Abstract
Tribbles homolog 3 (TRIB3) is pseudokinase involved in intracellular regulatory processes and has been implicated in several diseases. In this article, we report that human TRIB3 promoter contains a 33-bp variable number tandem repeat (VNTR) and characterize the heterogeneity and function of this genetic element. Analysis of human populations around the world uncovered the existence of alleles ranging from 1 to 5 copies of the repeat, with 2-, 3- and 5-copy alleles being the most common but displaying considerable geographical differences in frequency. The repeated sequence overlaps a C/EBP-ATF transcriptional regulatory element and is highly conserved, but not repeated, in various mammalian species, including great apes. The repeat is however evident in Neanderthal and Denisovan genomes. Reporter plasmid experiments in human cell culture reveal that an increased copy number of the TRIB3 promoter 33-bp repeat results in increased transcriptional activity. In line with this, analysis of whole genome sequencing and RNA-Seq data from human cohorts demonstrates that the copy number of TRIB3 promoter 33-bp repeats is positively correlated with TRIB3 mRNA expression level in many tissues throughout the body. Moreover, the copy number of the TRIB3 33-bp repeat appears to be linked to known TRIB3 eQTL SNPs as well as TRIB3 SNPs reported in genetic association studies. Taken together, the results indicate that the promoter 33-bp VNTR constitutes a causal variant for TRIB3 expression variation between individuals and could underlie the results of SNP-based genetic studies.
Author summary
The current article is devoted to a previously undescribed genetic element residing in the promoter region of human TRIB3 gene and consisting of a variable number of tandemly repeated 33-bp segments. We found that human genome contains from 1 to 5 copies of the repeat and that the copy number distribution varies in human populations around the world. Importantly, an increased number of copies associated with increased expression of the TRIB3 gene in many tissues of the body, giving rise to variation in TRIB3 gene expression between individuals. Mechanistically, the 33-bp segment possesses the ability to activate transcription, and it becomes more potent as it is repeated. The 33-bp segment is repeated in Neanderthals and Denisovans, but not in great apes or other mammals, suggesting that the expansion of the 33-bp unit in the TRIB3 promoter occurred after the split of the hominin and chimpanzee lineages. The TRIB3 protein participates in multiple biological processes as a regulatory factor and is linked to several diseases. Thus, the importance of our study is to shed light on a new mechanism determining TRIB3 expression level variation between individuals, and, more generally, to highlight how tandem repeat variations can serve as regulatory variants.
Introduction
The pseudokinase Tribbles homolog 3 (TRIB3) is known to interact and modulate the function of several transcription factors, protein kinases, ubiquitin ligases and other proteins (for review, see [1]). Through these interactions, TRIB3 has been reported to participate in processes such as the regulation of cellular stress response and cell death [2–5], megakaryocytopoiesis [6], metabolic adaptation to nutrient-limiting conditions [7], and to mediate cancer cell sensitivity to chemotherapeutics [8–13]. Moreover, TRIB3 has been implicated in the pathogenesis of several diseases, including type 2 diabetes and its complications [14–18], Parkinson’s disease [19, 20] and several types of cancer [21–23]. Recently, two novel associations relating TRIB3 genetic variants to disease have been described. Lorenzi et al. [24] identified a link between TRIB3 and the stereotypical pattern of gray matter loss in Alzheimer’s disease, and Yamada et al. [25] identified TRIB3 as a susceptibility locus for ischemic stroke in an exome-wide association study. Therefore, there is a considerable interest to improve our understanding of mechanisms which govern human TRIB3 expression and its variation between individuals.
Variable number tandem repeats (VNTRs) are defined as continuously located repeated DNA sequence motifs with the unit size ≥7 bp. VNTRs evolve as a result of recombination or replication slippage events and there are about ten thousand polymorphic VNTRs in the human genome [26]. It has been found that the unit copy number polymorphism of these loci can have a significant impact on the properties of genes, including gene expression level [27, 28]. Due to the repetitive nature of VNTRs, the analysis of their structure by short-read high-throughput sequencing is complicated [29]. In the current article, we report that the human TRIB3 promoter contains a 33-bp VNTR and characterize this genetic element using a k-mer counting approach to analyze short-read whole genome sequencing (WGS) data as well as search for sequencing reads spanning the VNTR region for direct allele detection. As a result of the analyses, we uncover that alleles with a higher number of the 33-bp repeat copies generate higher levels of TRIB3 expression and that variation in the copy number of the 33-bp repeat could represent the causal variant for a number of single nucleotide polymorphisms (SNP-s) that have been linked to variation in TRIB3 expression level in various tissues of the body.
Results
A VNTR with repeat length of 33 bp resides in the promoter of the human TRIB3 gene
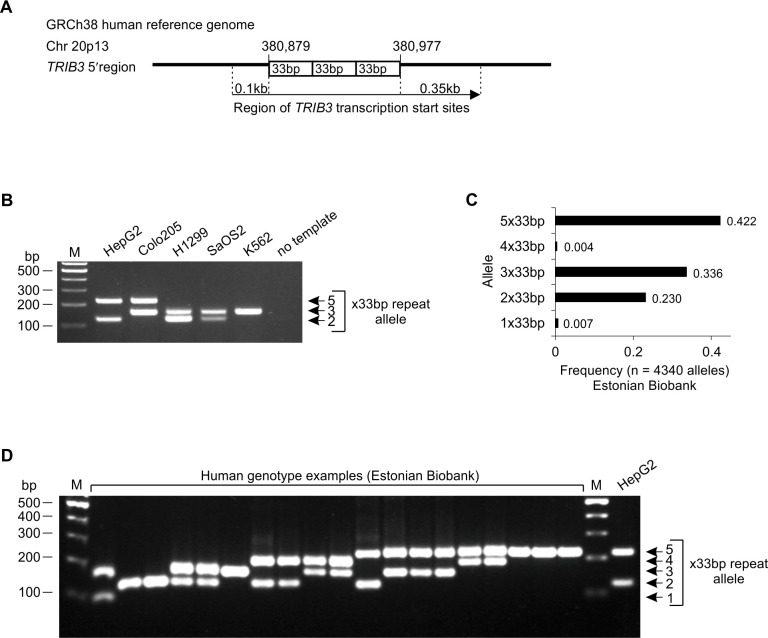
During previous studies of human TRIB3 gene, we observed that the 5’ region of the gene contains a variable number of tandemly repeated segments with the length of 33 bp [30]. TRIB3 transcripts are initiated at several sites extending from about 0.1 kb upstream to 0.35 kb downstream of the 33-bp repeats [30, 31]; thus, the repeats overlap with the promoter region of the gene (Fig 1A).
Fig 1. Characterization of the allelic heterogeneity of the 33-bp VNTR in the human TRIB3 promoter.
(A) A schematic of the TRIB3 gene 5’ region showing the location of VNTR, containing 3 copies of the 33-bp repeat as in the human reference genome (GRCh38). (B) PCR analysis of the TRIB3 promoter 33-bp repeat copy number in human cell lines. The DNA of hepatocellular carcinoma (HepG2), colorectal adenocarcinoma (Colo205), non-small cell lung carcinoma (H1299), osteosarcoma (SaOS2) and chronic myelogenous leukemia (K562) was amplified using primers flanking the VNTR and the products were separated by agarose gel electrophoresis. (C) Human TRIB3 promoter 33-bp VNTR allele frequencies in Estonian Biobank participants, based on whole genome sequencing data. (D) PCR analysis of the 33-bp repeat copy number in a selection of Estonian Biobank participants. The DNA was analyzed as in panel B, and HepG2 is shown as a control. Lane M in panels B and D contains a molecular weight marker (100 bp DNA ladder).
The GRCh38 human reference genome contains 3 copies of this repeating element, spanning positions 380,879 to 380,977 of chromosome 20. To assess the copy number heterogeneity of this repeat, a PCR amplicon using primers flanking the VNTR was designed. Analysis of genomic DNA from commonly used human cell lines reveals several examples of heterozygosity at the 33-bp repeat, with alleles of 2 and 5 copies present in HepG2, 3 and 5 copies in Colo205, and 2 and 3 copies in H1299 and SaOS2; in the case of K562 cells, only the 3-copy allele is detectable (Fig 1B).
To investigate the copy number variation of the TRIB3 33-bp repeats in a general population, we utilized sequencing data and DNA samples from the biobank of the Estonian Genome Center of University of Tartu (EGCUT) [32]. A 150-bp read length WGS data set comprised of >2000 individuals from the Estonian population was used to estimate allele frequencies [33]. From the 150 bp reads, alleles up to 4x33bp were called by searching for spanning reads; since alleles with ≥5 copies of the 33-bp repeat cannot be fully spanned with 150 bp read length, partial spanning was accepted and any reads supporting >4x33bp were designated 5x33bp. The results reveal that in the Estonian population, alleles with 2, 3 or 5 copies of the repeating element all appear to be quite frequent, with 5x33bp being the most common (allele frequency 0.42), followed by 3x33bp (0.34) and 2x33bp (0.23) (Fig 1C). This analysis further uncovered that alleles with 1 or 4 copies of the repeating element also occur, albeit at an allele frequency of less than 1% (Fig 1C).
