8 Herein we sought to explore parallels between glaucoma and another common age-related neurodegenerative disease, Alzheimer disease (AD), which is characterized by accumulation of amyloid-β plaques, neurofibrillary tangles, neuronal loss, and inflammation.9,10
The strongest risk factor for late-onset AD is apolipoprotein E (APOE), the major lipoprotein in the brain. APOE has three variants in humans (ε2, ε3, and ε4), which differ by two amino acid residues. The ε3 allele is the most common and is considered the baseline for AD risk; ε4 raises risk of AD relative to ε3, whereas ε2 is protective.11,12 More recently, rare variants of the transmembrane receptor TREM2 (triggering receptor expressed on myeloid cells 2) have also been identified as risk factors for AD13–16 and Nasu-Hakola disease, a neurodegenerative disease characterized by early dementia and bone cysts with fractures.17 Interestingly, in the brain, TREM2 is only expressed by myeloid cells (resident microglia and peripherally derived monocytes/macrophages),18 highlighting the importance of this cell type in the pathogenesis of AD.19–22 While APOE is expressed more broadly, recent work has found that APOE is upregulated in microglia in mouse models of neurodegenerative disease, including AD, ALS, and multiple sclerosis.23–25 Furthermore, TREM2 and APOE have been identified as key regulators of the microglial molecular phenotype associated with neurodegeneration (the so-called microglial neurodegenerative or disease-associated microglia molecular signature).23–25 Given that microglia have also been implicated in the pathogenesis of glaucoma26–30 and that APOE is upregulated in the retina and the aqueous humor of patients with glaucoma,31,32 we hypothesized that APOE and TREM2 may have genetic associations with POAG as well.
The genetic association between APOE and POAG has been previously examined in a series of small and likely underpowered studies that identified conflicting results.33–42 Several meta-analyses found no association between APOE and glaucoma,43,44 while others reported an association between the APOE ε4/ε4 genotype and POAG in Asians.45,46 Another large meta-analysis reported an association between a single-nucleotide polymorphism (SNP) in the promoter region of APOE (rs449647) and glaucoma47; this SNP, however, is not associated with AD.48 Interestingly, APOE ε4 is associated with decreased risk for another common neurodegenerative disease of the eye, age-related macular degeneration (AMD),49–52 opposite of its effect on AD. To our knowledge, the genetic association between TREM2 and POAG has not been previously explored.
In this study, we sought to examine the associations between APOE and TREM2 with POAG in a relatively large data set that includes subgroups with high-tension glaucoma (HTG) and normal-tension glaucoma (NTG). We find that APOE ε4 is associated with reduced risk of POAG, especially NTG. Three TREM2 rare variants associated with AD were not significantly associated with POAG, while a rare TREM2 variant (not implicated in AD) may contribute to HTG risk.
Methods
Study Participants
This study adhered to the tenets of the Declaration of Helsinki and has been reviewed and approved by the Institutional Review Boards of the Massachusetts Eye and Ear Infirmary, Harvard School of Public Health, the Brigham and Women's Hospital, University of Pittsburgh, Johns Hopkins University, Duke University, University of West Virginia, University of Miami, University of Michigan, Stanford University, Marshfield Clinic, and the University of California, San Diego. Informed consent was obtained from the participants after explanation of the nature and possible consequences of the study.
Two case-control genome-wide association study (GWAS) data sets were used for this study: the Mass Eye and Ear Infirmary (MEEI) component of the Glaucoma Genes and Environment (GLAUGEN) GWAS53 and the National Eye Institute Glaucoma Human Genetics Collaboration (NEIGHBOR) GWAS.54 Detailed information on these data sets has been described previously.4,53,54 Briefly, the MEEI component of the GLAUGEN data set includes 486 POAG cases and 344 controls, and the NEIGHBOR data set includes 2120 POAG cases and 2262 controls. Average age of enrollment was 64.5 ± 11.0 years for MEEI GLAUGEN controls and 62.0 ± 11.2 years for the MEEI GLAUGEN cases, as well as 68.9 ± 11.4 years for the NEIGHBOR controls and 66.6 ± 13.7 years for the NEIGHBOR cases. The MEEI GLAUGEN cases and controls were 58.4% and 59.8% female, respectively, while the NEIGHBOR cases and controls were 54.1% and 56.5% female, respectively.
A harmonized definition of POAG was adopted across these data sets based on the following criteria: (1) open anterior segment angles, (2) reproducible glaucomatous visual field loss on reliable tests or (3) an eye with cup-disc ratio of at least 0.7 with one visual field showing glaucomatous loss, and (4) no identifiable secondary cause for optic nerve disease. Elevated intraocular pressure (IOP) was not a criterion for POAG definition, but if present, there had to be no secondary causes on anterior segment examination.
