Abstract
Selective activation of p53 target genes in response to various cellular stresses is a critical step in determining the ability to induce cell-cycle arrest or apoptosis. Here we report the identification of the microRNA miR-22 as a p53 target gene that selectively determines the induction of p53-dependent apoptosis by repressing p21. Combinatorial analyses of the AGO2 immunocomplex and gene expression profiles identified p21 as a direct target of miR-22. Induction of p21 was inhibited by miR-22 after exposure to the genotoxic agent Adriamycin (doxorubicin; Bedford Laboratories), sensitizing cells to p53-dependent apoptosis. Interestingly, the activation of miR-22 depended on the intensity of the stresses that induced cells to undergo apoptosis in the presence of p21 suppression. Our findings define an intrinsic molecular switch that determines p53-dependent cellular fate through post-transcriptional regulation of p21.
Introduction
The p53 tumor suppressor network plays a crucial role in the prevention of malignant transformation in normal cells by maintaining the integrity of signaling pathways in response to various oncogenic stresses, including DNA damage, acute activation of oncogenes, and hypoxic conditions (1). The outcome of p53 activation in response to cellular stresses ranges from the induction of cell-cycle arrest for DNA repair to apoptosis for the complete elimination of damaged cells (2–4). The commitment to one of these alternative cellular fates depends on the set of p53 target genes induced by different stresses. Induction of cell-cycle arrest is mediated by the activation of the cyclin-dependent kinase inhibitor CDKN1A (hereafter referred to as p21), whereas apoptosis is induced by the activation of pro-apoptotic genes, including NOXA (5), PUMA (6), and BAX (7) that encode the regulators of intrinsic apoptosis pathways.
Post-translational modifications of p53 are involved in the selective activation of its various target genes leading to apoptosis (8, 9). Phosphorylation of p53 at serine 46 (Ser46), mediated by HIPK2 (10), regulates apoptotic pathways through the activation ofp53AIP1 (11). Furthermore, acetylation ofp53 at lysine 120 (K120) by Tip60 is essential for the expression of PUMA (12). Ongoing work focuses on the elucidation of p53 function and its regulation as a transcriptional factor.
Recently, the regulation of gene expression by small noncoding RNAs, including microRNAs (miRNA), has been reported to play crucial roles in the maintenance of homeostasis in a wide range of cellular processes, including differentiation, control of cell proliferation, and stress responses (13–15). The important feature of miRNAs is the targeting of multiple cellular mRNAs, resulting in the efficient activation or repression of intracellular or intercellular signaling networks at specific times during animal development. miRNA dysfunction therefore causes defects in the integration of signaling networks essential for the maintenance of cellular homeostasis.
miRNA dysfunction has been suggested as a dominant cause of the onset of human disorders, especially cancers. Indeed, aberrant expression of miRNA genes was observed in almost all types of human cancers (16, 17). As a consequence of miRNA dysfunction, cancer cells acquire properties that favor the activation of oncogenic pathways or the repression of tumor-suppressive networks, contributing to cancer progression and metastasis (18–22). MiR-21 was shown to repress PTEN, activating the phosphoinositide 3-kinase (PI3K)-AKT pathway and reflecting its oncogenic role (23). By contrast, miR-34a was identified as a p53-regulated tumor-suppressive miRNA in human colon cancer and shown to induce p53-dependent apoptosis or premature senescence, forming a positive feedback loop with p53 (24–28). The function of miRNAs as oncogenes or tumor suppressor genes is therefore well known, and it implies that the incorporation of miRNA species as critical components of intracellular signaling pathways is crucial for the reconstitution of integrated cancer-related networks necessary to fully clarify the molecular basis of carcinogenesis.
To analyze the connection between miRNAs and signaling networks, a functional genetic screening method named “dropout assay” was recently established using a lentivirus miRNA expression library and a home-made microarray to quickly and efficiently isolate tumor-suppressive miRNAs (29). In the present study, an in vitro functional genetic screen and comprehensive genomic screens of clinical samples were used to identify tumor suppressor miRNAs in colon carcinogenesis, with the resulting identification of miR-22 as a tumor suppressor gene. A p53-miR-22-p21 axis was identified as a crucial regulatory component involved in the determination of p53-dependent apoptosis. Our results suggest that miR-22 is an intrinsic molecular switch or sensor for the determination of p53-dependent cellular fate in response to distinct stresses, and miR-22 dysfunction could affect the anticancer barrier against various oncogenic insults.
Materials and Methods
Cell culture
HCT 116 (HCT 116 p53+/+) and HCT 116 p53−/− (30) were kindly provided by Dr. Bert Vogelstein (The Johns Hopkins University, Baltimore, MD). These cell lines were authenticated by morphologic inspection, and mycoplasma testing using PCR. The activation of p53 pathways was confirmed by checking the induction of p53 target genes after exposure to DNA damage before starting the experiments. The SW480 colon cancer cell line was obtained from the American Type Culture Collection and authenticated as described above. Mutation of TP53 was confirmed by sequencing. These cell lines were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% heat inactivated FBS in humidified air with 5% CO2.
Clinical samples
Paired surgical specimens of primary human colon cancers and surrounding noncancerous colon tissue counterparts were obtained from patients treated at the Teikyo University Hospital (Mizonokuchi, Kanagawa, Japan) with documented informed consent in each case. Institutional review board approval for the analysis of clinical samples was obtained at each institute.
Functional miRNA dropout screening
Functional dropout screening to identify tumor suppressor miRNAs was carried out according to our recent publication (29). HCT 116 cells were transduced with a pooled lentivirus miRNA expression library (SBI) at a multiplicity of infections (MOI) of 3. Cells were incubated in complete medium for 3 days (P1) and subjected to sequential passages every 3 days. After 9 passages, genomic DNA was prepared from P1, P5, and P9 cells and subjected to array CGH analysis using a home-made microarray.
Quantitative real-time PCR
For quantitative expression analysis of miRNAs, total RNAs from colon cancer patients were reverse-transcribed by Multiscribe RT and miRNA-specific miRNA primers (ABI), and quantitative real-time PCR (qRT-PCR) was carried out by using a TaqMan microRNA assay kit (ABI). The comparative cycle threshold (Ct) method was applied to quantify the expression levels of miRNAs. Relative expression levels were calculated by the 2-ΔΔCt method. U48 small nuclear RNA was used as an internal standard.
Chromatin immunoprecipitation sequencing
HCT 116 cells were treated with 5-fluorouracil (5-FU; 0.375 mmol/L) for 9 hours, and chromatin immunoprecipitation (ChIP) was conducted by using anti-p53, antimonomethylated or antitrimethylated histone H3 K4, or antitrimethylated histone H3 K36 antibodies. ChIP-isolated DNA was subjected to the sequencing using an Illumina platform.