A selection of individuals carrying the rare (1 or 4 repeats) alleles of the 33-bp repeat based on the WGS were subjected to follow-up PCR analysis, along with examples of individuals carrying the common alleles (2, 3 or 5 repeats). The results of PCR product gel electrophoresis confirmed the occurrence of the 1x33bp and 4x33bp alleles in these individuals (Fig 1D). We additionally sought to uncover potential alleles with >5 copies of the 33-bp repeat by using a k-mer counting technique (described below) on the EGCUT WGS data set; however, PCR-based follow-up of 11 highest ranking individuals (33-bp copy number estimates ranging from 6.13 to 6.69) revealed all of them to be 5x33bp homozygotes (S1 Fig). Similarly, searching for sequencing reads containing a full coverage of four 33-bp repeats and, at either end of that, partial matches for additional 33-bp copies (indicative of a hypothetical ≥6x33bp allele) did not uncover any individuals from the EGCUT WGS data set.
Evolutionary aspects of the human TRIB3 promoter 33-bp repeats: the highly conserved 33-bp segment is repeated also in Neanderthal and Denisovan hominins, but exists in a single copy in other mammalian genomes
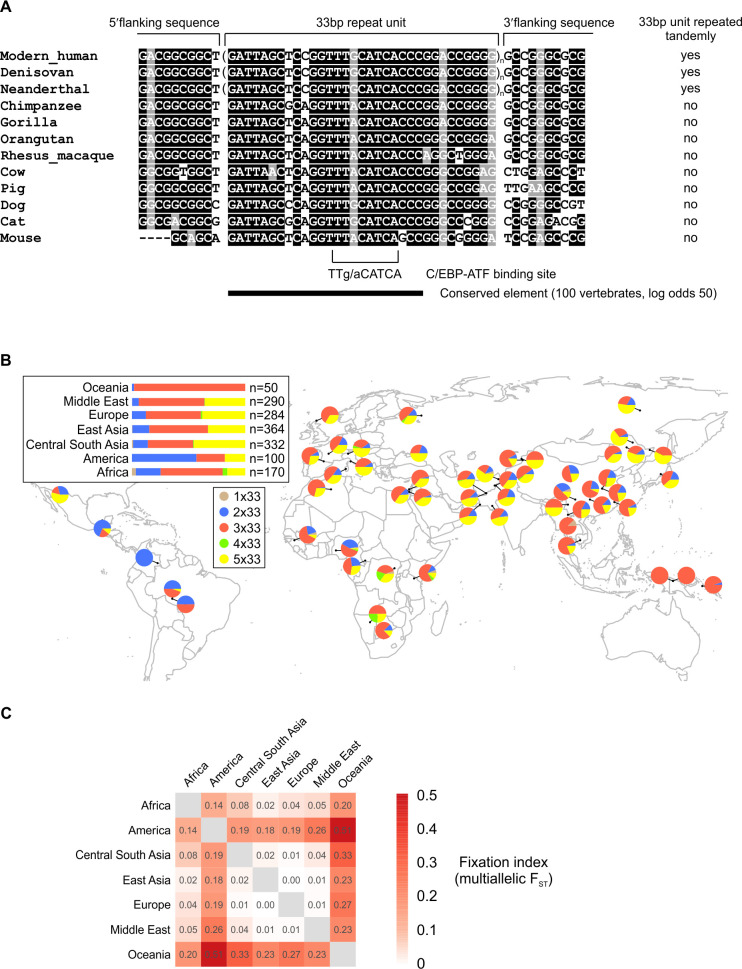
The inspection of mammalian genomes reveals that a segment identical or very similar to the 33-bp repeat element of human TRIB3 promoter is present in the 5’ region of TRIB3 genes from species representing diverse mammalian orders, such as primates, artiodactyls (cow and pig), carnivorans (dog and cat) and rodents (mouse) (Fig 2A). In particular, sequence conservation is strong in the first half of the 33bp segment, which is noted as a constrained element with a log odds score of 50 by PHAST analysis of 100 vertebrate genomes [34, 35].
Fig 2. Evolutionary conservation of the TRIB3 promoter 33-bp VNTR area across mammalian species and the geographical distribution of the TRIB3 33-bp VNTR alleles in human populations.
(A) Nucleotide sequence alignment of the human TRIB3 promoter 33-bp VNTR repeat unit and flanking areas with the corresponding sequence in various mammalian genomes. Only one copy of the repeat is shown for modern human and Denisovan and Neanderthal hominins. Alignment was produced using Clustal Omega and Boxshade, with identical nucleotides shown in black and conserved purines in grey. The location of the C/EBP-ATF binding site is highlighted, overlapping a highly conserved sequence element. (B) A map showing the allele frequency distribution of the human TRIB3 promoter 33-bp VNTR repeat in various populations around the world. The results are based on data from the Human Genome Diversity Project (HGDP). (C) Multi-allelic fixation index (FST) for TRIB3 33-bp repeat copy number alleles in HGDP superpopulations.
Remarkably, the high-coverage genomes of Neanderthal and Denisovan hominins [36, 37] reveal that the TRIB3 5’ regions of these extinct archaic humans contain an allele with at least three copies of the 33-bp repeat, while the TRIB3 5’ regions of the great apes chimpanzee, gorilla and orangutan contain only single copy of this 33-bp segment, similarly to the other mammalian orders (Fig 2A). Thus, the data suggests that the tandem repeat expansion of the 33-bp segment seen in the human TRIB3 promoter occurred after the divergence of the hominins and the great apes but before the divergence of the three branches of hominins. The scarcity of Neanderthal and Denisovan genomic data does not allow a more detailed analysis of these populations. Interestingly, the currently available high-coverage Denisovan genome contains a single nucleotide variation (SNV) in position 10 of the third copy of the 33-bp repeats (heterozygous, C/A). A matching SNV has been reported in modern humans as well, however it is a highly rare variant (rs867414698; gnomAD study-wide alternative allele frequency 0.0021 [38]).
As the genetic variation in modern humans tends to cluster with geographic origin, we also assessed the WGS data generated by Human Genome Diversity Project (HGDP) [39] for TRIB3 33-bp repeat alleles, using the approach described above for EGCUT data. The results, based on approx. 800 individuals spanning 54 geographically distinct world populations, reveal considerable variation in allele frequencies (Fig 2B). In many Asian, European and Middle Eastern populations, the 3x33bp and 5x33bp alleles are both quite frequent and can rank as the most common allele in a population (Fig 2B). The 2x33bp allele appears at a particularly high frequency in some American populations, while the Oceanic populations show nearly exclusively the 3x33bp allele (Fig 2B). For the 1x33bp and 4x33bp alleles, African populations display a noticeably higher incidence compared to populations from other continents (Fig 2B). To evaluate how much of the copy number variation in the TRIB3 33-bp repeat can be explained by population structure, we calculated the multi-allelic fixation index FST, as defined in [40], for pairwise comparisons between the seven superpopulations in HGDP. The results show that there is considerable population differentiation, particularly in Oceania and America relative to the other superpopulations (Fig 2C).
TRIB3 promoter variants containing more copies of the 33-bp repeat show increased transcriptional activity
The conservation of the TRIB3 promoter 33-bp repeat sequence throughout diverse mammalian orders suggests functional importance. In the center of the 33-bp segment, at positions 14 to 21, resides a motif TT(G/A)CATCA, which corresponds to a transcriptional regulatory motif termed the C/EBP-ATF response element (CARE) [30] (Fig 2A). CARE-s mediate gene activation in response to cellular stresses such as endoplasmic reticulum stress, oxidative stress, nutrient deficiency and viral infection [41, 42]. A key transcription factor for driving CARE-dependent gene expression is considered to be Activating Transcription Factor 4 (ATF4), acting as a heterodimer with various C/EBP factors [43].
