Abstract
Manipulation of PPAR activity is often a valuable approach toward elucidation of the cellular effects of PPARs. The activity of specific PPARs can be decreased using chemical inhibitors, but these approaches can be affected by nonspecific interactions or cell toxicity. Alternative approaches include targeting PPAR gene expression or activity through molecular biology strategies. Here, we describe the targeting of PPARγ through dominant-negative and siRNA-mediated knockdown constructs.
Keywords: Peroxisome proliferator-activated receptor, Dominant-negative, Adenovirus, siRNA
1. Introduction
Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors that regulate expression of target genes (1). Paramount to the elucidation of biological effects of PPARs is the demonstration that loss-of-function of the particular PPAR results in perturbation of the effect. A common approach to reduce PPAR activity is the use of chemical inhibitors that block ligand binding and activation of PPAR transcriptional activity. While useful in many systems, chemical inhibitors can have nonspecific and cytotoxic effects that can perturb the results. Therefore, alternative approaches are often required, such as those involving the use of dominant-negative mutants or reduced PPAR expression.
The transcriptional activity of PPARs relies on ligand binding, association with coactivator or corepressor proteins, and DNA binding to target gene promoter regions. Numerous mutant forms of PPARγ have been produced that result in dominant-negative forms of PPARs, as a result of impaired binding to coactivators or constitutive binding to corepressors, or inability to form a retinoid X- receptor-DNA complex (2–4). While dominant-negative contructs are of tremendous value in pinpointing the cellular effects of PPARs, many cell lines, especially primary cells, are difficult to transfect. Therefore, retroviral delivery of constructs can be a useful approach toward the use of dominant negative PPARs. Viral particles (such as adenovirus and lentivirus) carrying transcripts of interest are capable of infecting a wide number of cells without the need for lipofection or electroporation. Such approaches have been successfully employed frequently for PPARγ (2, 4–9), as well as PPARα (10) and PPARβ/δ (11, 12). Here, we describe adendovirus-mediated delivery of dominant-negative PPARγ (8) to reduce PPARγ transcriptional activity (13, 14).
The use of small inhibitory RNA (siRNA) to reduce the expression of target proteins is now a commonplace approach to the study of protein function. The method involves the design of small (19–23 bp) fragments of double-stranded RNA that bind to a protein complex known as RNA-induced silencing complex (RISC) (15). The RISC complex then binds to the target mRNA to form a triplex structure, followed by degradation of the mRNA complex by the nuclease Argonaute (16). Selected siRNA constructs are introduced into target cells using transfection reagents or electroporation approaches. Alternatively, the siRNA constructs can be delivered as short hairpin RNA (shRNA) inserted into plasmid or viral vectors for expression in cells. Because of possible global effects on expression caused by the response, it is critical to include controls using siRNA contructs targeting an unrelated protein (often housekeeping genes such as GAPDH) or nontargeting siRNA that will trigger the RISC machinery, target no mRNA species. Many suppliers now provide predesigned pools of 3–4 different siRNA constructs against a particular target to maximize the knockdown while minimizing off-target effects (17). We describe below the use of an siRNA pool (On-Target Plus, Thermo Scientific, Lafayette, CO) introduced into cells by an electroporation approach (Nucleofector, Lonza, Walkersville, MD), which we have used successfully to knockdown PPARγ (13, 14).
A number of methods can be employed to determine the effect of the above approaches on PPAR activity and expression. Because the activity of PPARs is employed at the protein level, the transcriptional activity or protein levels of PPARs should be determined whenever possible. However, the transcript level can provide a rapid means of assessing siRNA-mediated knockdown of PPAR expression for higher throughput screening methods. We describe below means to carry all of these determinations.
2. Materials
2.1. Adenovirus Amplification, Purification, and Infection
Healthy, low-passage HEK-293 cells (American Type Culture Collection, Manassas, VA).
Modified Eagle’s Medium (MEM), supplemented with 10% fetal bovine serum.
Dominant-negative, wild-type, and control β-galactosidase (β-Gal) adenovirus stock.
Dry ice/ethanol bath.
Adenovirus Mini Purification Virakit (Virapure, San Diego, CA). Includes columns and loading, wash, and elution buffers.
