Abstract
Purpose:
Cadmium (Cd) is reported to be associated with carcinogenesis. The molecular mechanisms associated with Cd-induced prostate cancer (PCa) remain elusive.
Materials and Methods:
RWPE1, PWR1E and DU 145 cells were used. RT2 Profiler array, real-time-quantitative-PCR, immunofluorescence, cell cycle, apoptosis, proliferation and colony formation assays along with Gene Set Enrichment Analysis (GSEA) were performed.
Results:
Chronic Cd exposure of non-malignant RWPE1 and PWR1E cells promoted cell survival, proliferation and colony formation with inhibition of apoptosis. Even a two-week Cd exposure of PCa cell line (DU 145) significantly increased the proliferation and decreased apoptosis. RT2 profiler array of 84 genes involved in the Erk/MAPK pathway revealed induction of gene expression in Cd-RWPE1 cells compared to RWPE1. This was confirmed by individual TaqMan gene expression analysis in both Cd-RWPE1 and Cd-PWR1E cell lines. GSEA showed an enrichment of the Erk/MAPK pathway along with other pathways such as KEGG-ERBB, KEGG-Cell Cycle, KEGG-VEGF, KEGG-Pathways in cancer and KEGG-prostate cancer pathway. We randomly selected upregulated genes from Erk/MAPK pathway and performed profile analysis in a PCa data set from the TCGA/GDC data base. We observed upregulation of these genes in PCa compared to normal samples. An increase in phosphorylation of the Erk1/2 and Mek1/2 was observed in Cd-RWPE1 and Cd-PWR1E cells compared to parental cells, confirming that Cd-exposure induces activation of the Erk/MAPK pathway.
Conclusion:
This study demonstrates that Erk/MAPK signaling is a major pathway involved in Cd-induced malignant transformation of normal prostate cells. Understanding these dominant oncogenic pathways may help develop optimal therapeutic strategies for PCa.
Keywords: Cadmium toxicity, prostate cancer, Erk/MAPK pathway
Introduction
Cadmium (Cd) occurs naturally in the earth’s crust at a concentration of 0.1–0.5 ppm and is commonly associated with zinc, lead, and copper ores(Bako et al., 1982; van der Gulden et al., 1995; Chan and Giovannucci, 2001; Amzal et al., 2009; Luevano and Damodaran, 2014; Rani et al., 2014; Byber et al., 2016; Mezynska and Brzoska, 2018). It is also a natural constituent of ocean water with average levels between <5 and 110 ng/L. Higher levels have been reported near coastal areas and in marine phosphates and phosphorites. Cd is widely distributed in the environment as a result of anthropogenic activity and is currently one of the most extensive toxic pollutants of occupational and environmental concern (Bako et al., 1982; van der Gulden et al., 1995; Chan and Giovannucci, 2001; Amzal et al., 2009; Luevano and Damodaran, 2014; Rani et al., 2014; Byber et al., 2016; Mezynska and Brzoska, 2018). The most dangerous characteristic of Cd is its life time accumulation due to its elimination half-life of 10–30 years (Bako et al., 1982; van der Gulden et al., 1995; Chan and Giovannucci, 2001; Amzal et al., 2009; Luevano and Damodaran, 2014; Rani et al., 2014; Byber et al., 2016; Mezynska and Brzoska, 2018). For non-occupationally exposed populations, Cd exposure normally results from tobacco consumption and diet choices. Cd is selectively taken up from soil by certain edible food items such as grains, potatoes and vegetables (Amzal et al., 2009). Occupational Cd exposure occurs when dust and fumes are inhaled in Cd emitting industries and metal mines (NTP, 2016). Following absorption by either the lung or intestinal epithelium, Cd enters the systemic circulation and exists as a mixture of free cations and metal compounds in blood (Zalups and Ahmad, 2003). Cd exposure is associated with diverse toxic effects including nephrotoxicity, teratogenicity, endocrine, reproductive and cardiovascular toxicities (Rani et al., 2014; Byber et al., 2016; Jacobo-Estrada et al., 2017). Cd has been implicated in cancer development and has been classified as a type I carcinogen by the International Agency for Cancer Research (IARC, 1993; Giaginis et al., 2006). Some epidemiological studies have demonstrated significant correlation between Cd exposure and prostate cancer risk (Ju-Kun et al., 2016).
Prostate cancer is the most frequent malignancy in men, with an estimated 191,930 new cases in the USA in 2020 (Siegel et al., 2020). In addition, prostate cancer is the second most fatal cancer in men after lung and bronchus tumors, resulting in 33,330 estimated deaths in USA during 2020 (Siegel et al., 2020) The number of identified cases of prostate cancer has substantially increased since the introduction of serum PSA screening and the development of prostate biopsy in the late 1980s (Potosky et al., 1995). Prostate cancer is generally a slowly progressing disease and, in many instances, will not progress from an indolent to an aggressive stage. Hence, active surveillance has emerged as an alternative to immediate treatment for men with low-risk disease. This management strategy is intended to decrease overtreatment and its related adverse effects but has raised the complex issue of patient selection. In this evolving landscape, the identification of environmental factors that might promote prostate cancer progression towards more aggressive stages has received much interest. The etiology of prostate cancer development has been found to be associated with a multitude of causative risk factors that include obesity, androgen, dietary fat, exposure to trace elements and cadmium (Chan and Giovannucci, 2001; Venkateswaran and Klotz, 2010; Allott et al., 2013). Several epidemiological and meta-analysis reports suggest a positive association between Cd exposure and prostate cancer risk (Bako et al., 1982; Elinder et al., 1985; Garcia Sanchez et al., 1992; van der Gulden et al., 1995; Vinceti et al., 2007; Julin et al., 2012; Cheung et al., 2014; Ju-Kun et al., 2016; Rapisarda et al., 2018). Several mechanisms have been reported to be responsible for Cd carcinogenesis including, induction of oxidative stress (Valverde et al., 2001), autophagy (Pal et al., 2017), suppression of DNA repair (Hartwig and Schwerdtle, 2002; Lutzen et al., 2004), alteration of DNA methylation (Pierron et al., 2014) inhibition of apoptosis and protooncogene activation (Asara et al., 2013), inactivation of tumor suppressor genes and cell adhesion disruption (Hartwig, 2010) and recently by induction of zinc finger of the cerebellum 2 (ZIC2) expression in benign prostatic hyperplasia 1 (BPH1) cells (Chandrasekaran et al., 2020). Long term exposure of nonmalignant human prostate epithelial cells (RPWE-1) to Cd in culture transformed them to malignant phenotypes (Achanzar et al., 2001; Luevano and Damodaran, 2014). However, the molecular mechanisms associated with malignant transformation of the normal prostate epithelial cells remain elusive.
The present study was undertaken to investigate the molecular mechanisms of Cd induced tumorigenicity in non-malignant prostate epithelial cells (RWPE1 and PWR1E). We exposed these cells to 10uM Cd in culture for almost a year following a previously published report (Achanzar et al., 2001). These transformed cells were named as Cd-RWPE1 and CdPWR1E. We performed a detailed analysis of the Erk/MAPK signaling pathway in these cells compared to their parental cells. This study provides evidence that the Erk/MAPK signaling is one of the dominant molecular fingerprint for the carcinogenic mode of action of Cd in inducing prostate cancer.