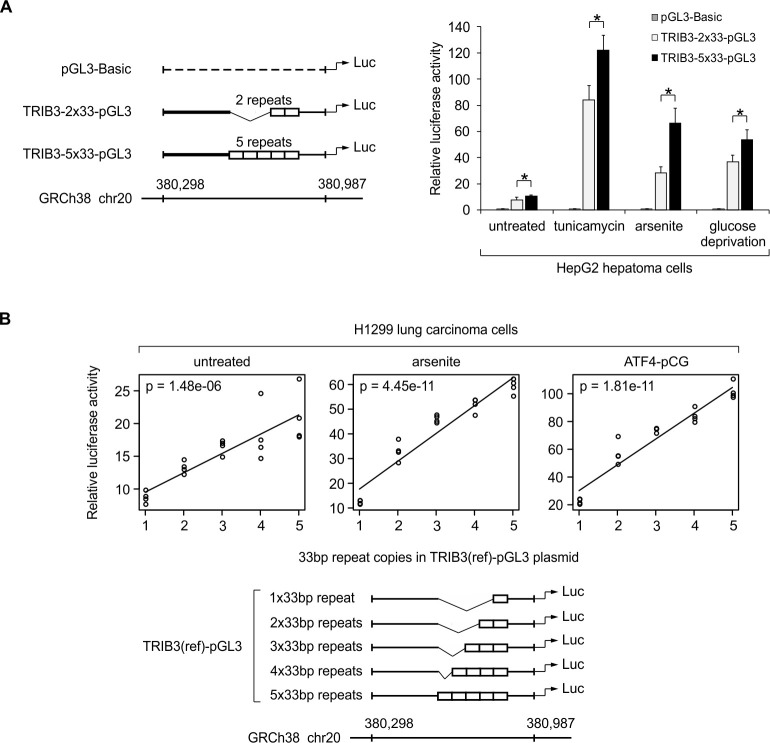
To study whether the copy number of 33-bp repeats affects TRIB3 promoter activity, we generated luciferase reporter constructs which differ by the number of 33-bp repeats. The constructs contain human TRIB3 gene segments up- and downstream of the 33-bp repeats (580 bp and 10 bp, respectively). For Fig 3A, the plasmids contain either two copies (plasmid TRIB3-2x33-pGL3) or five copies of the 33-bp repeat (TRIB3-5x33-pGL3), and both plasmids differ from the GRCh38 reference genome by six common SNPs that are listed in the Materials and Methods. The plasmids were transfected into HepG2 human hepatoma cells and the cells were either left untreated or treated with tunicamycin, an inducer of endoplasmic reticulum stress, with arsenite, an inducer of oxidative stress, or with glucose-free growth medium, to induce nutrient deficiency.
Fig 3. Human TRIB3 promoter variants containing more copies of the 33-bp repeat demonstrate elevated transcriptional activity in luciferase reporter assays.
(A) Results from HepG2 cells transfected with human TRIB3 promoter reporter constructs containing two (TRIB3-2x33-pGL3) or five (TRIB3-5x33-pGL3) copies of the 33-bp repeat. Cells were either left untreated or treated with tunicamycin (2 μg/ml, 10 h) or arsenite (20 μM, 10 h), or subjected to glucose deprivation (10 h). Luciferase activities are shown as mean ± SD from 4 independent transfections. *P < 0.05 comparing allelic variants using two-tailed t test followed by Holm–Bonferroni correction. (B) Reporter constructs carrying 1 to 5 copies of the 33-bp repeat were transfected into H1299 cells and their transcriptional activity was assayed under control condition (untreated), oxidative stress (30 μM arsenite, 10 h), and with co-transfection of an ATF4 overexpression plasmid (ATF4-pCG). Four independent transfections were performed, and the relationship between 33-bp copy number and luciferase activity was modeled using linear regression. For both panels, luciferase activities are presented relative to the activity of empty reporter vector (pGL3-Basic without TRIB3 fragment) under the same conditions. The genomic coordinates (GRCh38 human reference genome) of the region studied in the reporter assay are indicated below the plasmids.
As shown in Fig 3A, the plasmids containing the TRIB3 promoter fragments are strongly stress-responsive, upregulating the expression of reporter gene in all three treatments. The largest increase is observed in tunicamycin-treated cells, while arsenite exposure and glucose starvation result in slightly lower activation of reporter (Fig 3A). Importantly, in the cells transfected with plasmid TRIB3-5x33bp-pGL3 and exposed to stressors (tunicamycin, arsenite or glucose-free medium), the reporter activity is significantly higher than in the cells transfected with TRIB3-2x33-pGL3 and exposed to the same treatments (Fig 3A). Additionally, the 5-repeat allele also demonstrates increased luciferase activity compared to the 2-repeat allele in unstressed cells (Fig 3A).
To compare the full series of discovered copy number variants of the 33-bp repeat, a second set of luciferase reporter plasmids was prepared (Fig 3B), containing 1–5 copies of the TRIB3 33-bp repeat surrounded by flanking sequences that are identical to the GRCh38 reference genome (e.g., regarding SNPs in the region). To broaden the scope of the study, these plasmids were tested using the H1299 human non-small cell lung carcinoma cell line and the overexpression of ATF4 was included as a treatment. As depicted in Fig 3B, in the control cells as well as in the cells subjected to arsenite stress or overexpression of ATF4, the strength of the luciferase reporter signal positively and significantly correlates with the number of 33-bp repeats in the transfected reporter plasmid. Notably, increasing the 33-bp copies from 1 to 5 leads to approximately 5-fold increase in transcriptional activity under conditions that stimulate CARE activity (arsenite and ATF4 overexpression), indicating that transcriptional output increases quite steeply as a function of the 33-bp copy number. Thus, the results demonstrate that increased copy number of the 33-bp repeat results in elevated TRIB3 promoter activity.
Evidence for an in-cis regulatory effect of the TRIB3 33-bp repeat towards TRIB3 gene expression level from paired RNA-Seq and WGS data: k-mer-based repeat copy number estimation
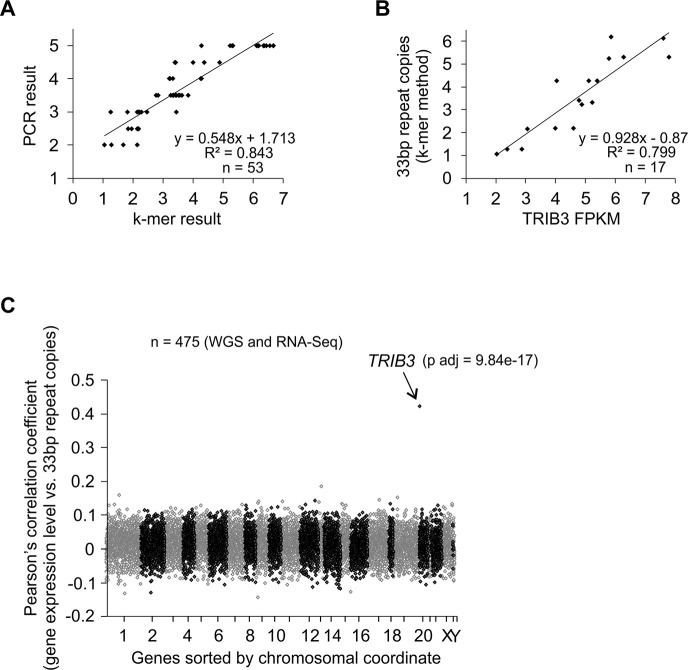
To study the effect of TRIB3 33-bp repeat copy number variation on the gene expression level in the human body, we turned to data sets with RNA-Seq and WGS data from the same individuals. To effectively use WGS data with read lengths <150 bp (e.g., 100 bp, in which case a read cannot reliably span even 3x33 bp), we switched to a k-mer counting based approach for quantifying the TRIB3 33-bp repeat copy number. The k-mer method estimates the average TRIB3 33-bp repeat copy number across each individual’s two alleles by querying the WGS reads for the occurrence of 25-mer oligonucleotides derived from the TRIB3 33-bp segment sequence, normalizing the counts to those of 25-mers derived from sequences neighboring the 33-bp repeat region.
We first analyzed individuals from the EGCUT cohort, where blood RNA-Seq has been performed for a subset of individuals analyzed by WGS [32, 44]. Analysis using PCR-validated individuals reveals a good agreement between the 33-bp repeat copy number values obtained from PCR and WGS k-mer counting across the range of possible values (Fig 4A). Further, for the PCR-validated individuals with RNA-Seq available, TRIB3 mRNA expression level from blood reveals a positive correlation to k-mer based 33bp copy number estimate (Fig 4B). Therefore, we extended this analysis to n = 475 blood samples and all expressed genes as detected by RNA-Seq. The results depicted in Fig 4C reveal that out of all expressed genes, the copy number of the TRIB3 33-bp repeat shows the strongest correlation with the expression level of TRIB3 itself, yielding a significant and positive correlation (Pearson's r = 0.42, corrected p = 9.8e-17), with no other genes demonstrating a correlation of |r|>0.2.
Fig 4. TRIB3 33-bp repeat copy number is positively correlated with the expression level of TRIB3 in genome-wide gene expression profiling.