Phosphate buffered saline 10× solution (PBS): 1.4 M NaCl, 27 mM KCl, 81 mM Na2HPO4, 15 mM KH2PO4. Dissolve 80 g NaCl, 2 g KCl, 11.5 g Na2HPO4 (anhydrous), 2 g KH2PO4 (anhydrous) in ~800 mL of deionized, endotoxin-free water. Adjust pH to 7.4 if necessary, bring to 1 L final volume. Sterile filter. Dilute with endotoxin-free water to 1× before use.
100% ice-cold methanol fixative.
PBS +1% BSA: To 100 mL1× PBS, add 1 g bovine serum albumin (BSA). Stir until dissolved, sterile filter, and store at 4°C.
Adeno-X Rapid Titer kit (Clontech, Mountain View, CA). Includes mouse anti-hexon primary antibody, horseradish peroxidase (HRP) conjugated rat anti-mouse secondary antibody, DAB substrate, and peroxidase solution.
Experimental cells for adenoviral infection.
Fixative for β-galactosidase assay: 0.05% glutaraldehyde in 1× PBS. Prepare fixative fresh on the day of use by diluting 100 μL 25% glutaraldehyde solution (Sigma Chemical, St. Louis, MO) with 1× PBS.
X-Gal stock solution: 40 mg/mL X-gal (5-bromo-4-chloro-3-indolyl-b-galactopyranoside) in dimethylformamide. Protect from light. Store at −20°C.
X-Gal buffer: 2 mM MgCl2, 35 mM potassium ferrocyanide, 35 mM potassium ferricyanide, 0.02% Nonidet P-40, 1× PBS.
X-Gal staining solution: Make fresh the day of use. Dilute X-Gal stock solution 1:40 in X-Gal buffer.
2.2. Knockdown of PPAR Expression with siRNA
Cultured cells of interest in log phase growth.
Nucleofector kit appropriate for cells of interest. Includes nucleofection solution and cuvettes.
Nucleofector II electroporation instrument.
Stock PPAR and control nontargeting siRNA solutions (20 nM).
Fluorescent siRNA for nucleofection efficiency (optional).
2.3. PPRE Luciferase Assay
PPRE reporter plasmid (e.g., ACO-PPRE-luc (18)).
Renilla luciferase reporter plasmid (e.g., pRL-TK, Promega, Madison WI).
Transfection reagent appropriate for cell line of interest.
Dual-Glo luciferase assay (Promega, Madison, WI).
3. Methods
3.1. Adenoviral Delivery of Dominant-Negative PPAR Constructs
3.1.1. Amplification and Purification of Adenoviral Vectors
Seed low-passage HEK-293 cells (see Note 1) onto a 100 mm2 culture dish and grow to ~90% confluency. Infect the cells with experimental and control adenoviruses separately (see Note 2). The amount of adenovirus to use will depend on the nature and purity of the individual preparations. A general rule of thumb is 150–300 μL of crude virus, or 30–100 μL of purified virus.
Culture for 2–4 days, checking daily for signs of the cytopathic effect (CPE). Initially, the CPE is detectable as a loss of attached cells in a grape-shaped area, usually in the center of the dish. When 80–90% of the cells have detached, collect the suspended and attached cells by pipetting up and down over the growth surface until all cells are suspended.
Pellet the cells at 1,000 × g for 5–10 min. Remove and discard all but 1 mL of the supernatant. Resuspend the cell pellet in the remaining 1 mL of medium.
Lyse the cells by freezing the cell suspension in a dry ice/ethanol bath, followed by thawing in a 37°C water bath. Vortex to mix the cells. Repeat for a total of three freeze-thaw cycles. Centrifuge the suspension at 10,000 × g for 10 min, and collect and save the supernatant. The clarified crude lysate can be stored at −80°C, or processed to purification as below
The crude lysate is further purified using the Adenovirus Mini Purification Virakit (Virapure, San Diego, CA). Each purification column can purify up to one half the total crude lysate collected from a 100 mm2 culture. All centrifugation steps are carried out at 2,200 × g for 5 min.
Condition the purification column by applying 0.4 mL loading buffer and centrifuge. Discard the flowthrough. Load up to 0.4 mL of crude lysate (see step 4 above) and centrifuge. Discard the flowthrough. If necessary, repeat the loading and centrifugation steps with additional lysate, up to half of the original crude lysate per column.