Sixty-seven percent of cases had a history of elevated IOP (≥22 mm Hg) measured in a clinical setting (typically between the hours of 8:00 am and 5:00 pm) and were classified as HTG. Cases with IOP <22 mm Hg (without treatment) measured in the clinic at the time of study enrollment were classified as NTG. Cases undergoing IOP-lowering therapy at the time of enrollment were included in the HTG group if they had a documented history of IOP >22 mm Hg prior to treatment, and cases undergoing IOP-lowering therapy at the time of enrollment were included in the NTG group if they did not have recorded pressures >22 mm Hg before treatment. Pretreatment IOP measurements were not available for all cases.
Genotyping
The MEEI and NEIGHBOR case control data sets were originally genotyped as part of the GLAUGEN and NEIGHBOR GWAS studies as previously described.4,53,54 Subsequently, the genotype data were imputed to the Haplotype Reference Consortium panel55 using the Michigan Imputation Server.56 APOE genotypes (including alleles ε2, ε3, and ε4) were determined from haplotypes of rs429358 and rs7412 (T-T, T-C, and C-C, respectively), the two SNPs known to define APOE alleles. Imputation scores were high for both SNPs (r2 > 0.93). Haplotypes are unambiguous from the unphased genotypes except for the double heterozygote, rs429358 C/T-rs7412 C/T, which was called ε2/ε4 rather than as ε1/ε3, since the ε1 allele is extremely rare.
Fourteen TREM2 rare variants (minor allele frequency [MAF] <1%, call rate ≥98%) were extracted from Illumina HumanExome BeadArray (Illumina, Inc., San Diego, CA) genotype data for the NEIGHBORHOOD and GLAUGEN data sets. We focused on TREM2 rare variants as those have previously been associated with AD.13–15 Genotyping was completed at the Center for Inherited Disease Research. The Illumina Genome Studio (Illumina) and PLINK57 were used for all quality controls (QC) steps except where noted. Basic QC for samples included screens for call rate (≥98.5%) and high (≥95%) concordance with a previous Illumina 660K panel run on the same sample54 where available (about 80% of samples). We verified recorded sex in the clinical records with genotyped sex by two criteria: mean fluorescence intensity on the X and Y chromosomes, plus genotype heterozygosity on the X chromosome and call rate on the Y, allowing male and female samples to have heterozygous X-linked and successful Y-linked genotypes, respectively. We tested samples for pairwise relationships and unexpected duplication using KING.58
We verified European ancestry from the first two principal components derived from genotypes at 9000 ancestry-informative markers by means of the SNPweights program,59 including representative HapMap CEU, YRI, CHB, and JPT samples as reference populations. Moreover, we conducted a principal components analysis over 52,040 independent (pairwise r2 < 0.1), common (MAF ≥0.005) SNPs using the smartpca program in EIGENSOFT60 to detect finer population structure. Of the first 20 principal components, the first, sixth, and eighth were significantly associated (P < 0.05 by logistic regression) with POAG status.
Initial QC screens for markers included call rate (≥98%) and consistency with Hardy-Weinberg proportions (P > 10−6 by Fisher exact test). We screened markers for differences in allele frequency between whole-genome-amplified DNA samples and all other samples by the Fisher exact test and removed from analysis all markers with P < 0.0001. All pseudoautosomal, Y-linked, and mitochondrial SNPs were subject to review in Illumina Genome Studio (Illumina, Inc.) and, if necessary, were reclustered by hand. We also confirmed genotype clustering for rare (MAF <0.02) variants from fluorescence intensity data by means of zCall, run with a stringent z score threshold of z = 21 for calling heterozygous genotypes. Every rare SNP with two or more additional heterozygous calls by zCall than by GenCall was reviewed in Genome Studio, and if necessary, cluster locations were adjusted manually.
Analyses
APOE Alleles.
Each of the three detectable APOE alleles was tested for association with glaucoma using logistic regression with age and sex as covariates. Results for ε2 and ε4 were compared to ε3 as the reference. A likelihood ratio test (LRT) was run to compare the logistic regression models that include age and sex and with or without the number of ε2 and ε4 alleles to assess the overall significance of adding APOE genotypes. APOE genotype frequencies were compared among POAG, HTG, and NTG cases and controls using the Pearson χ2 test.
TREM2 Rare Variants
Association between individual variants and POAG case/control status was assessed using logistic regression, including as covariates age at exam, sex, and three principal components observed to be significantly associated with POAG.