(A) Validation of a k-mer counting-based method for the estimation of TRIB3 33-bp repeat copy number from whole genome sequencing data. The k-mer based copy number estimates are compared to the results of PCR product gel analysis from the same individuals. The PCR results are shown as the average repeat copy number from the diploid genotype of each individual. (B) Correlation of TRIB3 33-bp copy number to TRIB3 gene expression in the subset of individuals with PCR-validated 33-bp repeat genotypes. Individuals from panel A that further had blood RNA-Seq data available are included. (C) Across all genes, the copy number of the TRIB3 33-bp repeat is correlated specifically with the expression level of TRIB3 mRNA. Correlation between gene expression and the copy number of the TRIB3 33-bp repeat was analyzed for all genes expressed in blood, as determined by RNA-Seq (total 13896 genes). For the panels of this figure, the genotyping samples and the DNA and RNA sequencing data originate from the Estonian Biobank.
The copy number of TRIB3 promoter 33-bp repeats positively correlates with TRIB3 mRNA expression level in many tissues
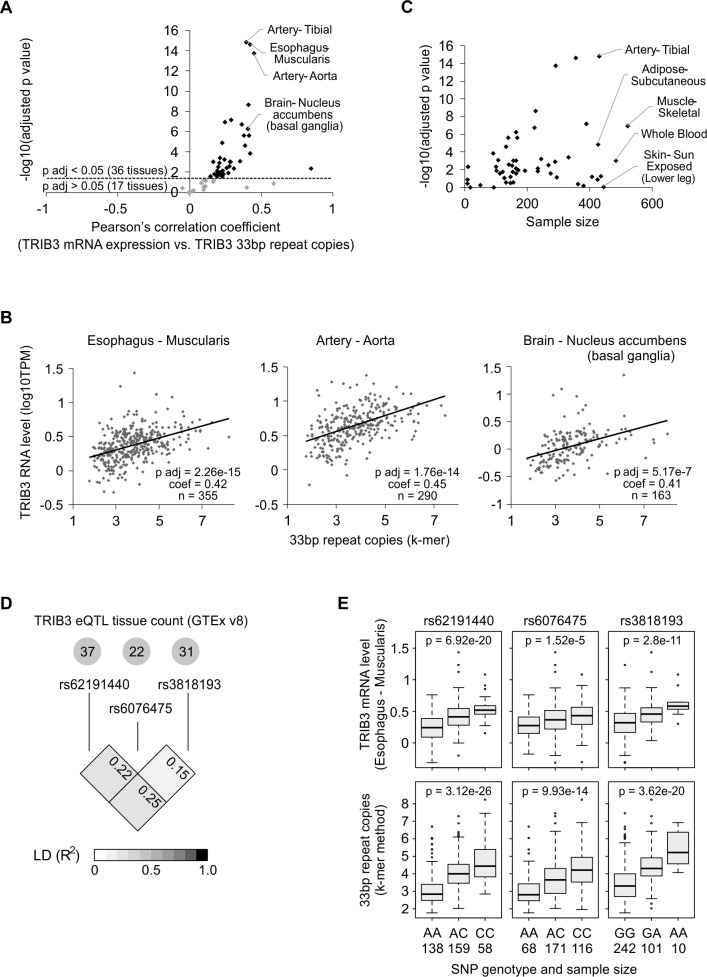
To assess whether the correlation between TRIB3 33-bp repeat copy number and TRIB3 mRNA level also occurs in other human tissues besides blood, we used the Genotype-Tissue Expression (GTEx) v8 data set [45]. All tissues with ≥5 samples with RNA-Seq and WGS were included, yielding 53 different tissues for analysis. Out of these, a total of 36 tissues revealed a significant (p adjusted < 0.05) correlation between genomic TRIB3 33-bp repeat copy number and TRIB3 mRNA expression in that tissue (Fig 5A, S1 Table), indicating a broad tissue distribution for the effect. Notably, in all tissues where a significant correlation was found, the direction of the correlation was positive (Fig 5, S1 Table). Tissues that yielded the most highly statistically significant results include arterial tissues and esophagus muscularis, characterized by high smooth muscle content; however, other tissues not rich in smooth muscle also ranked highly, such as nucleus accumbens of the brain (Fig 5B). The number of RNA-Seq samples available for each tissue varied greatly; however, some tissues with considerably large sample sizes still show a lack of association, implicating mechanisms other than the 33-bp repeat copy number as major determinants of TRIB3 expression level in those tissues (Fig 5C).
Fig 5. Variation in the TRIB3 33-bp repeat copy number is linked to TRIB3 expression level in various tissues of the body.
(A) Correlation between TRIB3 33-bp repeat copy number and TRIB3 gene expression level in 53 tissues from the Genotype-Tissue Expression (GTEx) v8 data set. The analysis included all tissues with ≥5 donors with RNA-Seq and whole genome sequencing from the same donor. The k-mer counting method was used to estimate the TRIB3 33-bp repeat copy number from genome sequencing reads. False discovery rate was used to correct the p values. (B) Scatter plots from selected tissues from panel A, showing the correlation between TRIB3 33-bp repeat copy number and TRIB3 mRNA expression at the level of individuals donors. (C) The strength of the association between 33-bp repeat copy number and TRIB3 expression level in different tissues (the correlation p value) does not appear to be driven strictly by the available sample size. All tissues from panel A are shown, and a selection of tissues with >400 samples are labeled. (D) Linkage disequilibrium results for SNPs that are TRIB3 eQTLs in multiple tissues. The eQTL tissue counts were obtained from the GTEx v8 data release. LD was calculated from GTEx donor genotypes. (E) Known SNP eQTLs for TRIB3 expression level demonstrate associations with the TRIB3 33-bp repeat copy number. Statistical significance was calculated by linear regression for the SNP allele effect.
The GTEx project has provided a wealth of SNP expression quantitative trait loci (eQTL) data for TRIB3 expression. We filtered the GTEx v8 eQTL SNPs for TRIB3 expression for SNPs that would be eQTLs in multiple tissues (≥20) and that would be largely independent of each other in terms of genetic linkage. This resulted in a set of 3 SNPs, rs62191440, rs6076475 and rs3818193, which are GTEx v8 eQTLs for TRIB3 in 37, 22 and 31 tissues, respectively, and have LD values of R2 < 0.3 between each other (Fig 5D). To assess whether the effects of these SNPs might be linked to variations in TRIB3 33-bp repeat copy number, we selected all GTEx v8 individuals that had SNP genotypes, WGS and esophagus muscularis RNA-Seq available for analysis. The results depicted in Fig 5E reveal that for these TRIB3 eQTL SNPs, individuals carrying the SNP allele associated with increased TRIB3 mRNA expression tend to also have a higher estimated TRIB3 33-bp repeat copy number, indicating that variation in the TRIB3 promoter VNTR could underlie the eQTL associations that have been discovered for these SNPs.
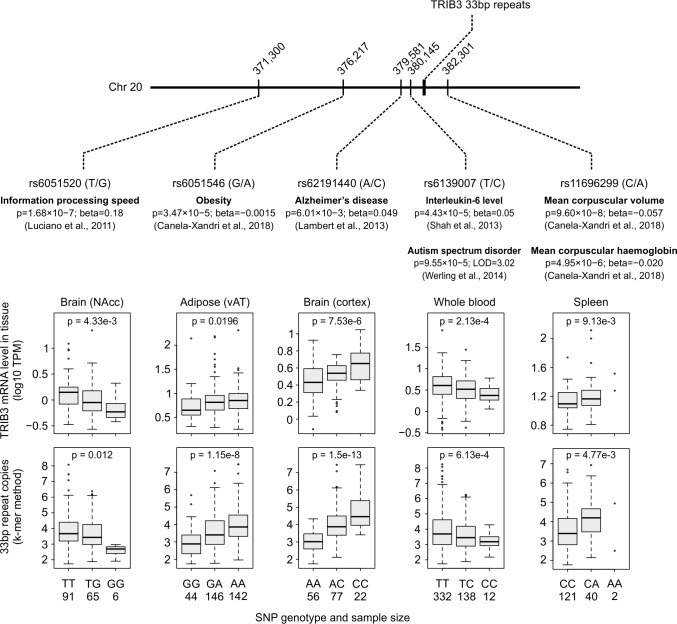
We searched literature for SNPs that are located in the vicinity of the TRIB3 33-bp repeat region and that have been noted as significant or suggestive in genetic association studies, including GWAS [46], UK Biobank PheWAS [47], linkage [48] and other types of genetic association studies [47, 49–51]. This uncovered 5 SNPs that have been associated with phenotypes, including several blood and nervous system traits or diseases, in genome-wide or more tightly focused studies (Fig 6). Analysis of the 5 association study lead SNPs in GTEx WGS data reveals positive as well as negative associations between the SNP alternative allele and the copy number of the TRIB3 33-bp repeat (Fig 6), indicating that the lead SNPs could be tagging various alleles of the 33-bp repeat. Further, GTEx RNA-Seq data from phenotype-relevant tissues reveals that these SNPs are linked to TRIB3 gene expression with directionality that is in line with their association to TRIB3 33-bp copy number (Fig 6), indicating that 33-bp repeat alleles could underlie the effects of these SNPs on TRIB3 expression level in tissues relevant to the reported trait or disease.