Wash the column by applying 0.4 mL of wash buffer to the column and centrifuging. Discard the flowthrough, and repeat the wash procedure two more times.
Place the column into a fresh sterile microcentrifuge tube. Add 0.4 mL of elution buffer to the column and centrifuge. The flowthrough contains the purified virus. Store the virus at −80°C, or proceed to determination of viral titer (see Note 3).
3.1.2. Determination of Viral Titer
Prepare serial dilutions of the purified viral stock in sterile PBS. The dilutions should range from 102–107 -fold dilutions, for a total of six viral concentrations.
Seed low-passage log phase (recently passaged) HEK-293 cells into a 12-well plate at 5 × 105 cells per well in 1 mL of medium. Add 100 μL PBS (vehicle control) to one well, and 100 μL of the 102 -fold dilution to a second well. Add 100 μL/well of each remaining diluted virus, two wells per dilution. Allow the cells to incubate in a culture hood for 48 h.
Carefully aspirate the media from the cells. Exercise caution, because the infected cells will detach easily. Leave the plate open in a laminar flow hood for 5 min to dry.
Fix the cells by gently adding 100% ice-cold methanol to each well along the well walls using a pipet. Incubate the plates at −20°C for 10 min. Aspirate methanol and wash the wells three times with 0.5 mL PBS + 1%BSA.
Add 250 μL of diluted mouse anti-hexon antibody (1:1,000 in PBS + 1% BSA) to each well. Incubate for 1 h at 37°C on an orbital shaker at low speed. Aspirate the anti-hexon antibody, and rinse the wells three times with 0.5 mL of PBS + 1% BSA. Add 250 μL of the secondary antibody (HRP-conjugated rat anti-mouse, 1:500 in PBS + 1% BSA), and incubate for 1 h at 37°C on an orbital shaker at low speed.
Prepare the 1× DAB working solution by diluting 600 μL DAB substrate solution with 6 mL peroxidase buffer. Bring working DAB solution to room temperature.
Remove the secondary antibody by aspiration, and rinse the wells three times with 0.5 mL of PBS + 1% BSA. Add 250 μL of 1× DAB working solution to each well, and incubate the plate at room temperature for 10 min. Protect the plate from light during the incubation. Remove the DAB solution by aspiration, and then add 1 mL PBS to each well.
Using an inverted microscope, observe a random field of cells under a 20× objective. Hexon-positive (infected) cells will appear as dark brown or black cells. The vehicle control well should have no hexon-positive cells. The wells with the higher concentration of virus may show morphological signs of cell death, and all cells may be hexon-positive. Looking at all of the wells, find the viral dilutions that resulted in 10% or fewer hexon-positive cells. Count the number of hexon-positive cells per field, a minimum of five fields per well. Calculate the mean positive cells per field for each well.
- Calculate viral titer in infectious units (ifu)/mL for each well using the following formula:
where 573 is the number of fields per well in a 12-well plate. The mean of the results from all of the counted wells is the ifu/mL for the viral preparation.
3.1.3. Determination of the Optimal Infectious Rate for the Target Cells Using a β-Gal Virus
Seed healthy, log phase cells in a 24-well plate at an appropriate density to reach 70–80% confluency at the time of infection, in a volume of 1 mL per well (see Note 4).
Dilute β-Gal adenovirus to appropriate concentrations to deliver 1–100 MOI (see Note 5). The diluted values should be tenfold higher than the final amount. Add 100 μL diluted virus to the cells. Incubate the cells for 4 h in a cell culture incubator. Remove the medium and replace with growth medium.
After 24–48 h, carefully remove the culture medium (see Note 6). Fix the cells in 0.05% glutaraldehyde for 5 min at room temperature. Add 1 mL per well PBS, aspirate. Add 1 mL PBS, incubate 10 min at room temperature. Aspirate, and then wash once more with 1 mL PBS.
Aspirate the final wash, and add 0.5 mL X-Gal staining solution. Incubate at 37°C for 1 h to overnight. Monitor frequently for development of blue color.
Choose the adenoviral MOI that produced good expression of β-Gal but with the least cytotoxicity (e.g., cell detachment). This adenoviral dilution will be the desired MOI for infection with the PPAR adenoviruses.
3.1.4. Infection of Cells with Wild-Type and Mutant PPARγ Adenoviruses
Calculate the infection strategy for the experiment. For experiments comparing different adenoviral constructs, the total amount of virus used for infection must be the same. A control virus (e.g., β-Gal) can be used to adjust the total (see Note 7).
