Abstract
Plastid genomes (plastomes) of land plants have a conserved quadripartite structure in a gene-dense unit genome consisting of a large inverted repeat that separates two single copy regions. Recently, alternative plastome structures were suggested in Geraniaceae and in some conifers and Medicago the coexistence of inversion isomers has been noted. In this study, plastome sequences of two Cyperaceae, Eleocharis dulcis (water chestnut) and Eleocharis cellulosa (gulf coast spikerush), were completed. Unlike the conserved plastomes in basal groups of Poales, these Eleocharis plastomes have remarkably divergent features, including large plastome sizes, high rates of sequence rearrangements, low GC content and gene density, gene duplications and losses, and increased repetitive DNA sequences. A novel finding among these features was the unprecedented level of heteroplasmy with the presence of multiple plastome structural types within a single individual. Illumina paired-end assemblies combined with PacBio single-molecule real-time sequencing, long-range polymerase chain reaction, and Sanger sequencing data identified at least four different plastome structural types in both Eleocharis species. PacBio long read data suggested that one of the four E. dulcis plastome types predominates.
Keywords: chloroplast, plastid genome, rearrangement, homologous recombination, RDR, repeat
Introduction
Plastids are one of the most essential features in photosynthetic eukaryotes. Plastid genomes (plastomes) of land plants have a conserved quadripartite structure in a gene-dense unit genome consisting of large and small single copy (LSC and SSC; ∼80 and 20 kb) regions separated by a large inverted repeat (IRA and IRB; ∼25 kb) (Wicke et al. 2011; Ruhlman and Jansen 2014). Plastomes of most land plants range from 120 to 170 kb in size with a median size and GC content of 154 kb and 37.6%, respectively (Wicke et al. 2011; Weng et al. 2014; Park et al. 2018). After massive transfer of plastid DNA to the host nucleus during symbiogenesis, most plastomes retain genes encoding ∼80 proteins, 30 tRNAs, and 4 rRNAs. Although variation in plastome size, structure, and gene content is uncommon in most photosynthetic taxa, extensive variation has been documented in several angiosperm lineages (Ruhlman and Jansen 2018).
Expansion, contraction, and loss of the IR are the main factors causing plastome size variation, and considerable variation in IR extent has been observed in diverse lineages, including Geraniaceae (Guisinger et al. 2011; Blazier, Jansen, et al. 2016; Zhang et al. 2016; Weng et al. 2017; Ruhlman and Jansen 2018), Fabaceae (Dugas et al. 2015; Schwarz et al. 2015), Ericaceae (Fajardo et al. 2013; Martínez-Alberola et al. 2013), Berberidaceae (Ma et al. 2013), Trochodendraceae (Sun et al. 2013), Plantaginaceae (Zhu et al. 2016), Cactaceae (Sanderson et al. 2015; Solórzano et al. 2019), Annonaceae (Blazier, Ruhlman, et al. 2016), Campanulaceae (Knox 2014; Cheon et al. 2017), Papaveraceae (Park et al. 2018), and Passifloraceae (Rabah et al. 2019; Shrestha et al. 2019). The plastome of Pelargonium transvaalense is ∼243 kb in size due to a remarkably expanded IR (∼88 kb), leaving relatively small LSC (∼60 kb) and SSC (∼7 kb) regions (Weng et al. 2017). Similar to plastome size variation, examples of gene and intron losses have been documented in several angiosperm families. The loss of the entire suite of ndh genes in Erodium (Blazier et al. 2011), Orchidaceae (Chang et al. 2006; Wu et al. 2010; Delannoy et al. 2011; Barrett et al. 2014; Kim et al. 2015; Ruhlman et al. 2015), Alismatales (Peredo et al. 2013), and Carnegiea (Sanderson et al. 2015) and loss of accD, clpP, rpl20, rps7, and ycf1 genes and the introns of atpF, rpoC1, and clpP in Passiflora (Jansen et al. 2007; Rabah et al. 2019; Shrestha et al. 2019) are examples. The monocot family Poaceae (Poales) also experienced gene and intron losses, such as accD, ycf1, and ycf2 genes and the introns of clpP and rpoC1 (Hiratsuka et al. 1989; Maier et al. 1995; Morris and Duvall 2010). These changes do not occur in basal lineages of Poales (Bromeliaceae and Typhaceae) (Stevens 2001), which retain the structure and content of conserved plastomes (Guisinger et al. 2010; Redwan et al. 2015; Poczai and Hyvönen 2017).
Although most plastomes contain very small amounts of repetitive DNA apart from the canonical IR, large numbers of repeats have been identified in several eudicot and monocot lineages with highly rearranged plastomes (Lee et al. 2007; Cai et al. 2008; Haberle et al. 2008; Guisinger et al. 2011; Zhang et al. 2011; Weng et al. 2014). In some of these, a significant positive correlation between the degree of genomic rearrangements and the number of repeats was detected (Guisinger et al. 2011; Weng et al. 2014). Repetitive DNA in plastomes plays a central role in genomic rearrangements and sequence divergence via illegitimate recombination and slipped-strand mispairing (Asano et al. 2004; Rogalski et al. 2006; Timme et al. 2007; Gray et al. 2009; Zhang et al. 2011). Although the large canonical IR is well known to reverse the polarity of SC regions through recombination (Kolodner and Tewari 1979; Bendich 2004), other inversions have been associated with short IRs (Kim and Lee 2005; Schwarz et al. 2015; Rabah et al. 2019). The typical IR does not occur in conifer plastomes but lineage-specific, short IRs are associated with large inversions. Since Tsumura et al. (2000) discovered inversion isomers associated with ∼1.2-kb IR in Pinaceae, examples of repeat associated isomeric plastomes within a single species or individual have been documented in several gymnosperm lineages including Pinaceae, Cupressaceae, Podocarpaceae, and Taxaceae (Wu et al. 2011; Guo et al. 2014; do Nascimento Vieira et al. 2016; Hsu et al. 2016; Qu et al. 2017; Fu et al. 2019). In angiosperms, two distinct plastome configurations that differed in the orientation of a ∼45-kb segment were detected among accessions of the legume Medicago truncatula with verification using DNA gel blot analysis (Gurdon and Maliga 2014). Recently, with the aid of PacBio long reads, alternative plastome arrangements were identified within the plastome of a single individual of Monsonia emarginata (Geraniaceae), and these structural variants were associated with large repeats (Ruhlman et al. 2017).
Approximately 4,000 land plant plastomes have been sequenced and are publicly available (accessed on October 2, 2019 from NCBI). However, taxon sampling is biased in favor of certain major groups, limiting the knowledge of plastome organization and evolution in many lineages. For example, despite 616 sequenced plastomes of the order Poales available in NCBI, just eight belong to the non-Poaceae lineages Bromeliaceae, Typhaceae, Eriocaulaceae, Cyperaceae, and Joinvilleaceae (Guisinger et al. 2010; Redwan et al. 2015; Wysocki et al. 2016). Among 14 Poales families (APG IV 2016), Cyperaceae (sedges) is the second largest with more than 5,500 species and a cosmopolitan distribution. However, little is known about plastome organization and evolution within the family, which is represented by three plastomes in NCBI that have not yet been published. In this study, plastome sequences of two Eleocharis species were completed. In contrast to basal Poales, Eleocharis plastomes exhibited multiple structural types within a single individual, unprecedented levels of genome rearrangement and repetitive DNA, and extensive gene loss and duplication.