Materials and Methods
Cell lines and culture
Human normal prostate epithelial cells RWPE1 and PWR1E and prostate cancer cell line DU 145 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). These human-derived cell lines were authenticated by DNA short-tandem repeat analysis by ATCC. RWPE1 and PWR1E cells were cultured in keratinocyte growth medium supplemented with 5 ng/mL human recombinant epidermal growth factor, 0.05 mg/mL bovine pituitary extract (Gibco/Invitrogen, Carlsbad, CA), 1x antibiotic/antimycotic solution. The prostate cancer cell line DU 145 was cultured as a monolayer in RPMI medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 50 ug/mL penicillin, 50 ug/mL streptomycin (Invitrogen, Carlsbad, CA). All cell lines were maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37˚C.
Chemicals, reagents and antibodies
CdCl2 was obtained from Sigma Chemical Co., St. Louis, MO. cDNA synthesis kits were purchased from Biorad. For real-time quantitative PCR (qRT-PCR) analysis, TaqMan primer assays and kits were purchased from ThermoFisher Scientific Inc. South San Francisco, CA. Erk/MAPK pathway array (Cat. # PAHS-061Z), a first strand cDNA synthesis kit and reagents for the array were obtained from Qiagen, Redwood city, CA. Antibodies for phosphor-Mek 1/2 (CST #9154P), phospho-p44/42 (CST #4370P) with anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor® 488 Conjugate) as secondary antibody (CST #4412) were used.
RNA extraction
Total RNA was extracted from confluent (70–80%) plates of cultured cells using a combination of TRIzol reagent (Invitrogen) and RNeasy columns (Qiagen, Redwood city, CA). On-column DNase digestion was also performed with RNase-Free DNase set (Qiagen, Redwood city, CA). RNA quality and concentration were assessed using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmingon, DE) spectrophotometer and were electrophoresed in agarose gels to check integrity. Extracted RNA was stored at –80°C.
RT2 profiler PCR array analysis
Expression profiling of the Erk/MAPK signaling pathway was performed using human RT2 profiler PCR array PAHS-061Z (Qiagen) based on SYBR-Green real time PCR. Three biological replicates were prepared from RWPE1 and Cd-RWPE1 cells and the isolated RNA from these replicates were pooled. cDNA was synthesized from the pooled RNA using RT2 first strand kit following the manufacturer’s instructions. SYBR-Green real-time PCR was performed and relative quantification changes in expression were measured by obtaining the threshold cycle. Data from the array was analyzed using the GeneGlobe Data Analysis Center (Qiagen). The real-time PCR module transforms threshold cycle (CT) values to calculated results for relative fold changes in expression normalized to housekeeping genes. The array includes a total of 84 Erk/MAPK pathway genes (Table 1), 5 housekeeping genes, an assay for determining genomic DNA contamination and three assays each for positive and negative PCR controls.
Table 1:
Differential expression of the Erk/MAPK pathway genes in qRT2 PCR array.
| Upregulated Genes: |
| Position | Gene Symbol | Fold Change |
|---|---|---|
| A08 | CCND1 | 2.61 |
| A10 | CCND3 | 2.6 |
| A11 | CCNE1 | 6.72 |
| B04 | CDKN1A | 3.73 |
| B05 | CDKN1B | 2.49 |
| B08 | CDKN2B | 2.88 |
| B10 | CDKN2D | 5.03 |
| C04 | E2F1 | 2.14 |
| C08 | ETS1 | 2.8 |
| C09 | ETS2 | 2.01 |
| D03 | JUN | 3.05 |
| D04 | KRAS | 2.51 |
| D05 | KSR1 | 2.79 |
| D07 | MAP2K1 | 2.01 |
| D09 | MAP2K3 | 2.86 |
| D12 | MAP2K6 | 2.5 |
| E02 | MAP3K1 | 2.09 |
| E03 | MAP3K2 | 2.07 |
| E11 | MAPK13 | 3.07 |
| E12 | MAPK14 | 2.41 |
| F03 | MAPK7 | 3.28 |
| F09 | MAX | 2.64 |
| G02 | MYC | 2.47 |
| G03 | NFATC4 | 5.5 |
| G06 | PRDX6 | 2.02 |
| G08 | RAF1 | 2.76 |
| G09 | RB1 | 2.06 |
| G12 | TP53 | 3.41 |
| Down-regulated Genes: |
| Position | Gene Symbol | Fold Change |
|---|---|---|
| E08 | MAPK10 | −3.32 |
Quantitative real time PCR
First strand cDNA was prepared from total RNA (1 ug) using the iScript Reverse Transcription Supermix (Biorad Lab. Inc., Hercules, CA). For real-time PCR, cDNA was amplified with Inventoried Gene Assay Products containing two gene-specific primers and one TaqMan MGB probe (6-FAM dye-labeled) using the TaqMan Universal Fast PCR Master Mix in a 7500 Fast Real Time PCR System (Applied Biosystems). Thermal cycling conditions included 95˚C for 20 seconds, 40 cycles of 95˚C for 3 seconds, and 60˚C for 30 seconds according to the TaqMan Fast Universal PCR Protocol. GAPDH was used as an endogenous control and vehicle control was used as a calibrator. The comparative Ct method was used to calculate the relative changes in gene expression in the 7500 Fast Real Time PCR System. The relative changes of gene expression were calculated using the following formula: Fold change in gene expression, 2-ΔΔCt = 2-where ΔCt = Ct (detected genes) – Ct (GAPDH) and Ct represents threshold cycle number. The experiments were repeated at least three times (biological replicates) with three technical replicates each times.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 15 minutes. Prior to overnight incubation with 1:100 fold diluted primary antibody, cells were blocked with blocking buffer (1× PBS/5% normal goat serum/0.3% Triton X-100) for 1 hour. After washing with PBS, cells were treated with 1:100 fold diluted secondary antibody for 2 hours and counterstained with 0.5 μg/mL of 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes. Cells were then mounted using Prolong Gold Antifade reagent and images were captured using Zeiss microscope (model: Axio Imager.D2). The primary antibodies used are pMek 1/2 (CST #9154P), phospho-p44/42 (CST #4370P) with anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor® 488 Conjugate) as secondary antibody (CST #4412).
Cell cycle, proliferation, colony formation and apoptosis assays
Fluorescence-activated cell sorting (FACS) analysis was done to test the effect of Cd exposure on cell cycle distribution and apoptosis of all cell lines. Equal numbers of cells were seeded and cultured for 24 hours. Cells were then harvested, washed with cold PBS, and stained with propidium iodide (PI-A) and Annexin-V-FITC using FITC-ANNEXIN V-PI-A KIT (BD Biosciences) for apoptosis and nuclear stain 4′,6-diamidino-2-phenylindole (DAPI) (BD Biosciences) for cell cycle analysis according to the manufacturer’s protocol. Stained cells were immediately analyzed by FACS (BD FACSVerse; BD Biosciences). Cell viability/proliferation was determined by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol. For cell viability assay, cells were seeded in 96-well microplates at a density of 5×103 cells per well. After 24 hours of culture, cell viability was determined by adding 20ul/well of the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol. Absorbance at 490 nm was measured with a kinetic microplate reader (Spectra MAX 190; Molecular Devices Co., Sunnyvale, CA).