Infect the cells as above, and incubate for 24 h (or an appropriate period depending on the cells) in a tissue culture incubator.
Assess the effect on PPARγ activity as described below.
3.2. Knockdown of PPAR Expression Using siRNA
The following protocol is for delivery of siRNA into HEK-293 cells by electroporation using the Nucleofector II instrument. The protocol can be easily adapted for use in many different cell lines and primary cells.
Passage healthy, low-passage cells 2 days before nucleofection, to maintain cells in log phase at the time of nucleofection. Make sure the cells are not confluent at the time of harvest and are sufficient for the experiment. For HEK-293 cells, nucleofection requires 1.2 × 106 cells per reaction (see Note 8).
Prepare the following ahead of nucleofection: Power on the nucleofector instrument and select the program Q-001. Prepare nucleofector solution V by adding 4.5 parts to 1 part supplement, and allow it to reach room temperature. For each nucleofection, 100 μL of supplemented solution V will be needed (see Note 8). Prewarm 1 mL of culture medium per nucleofection reaction into wells of a 6-well plate to 37°C in a cell culture incubator. Similarly, prewarm 500 μL culture medium per reaction to 37°C in an appropriately sized tube (see Note 9).
Harvest the cells by standard trypsinization and perform a cell count. Collect the required number of cells by gentle centrifugation. Aspirate the supernatant and resuspend the pelleted cells in 100 μL supplemented nucleofector solution. Do not store the cell suspension in nucleofector solution for more than 20 min (see Note 10).
For each reaction, add 100 μL of the cell suspension to 3–30 pmol of control or PPARγ siRNA (see Note 11). Transfer the cell suspension to a nucleofection cuvette, being sure that the solution covers the bottom surface of the cuvette. Insert the cuvette into the instrument and press the start button. Remove the cuvette immediately after the program has finished and add 500 μL of the prewarmed medium. Transfer the sample to a well of the pre-warmed plate, and incubate the plate in a tissue culture incubator (see Note 12).
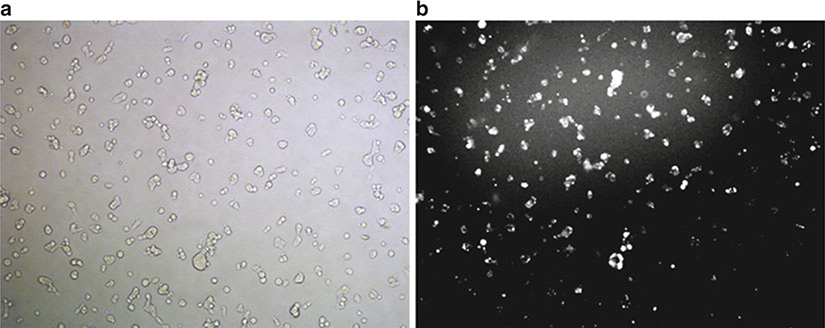
Assay for PPARγ knockdown as described below after 24–72 h. The efficiency of the siRNA transfer can be monitored using fluorescent siRNA (see Fig. 1).
Fig. 1.
Visualization of siRNA localization after a typical nucleofection reaction. HEK-293 cells were subject to nucleofection with fluorescent siRNA (siRNA-GLO Red, Thermo Scientific, Lafayette CO). After 24 h, the cells were visualized and bright field (a) and fluorescent (b) images were captured. The fluorescence reveals efficient delivery of siRNA, predominantly localized to the cytosol and nucleus.