Materials and Methods
Plant Materials and DNA Extraction
Corms of water chestnut (Eleocharis dulcis, accession # PI 106274) were provided by the U.S. National Plant Germplasm System in the United States Department of Agriculture and Gulf coast spikerush, Eleocharis cellulosa, was collected in west Texas (supplementary table S1, Supplementary Material online). Corms of E. dulcis and live plants of E. cellulosa were grown in the University of Texas at Austin (UT-Austin) greenhouses. Because culms (a hollow or pithy stalk or stem of grasses, sedges, and rushes) play the primary role in photosynthesis due to highly reduced leaves modified into a tubular sheath at the base of culms, total genomic DNA was extracted from newly emergent culms using the cetyl trimethylammonium bromide method of Doyle and Doyle (1987) with modifications that included the addition of 3% polyvinylpyrrolidone (PVP; Sigma, St. Louis, Mo, USA) and 3% beta-mercaptoethanol (Sigma, St Louis, MO). Following treatment with RNase A (ThermoScientific, Lafayette, CO) and phase separation with chloroform, DNA was recovered by ethanol precipitation, resuspended in DNase-free water, and stored at –20 °C for genome sequencing. DNA of E. dulcis but not E. cellulosa was extracted from a single individual. Voucher specimens of E. dulcis and E. cellulosa were deposited in the Billie L. Turner Plant Resources Center (TEX-LL) at UT-Austin (supplementary table S1, Supplementary Material online).
Genome Sequencing, Assembly, and Annotation
Genomic DNAs were sent to the Beijing Genomics Institute for library preparation and Illumina sequencing. Approximately 40 million 150-bp, paired-end (PE) reads were generated on the Illumina HiSeq X-Ten sequencing platform (Illumina, San Diego, CA) and reads were assembled de novo using Velvet version 1.2.08 (Zerbino and Birney 2008) at the Texas Advanced Computing Center. Multiple de novo assemblies were performed with a range of k-mer sizes (intervals of four from 61 to 117), three minimum depths of coverage (200×, 500×, and 1,000×), default insert size estimation, and scaffolding off. Plastid contigs from all assemblies were imported into Geneious v.9.1.8 (http://www.geneious.com) (Kearse et al. 2012) and all protein-coding genes were identified by BLAST against reference plastomes of Nicotiana tabacum (NC_001879), Typha latifolia (NC_013823), Ananas comosus (NC_026220), and Hypolytrum nemorum (NC_036036) and by GeSeq with MPI-MP chloroplast references (Tillich et al. 2017). Once draft plastomes were assembled, all plastid contigs were mapped to drafts to identify the IR boundaries and mismatches among contigs. To validate any ambiguities and determine the IR boundaries, Bowtie2 v.2 (Langmead and Salzberg 2012) was used to map the Illumina reads to draft plastomes from which IRa had been removed. Verification of protein-coding genes was manually performed in Geneious by aligning genes with their homologs from the plastome sequences listed above as well as from Spinacia oleracea (NC_002202) and Arabidopsis thaliana (NC_000932). Because the H. nemorum plastome is publicly available in NCBI but has not yet been published, annotations were carefully examined and manually modified following the method described above prior to use in comparative analyses. All transfer RNA genes were identified using tRNAscan-SE v.2.0 (Lowe and Chan 2016). In Eleocharis and H. nemorum plastomes, genes that were annotated with low similarity (25–50%) to reference genes were further examined and regarded as putatively missing or nonfunctional based on the following criteria used in Rabah et al. (2019): absence of an open-reading frame with a complete conserved domain as searched in conserved domain database (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and presence of internal stop codons that interrupted the reading frame or the conserved domain. Linear plastome maps were drawn with OGDRAW v. 1.2 (Lohse et al. 2013). For repeat visualization, circular maps were drawn using Circoletto (Darzentas 2010).
Whole-genome alignments were performed to examine the arrangement of locally colinear blocks (LCBs) of different plastome structural types of E. dulcis and E. cellulosa and to estimate the genomic rearrangements of Eleocharis plastomes compared with the basal Poales, T. latifolia, as a reference (supplementary fig. S1, Supplementary Material online) using progressiveMauve 2.3.1 in Geneious (Darling et al. 2010) with default parameters. One copy of the IR (IRa) was removed from plastomes prior to the Mauve alignment. The LCBs of E. dulcis, E. cellulosa, H. nemorum, A. comosus, and T. latifolia plastomes were manually numbered and annotated for genes within each LCB. Strand orientation was indicated with “–”, if the LCB was in the reverse orientation relative to the reference. Breakpoint (BP) and reversal distances were estimated using the web-based application CREx (Common Interval Rearrangement Explorer) (Bernt et al. 2007) with T. latifolia as a reference.
Long-Range Polymerase Chain Reaction and Sanger Sequencing
Nine oligonucleotide primers were designed for long-range polymerase chain reaction (PCR) using Primer 3 (supplementary table S2, Supplementary Material online) (Untergasser et al. 2012). Primer sequences were located in genes (petN, petD, rps2, rpoC2, rpoB, psbK, rpl20, rps8, and ndhJ) situated at the ends of syntenic blocks to confirm plastome assemblies. In addition, 11 primers were designed to perform nested PCR using amplicons of targeted regions (supplementary table S2, Supplementary Material online). Long-range PCR with multiple primer combinations was performed using the high-fidelity TaKaRa PrimeStar GXL DNA Polymerase (Takara Bio USA, Inc., Mountain View, CA). PCR reactions were performed in 12.5 µl, including 7 µl distilled water, 2.5 µl 5× PrimeSTAR GXL Buffer, 1 µl dNTP mixture, 0.5 µl DNA polymerase, 0.5 µl each primer, and 0.5 µl genomic DNA (∼50 ng). Each target was amplified using variable conditions depending on the size: initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 58–62 °C for 15 s, and extension at 68 °C for 50–150 s. PCR products were treated with Exonuclease I (New England BioLabs, Ipswich, MA) and Shrimp Alkaline Phosphatase (Fermentas, Glen Burnie, MD) to remove residual primers and PCR amplicons were Sanger sequenced at the Genome Sequencing and Analysis Facility at UT-Austin.
PacBio Sequencing
Genomic DNA of E. dulcis with a total mass of 13.1 μg was sent to Beijing Genomics Institute for 20-kb PacBio library construction and single-molecule real-time (SMRT) sequencing (Pacific Biosciences, Menlo Park, CA). PacBio long reads were generated from one SMRT cell on RSII system. Error correction of long reads was performed using the LSC v.2.0 tool (Au et al. 2012) with ∼46 million Illumina PE reads generated from DNA of the same E. dulcis individual. Error-corrected reads were mapped to complete plastome monomers and to genes at the end of each Illumina/Velvet-assembled syntenic block to detect reads that may support the presence of multiple structural types.
Repetitive DNA Sequence Analysis
Repeat analyses were performed to calculate the content of tandem and dispersed repeats. One copy of the IR (IRa) was removed from plastomes prior to repeat analyses. Dispersed repeats were detected by using the command line version BLAST v.2.8.1+ (Altschul et al. 1990) with a word size of 16 and percent identity of 80.0% and each Eleocharis, H. nemorum, A. comosus, and T. latifolia plastomes as both query and subject. BLAST results were carefully examined and duplicate repeats were eliminated. Tandem repeats were identified by using the web version of Tandem Repeats Finder v.4.09 with default parameters (Benson 1999). Total repeats were categorized into several size classes based on the length of BLAST hits. The proportion of total repeats and repeats by each size class was calculated for each plastome (length of repeats/length of plastome [without IRa] × 100).