Results
In the combined NEIGHBOR-MEEI data set, APOE ε4 was inversely associated with POAG overall (odds ratio [OR], 0.83; 95% confidence interval [CI], 0.74–0.94; P = 0.0022) and in both the HTG (OR, 0.81; 95% CI, 0.70–0.94; P = 0.0052) and NTG (OR, 0.71; 95% CI, 0.58–0.87; P = 0.0014) subgroups (Table 1). The LRT, considering all alleles jointly, confirmed these findings and demonstrated the most significant difference in overall allele frequencies with NTG (P = 0.0041). The APOE allele frequencies did not significantly differ among POAG cases and controls when grouped according to age (Fig. 1).
Table 1.
APOE Allelic Association With POAG, HTG, and NTG
| NEIGHBOR | ||||||
|---|---|---|---|---|---|---|
| Allele | POAG(2120 Cases, 2262 Controls) | HTG(978 Cases, 2262 Controls) | NTG(395 Cases, 2262 Controls) | |||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| ε2 | 0.96 (0.83–1.13) | 0.65 | 0.94 (0.77–1.15) | 0.57 | 1.09 (0.83–1.42) | 0.52 |
| ε4 | 0.84 (0.74–0.96) | 0.0078 | 0.81 (0.68–0.97) | 0.013 | 0.69 (0.54–0.88) | 0.0036 |
| LRT | 0.021 | 0.03 | 0.0099 | |||
| MEEI | ||||||
| POAG(486 Cases, 344 Controls) | HTG(320 Cases, 344 Controls) | NTG(166 Cases, 344 Controls) | ||||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| ε2 | 1.08 (0.74–1.60) | 0.68 | 1.18 (0.77–1.79) | 0.45 | 0.94 (0.54–1.59) | 0.82 |
| ε4 | 0.81 (0.60–1.10) | 0.18 | 0.81 (0.58–1.13) | 0.22 | 0.81 (0.53–1.20) | 0.30 |
| LRT | 0.39 | 0.38 | 0.55 | |||
| Combined data set | ||||||
| POAG(2606 Cases, 2606 Controls) | HTG(1298 Cases, 2606 Controls) | NTG(561 Cases, 2606 Controls) | ||||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| ε2 | 0.97 (0.84–1.12) | 0.70 | 0.98 (0.81–1.17) | 0.80 | 1.01 (0.80–1.28) | 0.92 |
| ε4 | 0.83 (0.74–0.94) | 0.0022 | 0.81 (0.70–0.94) | 0.0052 | 0.71 (0.58–0.87) | 0.0014 |
| LRT | 0.0067 | 0.02 | 0.0041 | |||
Figure 1.
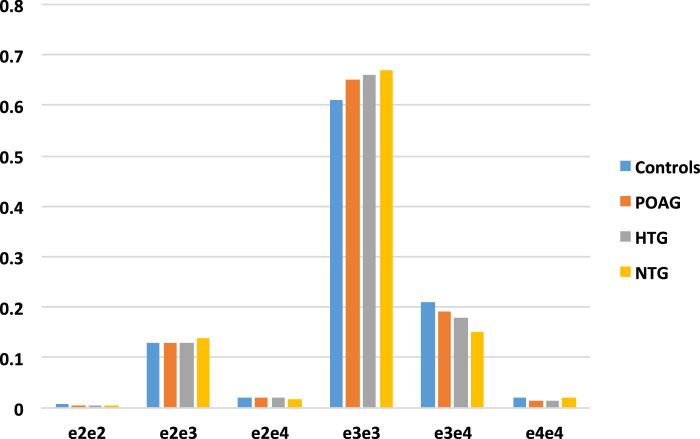
Distribution of APOE alleles according to age. Allele frequencies were not significantly different across age groups in either cases or controls (P > 0.2 from each of six pairwise comparisons).
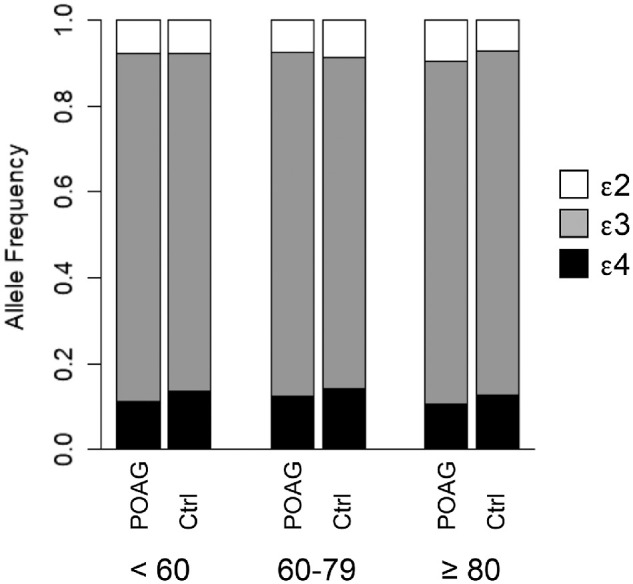
The distribution of APOE genotypes among POAG overall, HTG, and NTG cases and controls differed significantly, with the largest effect observed for the NTG case versus control comparison (P = 0.008; POAG, P = 0.02 and HTG, P = 0.085) (Fig. 2).