Fig 6. TRIB3 SNPs highlighted by genetic association studies are linked to 33-bp repeat copy number as well as TRIB3 mRNA expression in phenotype-relevant tissues.
SNPs localizing near the TRIB3 33-bp repeat and associated (or suggestive) for human phenotypes based on GWAS, PheWAS or other genetic association studies are shown. Using RNA-Seq and whole genome sequencing data from GTEx v8, the association between the lead SNP genotype and TRIB3 RNA expression level or TRIB3 33-bp repeat copy number was calculated using linear regression for the SNP allele effect. The k-mer counting method was used to estimate the TRIB3 33-bp repeat copy number from genome sequencing reads.
Discussion
In the current era of human genetic studies, powered mainly by short-read sequencing and SNP microarrays, VNTRs have been addressed comparatively less than other types of variants due to technological constraints. Nevertheless, more than a hundred thousand polymorphic tandem repeats have been described that localize within or near genes in the human genome, representing considerable potential to alter the protein coding or regulatory sequences of genes [52]. In the current article, we characterize copy number variation in a 33-bp tandem repeat localized to the promoter region of human TRIB3 and uncover that alleles with a higher copy number of the 33-bp repeating element lead to elevated TRIB3 gene expression level.
The human reference genome (GRCh38) contains 3 identical tandemly repeating elements of the TRIB3 33-bp VNTR. Alleles with 2 or 5 copies are also relatively frequent in many human populations. Crucially, this 33-bp region that has been repeated in humans contains a highly conserved sequence element, which likely accounts for the effect of 33-bp VNTR copy number variation on TRIB3 mRNA expression level. Previous functional studies of the human TRIB3 (as well as mouse Trib3) promoter sequences have identified that a C/EBP-ATF composite motif, which rests within the highly conserved part of the 33-bp repeat, represents a crucial transcription factor binding site for driving TRIB3 mRNA expression [30, 53]. A key factor shown to bind to this site is ATF4 [5, 30, 53], a bZIP transcription factor with several known roles in physiology and disease [54–56]. Thus, TRIB3 alleles with more copies of the 33-bp repeat can convey increased expression of TRIB3 by having more copies of the C/EBP-ATF binding site, a positive regulatory element, in their promoter. In support of this, we find that the overexpression of ATF4 is sufficient to induce TRIB3 promoter reporter constructs, and that the responsiveness to ATF4 (fold induction) increases steadily as the number of 33-bp repeats in the construct is increased step-wise from 1 to 5 copies, the range of natural variation.
Our finding that the great apes do not contain repeated copies of the 33-bp segment in the TRIB3 promoter, while the archaic hominin branches Neanderthal and Denisovan do, allows us to estimate the time window when the VNTR evolved. The ancestors of hominins and chimpanzees split about 6 million years ago [57], and present-day humans diverged from Neanderthal and Denisovan lineages about 800,000 years ago [36, 58]. Thus, the expansion of the 33-bp segment in TRIB3 promoter occurred within approximately 5 million years separating these two branching points of human evolution. All five of the copy number variants which we found in the current study are already present in the HGDP populations living in Africa, the ancestral home continent of modern humans, and the alleles 1x and 4x, the incidence of which is very low in most HGDP populations, are noticeably more frequent in Sub-Saharan Africa. Thus, the results suggest that all allelic variants ranging from 2x to 5x evolved before the migration of modern humans out of Africa, which, according to the earliest fossil evidence currently found, started about 200,000 years ago [59, 60].
Studies using knockout and overexpression models of mouse Trib3 have led to TRIB3 being implicated in several biological processes in vivo (reviewed in [1, 61]). Promisingly, some of these results line up with significant or suggestive phenotypes from human genetic association studies. For example, enforced overexpression of Trib3 in mouse adipose tissue has been reported to inhibit weight gain [62], and the human SNP rs6051546 alternative allele is associated with increased 33-bp repeat copy number, elevated TRIB3 mRNA level in adipose tissue, and suggestively (p = 3.47e-5) [47] with reduced obesity. Several mouse and cellular models have implicated increased TRIB3 in neurodegeneration and neuronal cell death [19, 63, 64], and in humans, SNP associations with information processing speed (p = 1.68e-7; rs6051520) [50] and Alzheimer’s disease (p = 6.01e-3; rs62191440) [49] correlate with the expected effects of TRIB3 33-bp repeat copy number on TRIB3 gene expression level in brain tissue. The SNP rs62191440 has received attention as a proxy for rs4813620 (R2 = 0.656 in 1000G EUR), which was reported by [24] to be associated with TRIB3 expression in human brain tissues, but [65] found that Alzheimer’s disease status may affect this association. Human genetic association studies also link TRIB3 to red blood cell parameters (p = 9.6e-8; rs11696299) [47], and we also uncovered a link to the 33-bp repeat copy count for this lead SNP. It has recently been reported that in mouse Trib3 plays a role in red blood cell production; however, the effect appears to be complex, with alternate consequences under steady-state and myeloablative challenge conditions [66].
The results of our study allow to propose that a tandemly repeated polymorphic 33-bp repeat element in the human TRIB3 gene gives rise to variation in TRIB3 expression levels between individuals by multiplying the number of C/EBP-ATF binding sites in the TRIB3 promoter. Further, this variation in the TRIB3 33-bp VNTR copy number appears to explain a number of SNP eQTLs for TRIB3 expression in various tissues and could possibly also underlie TRIB3 SNP effects from genetic association studies. Since the TRIB3 33-bp VNTR copy number is imperfectly associated with SNPs, direct genotyping of the alleles of the VNTR in large-scale study groups could allow for the discovery of stronger associations to human phenotypes.
Materials and methods
Ethics approval and consent to participate
The study was conducted in accordance with good ethical standards and was approved by the Research Ethics Committee of the University of Tartu (protocol number 289/T-10).
We used GTEx v8 (dbGap Accession phs000424.v8.p2) (Consent group: General Research Use) for the analyses described in this manuscript. The GTEx v8 DNA data were obtained from the GTEx Portal on September 27, 2019 and October 1–2, 2019, and the GTEx v8 RNA-seq data were obtained from the GTEx Portal on September 30, 2019.
Analysis of human TRIB3 promoter 33-bp repeat copy number by PCR
To study the human TRIB3 promoter 33-bp repeat copy number in the genomic DNA of human cell lines and EGCUT biobank human DNA samples, DNA was amplified by PCR using primers 5’-CCACTTCCGCTGCGAGTCTCGTG-3’ and 5’-CCCGAGGGCATCAGACGGCG-3’, which bind to the regions flanking the repeats. In addition to the repeats, the PCR products contain fragments up- and downstream of the repeats (23 bp and 33 bp, respectively). The PCR products were analyzed by electrophoresis in 2.5% agarose gel and, in several cases, Sanger sequencing was also carried out, which confirmed that the products contain the human TRIB3 33-bp repeats.
Human TRIB3 33-bp repeat copy number analyses from whole genome sequencing reads
Whole genome sequencing data was obtained from the Estonian Biobank [33], the Human Genome Diversity Project [39], and the GTEx project (v8 release) [45].
A k-mer counting approach was used to estimate the diploid allelic average copy number of the TRIB3 33-bp repeat from WGS reads. A list of 25-mer oligonucleotides was created from the genomic sequence of the TRIB3 33-bp repeat and its 5’ and 3’ flanking sequences using the glistmaker program of the GenomeTester4 package [67] with step of 1 bp, and k-mer frequencies were extracted from the list with the program glistquery from the same package. The following formula was then used for calculating the VNTR copy number: median from 10 repeat unit k-mers / max (median from 29 VNTR 5’ flank k-mers; median from 29 VNTR 3’ flank k-mers).
For direct detection of specific TRIB3 33-bp repeat alleles from 150 bp WGS reads, reads mapping to ±10 kb of the TRIB3 33-bp repeat were extracted for further analysis by text matching. Alleles with 1 to 4 copies of the 33-bp repeat were detected by searching for reads fully spanning the repeat region and additionally covering 6 bp of the 5’ and 3’ flanking regions, with a maximum allowed edit distance of 2 bp compared to the expected sequence. As the 5x33bp allele cannot be spanned by a 150 bp read, reads with partial coverage of 5 copies of the 33-bp repeat element were accepted, requiring a minimum repeat element fragment of 6 bp and a maximum edit distance of 2 bp. For each sample, alleles with <2 detection counts were excluded. Samples showing more than two alleles were excluded. To calculate multi-allelic FST, the approach in [40] was followed.