Human mRNA expression data
The expression levels of mRNAs in a human prostate adenocarcinoma sample cohort (n=534) were obtained from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). Human PCa (n=483) and normal (n=51) tissue cohorts from databases were analyzed using Mann–Whitney U-test after gene expression data was normalized by scaling the values of reads per million in order to eliminate bias of the differences between individual patients.
Gene set enrichment analysis
To compare the gene expression signatures between RWPE1 and Cd-RWPE1 cells, the expression data from QIAGEN qPCR array analysis was subjected to Broad Institute, “Gene Set Enrichment Analysis (GSEA) version 3.0”. The enrichment score (ES) and normalized enrichment score (NES) were computed by using parental RWPE1 as control.
Statistical analysis
Gene Globe data analysis software (Qiagen) was used to analyze the Erk/MAPK pathway gene expression profile in Cd-RWPE1 compared to RWPE1 samples. Three biological replicates for each group were pooled together to perform the qPCR RT2 array analysis. The CT cut-off was set to 35. Fold regulation cut off was set to 2. Fold-Change (2^ (- Delta Delta CT)) is the normalized gene expression (2^(- Delta CT)) in the Cd-RWPE1 sample divided by the normalized gene expression (2^ (- Delta CT)) in the parental RWPE1 sample that served as the control. The scatter plot was used to compare the normalized expression of every gene on the array between the Cd-RWPE1 and RWPE1 samples and the visualization was represented by a heat map. Statistical analyses were performed with GraphPad Prism 5 and/or MedCalc version 10.3.2. All quantified data represents an average of at least triplicate samples or as indicated. Error bars represent standard deviation or standard error of the mean as indicated in figure legends. All tests were performed two tailed and p-values <0.05 were considered statistically significant.
Results
Cadmium exposure modulates Erk/MAPK pathway genes
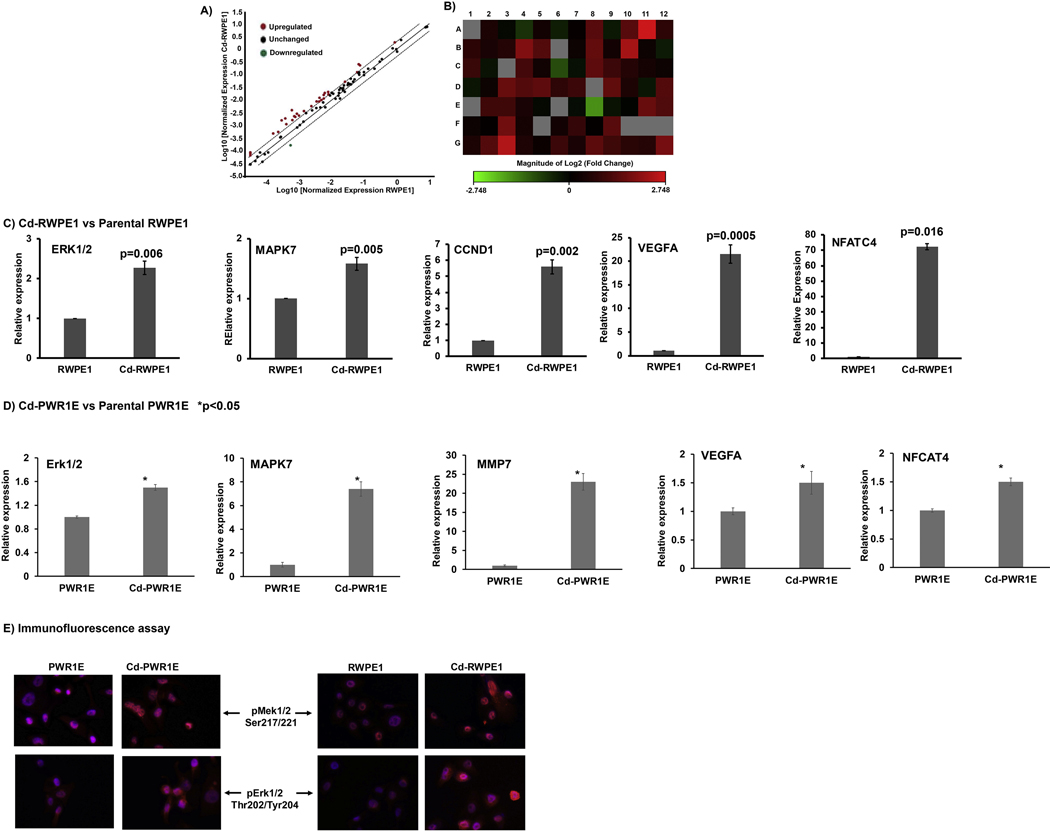
We exposed two non-malignant human prostate epithelial cells (RWPE1 and PWR1E) cells to 10uM Cd in culture for almost a year following a previously published report (Achanzar et al., 2001). Cd exposed cells Cd-RWPE1 and Cd-PWR1E showed increased cell growth and proliferation compared to normal parental controls (RWPE1 and PWR1E). Since ERK/MAPK pathway is the major signaling pathway involved in cell growth and proliferation, we sought to investigate this pathway in detail as a molecular driver of Cd-induced malignant transformation of normal prostate cells to prostate cancer. We performed ERK/MAPK pathway specific RT2 profiler array analysis. The Erk/MAPK array includes a total of 84 genes (Supplemental Table 1), 5 housekeeping genes, one assay for determining genomic DNA contamination along with 3 assays each for positive and negative PCR controls. For the analysis the Ct cut-off was set to 35. The scatter plot analysis showed that many genes in the Erk/MAPK pathway are upregulated in Cd-RPWE1 cells when compared to normal RWPE1 cells (Figure 1A). Relative fold change in expression normalized to housekeeping genes indicated that most of these genes were upregulated in Cd-RWPE1 cells compared to normal RWPE1. The cut-off for fold change was 2. The relative fold upregulation ranged from 2 to 7 fold (Table 1). Among the genes only MAPK10, a proapoptotic gene (Ying et al., 2006), was found to be downregulated (3.32 fold). Heat map shows the overall fold-regulation of genes involved in the Erk/MAPK signaling pathway (Figure 1B). All gene names for the heat map and relative fold regulation along with array location are shown in Supplemental Table 2.
Figure 1.
PCR array analysis of the Erk/MAPK signaling pathway. Scatter plot comparison of normalized gene expression between RWPE1 and Cd-RWPE1 cells. The central line indicates unchanged gene expression. Data points in red are the upregulated genes and in green are downregulated genes of the Erk/MAPK pathway (A). Heat Map visualization of the fold changes in expression in 84 genes of the Erk/MAPK pathway in Cd-RWPE1 compared to RWPE1 cells (B). The gene names and fold regulation along with the location on array are indicated in Supplemental Table 2. Quantitative real-time PCR analysis of selected Erk/MAPK pathway genes utilizing TaqMan gene expression assays in Cd-RWPE1 vs. parental normal RWPE1 (C) and Cd-PWR1E vs. parental normal PWR1E (D). Immunofluorescence assay (E). Error bars ±SD.