Figure 2.
APOE genotype distribution among POAG, HTG, and NTG cases and controls. P values for distribution compared to controls are POAG, 0.02; HTG, 0.085; and NTG, 0.008.
To determine whether the ε4 allele is associated with delayed age of disease onset, we compared the mean age of diagnosis for the ε3ε3 POAG cases with the mean age of diagnosis for the ε3ε4 POAG cases. While the mean (65.8 vs. 64.9 years) and median (67.3 vs. 65.6 years) ages of onset were indeed greater in the e3/e4 cases than in e3/e3 cases, the difference was not significant (P = 0.14 by t test, P = 0.13 by Wilcoxon rank-sum test), likely due to limited sample size (463 e3/e4 POAG cases).
TREM2 is known to interact with APOE, and rare TREM2 coding variants have been associated with AD.13–15 To determine whether any rare TREM2 coding variants are associated with POAG, we extracted NEIGHBOR and MEEI case-control association data for variants genotyped using the Human Exome bead array. Seven rare (MAF <1%) TREM2 missense variants were identified, and one variant (A105V) was identified in three HTG cases and in no controls (P = 0.03) (Table 2). Comparing the HTG allele frequency to the European Caucasian population in population database the Genome Aggregation Database v.2.1.1 (gnomAD)67 provided support for enrichment in HTG cases (P = 0.008). The remaining TREM2 variants, including variants known to contribute to AD risk, did not demonstrate comparable enrichment (P > 0.05).
Table 2.
TREM2 Rare Variants From the Human Exome Array
| rsID | Variant | CADD | PP | SIFT | gnomAD(MAF) | Controls(MAF) | POAG(MAF) | HTG(MAF) | NTG(MAF) | P *Case/Control | P † GnomAD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs200392967 | D39E | 23.4 | PS | D | 1.3E-4 | 1/6381 (1.5E-4) | 0/5628 (0.00) | 0/2866 (0.00) | 0/1158 (0.00) | 0.99 | 0.99 |
| rs143332484 | R62H‡ | 11.11 | B | T | 0.012 | 69/6347 (0.011) | 49/5623 (0.009) | 30/2836 (0.011) | 10/1148 (0.009) | 0.24 | 0.06 |
| rs142232675 | D87N‡ | 22.8 | PD | T | 0.002 | 4/6412 (6.2E-4) | 4/5668 (7.1E-4) | 0/2866 (0.00) | 1/1157 (8.6E-4) | 0.86 | 0.008 |
| rs145080901 | A105V | 24.2 | PD | D | 1.3E-4 | 0/6416 (0.00) | 3/5669 (5.3E-4) | 3/2863 (1.0E-3) | 0/1158 (0.00) | 0.07 (POAG)0.03 (HTG) | 0.05(POAG)0.008(HTG) |
| rs149622783 | R136Q | 1.841 | B | T | 1.3E-4 | 2/6414 (3.1E-4) | 3/5669 (5.3E-4) | 1/2865 (3.5E-4) | 1/1157 (8.6E-4) | 0.56 | 0.05 |
| rs79011726 | E151K | 23.2 | B | T | 1.8E-4 | 3/6413 (4.7E-4) | 1/5671 (1.8E-4) | 0/2866 (0.00) | 0/1158 (0.00) | 0.29 | 0.99 |
| rs2234255 | H157Y‡ | 23.1 | PS | D | 2.9E-4 | 3/6413 (4.7E-4) | 2/5670 (3.5E-4) | 0/2866 (0.00) | 1/1157 (8.6E-4) | 0.76 | 0.67 |
B, benign; CADD, combined annotation-dependent depletion score; D, damaging; PD, probably damaging; PP, polyphen2; PS, possibly damaging; SIFT, sorting intolerant from tolerant score; T, tolerated.
P value for logistic regression using cases and controls. Cases are POAG overall except for A105V, where the results are provided for both POAG overall and HTG.
Comparison of POAG cases to the European population distribution in GnomAD using the Fisher exact test. For A105V, the results are provided for both POAG and HTG.