AGO2-IP on ChIP analysis
The AGO2-IP on ChIP assay was carried out according to a previous report with minor modifications (31). In brief, HCT 116 cells stably expressing HA-AGO2 were transfected with either miR-22 (Pre-miR precursor molecule, Ambion) or miR-NC (Pre-miR miRNA Precursor Molecules Negative Control #2, Ambion) for 24 hours, and immunoprecipitated using anti-HA agarose beads. AGO2-bound RNA was eluted in boiling water, and the Trizol-LS reagent was added to extract total RNAs. AGO2-bound total RNAs were cleaned further using an RNeasy column and subjected to microarray analysis.
Reporter plasmid construction and luciferase assay
Amplification of the 3’ UTR of p21 mRNA was carried out by PCR from HCT 116 genomic DNA using a primer set (Supplementary Table S1). The DNA fragment was fused to the 3’ end of a firefly luciferase reporter gene in a pmirGLO dual luciferase vector (Promega). Site-directed mutagenesis of a miR-22 target site ofp21 mRNA was carried out by using a PrimeSTAR Max high fidelity DNA polymerase using the pmirGLO-p21 3’UTR plasmid as a template. HCT 116 cells, seeded at 5 × 104 cells/mL, were cotransfected with 200 ng of reporter plasmid and 10 nmol/L of either miR-22 or miR-NC using Lipofecta-mine 2000. After incubation for 24 hours, luciferase activities were determined by using a dual luciferase assay kit (Promega). Luciferase activity was normalized by Renilla luciferase activity as an internal standard.
Immunoblot analysis
Cells were lysed in lysis buffer consisting of 25 mmol/L Tris-HCl (pH7.5), 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, 0.1% SDS and 1x proteinase inhibitor cocktail, and equal concentrations of protein samples were loaded on a 10% to 20% polyacrylamide gradient gel (ATTO). After electrophoresis, proteins were transferred to a PVDF membrane, and immunoblot analysis was conducted by the standard method.
Supplementary information
Supplementary information includes extended Materials and Methods, 8 figures, and 4 tables.
Results
Identification of miR-22 as a candidate tumor suppressor miRNA by functional genetic and comprehensive genomic screens
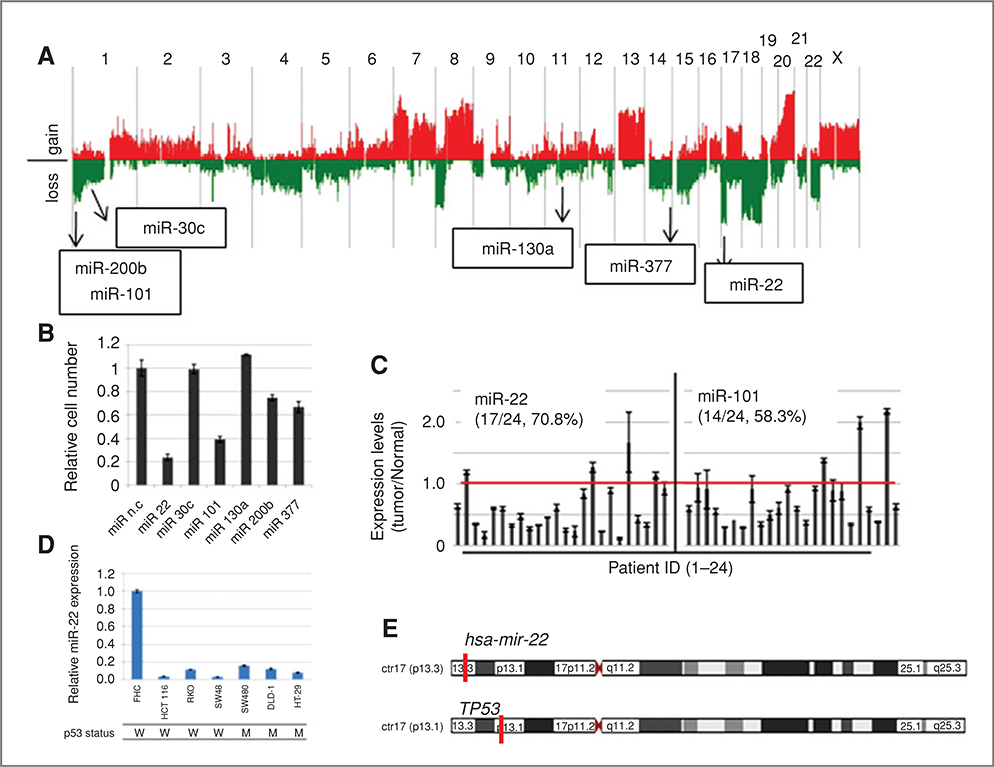
A screening method for the efficient identification of tumor suppressor miRNAs in colon cancer was established and is depicted in Supplementary Fig. S1A. Tumor suppressor miRNAs were defined as follows; (i) repressor of cell proliferation, (ii) expression in normal colon tissue, (iii) high-frequency loss of their chromosomal positions, and (iv) downregulation in human colon cancers. Following these criteria, a functional genetic screening, namely a “dropout assay,” was conducted using a lentivirus miRNA expression library (29) to isolate repressors of cell proliferation in a colon cancer cell line (Supplementary Fig. S1B). HCT 116 cells were transduced with a pooled lentivirus library containing 454 miRNA species and propagated for 3 weeks with sequential passages. Genomic DNA from the first passage (P1), fifth passage (P5), and ninth passage (P9) cell populations was prepared, and copy numbers of each miRNA clone in these cells were compared by array CGH analysis using a home-made microarray (Supplementary Fig. S1B). A total 55 miRNA clones were reproducibly dropped out in a culture time-dependent manner (Supplementary Fig. S1C and Table S2). Among these dropout clones, 24 miRNAs were confirmed for their expression in normal tissue (Supplementary Fig. S1D). Furthermore, we carried out array CGH analysis (aCGH) to examine autosomal copy number aberrations using 24 colon cancer patients and finally identified 6 miRNA clones whose genes show hemizygous deletions in cancers with a high frequency (>30%), as candidates for tumor suppressor gene in colon cancer (Fig. 1A and Supplementary Fig. S1E). Two of them, miR-22 and miR-101, showed strong inhibition of cell proliferation in HCT 116-p53+/+ cells by MST assay (Fig. 1B). As shown in Fig. 1C, miR-22 and miR-101 showed reduced expression in 70.8% and 50.3% of colon cancer cases, respectively, when compared with their normal counterparts. MiR-22 also showed significant downregulation in 6 colon cancer cell lines in comparison with FHC cells derived from normal colon epithelium (Fig. 1D), which was not observed for miR-101 (data not shown). Interestingly, CGH analysis showed deletion of the miR-22 locus without loss or mutation of TP53 localized to the 6Mb centromeric region of the miR-22 gene in 2 colon cancer patients, and 3 other cases showed a significant reduction of miR-22 expression (Fig. 1E and Table 1). Furthermore, in a copy number assay using another set of colon cancer samples, 5 of 36 cases showed hemizygous deletion of miR-22 locus with intact copy of TP53 (Supplementary Fig. S2).