3.3. Assessing PPARγ Activity and Expression
3.3.1. PPAR Response Element Luciferase Reporter Assay
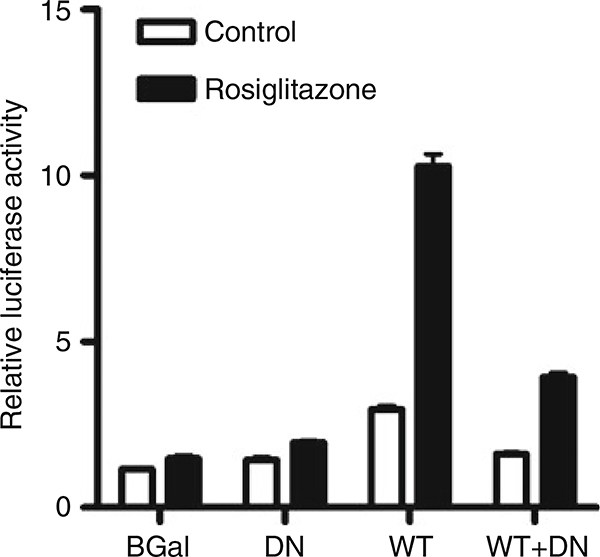
Activation of PPARs results in increased expression of genes regulated by PPAR response elements (PPREs). A number of reporter plasmids have been described using multiple copies of PPREs upstream of luciferase genes (18–21). The effect on transcription can be easily measured by measurement of luciferase activity in the treated cells (see Fig. 2). The procedure below is a description of the assay we have used using a reporter plasmid firefly luciferase driven by the acyl-CoA oxidase PPRE (ACO-PPRE-luc, (21)) reporter plasmid with a Renilla luciferase reporter (pRL-TK, Promega, Madison, WI) as a transfection control.
Fig. 2.
Determination of PPARγ transcriptional activity in cells infected with wild-type (WT) and dominant-negative (DN) adenoviruses. Cells were infected with control (β-Gal), DN, WT, or both DN and WT adenoviruses at an MOI of 1.0 each. Control (β-Gal) adenovirus was used to adjust the total viral load to 2.0 MOI when necessary. After 4 h, the cells were transfected with ACO-PPRE-luc and pRL-TK reporter plasmids, and incubated for 24 h. Cells were treated with rosiglitazone (1 μM) or vehicle, and 24 h later firefly and Renilla luciferase activities were measured. Rosiglitazone caused a marked increase in PPARγ activity in the WT cells, which was reduced in presence of the DN mutant.
Using appropriate conditions for the cell line of interest, transfect cells with ACO-PPRE-luc and pRL-TK at a ratio of 5:1–20:1, respectively, depending on the cell line. The transfection can take place before or after introduction of the adenovirus or siRNA, or concomitantly with nucleofection of the siRNA (see Note 13). Place the cells in a cell culture incubator for 24 h.
Treat the cells with an appropriate PPAR activator (such as a thiazolidinedione for PPARγ, or clofibrate for PPARα) for 24 h.
Measure the firefly and Renilla luciferase activities using the Dual-Glo Luciferase Assay kit (Promega, Madison, WI) (see Note 14). Divide the firefly luciferase activity by the Renilla luciferase activity for each well to determine the relative luciferase (PPRE) activity. Successful dominant-negative inhibition or knockdown in the PPAR of interest should result in decreased relative luciferase activity compared to the appropriate controls.
3.3.2. Western Blotting
Western blotting is a standard protocol in virtually any biomedical research laboratory, and therefore the specific protocol will not be presented here. In general, from 24 to 72 h after application of the siRNA constructs, cell lysates are subject to western blotting to assess the protein levels of the PPAR of interest. In addition, this method can be used to assess the efficiency of viral infection of a PPAR transcript, as many of these retain antigenic properties for recognition by PPAR antibodies. The advantage of this approach is that it provides direct assessment of the protein level of the PPAR of interest. The disadvantage of this procedure is that, unlike the PPRE reporter assay, it provides no information about PPAR transcriptional activity.
3.3.3. Quantitative (Real-Time) PCR
This procedure determines the level of an mRNA of interest relative to that of a control (housekeeping) gene. This procedure has become fairly standard in research laboratories, and so specific procedures will not be provided here. However, because the changes in PPAR activity occur at the protein level, and because changes in protein levels are not always reflected by transcript levels, we do not commonly use this approach in out laboratory. The advantage of this approach is that it is a more rapid, high throughput procedure than PPRE activity assays or western blotting, and therefore is useful for screening potential treatments.
Acknowledgments
Supported by funding through The Department of Veterans Affairs Biomedical Laboratory Research and Development Program, NIAAA (AA015509), and the Bly Memorial Research Fund (RGB), and by a University of Nebraska Medical Center Graduate College Fellowship (SS).
Footnotes
Most adenoviruses lack critical proteins to render them replication-incompetent. The HEK-293 cells are used because they express proteins that allow the viruses to replicate for amplification. Other cell lines can be used for this purpose, such as 911 or GH329.