RNA-Seq and SNP genotype data and processing
RNA-Seq data for all tissues and all samples in GTEx v8, pre-processed by GTEx, was downloaded from the project home page [45]. All RNA-Seq samples annotated by GTEx as passing quality control were kept, and all tissues with ≥5 donors having RNA-Seq and WGS data available were kept. No further exclusion of outlier samples was applied in the calculations. Pre-processed RNA-Seq data for blood cells was also obtained for participants of the Estonian Biobank [44]. The depth-normalized RNA-Seq results (TPM for GTEx; FPKM for Estonian Biobank) were log-transformed, and Pearson’s product-moment correlation (coefficient r) was used to assess correlation between RNA expression level and 33-bp repeat copy number. False discovery rate was used for the correction of p values for multiple testing.
For SNP genotypes of GTEx donors, the genotyping calls from the GTEx project were used. Association between SNP genotype and RNA expression level or 33-bp repeat copy number was calculated by linear regression model for the SNP allele effect. For the calculation of linkage disequilibrium, the R package LDheatmap was used (version 0.99–7) [68].
Luciferase reporter plasmid generation
To prepare reporter plasmids which contain approximately 0.7 kb of the human TRIB3 5’-flanking region (GRCh38 human reference genome chr 20 positions 380,298 to 380,987) with different copy numbers of the 33-bp repeat, the following two-step cloning procedure was carried out. Human DNA samples (received from EGCUT) were amplified by PCR using primers 5’-CCACTTCCGCTGCGAGTCTCGTG-3’ and 5’-CTCGGTACCTGAAGCTTCTGAACCACTTGC-3’, and the PCR products were cut with restriction enzymes Acc65I/MluI and subsequently inserted into Acc65I/MluI sites of the reporter vector pGL3-Basic (Promega). The inserts were analyzed for 33-bp repeat alleles and other genetic variants by Sanger sequencing and used as template for the PCR amplification with primers 5’-GCGGTTCCATCTTCCAGCGGATAG-3’ and 5’-CTCGGTACCCCGACAACAGTCACTGTTTG-3’, and the PCR products were cut with restriction enzymes Acc65I/BglII and inserted into Acc65I/BglII sites of the plasmid pGL3-Basic.
The luciferase reporter constructs TRIB3-2x33-pGL3 and TRIB3-5x33-pGL3 contain two or five copies of the 33-bp repeat, respectively. All other nucleotides of their inserts are identical: both constructs differ from the GRCh38 reference genome at a number of positions, all of which correspond to common SNPs (genotypes in the plasmids: rs575062197:delA, rs2092474:C, rs7267577:A, rs6115787:A, rs7261666:G, and rs7263534:C).
To prepare the series of luciferase reporter plasmids TRIB3(ref)-(1/2/3/4/5)x33-pGL3, carrying 1 to 5 copies of the 33-bp repeat with flanking sequences identical to the GRCh38 human reference genome (i.e., all SNPs in the region are represented by the reference allele), a genomic DNA fragment containing 3x33bp copies and matching exactly to the human reference genome chr 20 region 380,298–380,987 was used as a 33-bp repeat flanking sequence donor for fragments carrying 1, 2, 4 or 5 copies of 33-bp repeat.
Sanger sequencing was used to confirm the nucleotide sequences of all plasmids.
Dual-luciferase reporter assay
HepG2 and H1299 cells grown in 96-well plates were cotransfected in triplicate wells with 40 ng of firefly luciferase-encoding plasmid (either pGL3-Basic or reporter constructs containing the TRIB3 promoter region; described above) and 6 ng of Renilla luciferase-encoding plasmid (pRL-TK; Promega) using polyethylenimine (PEI-MAX 40,000; Polysciences Inc. #24765). Where indicated, 2.5 ng of the expression plasmid encoding human ATF4 (ATF4-pCG) [30], or the corresponding empty vector (pCG), was included in the transfection mixture. Before each transfection, fresh dilutions of reporter construct stocks were prepared, and DNA concentration was verified using NanoDrop 1000 (Thermo Scientific).
HepG2 cells were left to recover for 20 h after transfection and were subsequently incubated for 10 h in treatment medium (glucose-free DMEM (without sodium pyruvate; Gibco) supplemented with 10% dialyzed FCS (Sigma-Aldrich) and D-glucose at a final concentration of 4.5 g/l). Tunicamycin (2 μg/ml) and sodium arsenite (20 μM) (both obtained from Sigma-Aldrich) were added where indicated. To induce glucose deprivation, cells were washed twice with PBS and incubated in glucose-free treatment medium. H1299 cells were left to recover for 20 h after transfection and were subsequently either mock-treated or treated with 30 μM sodium arsenite in the cell growth medium (DMEM, without sodium pyruvate (Gibco) supplemented with 10% FCS (PAA)) for 10 h.
Firefly and Renilla luciferase activities were measured using a dual-luciferase assay (Promega) as described previously [3, 30]. The firefly luciferase activity was normalized to the Renilla luciferase activity, and the activities of the reporter constructs containing TRIB3 promoter region are presented relative to the results from the promoterless pGL3-Basic plasmid in the same experimental conditions.
Supporting information
The k-mer quantification result is shown below the gel. The PCR was performed as in Fig 1B and 1D, and the products generated from HepG2 cell line DNA are shown as a control. Lane M designates a 100 bp DNA ladder molecular weight marker.
(TIF)
All tissues with ≥5 samples of RNA-Seq and whole genome sequencing data from the same donor were included. The k-mer counting method was used to estimate the TRIB3 33-bp repeat copy number from genome sequencing reads, and TRIB3 gene expression was obtained from RNA-Seq data. Correlation was calculated using Pearson's product-moment correlation method. False discovery rate was used to correct the p values.
(DOCX)
(XLSX)
Acknowledgments
We are grateful to Dr. Reidar Andreson for helpful discussions and advice.
Data Availability
All relevant data are within the manuscript and its Supporting Information files, with the exception of individual-level genetic data of human donors. Access to the human genetic sequence data used in this manuscript can be obtained as follows. The GTEx datasets used for the analyses described in this manuscript were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap; accession number phs000424.v8.p2) using the procedure detailed at https://gtexportal.org/home/protectedDataAccess. The Human Genome Diversity Project (HGDP) datasets were obtained from The International Genome Sample Resource (https://www.internationalgenome.org/data-portal/data-collection/hgdp), as detailed at https://www.internationalgenome.org/data. The Estonian Biobank data were obtained under Request to the Estonian Committee on Bioethics and Human Research, using the procedure detailed at https://genomics.ut.ee/en/biobank.ee/data-access.