Results
Multiple Structural Types in Eleocharis Plastomes
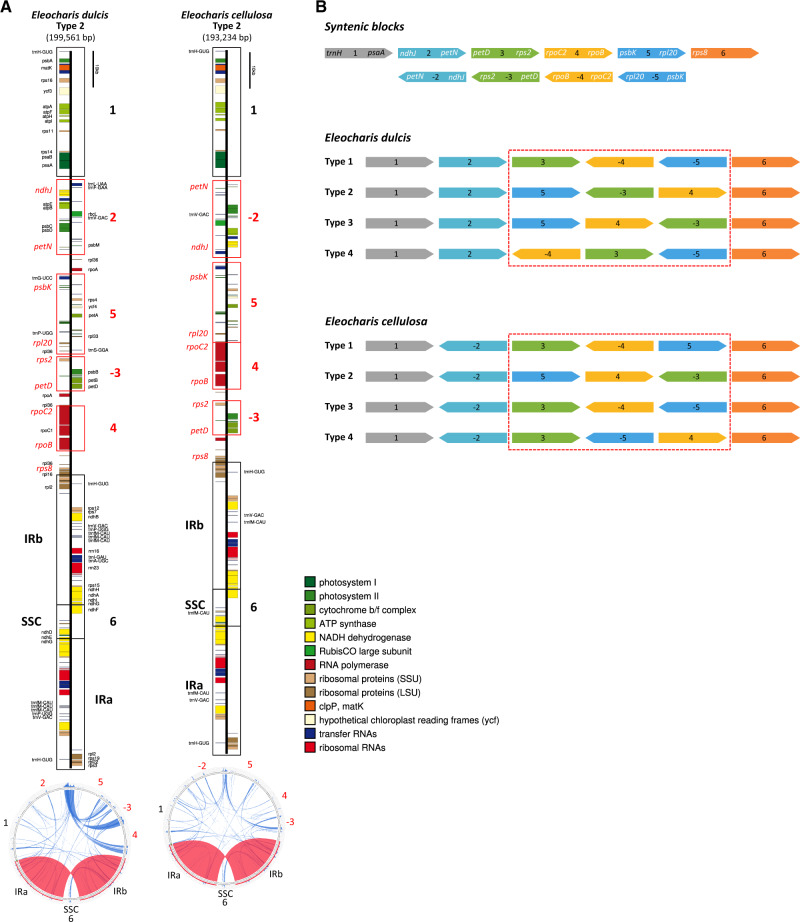
Plastome contigs assembled de novo from E. dulcis and E. cellulosa Illumina PE reads were very complex and suggested multiple arrangements of syntenic blocks (fig. 1B and supplementary figs. S2 and S3, Supplementary Material online). All possible arrangements of the syntenic blocks were assembled to yield the greatest number of unique draft plastome monomers. Careful examination of draft assemblies suggested four structural types that vary with respect to the arrangement of syntenic blocks in the LSC region.
Fig. 1.
—Schematic representations of Eleocharis dulcis and E. cellulosa plastomes. (A) Unit-genome maps and repetitive DNA content of E. dulcis and E. cellulosa plastomes. Completed Eleocharis plastome sequences were submitted to OGDRAW (Lohse et al. 2013) to generate physical maps and Circoletto (Darzentas 2010) to visualize repetitive DNA. Structural type and plastome size (in parentheses) are shown below the species name. Syntenic blocks (numbered 1–6) detected by progressiveMauve (Darling et al. 2010) are depicted by open boxes; the negative symbol (–) indicates reverse-oriented strands relative to the reference (Typha). Red boxes indicate blocks that that vary in order among different structural types, whereas blocks encompassed by black boxes maintain their positions across types. Genes indicated in red font are located near the end of each syntenic block and were employed as annealing sites for PCR confirmation of plastome arrangements. The vertical black bar at the top of each linear map is provided for scale. The circular representation of each species is below each linear map with syntenic block numbers shown. Dispersed repeats and IR are shown within the circular map in blue and red, respectively. (B) Each syntenic block is illustrated with a different color and follows the same numbering convention as in (A). Representative genes near the end of each syntenic block are shown at the top. Gene symbols in white font indicate those in long-range PCR and correspond to the red gene symbols in (A). Different syntenic block arrangements are highlighted in red boxes for both Eleocharis species. IR, inverted repeat; SSC, small single copy region.
Although the Illumina PE reads mapped to the four plastome types with ∼1,900× and 1,080× depth of coverage for E. dulcis and E. cellulosa (supplementary table S3, Supplementary Material online), respectively, these data were generated from 300- to 350-bp insert libraries and therefore could not confirm the presence of different plastome types. To address this and to confirm the putative multiple types, long-range PCR was performed for both species.
Long-range PCR amplification provided strong evidence for multiple structural conformations of Eleocharis plastomes (fig. 1B, table 1, and supplementary figs. S4 and S5, Supplementary Material online). All but 3 of the 20 unique junction regions among syntenic blocks 2–6 for each plastome type were amplified using multiple combinations of primers designed to anneal in nine genes located at the boundaries of each block (fig. 1B and supplementary table S2, Supplementary Material online). For E. dulcis, all junction regions were amplified and most of PCR amplifications yielded fragments of expected sizes, which ranged from ∼4.4 to ∼11 kb, confirming the presence of four plastome structural types (table 1). Although PCR of the 2–5 and (−3)–4 junctions exhibited two amplicons of different sizes, those with the stronger intensity were consistent with the expected sizes. Three fragments that were amplified from the 2–3 junction showed a similar intensity and one of them was congruent with the expected size. For E. cellulosa, all junctions for plastome types 1 and 4 were amplified with fragments of expected sizes except for the 5–6 and 4–6 junctions (table 1). Plastome types 2 and 3 were supported with PCR amplifications for 2 and 3 junctions. Unexpectedly, (−4)–5 and 5–6 junctions found only in E. cellulosa plastome types 1 and 4 were also amplified in E. dulcis (supplementary fig. S6 and table S4, Supplementary Material online). The sizes of each PCR amplicon from E. dulcis (∼8 kb and ∼4.8/∼3 kb) were very close to those from E. cellulosa (∼8.5 kb and ∼4.5 kb).
Table 1.