Figure 1.
A, result of copy number aberrations in 24 human colon cancer samples. Red and green indicate chromosomal gain and loss, respectively. Chromosomal positions of 6 identified miRNA genes are shown in the CGH result. B, cell proliferation assay. HCT 116 cells were transfected with each synthetic miRNA and incubated for 5 days. Cell viability was measured by MST assay. Error bars indicate SD in triplicate cultures. C, expression of miR-22 and miR-101 in human colon cancer patients. Expression levels of miR-22 and miR-101 were quantified by TaqMan microRNAqRT-PCR. The graphs show the relative expression levels of miR-22 and miR-101, calculated by adjusting their expression levels to matched normal counterparts in each cancer sample. The red line indicates the relative expression level of 1.0. D, expression of miR-22 in human colon cancer cell lines and normal colon-derived FHC cells. The genomic status of TP53 in cancer cell lines is indicated. E, chromosomal positions of miR-22 and TP53 genes on chromosome 17.
Table 1.
Genomic status of hsa-miR-22 and TP53 genes in 24 human colon cancers
| has-miR-22 (17p13.3) |
TP53(17p13.1) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| chrl7:1,563,947-1,564,081 |
Chrl7:7,520,037-7,531,588 |
||||||||
| Sample No | Patient ID | CNA | Expression (T/N) | CNA | Mutation | Exon | Codon | WT (A.A.) | Mut (A.A.) |
| 1 | 1002 | - | 0.635 | - | - | ||||
| 2 | 1004 | - | 1.184 | - | missense | 7 | 237 | ATG (M) | ATA (I) |
| 3 | 1008 | Loss | 0.587 | Loss | missense | 6 | 193 | CAT (H) | CGT(R) |
| 4 | 1010 | Loss | 0.172 | Loss | missense | 8 | 285 | GAG (E) | AAG (K) |
| 5 | 1011 | - | 0.602 | - | - | ||||
| 6 | 1013 | Loss | 0.001 | Loss | - | ||||
| 7 | 1014 | Loss | 0.321 | Loss | missense | 7 | 230 | GAA (E) | AAA (K) |
| 8 | 1015 | Gain | 0.470 | Gain | - | ||||
| 9 | 1016 | Loss | 0.272 | Loss | insertion (4) | 7 | 280 | ||
| 10 | 1017 | Loss | 0.327 | - | - | ||||
| 11 | 1018 | Loss | 0.455 | - | - | ||||
| 12 | 1019 | - | 0.614 | - | - | ||||
| 13 | 1022 | Loss | 0.248 | Loss | missense | 7 | 245 | GGC (G) | TGC (C) |
| 14 | 1023 | Loss | 0.227 | Loss | missense | 7 | 245 | GGC (G) | TGC (C) |
| 15 | 1024 | Loss | 0.835 | Loss | missense | 7 | 248 | CGG (R) | CAG (Q) |
| 16 | 1025 | Loss | 1.285 | Loss | missense | 5 | 175 | CGC (R) | CAC (H) |
| 17 | 1027 | Loss | 0.227 | Loss | missense | 7 | 248 | CGG (R) | CAG (Q) |
| 18 | 1028 | Loss | 0.888 | Loss | missense | 5 | 158 | CGC (R) | CAC (H) |
| 19 | 1029 | Loss | 0.114 | Loss | - | ||||
| 20 | 1033 | Loss | 1.668 | Loss | deletion (1) | 8 | 267 | ||
| 21 | 1035 | Loss | 0.429 | Loss | deletion (18) | 5 | 174 | ||
| 22 | 1036 | Loss | 0.339 | Loss | missense | 5 | 152 | CCG (P) | CTG (L) |
| 23 | 1037 | Loss | 1.139 | Loss | |||||
| 24 | 1039 | Loss | 0.922 | Loss | deletion (6) | 7 | 235 | AAC (N) | ATG (M) |
Two patients, sample numbers 10 and 11, showed hemizygous loss of the miR-22 gene locus with intact TP53. Three patients, sample numbers 1, 5, and 12, showed downregulation of miR-22 with intact TP53.
Induction of apoptosis by miR-22 in p53 wild-type colon cancer cells
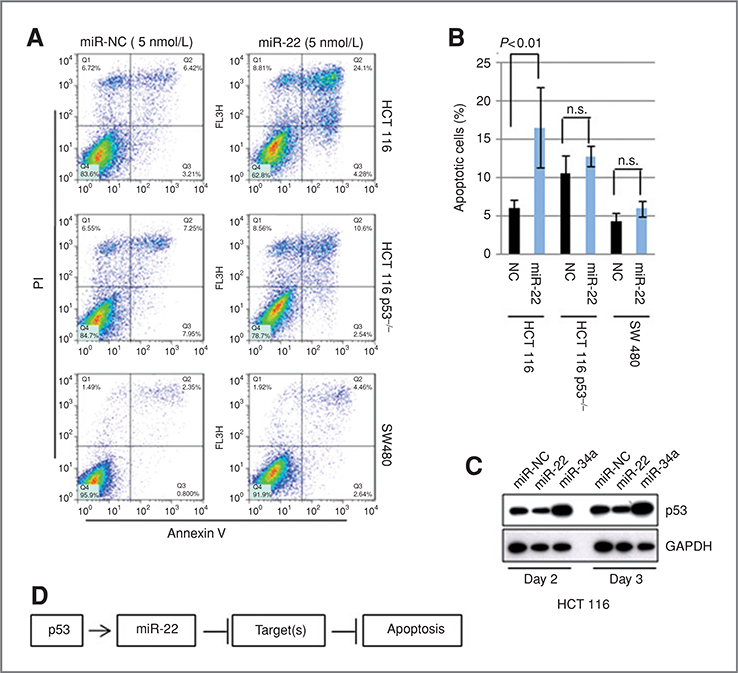
Cell proliferation assays using the HCT 116-p53+/+, HCT 116-p53−/−, and p53 mutant SW480 cell lines showed a significant repression of cell proliferation by miR-22 in 3 cell lines (Supplementary Fig. S3A). Interestingly, miR-22 induced apoptosis selectively in HCT 116-p53+/+ cells (Fig. 2A and B). In contrast, it caused cell-cycle arrest in HCT 116-p53−/− and SW480 cells (Supplementary Fig. S3B and C). These results indicate that miR-22 acts as a growth repressor in colon cancer cells, and that its ability to induce apoptosis depends on the TP53 status. Indeed, the expression profile of HCT 116 cells in the presence of miR-22 showed significant modulation of cellular p53 network (Supplementary Fig. S3D and Tables S3 and S4). Furthermore, the introduction of miR-22 into HCT 116 cells did not show upregulation or stabilization of p53, suggesting that miR-22 may function downstream of the p53 induced apoptotic pathways, and that its role in the induction of apoptosis could be mediated by the repression of p53 target genes (Fig. 2C and D).
Figure 2.