All procedures with adenoviral particles should be performed by fully trained personnel in accordance with the local Institutional Biosafety Committee. In general, work should be performed in a BSL2 biological safety cabinet. All items that contact the virus (plasticware, surfaces, etc.) should be disinfected with 10% bleach solution for at least 10 min. Although most adenoviral vectors have been rendered replication-incompetent, with extended use recombination events can occur to produce replication-competent virus, greatly increasing the hazardous potential. At each round of purification, the virus should be screened for replication-competent virus by standard approaches (22).
The effectiveness of some viral preparations will decrease with repeated freeze-thaw cycles. Therefore, it is recommended to store viral stocks in working aliquots.
Depending on the cell line, the presence of serum may greatly affect the efficiency of adenoviral infection. It is recommended to test serum at normal growth conditions (e.g., 10% serum), low-serum (e.g., 2%), and serum-free conditions to determine infection efficiency.
The optimal multiplicity of infection (MOI) will vary for each cell line, and is defined as the ifu per cell. Initial values should range from 1 to 100 MOI. For example, an MOI of 1.0 for a well containing 105 cells would require 105 ifu of virus. It is critical that the selected MOI does not cause cytotoxicity, and therefore the user must have a means of assessing cell death (e.g., cell morphology, lactate dehydrogenase leakage, etc.)
The time required for expression of the desired protein will vary depending on the cell line used. Efficient expression is detectable after 24 and 48 h in most cell lines, but shorter or longer times may be necessary.
When comparing the effects of different viral constructs, the total amount of virus must remain the same. Control or empty viruses can be used to adjust the total viral load. For example, if PPARγ activity is to be compared in cells overexpressing wt-PPARγ with various amounts of dn-PPARγ using an optimal MOI of 10, then cells could be infected with 5 ifu of wt-PPARγ + 5 ifu β-Gal, 5 ifu of wt-PPARγ + 5 ifu dn-PPARγ, or 10 ifu β-Gal.
The cell density and growth phase, nucleofection solution, oligonucleotide concentration, and nucleofector program are critical for the efficiency of the nucleofection procedure, and will vary depending on the cells used. An extensive list of optimized protocols for specific cells (both cell lines and primary cells), as well as a list of user-submitted protocols is available at the manufacturer’s website (www.lonzabio.com).
Cells will be affected differently by the presence of serum and antibiotics in the growth medium. In our hands, the cell viability is often greater if the cells are placed into antibiotic-free medium immediately after the nucleofection reaction.
Exposure to nucleofector solution for more than 20 min can have greatly detrimental effects on cell viability. If multiple nucleofection reactions are planned, the cell collection and resuspension should be staggered to minimize exposure to the solution.
The total amount of oligonucleotide is critical to the success of the experiment. For HEK-293 cells, 3–30 pmol is recommended (30 pmol is 1.5 μL of a 20 μM siRNA stock) to cell suspension. Optimal siRNA concentration should be determined empirically. In addition, co-nucleofection of siRNA with other oligonucleotides (e.g., plasmids) can be accomplished, but the total oligonucleotide amount is limited. See instructions for the specific cell lines. It is critical that appropriate controls are included with each experiment. These may include nontargeting siRNA, or knockdown of unrelated transcripts. Transfection efficiency can be monitored using fluorescently labeled siRNA.
Immediately after the nucleofection reaction, the display on the instrument should read “OK”. If an error message is displayed, refer to the instrument manual for an explanation. The most common causes of an error is failure to completely remove the cell growth medium after centrifugation, or failure to cover the bottom of the cuvette.
When using nucleofection, the reporter plasmids can often be introduced into the cells at the same time as the siRNA constructs, as long as the total oligonucleotide concentration does not exceed the usable level. In addition to Renilla luciferase, other kinds of reporter plasmids can be used for transfection normalization, such as fluorescent protein of β-Gal plasmids. For many cell lines, adherence to cell culture plasticware is decreased after transfection, and therefore cell cultureware coated with poly-d-lysine, collagen I, etc., can greatly improve performance.