Confirmation of array results by real-time quantitative PCR analysis
To confirm the array results, we performed qRT-PCR analysis for selected genes using the TaqMan gene expression assay system. Consistent with the array results, expression of genes such as Erk1 (p=0.006), MAPK7 (p=0.005) and CCND1 (p=0.002) were significantly higher in Cd-RWPE1 compared to the normal RWPE1 cells (Figure 1C). We further examined these genes in Cd-PWR1E compared to parental PWR1E cells. The results were consistent with that of Cd-RWPE1 cells (Fig. 1D). GAPDH was used as the internal control for qRT-PCR.
Cd exposure induces activation of Erk1/2 and Mek1/2
Phosphorylation of the Erk (p44/42) and Mek 1/2 represents the activation of the Erk/MAPK pathway. Hence, we performed immunofluorescence assay to determine the phospho-Mek 1/2 and phospho-Erk 1/2 at protein level in situ. Our results show that phosphorylation of both proteins increased in Cd-RWPE1 and Cd-PWR1E cells compared to normal parental RWPE1 and PWR1E cells (Fig. 1E).
Functional implication of Cd exposure in normal prostate epithelial cells
The Erk/MAPK signaling pathway is involved in the proliferation and survival of cancer cells. We observed an increase in pro-survival genes in both the array and qRT-PCR results. Thus, we sought to determine the effect of Cd exposure on cell proliferation, colony formation, cell cycle distribution and apoptosis in Cd-RWPE1 and Cd-PWR1E cells compared to their normal parental cells RPWE1 and PWR1E.
Cell Proliferation and colony formation
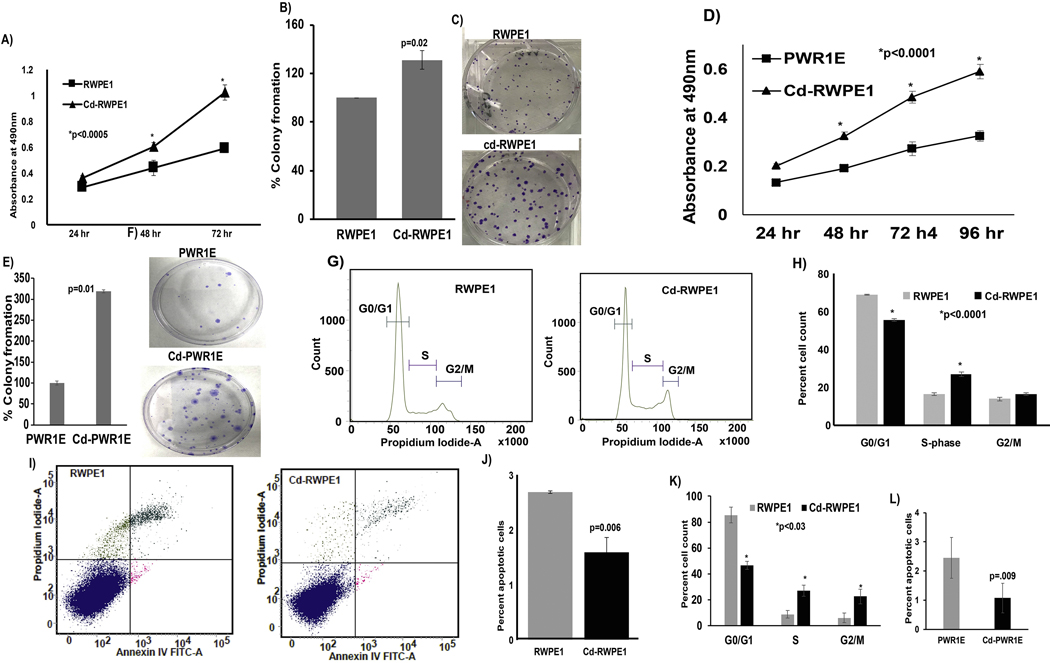
We observed that Cd-RWPE1 cells had a significant higher proliferation rate (p<0.0005) after seeding at 24, 48 or 72 hours compared to normal RWPE1 cells (Figure 2A). Cadmium exposure significantly induced colony formation (p=0.02) in Cd-RWPE1 cells compared to parental RWPE1 cells (Figure 2B–C). Similar results were observed in Cd-PWR1E cells with significantly increased proliferation (p<0.0001; Fig. 2D) and colony formation capability (p=0.01; Fig. 2E–F) compared to parental PWR1E. These results indicate the pro-survival effect of Cd exposure in normal prostate epithelial cells.
Figure 2.
Functional implication of Cd exposure in normal prostate epithelial cells. Cell proliferation assay shows increased proliferation and colony formation in Cd-RWPE1 compared to parental RWPE1 cells (A; B-C) and in Cd-PWR1E vs. parental normal PWR1E (D; E-F). FACS analysis for cell cycle distribution showed an increase in the S-phase population (C-D) and a decrease in the apoptotic cell fraction (E-F) in Cd-RWPE1 compared to parental RWPE1 cells. Similar effects were observed in Cd-PWR1E vs. normal parental PWR1E (K-L). Error bars ±SD.
Cell cycle and apoptosis analyses
Fluorescence-activated cell sorting (FACS) analysis revealed significantly higher number of Cd-RWPE1 cells (27%) in the proliferative S-phase of the cell cycle compared to parental RWPE1 cells (16%) (Figure 2G–H). This observation is consistent with significantly less Cd-RWPE1 cells (55%) in the G0/G1 phase compared to RWPE1 (69%), indicating that more Cd-RWPE1 cells had entered the actively dividing S-phase (Figure 2G–H). These results are consistent with the array gene expression results that showed up-regulation of cyclins (CCNE1, CCND3, CCND1) and cyclin dependent kinases (CDK2, CDK4, CDK6) (Table 1). FACS analysis for apoptosis was performed using an FITC-Annexin-V-propidium iodide-A (PI-A) kit. Viable cells (lower left quadrant) are FITC Annexin V and PI negative. Early apoptotic cells (lower right quadrant) are FITC Annexin V positive and PI negative. Late apoptotic cells (upper right quadrant) and necrotic (upper right quadrant) are Annexin V and PI positive. The movement of cells through these four stages provides a measure of apoptosis. The percentage of total apoptotic cells (early apoptotic + late apoptotic) in parental RWPE1 was double (~3%) that of Cd-RWPE1 cells (~1.5%) (Figure 2I–J). Consistent results were obtained in Cd-PWR1E cells compared to parental PWR1E (Fig 2K–L). These results indicate that Cd exposure decreased the capability of normal prostate epithelial cells to undergo apoptosis. We further examined the effect of two-week Cd exposure on prostate cancer cell line DU 145. Interestingly, Cd-exposed DU 145 (Cd-DU 145) showed increased proliferation (Supplemental Fig. 1A) and less apoptotic cell fraction (Supplemental Fig. 1B) compared to parental DU 145 cells. Disruption of apoptosis has been shown to play major role in tumor formation and malignant progression (Wyllie et al., 1999).
The above functional experiments indicate that increased proliferation and decreased apoptosis are key aspects of cadmium-induced malignant transformation of normal prostate epithelial cells RWPE1 and PWR1E.