A, fluorescence-activated cell sorting (FACS) analysis. HCT 116, HCT 116-p53−/−, and SW480 were transfected with 5 nmol/L of miR-22 or miR-NC, incubated for 3 days, and subjected to FACS analysis. B, quantification of apoptotic cells. Apoptotic cells were quantified by using 4 independent FACS experiments. Data indicate the mean value with SD. Statistical analysis was carried out by t test. C, p53 is not activated by miR-22. Cells, transfected with miR-22 or miR-NC, were incubated for 2 or 3 days, and subjected to immunoblotting. D, hypothesis of miR-22 function in the p53 network.
Identification of the miR-22 gene as a direct transcriptional target of p53
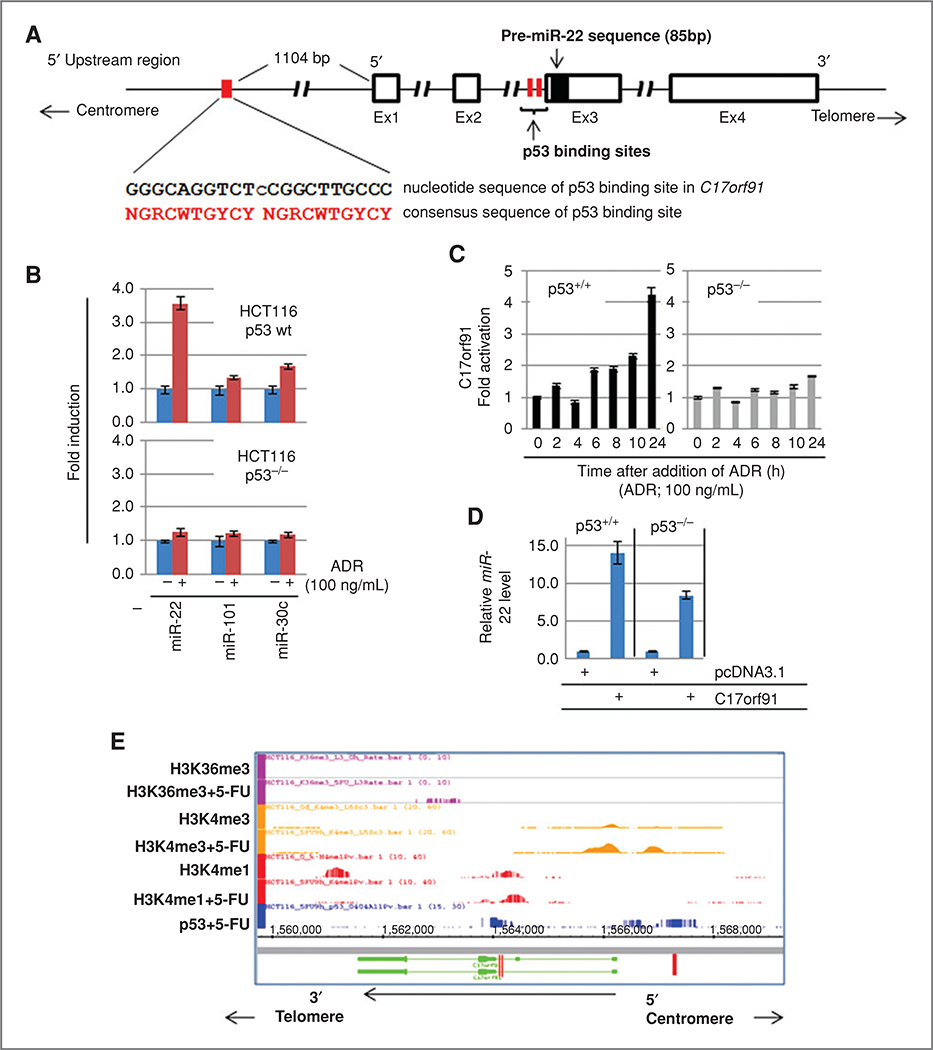
As shown in Fig. 3A, miR-22 is encoded within exon 3 of the C17orf91 gene, which is located on the minus strand of the 17p13.3 region of the human chromosome, and consensus sequence of p53 binding sites (p53BS) was identified at a 5’ upstream region and within the intron 2 of the C17orf91 gene (Fig. 3A and Supplementary Fig. S4). The expression of miR-22 was assessed in HCT 116-p53+/+ cells treated with 100 ng/mL of Adriamycin (ADR; doxorubicin, Bedford Laboratories), a genotoxic agent leading to activation of p53, for 24 hours. The result indicated that mature miR-22 was increased considerably by ADR treatment in HCT 116-p53+/+ cells, but not in HCT 116-p53−/− cells (Fig. 3B). The expression of C17orf91 was induced only in HCT 116-p53+/+ cells by ADR (Fig. 3C). Transcriptional activation of miR-22 was also found in HCT 116 cells after treatment with 5-FU, which was confirmed by qRT-PCR and reporter gene analyses (Supplementary Fig. S5A–C). Furthermore, introduction of a cDNA encoding C17orf91, cloned by using a gene-specific primer set (Supplementary Fig. S4), into cells clearly showed an increase of mature miR-22 in both p53 wild-type and p53−/− HCT 116 cells (Fig. 3D). These results suggest that miR-22 expression is regulated by p53 at the transcriptional level, not by p53-dependent processing during the maturation of the miRNA (32). Indeed, p53 binding on p53BS located at 5’ upstream and intron 2 of the miR-22 gene was significantly enhanced after exposure to 5-FU evidenced by p53 ChIP (Supplementary Fig. S5D and E). Furthermore, this was also confirmed by ChIP-sequencing (ChIP-Seq) analysis (Fig. 3E), indicating that miR-22 is a direct transcriptional target of p53. The concurrent increase in tri-methylation of lysine 4 of histone H3 (33) evidenced transcriptional activation of the miR-22 gene after exposure to 5-FU (Fig. 3E).
Figure 3.
A, genomic structure of miR-22 and its host gene, C17orf91. Genomic structure of C17ORF91 is indicated. Open boxes show exons and the region-encoded pre-miR-22 is indicated by a closed box. Red boxes are p53 binding site at 5’ upstream region and within intron 2. Consensus sequences of p53 binding sites located 5’ upstream of exon1 of C17orf91 are shown. B, induction of miR-22 expression after addition of the genotoxic agent ADR in p53-wild type (wt) and p53−/− HCT 116 cells. Cells were cultured in the presence or absence of ADR (100 ng/mL) for 24 hours. Mature-type miRNAs were measured by qRT-PCR. miR-101 and miR-30c, whose expression was not affected by p53, were used as negative controls. C, upregulation of C17orf91 by ADR. The cells were treated with ADR (100 ng/mL) for the indicated times, and C17orf91 was quantified by TaqMan qRT-PCR. D, upregulation of miR-22 by introduction of C17orf91 cDNA. Cells were transfected with an expression vector containing C17orf91 cDNA (Supplementary Fig. S4) for 48 hours. The expression of miR-22 was analyzed by qRT-PCR. E, ChIP-sequence analysis. Genomic region of C17orf91 indicates opposite direction as shown in (A) because of C17orf91 gene encoded on minus strand in the chromosome 17. HCT 116 cells were treated with a DNA-damaging agent, 5-FU, for 9 hours, and ChIP was carried out by using the indicated antibodies. Red boxes show p53BS located at 5’ upstream region and within intron 2. The direction of C17orf91 gene is indicated by arrows.