Verification of Multiple Structural Types in Eleocharis Plastomes
| Species | Type | Syntenic Block | Primer F | Primer R | Size (bp) | PCR | PCR Band Size (bp) | Sanger Seq. | PacBio Seq. |
|---|---|---|---|---|---|---|---|---|---|
| Eleocharis dulcis | 1 | 2 – 3 | E_petN | E_petD | 8,188 | + | 5,500/6,200/8,100 | + | |
| 3 – (−4) | E_rps2 | E_rpoB | 4,842 | + | ∼5,000 | +a | |||
| (−4) – (−5) | E_rpoC2 | E_rpl20 | 4,368 | + | ∼4,300 | +a | |||
| (−5) – 6 | E_psbK | E_rps8 | 8,550 | + | ∼8,500 | ||||
| 2 | 2 – 5 | E_petN | E_psbK | 11,127 | + | ∼11,000 b /∼6,200 | + | ||
| 5 – (−3) | E_rpl20 | E_rps2 | 4,381 | + | ∼4,300 | +a | + | ||
| (−3) – 4 | E_petD | E_rpoC2 | 5,261 | + | ∼5,000 b /∼3,000 | +c | + | ||
| 4 – 6 | E_rpoB | E_rps8 | 5,181 | + | ∼5,000 | +c | + | ||
| 3 | 2 – 5 | E_petN | E_psbK | 11,128 | + | ∼11,000 b /∼6,200 | + | ||
| 5 – 4 | E_rpl20 | E_rpoC2 | 4,368 | + | ∼4,300 | +a | |||
| 4 – (−3) | E_rpoB | E_rps2 | 4,842 | + | ∼5,000 | +a | |||
| (−3) – 6 | E_petD | E_rps8 | 5,610 | + | ∼5,500 | +d | |||
| 4 | 2 – (−4) | E_petN | E_rpoB | 4,869 | + | ∼4,000 | +c | ||
| (−4) – 3 | E_rpoC2 | E_petD | 5,257 | + | ∼4,300 | +c | + | ||
| 3 – (−5) | E_rps2 | E_rpl20 | 4,381 | + | ∼4,300 | +a | + | ||
| (−5) – 6 | E_psbK | E_rps8 | 8,550 | + | ∼8,500 | ||||
| Eleocharis cellulosa | 1 | (−2) – 3 | E_ndhJ | E_petD | 5,387 | + | ∼5,400 | n/a | n/a |
| 3 – (−4) | E_rps2 | E_rpoB | 4,816 | + | ∼5,000 | n/a | n/a | ||
| (−4) – 5 | E_rpoC2 | E_psbK | 9,000 | + | ∼8,500 | n/a | n/a | ||
| 5 – 6 | E_rpl20 | E_rps8 | 3,025 | + | ∼4,500 | n/a | n/a | ||
| 2 | (−2) – 5 | E_ndhJ | E_psbK | 8,326 | n/a | n/a | |||
| 5 – 4 | E_rpl20 | E_rpoC2 | 2,685 | + | ∼2,700 | n/a | n/a | ||
| 4 – (−3) | E_rpoB | E_rps2 | 4,846 | + | ∼4,500 | n/a | n/a | ||
| (−3) – 6 | E_petD | E_rps8 | 6,400 | n/a | n/a | ||||
| 3 | (−2) – 3 | E_ndhJ | E_petD | 5,387 | + | ∼5,000 | n/a | n/a | |
| 3 – (−4) | E_rps2 | E_rpoB | 4,816 | + | ∼4,500 | n/a | n/a | ||
| (−4) – (−5) | E_rpoC2 | E_rpl20 | 2,685 | + | ∼2,700 | n/a | n/a | ||
| (−5) – 6 | E_psbK | E_rps8 | 9,339 | n/a | n/a | ||||
| 4 | (−2) – 3 | E_ndhJ | E_petD | 5,387 | + | ∼5,000 | n/a | n/a | |
| 3 – (−5) | E_rps2 | E_rpl20 | 4,346 | + | ∼4,300 | n/a | n/a | ||
| (−5) – 4 | E_psbK | E_rpoC2 | 9,001 | + | ∼8,000 | n/a | n/a | ||
| 4 – 6 | E_rpoB | E_rps8 | 3,495/2,312 | + | ∼5,000 | n/a | n/a |
Note.—+ indicates that adjacencies of syntenic blocks were confirmed. F, forward; R, reverse; n/a, not available.
Entire junction was sequenced.
Band size in bold indicates the one with higher intensity.
Boundaries of junction were sequenced.
Almost entire junction was sequenced.
To further examine the E. dulcis plastome, the PCR amplicons for several junction regions were Sanger sequenced (table 1) and boundary sequences were compared with draft plastome assemblies from Illumina data. DNA sequences of seven junction boundaries consistently mapped to the expected regions in each plastome type. Among seven junctions, three, 3–(−4), (−4)–(−5), and 5–(−3), were fully sequenced by nested PCR using the PCR amplicons as templates with primers designed to anneal near the 3′ end (supplementary table S2, Supplementary Material online). One other region, (−3)–6, was almost fully sequenced.
Long read PacBio SMRT sequencing was performed for E. dulcis. A total of 68,167 raw PacBio reads with the mean size of 10,035 bp was generated. Following correction with Illumina sequences, the read count was reduced to 61,379 with the mean size of 9,139 bp and mean GC content of 35.7%. Mapping of long reads to each plastome type, emphasizing reads containing genes situated at the end of each syntenic block, showed that the majority of reads were consistent with the plastome type 2 except for four reads. Specifically, when corrected long reads were mapped to one end of syntenic block 2, where petN gene is located, 119 of 123 mapped reads were consistent with 2–5 syntenic block junction, but four reads that showed partial inconsistency with plastome type 2 had the sequences of 2–3 junction (adjacency pair of petN–petD genes) in plastome type 1, confirming the presence of multiple structural types in a single individual of E. dulcis.
Plastome Features
Minimal size variation was detected among the four plastome types in E. dulcis and E. cellulosa, ranging from 196,668 to 199,561 bp and 192,023 to 193,234 bp, respectively (supplementary table S5, Supplementary Material online), all of which were larger than the median plastome size (152 kb) for 3,656 angiosperms (accessed on October 2, 2019 from NCBI). Hypolytrum nemorum (NC_036036), representing a basal lineage of Cyperaceae, had a plastome size of 180,648 bp with the IR expanded to ∼ 38 kb and was also larger than the median angiosperm plastome size but smaller than Eleocharis. The IR and SSC regions of both Eleocharis species were similar in size (IR: ∼36 kb, E. dulcis and ∼35 kb, E. cellulosa; SSC: ∼10 kb) with only seven genes (ndhF, trnL-UAG, rpl32, trnT-UGU, ndhD, psaC, and ndhE genes) in the SSC (table 2). However, since the size variation in the IR and SSC among different plastome types could not be identified with the current data, minor size variation in LSC region (115,003–117,896 bp, E. dulcis and 111,670–112,881 bp, E. cellulosa) accounted for the overall size variation among four types identified within each species.
Table 2.