Characterization of gene expression by gene set enrichment analysis (GSEA)
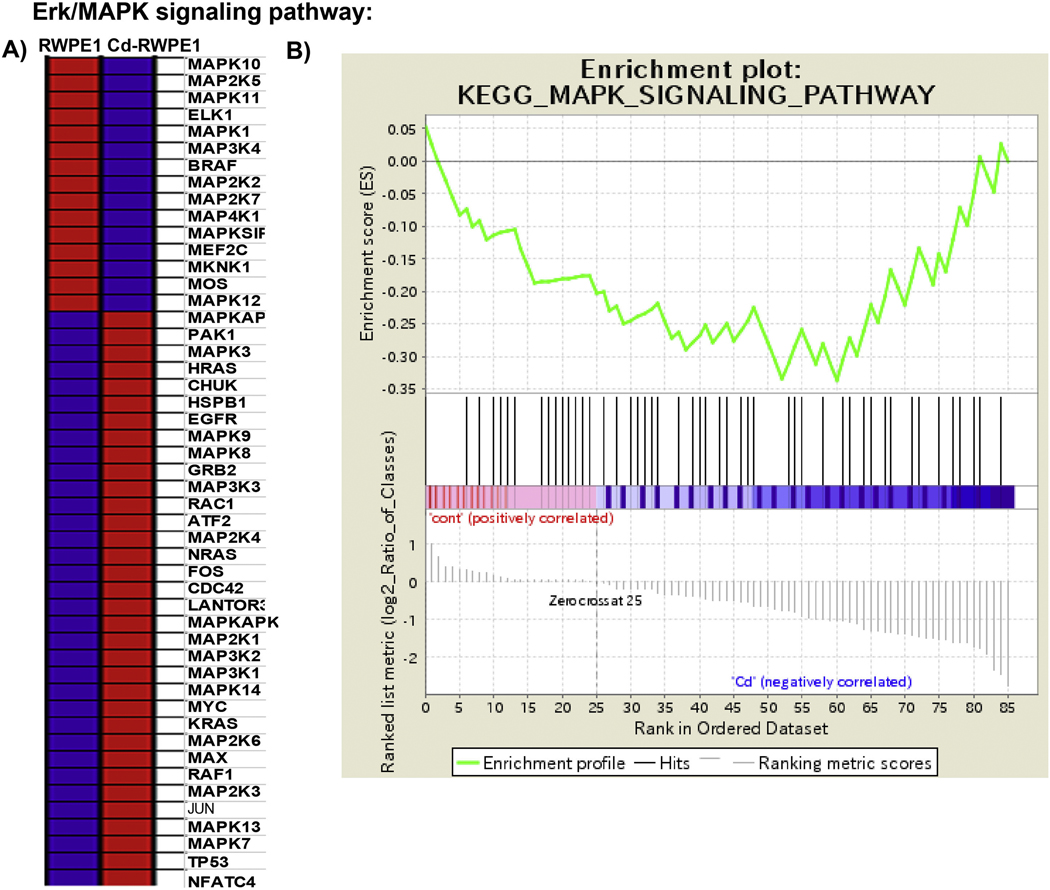
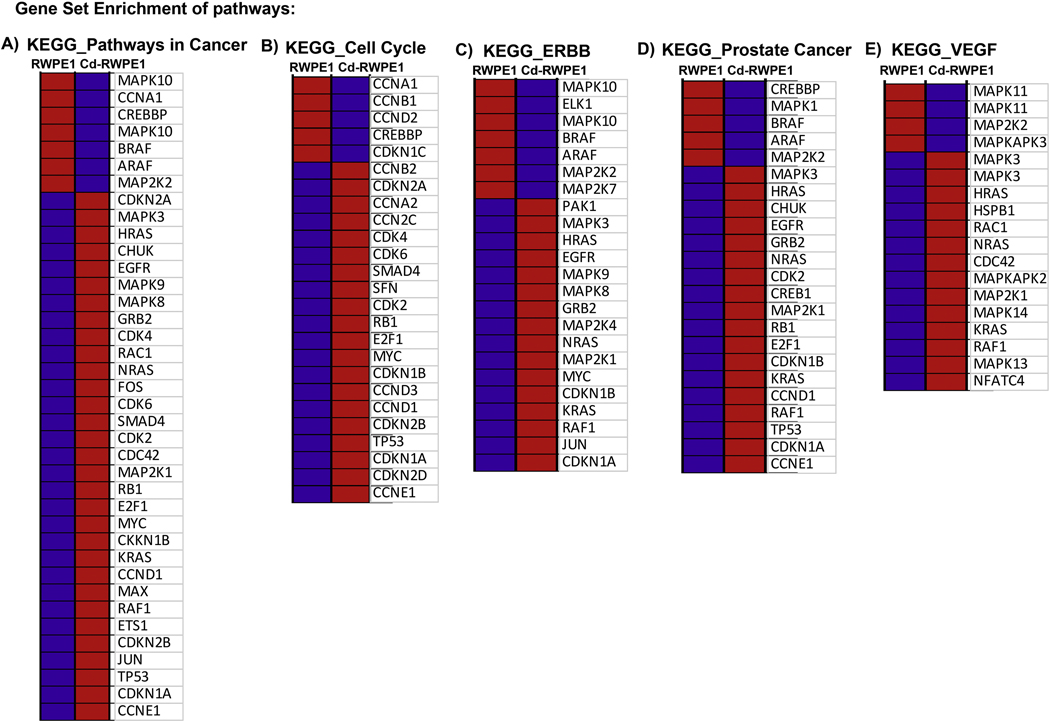
We performed GSEA analysis utilizing curated gene sets (C2) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping, whereby all the upregulated genes in Cd-RWPE1 compared to normal RWPE1 cells were compared with the KEGG pathway database to examine which pathways are likely to be correlated with our array results. In concordance with our finding in the qPCR array, KEGG MAPK signaling pathway was enriched in Cd-treated cells and negatively correlated with RWPE1 (enrichment score (ES) = −0.33, normalized enrichment score (NES) = −1) (Figure 3A–B). In addition, other related pathways such as KEGG ERBB pathway (ES= −0.34, NES= −1), KEGG Cell Cycle pathway (ES= −0.50, NES= −1), KEGG VEGF pathway (ES= −0.46, NES= −1), KEGG Pathways in cancer (ES= −0.43, NES= −1) and KEGG Prostate Cancer pathway (ES= −0.53, NES= −1) (Figure 4A–E; Supplemental Figure 2) were also enriched in Cd-treated cells compared to parental RWPE1. These results further confirm that the Erk/MAPK pathway is one of the major pathways involved in Cd induced malignant transformation of normal prostate epithelial cells.
Figure 3:
Gene Set Enrichment Analysis (GSEA) of the Erk/MAPK signaling pathway enriched in Cd-RWPE1 (denoted as “1” in the plot) compared to RWPE1 (denoted as “0” in the plot) as control. Differential expression of genes (A) correlated with the enrichment plot (B) of MAPK signaling pathway genes. Red-overexpression, Blue-lower expression. The gene lists counterpart to each gradient red and blue bar that is located at the middle of Enrichment plot and indicate the genes assigned in the plot.