Identification of p21 as a direct target of miR-22
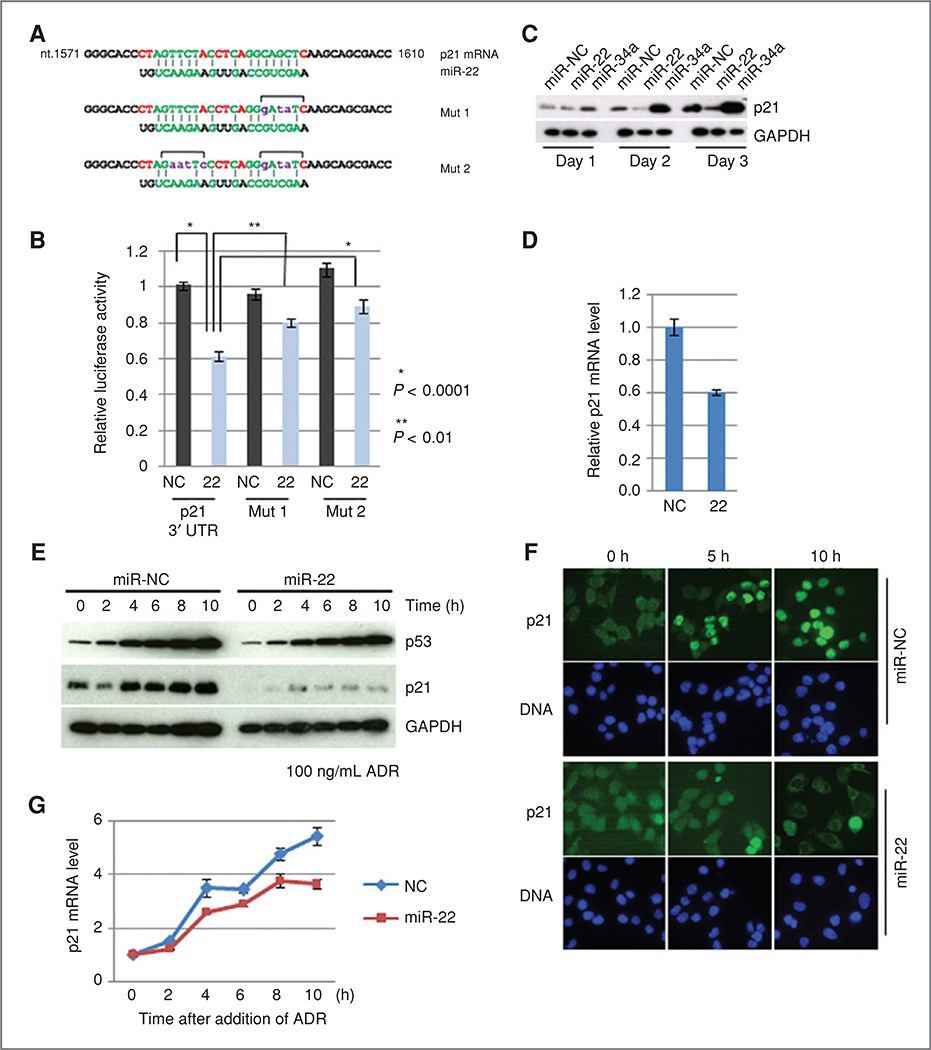
To identify the miR-22 target mRNAs involved in p53-dependent apoptosis, AGO2-immunoprecipitation (AGO2-IP) on ChIP analysis (31) was applied to screen mRNA species enriched in the AGO2 complex in a miR-22 dependent manner; an in silico database search was further carried out using candidate mRNAs. This strategy was expected to lead to the efficient identification of responsible miRNA targets. HCT 116 cells, stably expressing HA-AGO2, were transfected with miR-22, and the AGO2 complex was precipitated with anti-HA antibody, followed by the microarray analysis of the precipitated RNAs (Supplementary Fig. S6A). After calculation of the enrichment score (Supplementary Fig. S6B), 10 mRNAs were selected as miR-22-dependent AGO2-bound mRNAs, which included regulators of apoptosis and the cell cycle (Supplementary Fig. S6C and D). A search of the TargetScan database (34) using the top10 mRNAs revealed that only p21 was a potential target for miR-22. Indeed, p21 had a potential miR-22 target sequence, whose site was conserved among the other mammalian species (Fig. 4A and Supplementary Fig. S6E). The expression of a luciferase reporter gene fused with the 3’ UTR of p21 mRNA was suppressed by the introduction of miR-22 (Fig. 4B). This suppression was significantly reduced by the introduction of mutations into the miR-22 response sequence (Fig. 4B, Mut1 and Mut2), indicating that miR-22 represses p21 directly. Furthermore, ectopic expression of miR-22 in HCT 116-p53+/+ cells reduced p21 protein levels (Fig. 4C). Suppression of p21 mRNA levels was also observed by introduction of miR-22 (Fig. 4D). These results show that miR-22 controls p21 expression by both inhibition of translation and degradation of mRNA.
Figure 4.
A. sequence alignment of miR-22 and the 3’ UTR of p21 mRNA is indicated at the top. Mutant sequences used for the reporter gene assay are listed (Mut 1 and Mut 2). B, reporter gene assay. Error bar indicates SD (n = 6). C, expression level of p21 protein in the presence of miR-22. MiR-34a was used as positive control. D, expression levels of p21 mRNA in the presence of miR-22. Cells were transfected with miR-22, and incubated for 3 days. The relative expression levels of p21 mRNA were quantified by TaqMan assay. E, effect of miR-22 on the activation of p21 expression after exposure to ADR. HCT 116 cells were transfected with 5 nmol/L of either miR-22 or miR-NC and incubated for 48 hours, and further incubated in the presence of ADR for the indicated times. F, indirect immunocytochemistry. Cells were transfected as described above, and incubated in the presence of ADR for the indicated times. Cells were subjected to immunostaining. G, activation of p21 expression in the presence or absence of miR-22 after exposure to ADR. Cells were prepared as described in (E), and total RNAs were prepared from each time point. Relative expression levels of p21 mRNA were quantified by TaqMan assay.
As shown in Fig. 4E, miR-22 inhibited the ADR-induced upregulation of p21. Immunocytochemical analysis showed no nuclear accumulation of p21 in miR-22 introduced cells, even after a 10-hour ADR treatment (Fig. 4F). To show that this repression occurs at a post-transcriptional, but not at a transcriptional level, the ADR-induced increase in p21 mRNA was quantitatively assessed in the presence or absence of miR-22. As expected, transcriptional activation of p21 was observed with similar kinetics as the p53 response in both miR-NC and miR-22 introduced cells after ADR treatment (Fig. 4G). These observations suggest that miR-22 directly represses p21 expression via a post-transcriptional mechanism.