Summary of Major Features of Eleocharis, Hypolytrum, and Basal Poales Plastid Genomes
| Family |
Cyperaceae |
Bromeliaceae |
Typhaceae |
||
|---|---|---|---|---|---|
| Taxon | Eleocharis dulcis (Type 2) | Eleocharis cellulosa (Type 2) | Hypolytrum nemorum | Ananas comosus | Typha latifolia |
| Genome size (bp) | 199,561 | 193,234 | 180,648 | 159,636 | 161,572 |
| LSC (% of genome) | 117,896 (59.1) | 112,881 (58.4) | 95,644 (52.9) | 87,482 (54.8) | 89,140 (55.2) |
| SSC (% of genome) | 9,601 (4.8) | 10,311 (5.3) | 8,150 (4.5) | 18,622 (11.7) | 19,652 (12.2) |
| IR (% of genome) | 36,032 (18.1) | 35,021 (18.1) | 38,427 (21.3) | 26,766 (16.8) | 26,390 (16.3) |
| Total number of genes | 139 | 132 | 135 | 131 | 131 |
| Number of unique genes | 105 | 105 | 110 | 113 | 113 |
| Number of unique protein-coding genes (duplicated in IR) | 72 (14) | 72 (12) | 76 (12) | 79 (6) | 79 (6) |
| Number of unique tRNA genes (duplicated in IR) | 29 (10) | 29 (9) | 30 (8) | 30 (8) | 30 (8) |
| Number of unique rRNA genes (duplicated in IR) | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) |
| Number of genes with introns | 17 | 17 | 18 | 18 | 18 |
| GC content (%) | 32.6 | 32.8 | 34.9 | 37.4 | 36.6 |
| GC content of IR/LSC/SSC (%) | 37.6/30.2/25.5 | 37.8/30.4/25.8 | 38.5/32.6/28.1 | 42.7/35.4/31.4 | 42.4/34.4/30.5 |
| Genic DNA (% of genome, GC [%]) | 76,207 (38.2, 38.4) | 74,394 (38.5, 38.5) | 98,905 (54.8, 38.1) | 91,317 (57.2, 40.2) | 91,212 (56.4, 39.9) |
| Intergenic spacers (% of genome, GC [%]) | 123,354 (61.8, 29.1) | 118,840 (61.5, 29.3) | 81,743 (45.2, 31.1) | 68,611 (42.8, 33.7) | 70,541 (43.6, 32.4) |
| Gene density | 0.70 | 0.68 | 0.75 | 0.82 | 0.81 |
| Putative gene losses | accD, clpP, rpl23, rps18, ycf1, ycf2, infA, and trnT-GGU | accD, clpP, rpl23, rps18, ycf1, ycf2, infA, and trnT-GGU | accD, rps18, and infA | — | — |
| Putative gene duplications | rpoA (2) rpl36 (4), trnfM-CAU (6), trnH-GUG (3), trnP-UGG (3), and trnV-GAC (3) | rpl36 (2)* trnfM-CAU (3), trnH-GUG (3), and trnV-GAC (3) | trnV-GAC (3) | — | — |
Note.—The number in ( ) in putative gene duplications indicates the number of copies. Asterisk (*) on rpl36 gene indicates gene duplication in only plastome type 4 of E. cellulosa.
Plastomes of both Eleocharis species contained the same number of unique genes (105; 72 protein-coding genes, 29 tRNA, and 4 rRNA genes), which was fewer than H. nemorum (110; 76 protein-coding genes, 30 tRNA, and 4 rRNA genes) and the highly conserved basal Poales plastomes (113; 79 protein-coding genes, 30 tRNA, and 4 rRNA genes) due to putative gene losses. However, the total number of genes in Eleocharis was greater than basal Poales due to gene duplication events (table 2). The total number of genes was higher in E. dulcis (139) than E. cellulosa (132) due to more extensive duplications (table 2). The plastomes of both species experienced several putative gene losses including accD, clpP, rpl23, rps18, ycf1, ycf2, infA, and trnT-GGU. The plastome of H. nemorum also lacked accD, rps18 and infA and experienced duplication of trnV-GAC (table 2). Gene density (the number of genes per kb) in E. dulcis, E. cellulosa, and H. nemorum plastomes was 0.70, 0.68 and 0.75 and the percentage of the genome that contained coding regions was 38.2%, 38.5%, and 54.8%, respectively, substantially lower than basal Poales (0.82 and 57.2% for A. comosus and 0.81 and 56.4% for T. latifolia) (table 2).
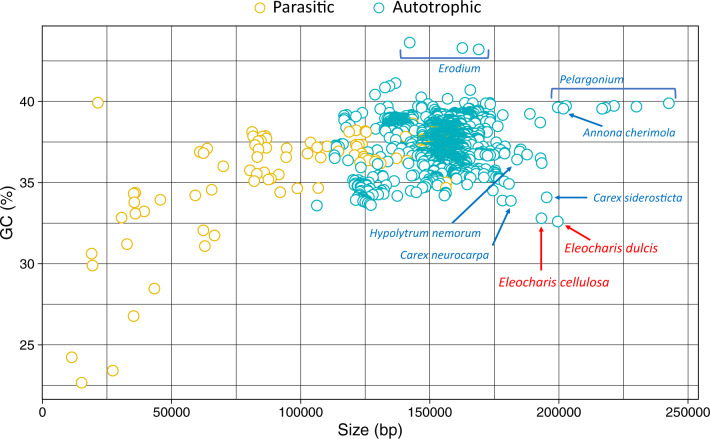
The GC content of E. dulcis and E. cellulosa plastomes was low at 32.6% and 32.9%, respectively, compared with the earliest diverging taxon of Cyperaceae, H. nemorum (34.9%). All Cyperaceae had GC values less than the basal Poales T. latifolia (36.6%) and A. comosus (37.4%) and the mean value (37.5%) of 3,656 angiosperm plastomes (fig. 2 and table 2). The IR GC content of E. dulcis (37.6%) and E. cellulosa (37.8%) plastomes was higher than the mean for angiosperm plastome IRs, whereas the GC content of LSC and SSC regions for E. dulcis (30.2% and 25.5%) and E. cellulosa (30.4% and 25.8%) was lower (table 2).
Fig. 2.
—Eleocharis plastomes exhibit atypical size and GC content. Plastome size and GC content of 3,656 angiosperms in the NCBI Genome database were plotted. Parasitic (yellow) and autotrophic (teal) species are indicated with different colors. The two Eleocharis plastomes are labeled with red font. Other Cyperaceae and larger plastomes, including Pelargonium and Annona cherimola, are indicated with teal font. The Erodium plastomes with high GC content are also represented with a teal label. bp, basepairs.
Overall, the organization of Eleocharis plastomes was substantially different from the highly conserved plastomes of basal Poales, Bromeliaceae and Typhaceae, whereas the basal taxon in Cyperaceae, H. nemorum, exhibited features intermediate between those of Eleocharis and basal Poales.
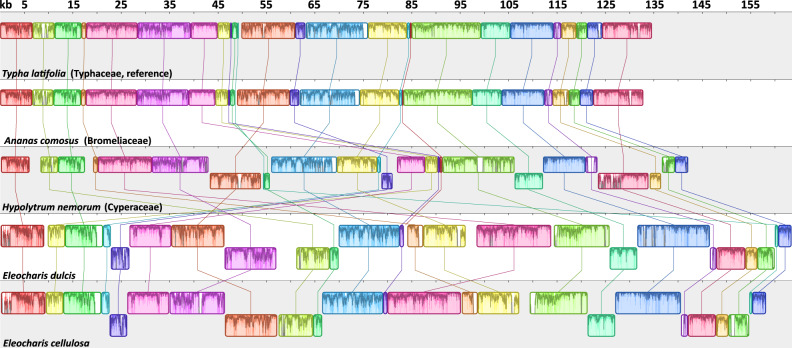
Whole-genome alignment using progressiveMauve (fig. 3 and supplementary table S6, Supplementary Material online) identified 25 LCBs from five complete plastomes (E. dulcis, E. cellulosa, H. nemorum, A. comosus, and T. latifolia). The two basal Poales plastomes were collinear, whereas Hypolytrum had BP and reversal distances of 11 and 7, respectively compared with basal Poales plastomes (table 3). The BP and reversal distances were 16 and 13 (E. dulcis) or 16 and 14 (E. cellulosa) between Hypolytrum and Eleocharis plastomes, respectively. The largest BP distance (25) was between E. dulcis and basal Poales plastomes, whereas the largest reversal distance (21) was between E. cellulosa and basal Poales. The BP and reversal distances were 5 and 4 between the two Eleocharis species, respectively. Among the four plastome types in each Eleocharis species, six LCBs (1–6 syntenic blocks) were identified excluding small LCBs of large repeats (supplementary fig. S3, Supplementary Material online). Plastomes of both Eleocharis species had different orders of LCBs (3–5 syntenic blocks) in the LSC region and in E. cellulosa LCB 2 was reversed compared with E. dulcis.