Figure 4:
Gene Set Enrichment Analysis of pathways enriched in Cd-RWPE1 compared to RWPE1 as control. KEGG_Pathways in cancer (A). KEGG_Cell Cycle (B). KEGG_ERBB (C). KEGG_Prostate Cancer (D). KEGG_VEGF (E). Enrichment plots are shown in Supplemental Figure 2. Red-overexpression, Blue-lower expression. The gene lists counterpart to each gradient red and blue bar that is located at the middle of Enrichment plot in Supplemental Figure 2A–E and indicate the genes were assigned in the plot.
Genes induced by Cd exposure are overexpressed in prostate cancer
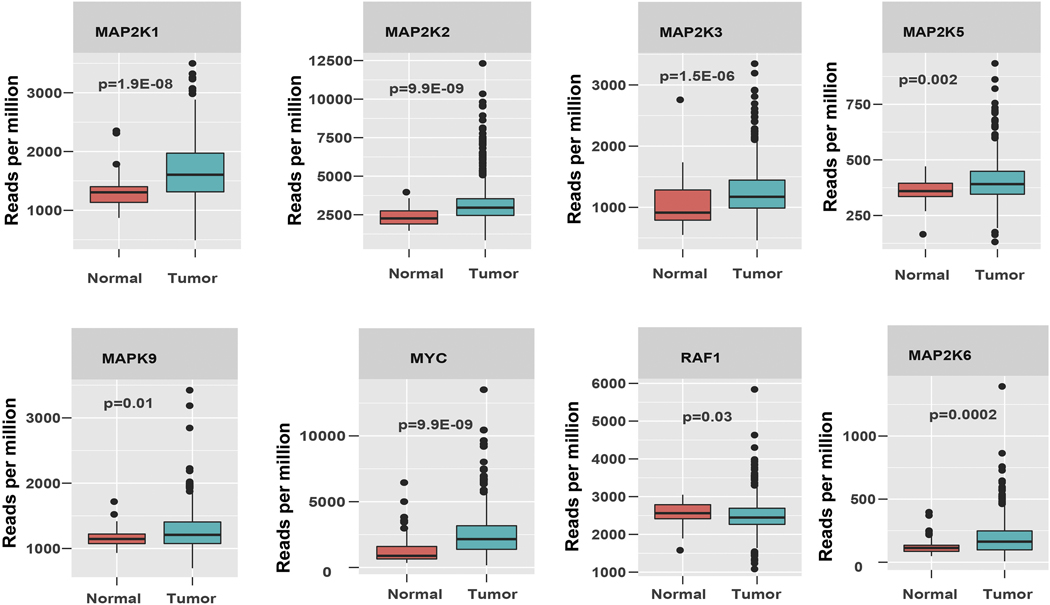
We randomly selected some genes that were enriched in Cd-RWPE1 compared to normal RWPE1 cells and profiled their expression in a large cohort of prostate adenocarcinoma (n= 534) available at TCGA/GDC data base. As expected the expression profiles of the selected genes [MAP2K1 (p=1.9E-08), MAP2K2 (p=9.9E-09), MAP2K3 (p=1.5E-06), MAP2K5 (p=0.002), MAP2K6 (p=0.0002), MAPK9 (p=0.01), MYC (p=9.9E-09) and RAF1(p=0.03)] were significantly higher in prostate cancer compared to normal prostate samples (Figure 5).
Figure 5:
TCGA data analysis. Box plot analysis of the Erk/MAPK signaling pathway genes in a large prostate adenocarcinoma data cohort (n=534) from TCGA data base.
Discussion
This study provides evidence about the Erk/MAPK signaling pathway being one of the major molecular pathways for the carcinogenic mode of action of Cd exposure in prostate cancer. In the USA, prostate cancer incidence and mortality rates have risen from year to year. Prostate cancer is currently the most commonly diagnosed cancer in American men with estimated new cases of 191,930 in 2020 (Siegel et al., 2020). It is the second-leading cause of cancer related deaths with an estimated 33,330 deaths for 2020 (Siegel et al., 2020). The etiology of prostate cancer remains elusive because of its complex, multifactorial nature, which includes evidence of genetic, dietary, hormonal, and environmental risk factors (Sommer et al., 1995; Hsing et al., 2000). Substantial evidence indicates that one such risk factor is exposure to carcinogenic metals (Luevano and Damodaran, 2014; Ju-Kun et al., 2016; Wei et al., 2017; Mezynska and Brzoska, 2018; Rapisarda et al., 2018). Several epidemiologic studies have reported an association between occupational or environmental Cd exposure and prostate cancer incidence (Bako et al., 1982; Elinder et al., 1985; Garcia Sanchez et al., 1992; van der Gulden et al., 1995; Vinceti et al., 2007; Julin et al., 2012; Cheung et al., 2014). In this study we exposed normal prostate epithelial cells (RWPE1 and PWR1E) to Cd similarly as guided by a previous publication (Achanzar et al., 2001) to perform in depth investigation of role of the Erk/MAPK pathway in this malignant transformation.
Erk/MAPK pathway mediated intracellular signaling is associated with a variety of cellular activities including cell proliferation, differentiation, survival, death, and transformation (McCubrey et al., 2006; Kholodenko and Birtwistle, 2009). In prostate cancer, the MAPK pathway was found to be significantly elevated in both primary and metastatic lesions (Mulholland et al., 2012) and its inhibition was highly effective in preventing the development of metastatic prostate cancer (Fu et al., 2003; Maroni et al., 2004; Mulholland et al., 2012). An increase in the activity of the MAPK pathway has been correlated with the progression of prostate cancer to more advanced disease (Gioeli et al., 1999; Clark et al., 2005). We observed that functionally, chronic Cd exposure of normal prostate epithelial cells induced tumorigenic characteristics indicated by higher survival and proliferation of Cd-RWPE1 and Cd-PWR1E cells compared to parental RWPE1 and PWR1E cells. Our PCR array showed an increase in the expression of pro-survival genes of the Erk/MAPK pathway. Interestingly, array results showed downregulation of the MAPK10 gene on Cd exposure. This gene is known to be pro-apoptotic in various cancers (Ying et al., 2006; Li and Luo, 2017). Consistent with these results we observed that Cd exposure induced apoptotic resistance in prostate epithelial cells as indicated by less apoptotic cells in Cd-RWPE1 compared to parental RWPE1. Apoptosis is a genetically controlled process, by which cells with irreparable damage self-destruct, facilitating their removal from the population (Wyllie, 1992). Disruption of apoptosis has been shown to play major role in tumor formation and malignant progression (Wyllie et al., 1999). Previous studies have shown that chronic cadmium exposure of prostate epithelial cells altered expression of important apoptotic regulators and derangement of the apoptotic process was a key aspect of their malignant transformation (Lowe and Lin, 2000; Achanzar et al., 2001). Our study highlights another dimension of Cd induced malignant transformation of normal prostate epithelial cells involving entire Erk/MAPK pathway.