Discussion
In this study, we have examined the genetic association of POAG with APOE and TREM2, two well-established risk factors for AD. We selected the APOE ε2, ε3, and ε4 alleles for evaluation because of the known and important contributions of these alleles to risk for other neurodegenerative diseases, specifically AD11,12 and age-related macular degeneration.49–51 Interestingly, we have found that APOE ε4, which is positively associated with AD, is inversely associated with POAG. This effect was most pronounced in NTG, consistent with APOE playing a direct role in regulating RGC degeneration rather than indirectly via IOP regulation. Although prior published reports on the association of APOE and glaucoma had small sample sizes and reported conflicting results, it is worth noting that two studies that also demonstrated a protective effect for APOE ε4 in glaucoma were larger than the others (with more than 300 enrolled participants) and also enriched in patients with NTG.33,34 Lam et al.34 demonstrated that APOE ε4 was protective in NTG patients of Chinese ancestry, while Mabuchi et al.33 found that ε4 was inversely associated with open-angle glaucoma in Japan, where the predominant form of glaucoma is the NTG subtype.61 We speculate that the strength of association noted between APOE ε4 and POAG will depend on the proportion of NTG patients enrolled in a given study, which may explain some of the variability in the published individual studies33–42 and meta-analyses.43–47
What is the mechanism by which APOE ε4 may be protective in glaucoma? APOE is expressed in a variety of cell types in the healthy retina and the optic nerve, including Müller glia and astrocytes.62 However, APOE is also upregulated in the neurodegeneration-associated microglia in the brain in a variety of neurodegenerative disease mouse models, including AD, ALS, and multiple sclerosis.23,24 Furthermore, APOE has been found to be a critical regulator of this microglial neurodegenerative phenotype that develops in response to apoptotic neurons.23 We speculate that APOE may be similarly upregulated in microglia in glaucoma, as microglial reactivity has been found to contribute to glaucoma pathogenesis.26–30
Another interesting parallel can be made between our findings and the association of APOE with AMD, where APOE ε4 has also been found to be inversely associated with disease.49–52 The reason why the same APOE allele has an opposing relationship in ocular neurodegenerative diseases and AD is presently poorly understood. A study by Levy et al.63 has shown that in a mouse model of AMD, mice with human APOE ε4 allele had lower levels of Monocyte chemoattractant protein-1 (MCP-1/CCL2) (a major monocyte attractant), less myeloid cell accumulation in the subretinal space, and decreased photoreceptor degeneration. Therefore, microglia with the APOE ε4 allele appear to be less reactive, which may be helpful in retinal neurodegenerations but harmful in AD, which is characterized by toxic Aβ plaques and tau deposits that need to be contained by the immune system. Furthermore, there could be additional mechanisms by which APOE plays a role in AD pathogenesis that are unrelated to its effect on the microglial transcriptional phenotype. For instance, APOE stimulates production of the Aβ precursor Amyloid precursor protein (APP) and thus directly contributes to Aβ plaque formation, with ε4 allele being most potent at producing this effect.64
We also investigated the association between glaucoma and TREM2, a transmembrane receptor expressed by myeloid cells that has recently been associated with AD.20–22 We have found that rare variants previously associated with AD interrogated by our exome chip (R62H,13,14 D87N,15 and H157Y14,16) were not associated with POAG, which further underscores different underlying pathogenic mechanisms in glaucoma and AD. Notably, unlike APOE ε4, TREM2 rare variants were also not associated with NTG. Although both APOE and TREM2 have been implicated in the regulation of the microglial neurodegeneration-associated phenotype, single-cell RNAseq analysis in a mouse model of AD has found that TREM2 and APOE may regulate different subpopulations of neurodegeneration-associated microglia.24 Whether TREM2 plays a role in regulating microglial molecular signature in the retina will merit further investigation.
An unexpected finding of our study was the association of one rare variant of TREM2, A105V, with HTG, although the number of cases was quite small. This variant has a high pathogenicity score based on in silico analyses, is not associated with AD or Nasu-Hakola disease, and is located in a different region of the TREM2 receptor than the AD-associated rare variants. Given the association with HTG only, we speculate that TREM2 may be involved in IOP regulation. In addition to microglia, TREM2 is also expressed by peripheral myeloid cells (monocytes and macrophages).18 Notably, macrophages are abundantly present in the conventional outflow pathway65 and have been implicated in IOP regulation after selective laser trabeculoplasty.66 Further research will be necessary to validate our genetic findings and explore the possible role of TREM2 in IOP regulation.
There were several limitations of our study. First, we do not have whole-exome sequencing data for TREM2, and therefore additional TREM2 rare variants may be associated with glaucoma that were not present on our chip. Second, although our data set is the largest of its kind, its statistical power for association of rare variants is limited. Finally, while we did show a consistent effect for APOE ε4 association with POAG in two independent data sets (NEIGHBOR and MEEI), additional data sets with a sufficient number of NTG cases are currently not available for further replication.