Funding Statement
This work was supported by the Estonian Research Council grants PUT1701, PRG555, PRG687 and institutional grant IUT34-11 and the EU ERDF grant No. 2014-2020.4.01.15-0012 (Estonian Center of Excellence in Genomics and Translational Medicine). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health (commonfund.nih.gov/GTEx). Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\Leidos Biomedical Research, Inc. subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to the The Broad Institute, Inc. Biorepository operations were funded through a Leidos Biomedical Research, Inc. subcontract to Van Andel Research Institute (10ST1035). Additional data repository and project management were provided by Leidos Biomedical Research, Inc.(HHSN261200800001E). The Brain Bank was supported supplements to University of Miami grant DA006227. Statistical Methods development grants were made to the University of Geneva (MH090941 & MH101814), the University of Chicago (MH090951,MH090937, MH101825, & MH101820), the University of North Carolina - Chapel Hill (MH090936), North Carolina State University (MH101819),Harvard University (MH090948), Stanford University (MH101782), Washington University (MH101810), and to the University of Pennsylvania (MH101822). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Örd T, Örd T. Mammalian Pseudokinase TRIB3 in Normal Physiology and Disease: Charting the Progress in Old and New Avenues. Curr Protein Pept Sci. 2017;18(8):819–42. 10.2174/1389203718666170406124547 MEDLINE:28393700. [DOI] [PubMed] [Google Scholar]
- 2.Jousse C, Deval C, Maurin A-C, Parry L, Cherasse Y, Chaveroux C, et al. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J Biol Chem. 2007;282(21):15851–61. 10.1074/jbc.M611723200 MEDLINE:17369260. [DOI] [PubMed] [Google Scholar]
- 3.Örd D, Meerits K, Örd T. TRB3 protects cells against the growth inhibitory and cytotoxic effect of ATF4. Exp Cell Res. 2007;313(16):3556–67. 10.1016/j.yexcr.2007.07.017 MEDLINE:17707795. [DOI] [PubMed] [Google Scholar]
- 4.Örd T, Örd D, Adler P, Vilo J, Örd T. TRIB3 enhances cell viability during glucose deprivation in HEK293-derived cells by upregulating IGFBP2, a novel nutrient deficiency survival factor. Biochim Biophys Acta Mol Cell Res. 2015;1853(10 Pt A):2492–505. 10.1016/j.bbamcr.2015.06.006 MEDLINE:26094770. [DOI] [PubMed] [Google Scholar]
- 5.Örd D, Örd T, Biene T, Örd T. TRIB3 increases cell resistance to arsenite toxicity by limiting the expression of the glutathione-degrading enzyme CHAC1. Biochim Biophys Acta Mol Cell Res. 2016;1863(11):2668–80. 10.1016/j.bbamcr.2016.08.003 MEDLINE:27526673. [DOI] [PubMed] [Google Scholar]
- 6.Butcher L, Ahluwalia M, Örd T, Johnston J, Morris RH, Kiss-Toth E, et al. Evidence for a role of TRIB3 in the regulation of megakaryocytopoiesis. Sci Rep. 2017;7(1):6684 10.1038/s41598-017-07096-w MEDLINE:28751721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Örd T, Örd D, Örd T. TRIB3 limits FGF21 induction during in vitro and in vivo nutrient deficiencies by inhibiting C/EBP-ATF response elements in the Fgf21 promoter. Biochim Biophys Acta Gene Regul Mech. 2018;1861(3):271–81. 10.1016/j.bbagrm.2018.01.014 MEDLINE:29378327. [DOI] [PubMed] [Google Scholar]
- 8.Erazo T, Lorente M, Lopez-Plana A, Munoz-Guardiola P, Fernandez-Nogueira P, Garcia-Martinez JA, et al. The New Antitumor Drug ABTL0812 Inhibits the Akt/mTORC1 Axis by Upregulating Tribbles-3 Pseudokinase. Clin Cancer Res. 2016;22(10):2508–19. 10.1158/1078-0432.CCR-15-1808 WOS:000375839200023. [DOI] [PubMed] [Google Scholar]
- 9.Felip I, Moiola CP, Megino-Luque C, Lopez-Gil C, Cabrera S, Sole-Sanchez S, et al. Therapeutic potential of the new TRIB3-mediated cell autophagy anticancer drug ABTL0812 in endometrial cancer. Gynecol Oncol. 2019;153(2):425–35. 10.1016/j.ygyno.2019.03.002 MEDLINE:30853360. [DOI] [PubMed] [Google Scholar]
- 10.López-Plana A, Fernández-Nogueira P, Muñoz-Guardiola P, Solé-Sánchez S, Megías-Roda E, Pérez-Montoyo H, et al. The novel pro-autophagy anticancer drug ABTL0812 potentiates chemotherapy in adenocarcinoma and squamous Non-Small Cell Lung Cancer. Int J Cancer. 2020. 10.1002/ijc.32865 [DOI] [PubMed] [Google Scholar]
- 11.Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119(5):1359–72. 10.1172/jci37948 WOS:000265843400034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salazar M, Lorente M, Garcia-Taboada E, Hernandez-Tiedra S, Davila D, Francis SE, et al. The pseudokinase tribbles homologue-3 plays a crucial role in cannabinoid anticancer action. Biochim Biophys Acta Mol Cell Biol Lipids. 2013;1831(10):1573–8. 10.1016/j.bbalip.2013.03.014 MEDLINE:23567453. [DOI] [PubMed] [Google Scholar]
- 13.Salazar M, Lorente M, Garcia-Taboada E, Perez Gomez E, Davila D, Zuniga-Garcia P, et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015;22(1):131–44. 10.1038/cdd.2014.133 MEDLINE:25168244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borsting E, Patel SV, Decleves AE, Lee SJ, Rahman QM, Akira S, et al. Tribbles Homolog 3 Attenuates Mammalian Target of Rapamycin Complex-2 Signaling and Inflammation in the Diabetic Kidney. J Am Soc Nephrol. 2014;25(9):2067–78. 10.1681/ASN.2013070811 WOS:000341059200021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du KY, Herzig S, Kulkarni RN, Montminy M. TRB3: A tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300(5625):1574–7. 10.1126/science.1079817 WOS:000183333100056. [DOI] [PubMed] [Google Scholar]
- 16.Liew CW, Bochenski J, Kawamori D, Hu J, Leech CA, Wanic K, et al. The pseudokinase tribbles homolog 3 interacts with ATF4 to negatively regulate insulin exocytosis in human and mouse beta cells. J Clin Invest. 2010;120(8):2876–88. 10.1172/JCI36849 WOS:000280492100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prudente S, Hribal ML, Flex E, Turchi F, Morini E, De Cosmo S, et al. The functional Q84R polymorphism of mammalian Tribbles homolog TRB3 is associated with insulin resistance and related cardiovascular risk in Caucasians from Italy. Diabetes. 2005;54(9):2807–11. 10.2337/diabetes.54.9.2807 MEDLINE:16123373. [DOI] [PubMed] [Google Scholar]
- 18.Prudente S, Baratta R, Andreozzi F, Morini E, Farina MG, Nigro A, et al. TRIB3 R84 variant affects glucose homeostasis by altering the interplay between insulin sensitivity and secretion. Diabetologia. 2010;53(7):1354–61. 10.1007/s00125-010-1749-1 MEDLINE:20393693. [DOI] [PubMed] [Google Scholar]
- 19.Aimé P, Sun X, Zareen N, Rao A, Berman Z, Volpicelli-Daley L, et al. Trib3 Is Elevated in Parkinson's Disease and Mediates Death in Parkinson's Disease Models. J Neurosci. 2015;35(30):10731–49. 10.1523/JNEUROSCI.0614-15.2015 MEDLINE:26224857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aimé P, Karuppagounder SS, Rao A, Chen Y, Burke RE, Ratan RR, et al. The drug adaptaquin blocks ATF4/CHOP-dependent pro-death Trib3 induction and protects in cellular and mouse models of Parkinson's disease. Neurobiol Dis. 2020;136 10.1016/j.nbd.2019.104725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua F, Li K, Yu JJ, Lv XX, Yan J, Zhang XW, et al. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;6 10.1038/ncomms8951 WOS:000360346700006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izrailit J, Berman HK, Datti A, Wrana JL, Reedijk M. High throughput kinase inhibitor screens reveal TRB3 and MAPK-ERK/TGFbeta pathways as fundamental Notch regulators in breast cancer. Proc Natl Acad Sci U S A. 2013;110(5):1714–9. 10.1073/pnas.1214014110 MEDLINE:23319603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li K, Wang F, Cao W-B, Lv X-X, Hua F, Cui B, et al. TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARalpha and Inhibition of p53-Mediated Senescence. Cancer cell. 2017;31(5):697–710.e7. 10.1016/j.ccell.2017.04.006 MEDLINE:28486108. [DOI] [PubMed] [Google Scholar]
- 24.Lorenzi M, Altmann A, Gutman B, Wray S, Arber C, Hibar DP, et al. Susceptibility of brain atrophy to TRIB3 in Alzheimer's disease, evidence from functional prioritization in imaging genetics. Proc Natl Acad Sci U S A. 2018;115(12):3162–7. 10.1073/pnas.1706100115 MEDLINE:29511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada Y, Kato K, Oguri M, Horibe H, Fujimaki T, Yasukochi Y, et al. Identification of nine genes as novel susceptibility loci for early-onset ischemic stroke, intracerebral hemorrhage, or subarachnoid hemorrhage. Biomed Rep. 2018;9(1):8–20. 10.3892/br.2018.1104 WOS:000440677300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulovari A, Li RY, Audano PA, Porubsky D, Vollger MR, Logsdon GA, et al. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc Natl Acad Sci U S A. 2019;116(46):23243–53. 10.1073/pnas.1912175116 WOS:000496506600056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85(2):258–63. 10.1016/j.ygeno.2004.11.003 WOS:000226843300011. [DOI] [PubMed] [Google Scholar]
- 28.Song JHT, Lowe CB, Kingsley DM. Characterization of a Human-Specific Tandem Repeat Associated with Bipolar Disorder and Schizophrenia. Am J Hum Genet. 2018;103(3):421–30. 10.1016/j.ajhg.2018.07.011 WOS:000443819500011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaisson MJP, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet. 2015;16(11):627–40. 10.1038/nrg3933 WOS:000362909300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Örd D, Örd T. Characterization of human NIPK (TRB3, SKIP3) gene activation in stressful conditions. Biochem Biophys Res Commun. 2005;330(1):210–8. 10.1016/j.bbrc.2005.02.149 MEDLINE:15781252. [DOI] [PubMed] [Google Scholar]
- 31.Örd T, Örd D, Kõivomägi M, Juhkam K, Örd T. Human TRB3 is upregulated in stressed cells by the induction of translationally efficient mRNA containing a truncated 5'-UTR. Gene. 2009;444(1–2):24–32. 10.1016/j.gene.2009.06.001 MEDLINE:19505541. [DOI] [PubMed] [Google Scholar]