Sensitization of p53-dependent apoptosis by miR-22
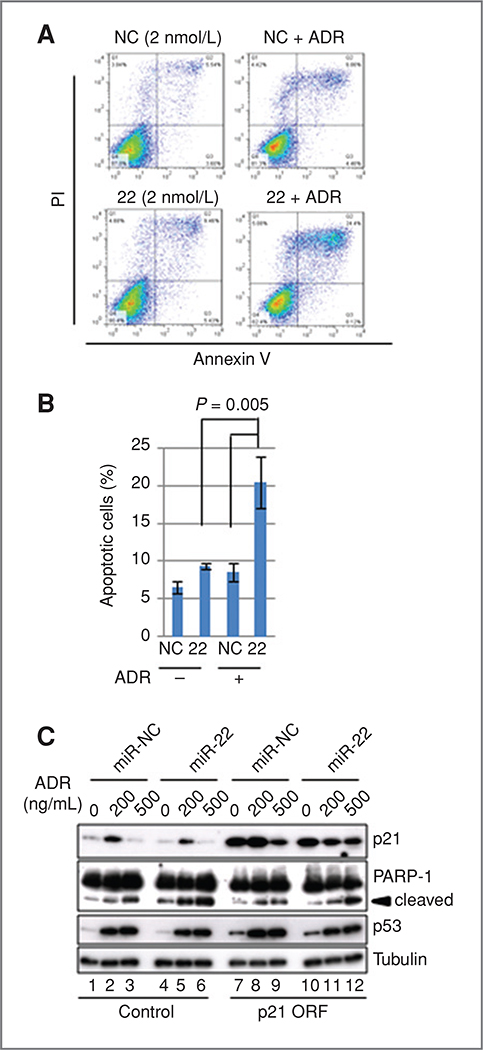
p21 is known to be a key regulator of cell-cycle arrest after the activation of p53, and also an inhibitor of apoptosis (35). Thus, we analyzed the effect of miR-22 levels on the p53-dependent apoptosis. HCT 116-p53+/+ cells were transfected with either miR-22 or miR-NC, and apoptotic cells were quantified by FACS in the presence or absence of ADR. As shown in Fig. 5A, cells transfected with miR-NC showed a slight increase of the Annexin V and PI double-positive fraction after 12-hour exposure to 100 ng/mL of ADR (Fig. 5A, top right and B). The introduction of low amounts (2 nmol/L) of miR-22 slightly enhanced the induction of apoptosis compared with those with miR-NC in the absence of ADR (Fig. 5A, bottom left, and B). The addition of ADR caused a marked increase of apoptotic cells in miR-22-transfected cells (Fig. 5A, bottom right, and B), indicating that miR-22 sensitizes cells to p53-dependent apoptosis induced by DNA damage. Next, we analyzed the effect of p21 protein levels on miR-22-induced apoptosis. MiR-22 caused significant repression of p21 upregulation by ADR treatment for 24 hours (Fig. 5C, lanes 2 and 5, and Supplementary Fig. S7A). The introduction of p21 ORF showed the reduction of apoptosis, evidenced by the decrease in PARP-1 cleavage (36), in cells transfected with miR-22 (Fig. 5C, lanes 5 and 11). This was reproducibly detected (Supplementary Fig. S7B).
Figure 5.
A, sensitization to p53-dependent apoptosis by miR-22. HCT116 cells, transfected with either 2 nmol/Lof miR-22 or miR-NC, were incubated in the presence or absence of 100 ng/mL ADR for 12 hours. Apoptotic cells were determined by FACS. B, quantification of apoptotic cells using 3 independent FACS experiments as described in (A). Data show mean with SD. C, expression of p21 protein reduced miR-22-induced sensitization of apoptosis. HCT 116 cells were transduced with either control or p21 ORF lentivirus. After selection, cells transfected with either miR-NC or miR-22 were treated with indicated concentration of ADR for 24 hours and cleaved PARP-1 was detected by immunoblotting.
These results suggest that endogenous levels of miR-22 are a cellular determinant for the induction of apoptosis through the repression of p21. On the other hand, p21 knockdown induced the cleavage of PARP-1 (Supplementary Fig. S7C). This was consistent with previous reports that p21 deficiency sensitizes cells to apoptosis (37, 38). However, p21 knockdown was not as prominent as is observed by miR-22 introduction. This strongly suggests that other factors, being also regulated by miR-22, could be involved in the sensitization of p53-dependent apoptosis by miR-22. Furthermore, inhibition of miR-22 by expression of an antisense miR-22 transcript causes the substantial decrease of S-phase cells, suggesting the cell-cycle arrest at G1 phase (Supplementary Fig. S7D).