In summary, our results demonstrate, for the first time, that APOE ε4 association in glaucoma is strongest in the NTG subgroup and that the APOE ε4 allele is inversely associated with NTG and also with POAG overall. This result helps clarify the overall contribution of APOE to glaucoma risk and suggests that prior conflicting results could have reflected varying numbers of NTG cases in tested cohorts. Understanding the underlying mechanism by which APOE ε4 is involved in the pathogenesis of glaucoma will necessitate further study in animal glaucoma models (for instance, in APOE ε2, ε3, and ε4 humanized mice, which are commercially available). Interestingly, TREM2 rare variants associated with AD did not contribute to POAG risk in our study. The association between a different rare variant of TREM2, A105V, with HTG is intriguing but at this point preliminary and warrants further study and validation in larger data sets.
Acknowledgments
Presented at the Association for Research in Vision and Ophthalmology (ARVO) meeting in Vancouver, Canada, April 29th 2019.
Supported by MEE P30 (NIH/NEI EY014104); NIH/NEI R01 EY022305 (JLW); NIH/NEI K12 EY016335 (MAM); NIH/NEI K08 EY030160 (MAM); American Glaucoma Society Young Clinician Scientist Award (MAM); Research to Prevent Blindness Career Development Award (MAM); NIH/NEI R01 EY015473 (LRP); NIH/NINDS R01 NS088137 (OB), R21 NS104609 (OB), R21 NS101673 (OB), NIH/NIA R01 AG051812 (OB), and R01 AG054672 (OB); NIH/NEI R01 EY027921 (OB); National Multiple Sclerosis Society (5092A1) (OB); National Health and Medical Research Council Australia (RG180378) (OB); Nancy Davis Foundation Faculty Award (OB); Cure Alzheimer's Fund Award (OB); and Amyotrophic Lateral Sclerosis Association Award (OB).
Disclosure: M.A. Margeta, None; S.M. Letcher, None; R.P. Igo Jr., None; J.N. Cooke Bailey, None; L.R. Pasquale, Verily (C), Nicox (C), Emerald Bioscience (C), Eyenovia (C), and Bausch+Lomb (C); J.L. Haines, None; O. Butovsky, None; J.L. Wiggs, None
References
- 1. Almasieh M, Levin LA.. Neuroprotection in glaucoma: animal models and clinical trials. Annu Rev Vis Sci. 2017; 3: 91–120. [DOI] [PubMed] [Google Scholar]
- 2. Rezaie T, Child A, Hitchings R, et al.. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002; 295: 1077–1079. [DOI] [PubMed] [Google Scholar]
- 3. Kawase K, Allingham RR, Meguro A, et al.. Confirmation of TBK1 duplication in normal tension glaucoma. Exp Eye Res. 2012; 96: 178–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey JN, Loomis SJ, Kang JH, et al.. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016; 48: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maruyama H, Morino H, Ito H, et al.. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010; 465: 223–226. [DOI] [PubMed] [Google Scholar]
- 6. Cirulli ET, Lasseigne BN, Petrovski S, et al.. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015; 347: 1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elden AC, Kim HJ, Hart MP, et al.. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010; 466: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee T, Li YR, Ingre C, et al.. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet. 2011; 20: 1697–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardy J, Selkoe DJ.. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002; 297: 353–356. [DOI] [PubMed] [Google Scholar]
- 10. Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. Sci Transl Med. 2011; 3: 77sr71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corder EH, Saunders AM, Strittmatter WJ, et al.. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993; 261: 921–923. [DOI] [PubMed] [Google Scholar]
- 12. Corder EH, Saunders AM, Risch NJ, et al.. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994; 7: 180–184. [DOI] [PubMed] [Google Scholar]
- 13. Jin SC, Benitez BA, Karch CM, et al.. Coding variants in TREM2 increase risk for Alzheimer's disease. Hum Mol Genet. 2014; 23: 5838–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sims R, van der Lee SJ, Naj AC, et al.. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat Genet. 2017; 49: 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guerreiro R, Wojtas A, Bras J, et al.. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013; 368: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang T, Hou JK, Gao Q, et al.. TREM2 p.H157Y variant and the risk of Alzheimer's disease: a meta-analysis involving 14,510 subjects. Curr Neurovasc Res. 2016; 13: 318–320. [DOI] [PubMed] [Google Scholar]
- 17. Paloneva J, Manninen T, Christman G, et al.. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002; 71: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003; 3: 445–453. [DOI] [PubMed] [Google Scholar]