- 32.Leitsalu L, Alavere H, Tammesoo ML, Leego E, Metspalu A. Linking a Population Biobank with National Health Registries-The Estonian Experience. J Pers Med. 2015;5(2):96–106. 10.3390/jpm5020096 WOS:000457944500005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitt M, Kals M, Parn K, Gabriel SB, Lander ES, Palotie A, et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur J Hum Genet. 2017;25(7):869–76. 10.1038/ejhg.2017.51 WOS:000403061300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou MM, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. 10.1101/gr.3715005 WOS:000231032000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–21. 10.1101/gr.097857.109 WOS:000273249500012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science. 2012;338(6104):222–6. 10.1126/science.1224344 WOS:000309712300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 2017;358(6363):655–8. 10.1126/science.aao1887 WOS:000414240500039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019:531210 10.1101/531210 [DOI] [Google Scholar]
- 39.Bergström A, McCarthy SA, Hui RY, Almarri MA, Ayub Q, Danecek P, et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367(6484):1339–+. 10.1126/science.aay5012 WOS:000522167400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slatkin M, Barton NH. A COMPARISON OF 3 INDIRECT METHODS FOR ESTIMATING AVERAGE LEVELS OF GENE FLOW. Evolution. 1989;43(7):1349–68. 10.1111/j.1558-5646.1989.tb02587.x WOS:A1989AX63000001. [DOI] [PubMed] [Google Scholar]
- 41.Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, et al. C/EBP gamma Is a Critical Regulator of Cellular Stress Response Networks through Heterodimerization with ATF4. Mol Cell Biol. 2016;36(5):693–713. 10.1128/mcb.00911-15 WOS:000372328600004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan JX, Zhang F, Sharkey J, Tang TA, Ord T, Kilberg MS. The C/ebp-Atf response element (CARE) location reveals two distinct Atf4-dependent, elongation-mediated mechanisms for transcriptional induction of aminoacyl-tRNA synthetase genes in response to amino acid limitation. Nucleic Acids Res. 2016;44(20):9719–32. 10.1093/nar/gkw667 WOS:000393817800023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan JX, Örd D, Örd T, Kilberg MS. Elevated ATF4 Expression, in the Absence of Other Signals, Is Sufficient for Transcriptional Induction via CCAAT Enhancer-binding Protein-activating Transcription Factor Response Elements. J Biol Chem. 2009;284(32):21241–8. 10.1074/jbc.M109.011338 WOS:000268564400017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepik K, Annilo T, Kukuskina V, Kisand K, Kutalik Z, Peterson P, et al. C-reactive protein upregulates the whole blood expression of CD59—an integrative analysis. PLoS Comput Biol. 2017;13(9). 10.1371/journal.pcbi.1005766 WOS:000411981000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. 10.1038/ng.2653 WOS:000319563900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D12. 10.1093/nar/gky1120 WOS:000462587400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50(11):1593–9. 10.1038/s41588-018-0248-z WOS:000448398000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werling DM, Lowe JK, Luo R, Cantor RM, Geschwind DH. Replication of linkage at chromosome 20p13 and identification of suggestive sex-differential risk loci for autism spectrum disorder. Mol Autism. 2014;5 10.1186/2040-2392-5-13 WOS:000334716000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452–U206. 10.1038/ng.2802 WOS:000327715800010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luciano M, Hansell NK, Lahti J, Davies G, Medland SE, Raikkonen K, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol. 2011;86(3):193–202. 10.1016/j.biopsycho.2010.11.008 MEDLINE:21130836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah T, Zabaneh D, Gaunt T, Swerdlow DI, Shah S, Talmud PJ, et al. Gene-Centric Analysis Identifies Variants Associated With Interleukin-6 Levels and Shared Pathways With Other Inflammation Markers. Circ Cardiovasc Genet. 2013;6(2):163–70. 10.1161/CIRCGENETICS.112.964254 WOS:000317741400005. [DOI] [PubMed] [Google Scholar]
- 52.Genovese LM, Geraci F, Corrado L, Mangano E, D'Aurizio R, Bordoni R, et al. A Census of Tandemly Repeated Polymorphic Loci in Genic Regions Through the Comparative Integration of Human Genome Assemblies. Front Genet. 2018;9 10.3389/fgene.2018.00155 WOS:000431218500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carraro V, Maurin AC, Lambert-Langlais S, Averous J, Chaveroux C, Parry L, et al. Amino Acid Availability Controls TRB3 Transcription in Liver through the GCN2/eIF2 alpha/ATF4 Pathway. Plos One. 2010;5(12). 10.1371/journal.pone.0015716 WOS:000285576900037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams CM, Ebert SM, Dyle MC. Role of ATF4 in skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2017;20(3):164–8. 10.1097/MCO.0000000000000362 WOS:000400991100004. [DOI] [PubMed] [Google Scholar]
- 55.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–96. 10.1146/annurev.genom.9.081307.164437 WOS:000259629000010. [DOI] [PubMed] [Google Scholar]
- 56.Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells. Trends Endocrinol Metab. 2017;28(11):794–806. WOS:000414064400004. 10.1016/j.tem.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MAM, et al. A Molecular Phylogeny of Living Primates. PLoS Genet. 2011;7(3). 10.1371/journal.pgen.1001342 WOS:000288996600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A Draft Sequence of the Neandertal Genome. Science. 2010;328(5979):710–22. 10.1126/science.1188021 WOS:000277357100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harvati K, Roding C, Bosman AM, Karakostis FA, Grun R, Stringer C, et al. Apidima Cave fossils provide earliest evidence of Homo sapiens in Eurasia. Nature. 2019;571(7766):500–+. 10.1038/s41586-019-1376-z WOS:000477016700058. [DOI] [PubMed] [Google Scholar]
- 60.Hershkovitz I, Weber GW, Quam R, Duval M, Grun R, Kinsley L, et al. The earliest modern humans outside Africa. Science. 2018;359(6374):456–9. 10.1126/science.aap8369 WOS:000423283200046. [DOI] [PubMed] [Google Scholar]
- 61.Eyers PA, Keeshan K, Kannan N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol. 2017;27(4):284–98. 10.1016/j.tcb.2016.11.002 WOS:000399431000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312(5781):1763–6. 10.1126/science.1123374 WOS:000238452800048. [DOI] [PubMed] [Google Scholar]
- 63.Saleem S, Biswas SC. Tribbles Pseudokinase 3 Induces Both Apoptosis and Autophagy in Amyloid-beta-induced Neuronal Death. J Biol Chem. 2017;292(7):2571–85. 10.1074/jbc.M116.744730 WOS:000395535100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zareen N, Biswas SC, Greene LA. A feed-forward loop involving Trib3, Akt and FoxO mediates death of NGF-deprived neurons. Cell Death Differ. 2013;20(12):1719–30. 10.1038/cdd.2013.128 MEDLINE:24212932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu GY, Jin SL, Hu Y, Jiang QH. Disease status affects the association between rs4813620 and the expression of Alzheimer's disease susceptibility gene TRIB3. Proc Natl Acad Sci U S A. 2018;115(45):E10519–E20. 10.1073/pnas.1812975115 WOS:000449459000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dev A, Asch R, Jachimowicz E, Rainville N, Johnson A, Greenfest-Allen E, et al. Governing roles for Trib3 pseudokinase during stress erythropoiesis. Exp Hematol. 2017;49:48–55. 10.1016/j.exphem.2016.12.010 WOS:000399861000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplinski L, Lepamets M, Remm M. GenomeTester4: a toolkit for performing basic set operations—union, intersection and complement on k-mer lists. Gigascience. 2015;4 10.1186/s13742-015-0097-y WOS:000365669500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin J-H, Blay S, McNeney B, Graham J. LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria Between Single Nucleotide Polymorphisms. J Stat Softw. 2006;16(Code Snippet 3):1–9. 10.18637/jss.v016.c03 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The k-mer quantification result is shown below the gel. The PCR was performed as in Fig 1B and 1D, and the products generated from HepG2 cell line DNA are shown as a control. Lane M designates a 100 bp DNA ladder molecular weight marker.
(TIF)
All tissues with ≥5 samples of RNA-Seq and whole genome sequencing data from the same donor were included. The k-mer counting method was used to estimate the TRIB3 33-bp repeat copy number from genome sequencing reads, and TRIB3 gene expression was obtained from RNA-Seq data. Correlation was calculated using Pearson's product-moment correlation method. False discovery rate was used to correct the p values.
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files, with the exception of individual-level genetic data of human donors. Access to the human genetic sequence data used in this manuscript can be obtained as follows. The GTEx datasets used for the analyses described in this manuscript were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap; accession number phs000424.v8.p2) using the procedure detailed at https://gtexportal.org/home/protectedDataAccess. The Human Genome Diversity Project (HGDP) datasets were obtained from The International Genome Sample Resource (https://www.internationalgenome.org/data-portal/data-collection/hgdp), as detailed at https://www.internationalgenome.org/data. The Estonian Biobank data were obtained under Request to the Estonian Committee on Bioethics and Human Research, using the procedure detailed at https://genomics.ut.ee/en/biobank.ee/data-access.