Fig. 3.
—Whole-plastome alignment of five Poales species. Newly completed plastomes of Eleocharis dulcis and E. cellulosa and publicly available basal Cyperaceae and Poales plastomes from NCBI (Hypolytrum nemorum, Ananas comosus, and Typha latifolia) were analyzed by progressiveMauve to identify LCBs with the Typha plastome as a reference. One copy of the inverted repeat was removed before the analysis and numerals at top indicate size in kilobases (kb). The corresponding LCBs among five plastomes are shaded and connected with a line of the same color. The histogram inside each block shows pairwise nucleotide sequence identity. LCBs that are flipped across the plane indicate an inverted strand.
Table 3.
Pairwise Comparison of Breakpoint and Reversal Distances for Eleocharis, Hypolytrum, and Basal Poales Plastomes
| Typha latifolia | Ananas comosus | Hypolytrum nemorum | Eleocharis dulcis | Eleocharis cellulosa | |
|---|---|---|---|---|---|
| Typha latifolia | — | ||||
| Ananas comosus | 0/0 | — | |||
| Hypolytrum nemorum | 11/7 | 11/7 | — | ||
| Eleocharis dulcis | 25/20 | 25/20 | 16/13 | — | |
| Eleocharis cellulosa | 24/21 | 24/21 | 16/14 | 5/4 | — |
Repetitive DNA in Eleocharis Plastomes
Repeat analyses revealed that Eleocharis plastomes contain abundant repetitive DNA with minor variation among different structural types (supplementary tables S7 and S8, Supplementary Material online). Along with Eleocharis, basal Cyperaceae and Poales taxa were included in repeat analyses. The amount and proportion of dispersed repeats in Eleocharis plastomes (39,752 bp, 24.3% for E. dulcis and 31,118 bp, 19.7% for E. cellulosa) were substantially greater than in H. nemorum (8.8%, 12,520 bp), which in turn were greater than A. comosus (1.1%, 1,495 bp) and T. latifolia (0.9%, 1,210 bp) (table 4).
Table 4.
Statistics of Dispersed and Tandem Repeats in Eleocharis, Hypolytrum, and Basal Poales Plastomes
| Family |
Cyperaceae |
Bromeliaceae |
Typhaceae |
||
|---|---|---|---|---|---|
| Species | Eleocharis dulcis Type 2 | Eleocharis cellulosa Type 2 | Hypolytrum nemorum | Ananas comosus | Typha latifolia |
| Genome size (IRa excluded) | 163,529 | 158,213 | 142,221 | 132,862 | 134,642 |
| GC % | 31.6 | 31.8 | 34 | 36.3 | 35.5 |
| Dispersed repeats (DRs) | |||||
| Length of DR | 39,752 | 31,118 | 12,520 | 1,495 | 1,210 |
| GC % of DR | 30.9 | 30.2 | 32.6 | 35.9 | 33.7 |
| GC % without DR | 31.8 | 32 | 33.9 | 36.3 | 35.5 |
| % of DR in genome | 24.3 | 19.7 | 8.8 | 1.1 | 0.9 |
| Tandem repeats (TRs) | |||||
| Length of TR | 5,864 | 2,833 | 6,638 | 2,057 | 3,270 |
| GC % of TR | 25.9 | 25.5 | 27.8 | 18.4 | 13.5 |
| GC % without TR | 31.8 | 31.9 | 34.3 | 36.6 | 36 |
| % of TR in genome | 3.6 | 1.8 | 4.7 | 1.5 | 2.4 |
| Total repeats | |||||
| Length of total repeats | 42,216 | 32,695 | 16,718 | 3,552 | 4,436 |
| GC % of total repeats | 30.4 | 29.9 | 31.1 | 25.7 | 19.1 |
| GC % without total repeats | 32 | 32.1 | 34.3 | 36.6 | 36 |
| % of total repeats in genome | 25.8 | 20.7 | 11.8 | 2.7 | 3.3 |
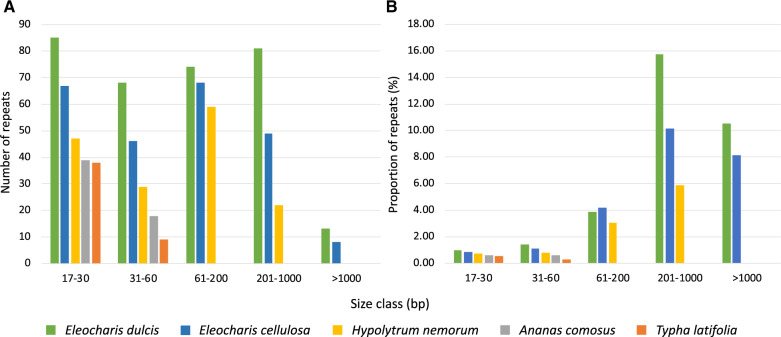
Dispersed repeats were grouped into five size classes (fig. 4). The E. dulcis plastome contained the largest number of dispersed repeats (321) ranging from 17 bp to ∼5 kb followed by E. cellulosa (238; 17 bp to ∼3 kb) and H. nemorum (157; 17 to ∼850 bp), whereas basal Poales had 57 and 47 repeats smaller than 60 bp (fig. 4 and supplementary table S8, Supplementary Material online). Repeats >1,000 bp were only detected in Eleocharis plastomes where they represented more than 10% and 8% of the E. dulcis and E. cellulosa genomes, respectively (fig. 4). The three Cyperaceae plastomes contain larger repeats in the range of 61–1,000 bp relative to other Poales, and these larger repeats constitute a substantial proportion of the plastome (fig. 4). In Eleocharis, many of the larger repeats in the >201 bp size range were concentrated between syntenic blocks 2–6 (fig. 1A).
Fig. 4.
—Repetitive DNA content in five Poales plastomes. (A) The number of dispersed repeats in different size classes. (B) The proportion of plastome that represents dispersed repeats in different size classes. bp, basepairs.
The greatest proportion of tandem repeats were identified in H. nemorum (6,638 bp; 4.7%) followed by E. dulcis (5,864 bp; 3.6%) (table 4). Unlike dispersed repeats, only E. dulcis plastome contained unusually large amounts of tandem repeats. Eleocharis cellulosa had fewer tandem repeats (2,833 bp; 1.8%) than T. latifolia (3,270 bp; 2.4%). Some tandem repeats were embedded in large dispersed repeats in Eleocharis plastomes, and there were instances where small dispersed repeats were located within tandem repeats. In the E. dulcis plastome, three copies of the largest tandem repeat (278 bp) with 99% sequence identity included trnfM-CAU in the IR. Most tandem repeats were found in intergenic spacers (IGSs) except for occurrences in the introns of several genes (rps16, ycf3, rpoC1, rpl16, and ndhB), including 16 copies of a 48-bp tandem repeat that share 84% sequence identity within the intron of rpoC2 causing an expansion of its size to 5,097 bp. Similar to E. dulcis, most tandem repeats in E. cellulosa were found in IGS regions. Three introns included tandem repeats, the introns of rps16, ndhB, and rpoC2. The rpoC2 gene (4,707 bp) was expanded by 12 copies of a 48-bp tandem repeat with 84% sequence identity within its intron. Three and seven copies of a 48- and 45-bp tandem repeat were identified within rpoC2 (4,707 bp) in H. nemorum. Overall, the exceptionally abundant repeats that enlarged IGS regions contributed to plastome size expansion in Eleocharis.