Cancer genomics has the potential to identify biologic determinants associated with any cancer. We explored this at the transcriptome level by performing Gene Set Enrichment Analysis (GSEA). Comparative transcriptomics was performed for all the differentially expressed genes in the Cd-RWPE1 array for pathway enrichment considering parental RWPE1 as control. In concordance with our finding in the qPCR array, we found enrichment of the MAPK pathway. In addition, our array data overlapped with other pathways that are implicated in prostate carcinogenesis. We observed enrichment of KEGG-ERBB, KEGG-Cell Cycle, KEGG-VEGF, KEGG-Pathways in cancer and KEGG-prostate cancer pathways were also enriched as the genes overlapped with that of Cd-RWPE1 compared to parental RWPE1 cells. Increased Erk was observed in human lung epithelial cells upon exposure to Cd (Jing et al., 2012). Low dose Cd also activated Erk signaling in endothelial cells (HUVECs) with an increase in VEGFR-2 (Kim et al., 2012). We observed an increase in Erk and Mek1/2 protein levels in situ in Cd-RWPE1 and Cd-PWR1E cells compared to their normal parental controls. Further, we randomly selected upregulated genes from Erk/MAPK pathway and performed profile analysis in a larger prostate adenocarcinoma data set (n=534) from the TCGA/GDC data base. We observed upregulation of these genes in prostate cancer compared to normal prostate samples. These data further strengthen our finding that the Erk/MAPK pathway is involved in Cd-induced malignant transformation of normal prostate epithelial cells.
This study provides evidence that enhanced Erk/MAPK signaling is one of the major molecular events involved in the transformation of Cd exposed carcinogenesis of normal prostate epithelial cells. In addition, Cd exposure increased the proliferative capability of prostate cancer cells as well. Functionally, Cd-induced Erk/MAPK signaling resulted in the induction of tumorigenic attributes in normal prostate epithelial cells. In the USA, prostate cancer incidence and mortality rates have risen, and it is the second leading cause of cancer deaths in men. Understanding the dominant oncogenic pathways involved in the transformation of normal prostate epithelial cells to malignant forms by cadmium will help in developing better therapeutic strategies to reduce the morbidity and mortality of prostate cancer.
Supplementary Material
Supplemental Figure 1: FACS analysis for cell cycle distribution showed an increase in the S-phase population (A) and a decrease in the apoptotic cell fraction (B) in prostate cancer cell line exposed to Cd (Cd-DU 145) compared to parental DU 145. Error bars ±SD.
Supplemental Figure 2: Gene Set Enrichment Analysis (GSEA) plots for enriched pathways. Cd-RWPE1 (denoted as “1” in the plot) compared to RWPE1 (denoted as “0” in the plot) as control.
Normal prostate epithelial RWPE1 and PWR1E cells were exposed to cadmium (Cd).
Chronic Cd exposure transformed these normal cells to malignant form.
Molecular mechanism involved was the Erk/MAPK signaling genes.
Activation of these genes was observed at transcription and translation levels.
Gene Set Enrichment Analysis showed enrichment of prostate cancer related pathways.
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript.
Funding
This study was supported by the Department of Veterans Affairs VA Merit Review 101 BX001123, Senior Research Career Scientist Award (Rajvir Dahiya, IK6-BX004473), and the National Institutes of Health / National Cancer Institute RO1CA199694, RO1CA196848.
Abbreviations:
- PCa
Prostate cancer
- Cd
Cadmium
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no potential conflicts of interest.
References:
- Achanzar WE, Diwan BA, Liu J, Quader ST, Webber MM, Waalkes MP, 2001. Cadmium-induced malignant transformation of human prostate epithelial cells. Cancer Res 61, 455–458. [PubMed] [Google Scholar]
- Allott EH, Masko EM, Freedland SJ, 2013. Obesity and prostate cancer: weighing the evidence. Eur Urol 63, 800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amzal B, Julin B, Vahter M, Wolk A, Johanson G, Akesson A, 2009. Population toxicokinetic modeling of cadmium for health risk assessment. Environ Health Perspect 117, 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asara Y, Marchal JA, Carrasco E, Boulaiz H, Solinas G, Bandiera P, Garcia MA, Farace C, Montella A, Madeddu R, 2013. Cadmium modifies the cell cycle and apoptotic profiles of human breast cancer cells treated with 5-fluorouracil. Int J Mol Sci 14, 16600–16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bako G, Smith ES, Hanson J, Dewar R, 1982. The geographical distribution of high cadmium concentrations in the environment and prostate cancer in Alberta. Can J Public Health 73, 92–94. [PubMed] [Google Scholar]
- Byber K, Lison D, Verougstraete V, Dressel H, Hotz P, 2016. Cadmium or cadmium compounds and chronic kidney disease in workers and the general population: a systematic review. Crit Rev Toxicol 46, 191–240. [DOI] [PubMed] [Google Scholar]
- Chan JM, Giovannucci EL, 2001. Vegetables, fruits, associated micronutrients, and risk of prostate cancer. Epidemiol Rev 23, 82–86. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran B, Dahiya NR, Tyagi A, Kolluru V, Saran U, Baby BV, States JC, Haddad AQ, Ankem MK, Damodaran C, 2020. Chronic exposure to cadmium induces a malignant transformation of benign prostate epithelial cells. Oncogenesis 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MR, Kang J, Ouyang D, Yeung V, 2014. Association between urinary cadmium and all cause, all cancer and prostate cancer specific mortalities for men: an analysis of national health and nutrition examination survey (NHANES III) data. Asian Pac J Cancer Prev 15, 483–488. [DOI] [PubMed] [Google Scholar]
- Clark DE, Errington TM, Smith JA, Frierson HF Jr., Weber MJ, Lannigan DA, 2005. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res 65, 3108–3116. [DOI] [PubMed] [Google Scholar]
- Elinder CG, Kjellstrom T, Hogstedt C, Andersson K, Spang G, 1985. Cancer mortality of cadmium workers. Br J Ind Med 42, 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, Keller ET, 2003. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst 95, 878–889. [DOI] [PubMed] [Google Scholar]
- Garcia Sanchez A, Antona JF, Urrutia M, 1992. Geochemical prospection of cadmium in a high incidence area of prostate cancer, Sierra de Gata, Salamanca, Spain. Sci Total Environ 116, 243251. [DOI] [PubMed] [Google Scholar]
- Giaginis C, Gatzidou E, Theocharis S, 2006. DNA repair systems as targets of cadmium toxicity. Toxicol Appl Pharmacol 213, 282–290. [DOI] [PubMed] [Google Scholar]
- Gioeli D, Mandell JW, Petroni GR, Frierson HF Jr., Weber MJ, 1999. Activation of mitogenactivated protein kinase associated with prostate cancer progression. Cancer Res 59, 279–284. [PubMed] [Google Scholar]
- Hartwig A, 2010. Mechanisms in cadmium-induced carcinogenicity: recent insights. Biometals 23, 951960. [DOI] [PubMed] [Google Scholar]