- 19. Jay TR, Miller CM, Cheng PJ, et al.. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer's disease mouse models. J Exp Med. 2015; 212: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeh FL, Hansen DV, Sheng M. TREM2, microglia, and neurodegenerative diseases. Trends Mol Med. 2017; 23: 512–533. [DOI] [PubMed] [Google Scholar]
- 21. Ulrich JD, Ulland TK, Colonna M, Holtzman DM. Elucidating the role of TREM2 in Alzheimer's disease. Neuron. 2017; 94: 237–248. [DOI] [PubMed] [Google Scholar]
- 22. Colonna M, Wang Y. TREM2 variants: new keys to decipher Alzheimer disease pathogenesis. Nat Rev Neurosci. 2016; 17: 201–207. [DOI] [PubMed] [Google Scholar]
- 23. Krasemann S, Madore C, Cialic R, et al.. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017; 47: 566–581e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keren-Shaul H, Spinrad A, Weiner A, et al.. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017; 169: 1276–1290e1217. [DOI] [PubMed] [Google Scholar]
- 25. Butovsky O, Weiner HL.. Microglial signatures and their role in health and disease. Nat Rev Neurosci. 2018; 19: 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neufeld AH. Microglia in the optic nerve head and the region of parapapillary chorioretinal atrophy in glaucoma. Arch Ophthalmol. 1999; 117: 1050–1056. [DOI] [PubMed] [Google Scholar]
- 27. Yuan L, Neufeld AH.. Activated microglia in the human glaucomatous optic nerve head. J Neurosci Res. 2001; 64: 523–532. [DOI] [PubMed] [Google Scholar]
- 28. Bosco A, Steele MR, Vetter ML. Early microglia activation in a mouse model of chronic glaucoma. J Comp Neurol. 2011; 519: 599–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chidlow G, Ebneter A, Wood JP, Casson RJ. Evidence supporting an association between expression of major histocompatibility complex II by microglia and optic nerve degeneration during experimental glaucoma. J Glaucoma. 2016; 25: 681–691. [DOI] [PubMed] [Google Scholar]
- 30. Bosco A, Inman DM, Steele MR, et al.. Reduced retina microglial activation and improved optic nerve integrity with minocycline treatment in the DBA/2J mouse model of glaucoma. Invest Ophthalmol Vis Sci. 2008; 49: 1437–1446. [DOI] [PubMed] [Google Scholar]
- 31. Mirzaei M, Gupta VB, Chick JM, et al.. Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci Rep. 2017; 7: 12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Inoue T, Kawaji T, Tanihara H. Elevated levels of multiple biomarkers of Alzheimer's disease in the aqueous humor of eyes with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2013; 54: 5353–5358. [DOI] [PubMed] [Google Scholar]
- 33. Mabuchi F, Tang S, Ando D, et al.. The apolipoprotein E gene polymorphism is associated with open angle glaucoma in the Japanese population. Mol Vis. 2005; 11: 609–612. [PubMed] [Google Scholar]
- 34. Lam CY, Fan BJ, Wang DY, et al.. Association of apolipoprotein E polymorphisms with normal tension glaucoma in a Chinese population. J Glaucoma. 2006; 15: 218–222. [DOI] [PubMed] [Google Scholar]
- 35. Al-Dabbagh NM, Al-Dohayan N, Arfin M, Tariq M. Apolipoprotein E polymorphisms and primary glaucoma in Saudis. Mol Vis. 2009; 15: 912–919. [PMC free article] [PubMed] [Google Scholar]
- 36. Vickers JC, Craig JE, Stankovich J, et al.. The apolipoprotein epsilon4 gene is associated with elevated risk of normal tension glaucoma. Mol Vis. 2002; 8: 389–393. [PubMed] [Google Scholar]
- 37. Junemann A, Bleich S, Reulbach U, et al.. Prospective case control study on genetic association of apolipoprotein epsilon2 with intraocular pressure. Br J Ophthalmol. 2004; 88: 581–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ressiniotis T, Griffiths PG, Birch M, Keers S, Chinnery PF. The role of apolipoprotein E gene polymorphisms in primary open-angle glaucoma. Arch Ophthalmol. 2004; 122: 258–261. [DOI] [PubMed] [Google Scholar]
- 39. Lake S, Liverani E, Desai M, et al.. Normal tension glaucoma is not associated with the common apolipoprotein E gene polymorphisms. Br J Ophthalmol. 2004; 88: 491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zetterberg M, Tasa G, Palmer MS, et al.. Apolipoprotein E polymorphisms in patients with primary open-angle glaucoma. Am J Ophthalmol. 2007; 143: 1059–1060. [DOI] [PubMed] [Google Scholar]
- 41. Saglar E, Yucel D, Bozkurt B, Ozgul RK, Irkec M, Ogus A. Association of polymorphisms in APOE, p53, and p21 with primary open-angle glaucoma in Turkish patients. Mol Vis. 2009; 15: 1270–1276. [PMC free article] [PubMed] [Google Scholar]
- 42. Yaylacioglu Tuncay F, Aktas Z, Ergun MA, Ergun SG, Hasanreisoglu M, Hasanreisoglu B. Association of polymorphisms in APOE and LOXL1 with pseudoexfoliation syndrome and pseudoexfoliation glaucoma in a Turkish population. Ophthalmic Genet. 2017; 38: 95–97. [DOI] [PubMed] [Google Scholar]