Transcriptional activation of miR-22 depending on the intensity of stresses
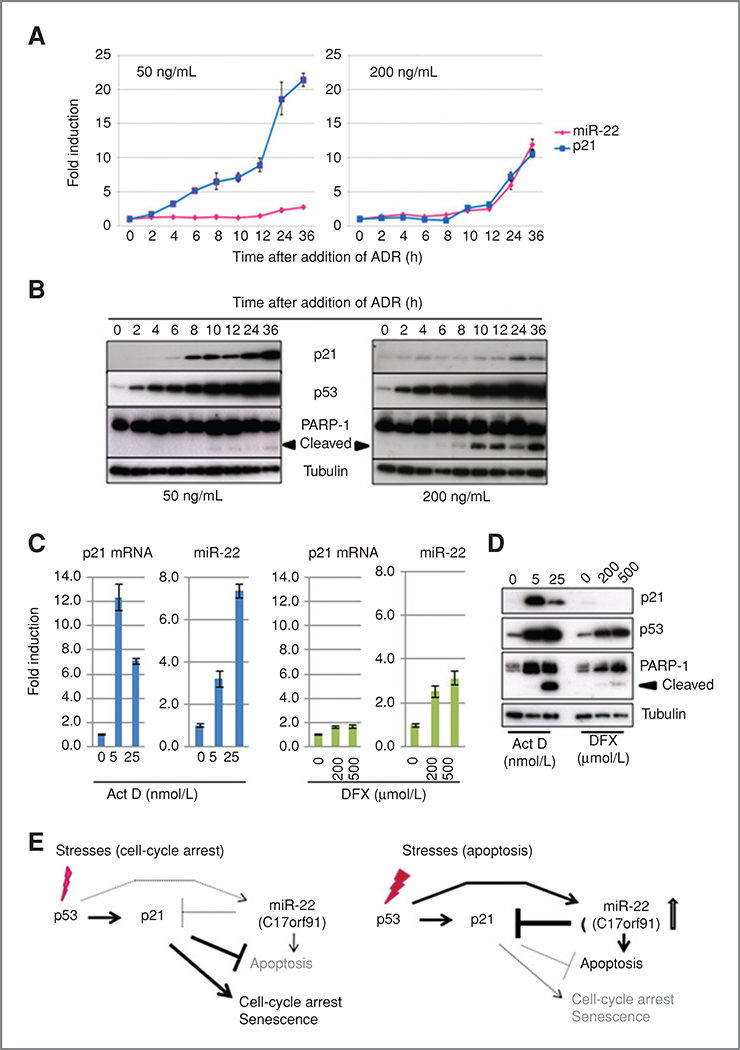
To examine whether the expression of miR-22 and p21 levels correlate with the induction of apoptosis in a physiologic setting, the kinetics of miR-22 and p21 mRNA expression was examined by treating cells with different doses of ADR. As expected, HCT 116 cells treated with 50 ng/mL of ADR showed cell-cycle arrest, but no apoptosis, with rapid increments of p21 at both mRNA and protein levels; upregulation of miR-22 was not observed, even after the ADR-mediated activation of p53 (Fig. 6A, left graph, and 6B, left). Under a high-dose exposure to ADR, in contrast, the expression levels of p21 mRNA and miR-22 increased from 8 hours after the addition of 200 ng/mL of ADR (Fig. 6A, right). Interestingly, p21 protein levels were not elevated significantly after 36 hours of incubation with ADR, despite the striking increase in p21 mRNA level (Fig. 6B, top right). The PARP-1 cleavage was observed at a similar kinetics with miR-22 expression (Fig. 6B). Similarly, a significant activation of miR-22 accompanying the repression of p21 protein and increase of PARP-1 cleavage was also observed in HCT 116 cells after exposure to high doses of actinomycin D (Act D), an inhibitor of RNA polymerases that activates p53 (ref. 39; Fig. 6C and D). ChIP analysis indicated the enhancement of p53 binding to p53BS in the miR-22 gene only after addition of high doses of Act D (Supplementary Fig. S8A and B). Interestingly, treatment with deferoxiamine, an inducer of HIF1a that stabilizes and activates p53 (40), did not upregulate miR-22 or p21 mRNA and did not induce apoptosis despite the activation of p53 (Fig. 6C and D). These results indicate that the activation of miR-22 regulated by p53 is dependent on the strength and type of stresses.
Figure 6.
A and B, kinetics of miR-22 and p21 increments after exposure to ADR. Time-dependent increments of p21 mRNA and miR-22 after exposure to different doses of ADR were quantified by RT-PCR. Relative expression of p21 and miR-22 was calculated by 2-ΔΔct using GAPDH as an internal standard (A). Protein levels of p21, p53, and cleaved PARP-1 were analyzed by immunoblotting in cells treated with different doses of ADR (B). C and D, activation of miR-22 expression by Act D. Cells were treated with specific concentration of either Act D or deferoxiamine for 24 hours, and p21 and miR-22 levels were determined as described above. E, 2 modes of action of miR-22 in the p53 network.
Discussion
In the present study, miR-22 was identified as a strong candidate for tumor suppressor gene in human colon cancers, and its role in the determination of p53-dependent cellular fate through the formation of the p53-miR-22-p21 axis was shown. This axis might be activated by specific stresses that require the elimination of damaged cells. The current findings provide a novel insight into the regulatory mechanism of cell fate determination by a specific molecule, miR-22, in response to various oncogenic stresses and in a p53-dependent manner.
As depicted in Fig. 6E, 2 modes of action of the p53-miR- 22-p21 axis were suggested in response to the different intensity of the stresses applied. In brief, p53 only activates p21 to induce cell-cycle arrest against weak stresses in the p53-p21 pathway. On the other hand, severe damage transcriptionally activates both p21 and miR-22, and miR-22 represses p21 expression through the inhibition of protein synthesis and enhancement of p21 mRNA degradation. Under severe damage conditions, apoptosis may be induced by entry into the cell cycle via direct repression of p21 by miR-22.
Antiapoptotic function of p21 has recently attracted attention for its oncogenic action, which is opposed to a traditional tumor suppressor function. Lack of p21 induces apoptosis through the accumulation of DNA damage in leukemic stem cells (41). Disruption of the p21 gene sensitized cancer cells to apoptosis after treatment with chemotherapeutic agents (37, 38). Recently, the small molecule RITA, an activator of p53, was shown to efficiently induce apoptosis through inhibition of p21 (42). Furthermore, a single recombinant adenovirus containing p53 cDNA and synthetic p21 shRNA also efficiently induced apoptosis in colon cancer cell lines (43). These findings indicate that the downregulation or inhibition of p21 after activation of p53 in stressed cells is one of the key factors as an anticancer mechanism by inducing the change of cellular phenotype from cell-cycle arrest to apoptosis, which could be the mechanism triggered by miR-22 as an intrinsic stress-response network.
MicroRNAs are known to repress multiple target mRNAs, leading to efficient shut down or activation of intracellular networks (44, 45). Indeed, introduction of miR-22 broadly and significantly modulates cellular networks in p53 wild-type HCT 116 cells (Supplementary Tables S3, S4, and Fig. S3D). Furthermore, high levels of expression of miR-22 alone clearly showed apoptosis without activation of p53 in HCT 116 cells (Fig. 2A), where p21 might not be a promising target of miR-22, suggesting that other miR-22 target genes also contribute to the induction of p53-dependent apoptosis.
In addition to the miR-22 function in p53 wild-type colon cancer cells, another interesting feature is that miR-22 expression induces cell-cycle arrest in p53 knockout and mutant cell lines (Supplementary Fig. S3B and C). We searched for a TargetScan database to obtain a list of potential miR-22 targets and conducted gene ontology analysis to identify genes whose repression theoretically induces cell-cycle arrest. These analyses indicated that several positive cell-cycle regulators, CDK6, CDK3, SIRT1, CDC25B, and HDAC4, are possible targets of miR-22. We analyzed the protein levels of CDK6 and SIRT1 by immunoblot analysis, and found no significant changes in their protein levels in the presence of miR-22 in SW480 cells. Then, we re-evaluated the data of AGO2-IP on ChIP analysis using HCT 116 cells. Interestingly, CDK3, CDC25B, and HDAC4 mRNAs were enriched in the AGO2 complex in a miR-22-dependent manner (data not shown). Although it is currently unclear that these mRNAs are directly downregulated by miR-22, miR-22 may induce p53 independent cell-cycle arrest through the repression of these genes.
The present data suggest a tumor-suppressive role of miR-22 in colon cancer cells. MiR-22 was also reported to be downregulated in estrogen receptor (ER)-positive breast cancers, and repression of ER expression by miR-22 suppressed cell proliferation (46, 47). On the other hand, miR-22 was recently suggested to have an oncogenic role through the direct silencing of PTEN and its upregulation in prostate cancer cell lines (48). These authors identified the transforming activity of miR-22 in mouse embryonic fibroblast cooperatively with c-Myc, and showed that the overexpression of miR-22 in the prostate cancer cell line DU145, harboring p53 mutations in both alleles, caused an enhancement of colony formation. The reported paradoxical function of miR-22 implies that miR-22 could act as a tissue-specific or context-dependent tumor suppressor gene.