References
- 1.Tontonoz P, Spiegelman BM (2008) Fat and beyond: the diverse biology of PPARγ. Annu Rev Biochem 77:289–312 [DOI] [PubMed] [Google Scholar]
- 2.Agostini M, Schoenmakers E, Mitchell C et al. (2006) Non-DNA binding, dominant-negative, human PPARγ mutations cause lipodystrophic insulin resistance. Cell Metab 4:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barroso I, Gurnell M, Crowley V et al. (1999) Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880–883 [DOI] [PubMed] [Google Scholar]
- 4.Masugi J, Tamori Y, Kasuga M (1999) Inhibition of adipogenesis by a COOH-terminally truncated mutant of PPARγ 2 in 3T3-L1 cells. Biochem Biophys Res Commun 264:93–99 [DOI] [PubMed] [Google Scholar]
- 5.Ferguson HE, Kulkarni A, Lehmann G et al. (2009) Electrophilic peroxisome proliferator-activated receptor-γ ligands have potent antifibrotic effects in human lung fibroblasts. Am J Respir Cell Mol Biol 41:722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurnell M, Wentworth J, Agostini M et al. (2000) A dominant-negative peroxisome proliferator-activated receptor γ (PPARγ) mutant is a constitutive repressor and inhibits PPARγ-mediated adipogenesis. J Biol Chem 275:5754–5759 [DOI] [PubMed] [Google Scholar]
- 7.Hata K, Nishimura R, Ikeda F et al. (2003) Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol Biol Cell 14:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park Y, Freedman B, Lee E et al. (2003) A dominant negative PPARγ mutant shows altered cofactor recruitment and inhibits adipogenesis in 3T3-L1 cells. Diabetologia 46:365–377 [DOI] [PubMed] [Google Scholar]
- 9.Wada K, Nakajima A, Katayama K et al. (2006) Peroxisome proliferator-activated receptor γ-mediated regulation of neural stem cell proliferation and differentiation. J Biol Chem 281:12673–12681 [DOI] [PubMed] [Google Scholar]
- 10.Semple RK, Meirhaeghe A, Vidal-Puig A et al. (2005) A dominant negative human peroxisome proliferator-activated receptor (PPAR) α is a constitutive transcriptional corepressor and inhibits signaling through all PPAR isoforms. Endocrinology 146:1871–1882 [DOI] [PubMed] [Google Scholar]
- 11.Bastie C, Luquet S, Holst D et al. (2000) Alterations of peroxisome proliferator-activated receptor δ activity affect fatty acid-controlled adipose differentiation. J Biol Chem 275:38768–38773 [DOI] [PubMed] [Google Scholar]
- 12.Nahle Z, Hsieh M, Pieka T et al. (2008) CD36-dependent regulation of muscle FoxO1 and PDK4 in the PPARδ/β-mediated Adaptation to Metabolic Stress. J Biol Chem 283:14317–14326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S, Bennett RG (2010) Relaxin signaling activates peroxisome proliferator-activated receptor gamma. Mol Cell Endocrinol 315:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh S, Bennett RG (2009) Relaxin family peptide receptor 1 activation stimulates peroxisome proliferator-activated receptor gamma. Ann N Y Acad Sci 1160:112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Weber K et al. (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199–213 [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Carmell M, Rivas F et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441 [DOI] [PubMed] [Google Scholar]
- 17.Reynolds A, Leake D, Boese Q et al. (2004) Rational siRNA design for RNA interference. Nat Biotechnol 22:326–330 [DOI] [PubMed] [Google Scholar]
- 18.Forman BM, Tontonoz P, Chen J et al. (1995) 15-Deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 83:803–812 [DOI] [PubMed] [Google Scholar]
- 19.Palmer CNA, Hsu M, Griffin H et al. (1995) Novel sequence determinants in peroxisome proliferator signaling. J Biol Chem 270: 16114–16121 [DOI] [PubMed] [Google Scholar]
- 20.Varanasi U, Chu R, Huang Q et al. (1996) Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J Biol Chem 271:2147–2155 [DOI] [PubMed] [Google Scholar]
- 21.Jiang C, Ting AT, Seed B (1998) PPARγ agonists inhibit production of monocyte inflammatory cytokines. Nature 391:82–86 [DOI] [PubMed] [Google Scholar]
- 22.Dion LD, Fang J, Garver RI (1996) Supernatant rescue assay vs. polymerase chain reaction for detection of wild type adenovirus-contaminating recombinant adenovirus stocks. J Virol Methods 56:99–107 [DOI] [PubMed] [Google Scholar]