Repetitive DNA also resulted in gene duplications in Eleocharis plastomes. Three copies of the trnfM-CAU were located in the 278-bp tandem repeat of the E. dulcis IR, resulting in six total copies of this sequence. Several other genes were multiplied, including four copies of rpl36 and two copies of rpoA in the LSC, and three copies of trnH-GUG, trnV-GAC, and trnP-UGG (two copies in the IRs). In E. cellulosa, whereas rpl36 was duplicated in a 593-bp dispersed repeat only in the LSC of plastome type 4, all four structural types had three copies of trnfM-CAU, trnH-GUG, and trnV-GAC (two copies in the IRs) located in various sized dispersed repeats (177–456 bp). In H. nemorum, one additional copy of trnV-GAC along with two in the IR was identified in the LSC within an 800-bp dispersed repeat with 98.5% sequence identity.
Discussion
Despite being the second largest family of Poales, plastome organization and evolution of Cyperaceae have not been examined. In this study, plastomes of two Eleocharis species, E. dulcis and E. cellulosa, were atypical with respect to size, gene content, GC content, gene order, and gene density compared with the highly conserved basal Poales plastomes. The most unusual feature was the presence of multiple plastome structural types within each Eleocharis species. Furthermore, plastome structural heteroplasmy was observed within single individuals and was associated with a remarkable abundance of repetitive DNA. The discussion mainly focuses on possible mechanisms that could generate different structural types in plastomes and comparisons to alternative plastome structures (isomeric forms) reported in a few distantly related seed plant lineages. The plastome organization of Eleocharis is also compared with basal Cyperaceae and other Poales.
Structural Heteroplasmy in Eleocharis Plastomes
In two species of Eleocharis, at least four different plastome structural types are present, and in one species, E. dulcis, this unprecedented structural heteroplasmy was identified in a single plant. Initial assemblies using Illumina short reads suggested multiple plastome types, a suggestion confirmed by long-range PCR, Sanger sequencing, and PacBio SMRT data. A few previous studies in conifers and Medicago (Fabaceae) detected isomeric plastomes associated with inversions mediated by lineage-specific short repeats (only for conifers) using a variety of approaches ranging from Southern hybridization to Illumina sequencing and PCR (Tsumura et al. 2000; Wu et al. 2011; Guo et al. 2014; do Nascimento Vieira et al. 2016; Hsu et al. 2016; Qu et al. 2017; Fu et al. 2019). Recently, PacBio long read data combined with Illumina read assembly demonstrated alternative gene order arrangements for the M. emarginata (Geraniaceae) plastome (Ruhlman et al. 2017).
Although four major plastome structural types were clearly evident in Eleocharis, long-range PCR data suggested the possibility of a higher degree of heteroplasmy as shown in supplementary table S4, Supplementary Material online. A similar suggestion was made by Ruhlman et al. (2017) for Monsonia. The possibility that the unexpected PCR results may be the result of amplifications of plastid DNA that has been transferred to nuclear or mitochondrial genomes cannot be ruled out. It is also possible that PCR artifacts arising from PCR-mediated recombination (Lahr and Katz 2009; Alverson et al. 2011) could explain the unexpected amplicons. However, consistent sizes between PCR amplicons for most junctions, Sanger sequencing, Illumina assemblies, and PacBio reads strongly support the unprecedented structural heteroplasmy in Eleocharis.
For decades, plastomes were considered circular with limited recombination. The presence of isomeric, presumed circular plastomes with different orientations of the single copy regions was attributed to intramolecular recombination between IR copies within a single unit genome and was referred to as “flip-flop” recombination (Palmer 1983, 1985; Brears et al. 1986; Stein et al. 1986). Two previous studies in Pinaceae adopted the flip-flop intramolecular recombination model to explain the different plastome isoforms (Tsumura et al. 2000; Wu et al. 2011), whereas other studies of conifers suggested a repeat-mediated homologous recombination mechanism based on a circular unit genome (Guo et al. 2014; do Nascimento Vieira et al. 2016; Hsu et al. 2016; Qu et al. 2017; Fu et al. 2019). These previous studies ignored the overwhelming evidence that the plastome exists as linear, branched and occasionally circular forms (Deng et al. 1989; Lilly et al. 2001; Oldenburg and Bendich 2004; Scharff and Koop 2006; Shaver et al. 2006). In fact, it has been shown that the different isomers of the SC regions occur by a recombination-dependent-replication (RDR) mechanism involving different unit genomes (Oldenburg and Bendich 2004; Maréchal and Brisson 2010). Thus, any discussion of the mechanism for generating multiple structural types of plastomes must consider RDR as the primary cause.
Ruhlman et al. (2017) proposed that RDR between different linear copies of plastomes generated alternative plastome arrangements associated with large IRs (>2 kb) in M. emarginata (see fig. 4 in Ruhlman et al. 2017). The unusually large number and size of repeats (up to ∼5 kb in E. dulcis and ∼3 kb in E. cellulosa) could account for the large number of plastome structural types by providing homologous sequence for the RDR pathway. Many of the large (>1 kb) and intermediate (201–1,000 bp) repeats in Eleocharis plastomes are located between syntenic blocks 3–5 and may be involved with repeat-mediated RDR within or between unit genomes. Structural heteroplasmy may be more prevalent than appreciated, especially in other lineages that have plastomes with highly rearranged gene orders and abundant larger repeats, such as Passiflora, Trifolium, Campanulaceae, and Geraniaceae.
The phylogenetic distribution of this phenomenon in Cyperaceae or other Poales is unknown due to limited taxon sampling in the family and order. The publicly available plastome of one of the basal taxa in Cyperaceae, H. nemorum, has not been published and no raw read data is available for analysis. Plastome sequences of two basal lineages, Bromeliaceae and Typhaceae, do not show structural heteroplasmy (Guisinger et al. 2010; Redwan et al. 2015; Poczai and Hyvönen 2017) and two other Poales lineages (Poaceae and Eriocaulaceae) do not exhibit multiple plastome types (Darshetkar et al. 2019; Orton et al. 2019). Additional sampling of plastome sequences in Cyperaceae and related families of cyperids is needed to explore the origin and extent of structural heteroplasmy in this clade.
Plastome Organization
In addition to remarkable structural heteroplasmy, Eleocharis plastomes have a number of unusual genomic characteristics compared with basal Poales, including size expansion, abundant repetitive DNA, higher degree of genome rearrangement, IR expansion, increased IGS, low GC content, low gene density, and numerous gene duplications and losses.