- Hartwig A, Schwerdtle T, 2002. Interactions by carcinogenic metal compounds with DNA repair processes: toxicological implications. Toxicol Lett 127, 47–54. [DOI] [PubMed] [Google Scholar]
- Hsing AW, Tsao L, Devesa SS, 2000. International trends and patterns of prostate cancer incidence and mortality. Int J Cancer 85, 60–67. [DOI] [PubMed] [Google Scholar]
- IARC, 1993. IARC monographs on the evaluation of carcinogenic risks to humans, Lyon, pp. 1972–1993. [Google Scholar]
- Jacobo-Estrada T, Santoyo-Sanchez M, Thevenod F, Barbier O, 2017. Cadmium Handling, Toxicity and Molecular Targets Involved during Pregnancy: Lessons from Experimental Models. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Liu LZ, Jiang Y, Zhu Y, Guo NL, Barnett J, Rojanasakul Y, Agani F, Jiang BH, 2012. Cadmium increases HIF-1 and VEGF expression through ROS, ERK, and AKT signaling pathways and induces malignant transformation of human bronchial epithelial cells. Toxicol Sci 125, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju-Kun S, Yuan DB, Rao HF, Chen TF, Luan BS, Xu XM, Jiang FN, Zhong WD, Zhu JG, 2016. Association Between Cd Exposure and Risk of Prostate Cancer: A PRISMA-Compliant Systematic Review and Meta-Analysis. Medicine (Baltimore) 95, e2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julin B, Wolk A, Johansson JE, Andersson SO, Andren O, Akesson A, 2012. Dietary cadmium exposure and prostate cancer incidence: a population-based prospective cohort study. Br J Cancer 107, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN, Birtwistle MR, 2009. Four-dimensional dynamics of MAPK information processing systems. Wiley Interdiscip Rev Syst Biol Med 1, 28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lim W, Ko Y, Kwon H, Kim S, Kim O, Park G, Choi H, Kim O, 2012. The effects of cadmium on VEGF-mediated angiogenesis in HUVECs. J Appl Toxicol 32, 342–349. [DOI] [PubMed] [Google Scholar]
- Li L, Luo Z, 2017. Dysregulated miR-27a-3p promotes nasopharyngeal carcinoma cell proliferation and migration by targeting Mapk10. Oncol Rep 37, 2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Lin AW, 2000. Apoptosis in cancer. Carcinogenesis 21, 485–495. [DOI] [PubMed] [Google Scholar]
- Luevano J, Damodaran C, 2014. A review of molecular events of cadmium-induced carcinogenesis. J Environ Pathol Toxicol Oncol 33, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzen A, Liberti SE, Rasmussen LJ, 2004. Cadmium inhibits human DNA mismatch repair in vivo. Biochem Biophys Res Commun 321, 21–25. [DOI] [PubMed] [Google Scholar]
- Maroni PD, Koul S, Meacham RB, Koul HK, 2004. Mitogen Activated Protein kinase signal transduction pathways in the prostate. Cell Commun Signal 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Lahair MM, Franklin RA, 2006. Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid Redox Signal 8, 1775–1789. [DOI] [PubMed] [Google Scholar]
- Mezynska M, Brzoska MM, 2018. Environmental exposure to cadmium-a risk for health of the general population in industrialized countries and preventive strategies. Environ Sci Pollut Res Int 25, 3211–3232. [DOI] [PubMed] [Google Scholar]
- Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H, 2012. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 72, 1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP, N.T.P., 2016. Cadmium and Cadmium Compounds. Report on Carcinogen. US Department of Health and Human Services, Public Health Service., pp. [Google Scholar]
- Pal D, Suman S, Kolluru V, Sears S, Das TP, Alatassi H, Ankem MK, Freedman JH, Damodaran C, 2017. Inhibition of autophagy prevents cadmium-induced prostate carcinogenesis. Br J Cancer 117, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron F, Baillon L, Sow M, Gotreau S, Gonzalez P, 2014. Effect of low-dose cadmium exposure on DNA methylation in the endangered European eel. Environ Sci Technol 48, 797–803. [DOI] [PubMed] [Google Scholar]
- Potosky AL, Miller BA, Albertsen PC, Kramer BS, 1995. The role of increasing detection in the rising incidence of prostate cancer. JAMA 273, 548–552. [PubMed] [Google Scholar]
- Rani A, Kumar A, Lal A, Pant M, 2014. Cellular mechanisms of cadmium-induced toxicity: a review. Int J Environ Health Res 24, 378–399. [DOI] [PubMed] [Google Scholar]
- Rapisarda V, Miozzi E, Loreto C, Matera S, Fenga C, Avola R, Ledda C, 2018. Cadmium exposure and prostate cancer: insights, mechanisms and perspectives. Front Biosci (Landmark Ed) 23, 1687–1700. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2020. Cancer statistics, 2020. CA Cancer J Clin 70, 7–30. [DOI] [PubMed] [Google Scholar]
- Sommer M, Gathof BS, Podskarbi T, Giugliani R, Kleinlein B, Shin YS, 1995. Mutations in the galactose-1-phosphate uridyltransferase gene of two families with mild galactosaemia variants. J Inherit Metab Dis 18, 567–576. [DOI] [PubMed] [Google Scholar]
- Valverde M, Trejo C, Rojas E, 2001. Is the capacity of lead acetate and cadmium chloride to induce genotoxic damage due to direct DNA-metal interaction? Mutagenesis 16, 265–270. [DOI] [PubMed] [Google Scholar]
- van der Gulden JW, Kolk JJ, Verbeek AL, 1995. Work environment and prostate cancer risk. Prostate 27, 250–257. [DOI] [PubMed] [Google Scholar]
- Venkateswaran V, Klotz LH, 2010. Diet and prostate cancer: mechanisms of action and implications for chemoprevention. Nat Rev Urol 7, 442–453. [DOI] [PubMed] [Google Scholar]
- Vinceti M, Venturelli M, Sighinolfi C, Trerotoli P, Bonvicini F, Ferrari A, Bianchi G, Serio G, Bergomi M, Vivoli G, 2007. Case-control study of toenail cadmium and prostate cancer risk in Italy. Sci Total Environ 373, 77–81. [DOI] [PubMed] [Google Scholar]
- Wei T, Jia J, Wada Y, Kapron CM, Liu J, 2017. Dose dependent effects of cadmium on tumor angiogenesis. Oncotarget 8, 44944–44959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AH, 1992. Apoptosis and the regulation of cell numbers in normal and neoplastic tissues: an overview. Cancer Metastasis Rev 11, 95–103. [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Bellamy CO, Bubb VJ, Clarke AR, Corbet S, Curtis L, Harrison DJ, Hooper ML, Toft N, Webb S, Bird CC, 1999. Apoptosis and carcinogenesis. Br J Cancer 80 Suppl 1, 34–37. [PubMed] [Google Scholar]
- Ying J, Li H, Cui Y, Wong AH, Langford C, Tao Q, 2006. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia 20, 1173–1175. [DOI] [PubMed] [Google Scholar]
- Zalups RK, Ahmad S, 2003. Molecular handling of cadmium in transporting epithelia. Toxicol Appl Pharmacol 186, 163–188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: FACS analysis for cell cycle distribution showed an increase in the S-phase population (A) and a decrease in the apoptotic cell fraction (B) in prostate cancer cell line exposed to Cd (Cd-DU 145) compared to parental DU 145. Error bars ±SD.
Supplemental Figure 2: Gene Set Enrichment Analysis (GSEA) plots for enriched pathways. Cd-RWPE1 (denoted as “1” in the plot) compared to RWPE1 (denoted as “0” in the plot) as control.