- 43. Song Q, Chen P, Liu Q. Role of the APOE epsilon2/epsilon3/epsilon4 polymorphism in the development of primary open-angle glaucoma: evidence from a comprehensive meta-analysis. PLoS One. 2013; 8: e82347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang W, Zhou M, Huang W, Chen S, Zhang X. Lack of association of apolipoprotein E (Apo E) epsilon2/epsilon3/epsilon4 polymorphisms with primary open-angle glaucoma: a meta-analysis from 1916 cases and 1756 controls. PLoS One. 2013; 8: e72644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Zhou YF, Zhao BY, Gu ZY, Li SL. Apolipoprotein E gene epsilon4epsilon4 is associated with elevated risk of primary open angle glaucoma in Asians: a meta-analysis. BMC Med Genet. 2014; 15: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liao R, Ye M, Xu X. An updated meta-analysis: apolipoprotein E genotypes and risk of primary open-angle glaucoma. Mol Vis. 2014; 20: 1025–1036. [PMC free article] [PubMed] [Google Scholar]
- 47. Chen M, Yu X, Xu J, et al.. Association of gene polymorphisms with primary open angle glaucoma: a systematic review and meta-analysis. Invest Ophthalmol Vis Sci. 2019; 60: 1105–1121. [DOI] [PubMed] [Google Scholar]
- 48. Xiao H, Gao Y, Liu L, Li Y. Association between polymorphisms in the promoter region of the apolipoprotein E (APOE) gene and Alzheimer's disease: a meta-analysis. EXCLI J. 2017; 16: 921–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiying M, Wenbo W, Wangyi F, Qinghuai L. Association of Apolipoprotein E polymorphisms with age-related macular degeneration subtypes: an updated systematic review and meta-analysis. Arch Med Res. 2017; 48: 370–377. [DOI] [PubMed] [Google Scholar]
- 50. Schmidt S, Klaver C, Saunders A, et al.. A pooled case-control study of the apolipoprotein E (APOE) gene in age-related maculopathy. Ophthalmic Genet. 2002; 23: 209–223. [DOI] [PubMed] [Google Scholar]
- 51. Baird PN, Guida E, Chu DT, Vu HT, Guymer RH. The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004; 45: 1311–1315. [DOI] [PubMed] [Google Scholar]
- 52. McKay GJ, Patterson CC, Chakravarthy U, et al.. Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum Mutat. 2011; 32: 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiggs JL, Kang JH, Yaspan BL, et al.. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011; 20: 4707–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wiggs JL, Yaspan BL, Hauser MA, et al.. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012; 8: e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCarthy S, Das S, Kretzschmar W, et al.. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016; 48: 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Das S, Forer L, Schonherr S, et al.. Next-generation genotype imputation service and methods. Nat Genet. 2016; 48: 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010; 26: 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen CY, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013; 29: 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 61. Iwase A, Suzuki Y, Araie M, et al.. The prevalence of primary open-angle glaucoma in Japanese: the Tajimi Study. Ophthalmology. 2004; 111: 1641–1648. [DOI] [PubMed] [Google Scholar]
- 62. Omodaka K, Nishiguchi KM, Yasuda M, et al.. Neuroprotective effect against axonal damage-induced retinal ganglion cell death in apolipoprotein E-deficient mice through the suppression of kainate receptor signaling. Brain Res. 2014; 1586: 203–212. [DOI] [PubMed] [Google Scholar]
- 63. Levy O, Lavalette S, Hu SJ, et al.. APOE isoforms control pathogenic subretinal inflammation in age-related macular degeneration. J Neurosci. 2015; 35: 13568–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang YA, Zhou B, Wernig M, Sudhof TC. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Abeta secretion. Cell. 2017; 168: 427–441 e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Margeta MA, Lad EM, Proia AD. CD163+ macrophages infiltrate axon bundles of postmortem optic nerves with glaucoma. Graefes Arch Clin Exp Ophthalmol. 2018; 256: 2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alvarado JA, Katz LJ, Trivedi S, Shifera AS. Monocyte modulation of aqueous outflow and recruitment to the trabecular meshwork following selective laser trabeculoplasty. Arch Ophthalmol. 2010; 128: 731–737. [DOI] [PubMed] [Google Scholar]
- 67. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, et al.. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020; 581: 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]