Chromosome 17p13.3, where the miR-22 gene resides, is well known to be a target for allelic loss, and loss of heterozygosity in 17p13.3 is often found independently of the TP53 mutation in human cancers, including lung and breast cancer (49, 50). Furthermore, an unknown tumor suppressor gene has been suggested to be present at this locus. The present data suggest that miR-22 is a candidate haploinsufficient-type tumor suppressor gene within this region, and its hemizygous loss or downregulation reduced apoptosis induction in response to stresses, even in cells retaining an intact TP53.
In summary, the data presented suggest a role for miR-22 as an intrinsic molecular switch in the p53 tumor suppressor network, functioning as a determinant of cell fate at a post-transcriptional level by inducing apoptosis via direct repression ofp21. This system might function in the p53-dependent activation of a specific anticancer barrier in response to various oncogenic stresses, and dysfunction of miR-22 might confer a chance of survival for damaged cells with tumorigenic potential.
Supplementary Material
Acknowledgments
We would like to thank Drs. Hirofumi Arakawa, Masato Enari, Hiroki Sasaki, Kazuhiko Aoyagi, and Ryo-u Takahashi at NCCRI for providing us with antibodies, the p21 expression vector, bioinformatics analysis, technical support, and helpful discussions.
Grant Support
This work is supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO), a Grant-in-Aid for 3rd Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labor and Welfare, Japan, a grant from Takeda Science Foundation (H. Nakagama), and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports & Technology of Japan (N. Tsuchiya). H. Ogata-Kawata is an awardee of the Research Resident Fellowship from the Foundation for Promotion of Cancer Research Japan for the 3rd Term Comprehensive 10-Year Strategy for Cancer Control.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323–31. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell 2009;137:413–31. [DOI] [PubMed] [Google Scholar]
- 3.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J 2003;22:4212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene 2005;24:2899–908. [DOI] [PubMed] [Google Scholar]
- 5.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000;288:1053–8. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 2001;7:673–82. [DOI] [PubMed] [Google Scholar]
- 7.MiyashitaT Reed JC. Tumorsuppressorp53 is a direct transcriptional activator of the human bax gene. Cell 1995;80:293–9. [DOI] [PubMed] [Google Scholar]
- 8.Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol 2000;20:3224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruse JP, Gu W. Modes of p53 regulation. Cell 2009;137:609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002;4:11–9. [DOI] [PubMed] [Google Scholar]
- 11.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000;102:849–62. [DOI] [PubMed] [Google Scholar]
- 12.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 2006;24:827–39. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V, Lee RC, Lavanway A,Williams PT, Jewell D. MicroRNAs and Other tiny endogenous RNAs in C. elegans. Curr Biol 2003;13:807–18. [DOI] [PubMed] [Google Scholar]
- 14.Plasterk RH. Micro RNAs in animal development. Cell 2006;124: 877–81. [DOI] [PubMed] [Google Scholar]
- 15.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008;9:219–30. [DOI] [PubMed] [Google Scholar]
- 16.Voorhoeve PM, Agami R. Classifying microRNAs in cancer: the good, the bad and the ugly. Biochim Biophys Acta 2007;1775:274–82. [DOI] [PubMed] [Google Scholar]
- 17.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanai-hara N, et al. MicroRNA expression profiles associated with prog-nosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008;299:425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Good-son S, et al. A microRNA polycistron as a potential human oncogene. Nature 2005;435:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–8. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007; 449:682–8. [DOI] [PubMed] [Google Scholar]
- 22.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 2008;40:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermeking H p53 enters the microRNA world. Cancer Cell 2007;12:414–8. [DOI] [PubMed] [Google Scholar]
- 25.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007;26:731–43. [DOI] [PubMed] [Google Scholar]
- 26.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A 2007;104:15472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 2008;105: 13421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumiya M, Okamoto K, Tsuchiya N, Nakagama H. Functional screening using a microRNA virus library and microarrays: a new high-throughput assay to identify tumor-suppressive microRNA. Carcinogenesis 2010;31:1354–9. [DOI] [PubMed] [Google Scholar]
- 30.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998;282:1497–501. [DOI] [PubMed] [Google Scholar]
- 31.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A 2007;104:19291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature 2009;460:529–33. [DOI] [PubMed] [Google Scholar]
- 33.Sims RJ 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev 2006;20:2779–86. [DOI] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20. [DOI] [PubMed] [Google Scholar]
- 35.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 1994;371:346–7. [DOI] [PubMed] [Google Scholar]
- 37.McDonald ER 3rd, Wu GS, Waldman T, El-Deiry WS. Repair defect in p21 WAF1/CIP1 —/— human cancer cells. Cancer Res 1996;56:2250–5. [PubMed] [Google Scholar]
- 38.Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene 1999;18:4808–18. [DOI] [PubMed] [Google Scholar]
- 39.Choong ML, Yang H, Lee MA, Lane DP. Specific activation of the p53 pathway by low dose actinomycin D: a new route to p53 based cyclotherapy. Cell Cycle 2009;8:2810–8. [DOI] [PubMed] [Google Scholar]
- 40.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 1998;392:405–8. [DOI] [PubMed] [Google Scholar]
- 41.Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature 2009;457:51–6. [DOI] [PubMed] [Google Scholar]
- 42.Enge M, Bao W, Hedstrom E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell 2009;15:171–83. [DOI] [PubMed] [Google Scholar]
- 43.Idogawa M, Sasaki Y, Suzuki H, Mita H, Imai K, Shinomura Y, et al. A single recombinant adenovirus expressing p53 and p21-targeting artificial microRNAs efficiently induces apoptosis in human cancer cells. Clin Cancer Res 2009;15:3725–32. [DOI] [PubMed] [Google Scholar]
- 44.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 2010;12:372–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajewsky N microRNA target predictions in animals. Nat Genet 2006;38 Suppl: S8–S13. [DOI] [PubMed] [Google Scholar]
- 46.Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 2009;29:3783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y, et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J 2010;277:1684–94. [DOI] [PubMed] [Google Scholar]
- 48.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 2010; 3:ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornelis RS, van Vliet M, Vos CB, Cleton-Jansen AM, van de Vijver MJ, Peterse JL, et al. Evidence for a gene on 17p13.3, distal to TP53, as a target for allele loss in breast tumors without p53 mutations. Cancer Res 1994;54:4200–6. [PubMed] [Google Scholar]
- 50.Konishi H, Takahashi T, Kozaki K, Yatabe Y, Mitsudomi T, Fujii Y, et al. Detailed deletion mapping suggests the involvement of a tumor suppressor gene at 17p13.3, distal to p53, in the pathogenesis of lung cancers. Oncogene 1998;17:2095–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.