In most angiosperms, increases in plastome size are caused by IR expansion as reported in several unrelated lineages, including Pelargonium (IR ∼ 88 kb; Chumley et al. 2006; Weng et al. 2014, 2017), Annona (IR ∼ 64.5 kb; Blazier, Ruhlman, et al. 2016), Lamprocapnos (IR ∼ 51 kb; Park et al. 2018), Asarum (IR ∼ 45 kb; Sinn et al. 2018), and Passiflora (IR ∼ 35 kb; Shrestha et al. 2019). Plastome size expansion in Eleocharis is mainly due to increased number and size of repeats and to a lesser extent IR expansion (tables 2 and 4). Increased plastome size in the basal member of Cyperaceae, H. nemorum, is also caused by these two phenomena. This is in contrast to other members of the Poales (Typha, Ananas, and Eriocaulon), which have typical IRs and much less repetitive DNA and size variation (Guisinger et al. 2010; Redwan et al. 2015; Darshetkar et al. 2019). Eleocharis plastomes have similar SSC/IR boundaries to H. nemorum but they have an independent IR expansion into the LSC. Despite IR expansion into both the SSC and LSC and increased number of repeats in the IGS regions of the IR in Eleocharis plastomes, the size of their IR is smaller than H. nemorum because they lack ycf1 and ycf2. Repeat content and IR expansion have contributed to the increase in plastome size of Eleocharis and both of these factors affect GC content. The IR has higher GC content than SC regions (table 2) so IR expansion should result in higher overall GC content in plastomes. However, Eleocharis plastomes had the lowest GC values (∼32.6%) among photosynthetic angiosperms despite their genome size expansion (fig. 2). Other Cyperaceae have a similar pattern, which is likely due to having more repeats in IGS regions, which are known to be GC poor (table 4, Cai et al. 2006). The prevalence of repeats in the IGS regions also contributed to the lower gene density in Eleocharis and Hypolytrum compared with basal Poales plastomes (table 2). Thus, unlike most other photosynthetic angiosperms, Eleocharis plastomes have experienced increased plastome size driven by IR expansion and accumulation of repeats, and low GC content and gene density.
Whole-genome alignment revealed extensive gene order changes in Eleocharis plastomes (fig. 3 and table 3) relative to T. latifolia. The exceptional number of gene order changes is likely related to the abundance of repeats, which facilitate repeat-mediated homologous recombination. The IR was previously suggested to play a role in plastome structural stability based on the presence of extensive rearrangements in IR-lacking groups in legumes (Palmer and Thompson 1982) and Cryptomeria (Hirao et al. 2008). However, many studies failed to find a correlation between plastome stability and the presence of the IR in several angiosperm lineages, including Campanulaceae (Haberle et al. 2008; Knox 2014), Oleaceae (Lee et al. 2007), Ericaceae (Fajardo et al. 2013; Martínez-Alberola et al. 2013), Plantago (Zhu et al. 2016), Geraniaceae (Chumley et al. 2006; Guisinger et al. 2011; Weng et al. 2014; Blazier, Jansen, et al. 2016), and Passifloraceae (Rabah et al. 2019; Shrestha et al. 2019). In Eleocharis and other lineages, the presence of dispersed repeats was more likely the primary factor facilitating plastome rearrangements.
Plastomes of basal Poales, Typha (Guisinger et al. 2010) and Ananas (Redwan et al. 2015), as well as Eriocaulon (Darshetkar et al. 2019) have a conserved gene content, similar to the ancestral angiosperm plastome (Ruhlman and Jansen 2014). However, Cyperaceae plastomes have numerous gene duplications and losses (table 2). Many gene duplications are the result of IR expansion into the SSC and LSC. The other duplicated genes are associated with the dispersed repeats. Gene losses are more extensive in Eleocharis than in H. nemorum (table 2). The missing Eleocharis genes have been pseudogenized or lost in several eudicot lineages, including Campanulaceae (Cosner et al. 1997; Haberle et al. 2008; Knox 2014; Cheon et al. 2017), Trifolium (Cai et al. 2008; Magee et al. 2010; Sabir et al. 2014), Ericaceae (Fajardo et al. 2013; Martínez-Alberola et al. 2013), Carnegiea (Sanderson et al. 2015), Silene (Erixon and Oxelman 2008; Sloan et al. 2012), Geraniaceae (Chumley et al. 2006; Guisinger et al. 2011; Weng et al. 2014, 2017; Blazier, Jansen, et al. 2016; Park et al. 2017), and Passifloraceae (Rabah et al. 2019; Shrestha et al. 2019). Plastomes of these groups have abundant repetitive DNA and highly rearranged gene order, a syndrome that is shared with Eleocharis. In monocots, Poaceae plastomes are structurally similar to basal Poales except for three inversions and plastome size reduction caused by several gene losses (Quigley and Weil 1985; Hiratsuka et al. 1989; Doyle et al. 1992; Maier et al. 1995; Michelangeli et al. 2003; Morris and Duvall 2010; Harris et al. 2013). The loss of accD, ycf1, ycf2, clpP, and rpl23 is homoplastic in Poales as the same losses have occurred independently in the distantly related families Poaceae and Cyperaceae (supplementary fig. S1, Supplementary Material online). More plastome sequencing in Cyperaceae lineages is needed to elucidate the evolution of clpP, ycf1, ycf2, and rpl23 in the family. Moreover, transcriptome data would allow the determination of the fate of missing genes in Eleocharis and other related Cyperaceae.
Conclusion
Analyses utilizing Illumina and PacBio SMRT sequences, long-range PCR, and Sanger sequencing data demonstrated unprecedented structural heteroplasmy within a single E. dulcis individual. At least four different plastome structural types are present in both Eleocharis species, and it is likely that this phenomenon is much more widespread in the family. The long-held notion that plastomes are highly conserved in structure within individuals and species must be reconsidered, especially for lineages with a prevalence of large dispersed repeats that are likely to exhibit structural heteroplasmy in plastomes.
Supplementary Material
Acknowledgments
This work was supported by Texas Ecolab program to C.L. and the S.F. Blake Centennial Professorship to R.K.J. and T.A.R. The authors thank the Billie L. Turner Plant Resources Center (TEX-LL) at UT-Austin for serving as a repository for voucher specimens, United States Department of Agriculture Germplasm Resources Information Network for providing corms, and the Texas Advanced Computing Center (TACC) at UT-Austin for access to supercomputers.
Data deposition: The data have been deposited at GenBank under accession MN967018 and MN985041.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD.. 2011. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One 6(1):e16404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20. [Google Scholar]
- Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K-I.. 2004. Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res. 11(2):93–99. [DOI] [PubMed] [Google Scholar]
- Au KF, Underwood JG, Lee L, Wong WH.. 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7(10):e46679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Bendich AJ. 2004. Circular chloroplast chromosomes: the grand illusion. Plant Cell 16(7):1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, et al. 2007. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics 23(21):2957–2958. [DOI] [PubMed] [Google Scholar]
- Blazier JC, Guisinger MM, Jansen RK.. 2011. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol. 76(3–5):263–272. [DOI] [PubMed] [Google Scholar]
- Blazier JC, Jansen RK, et al. 2016. Variable presence of the inverted repeat and plastome stability in Erodium. Ann Bot. 117(7):1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier JC, Ruhlman TA, et al. 2016. Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci Rep. 6(1):24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brears T, Schardl CL, Lonsdale DM.. 1986. Chloroplast genome organisation in sugar beet and maize. Plant Mol Biol. 6(3):171–177. [DOI] [PubMed] [Google Scholar]
- Cai Z, et al. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, et al. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67(6):696–704. [DOI] [PubMed] [Google Scholar]
- Chang C-C, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 23(2):279–291. [DOI] [PubMed] [Google Scholar]
- Cheon K-S, Kim K-A, Yoo K-O.. 2017. The complete chloroplast genome sequences of three Adenophora species and comparative analysis with Campanuloid species (Campanulaceae). PLoS One 12(8):e0183652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley TW, et al. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23(11):2175–2190. [DOI] [PubMed] [Google Scholar]
- Cosner ME, Jansen RK, Palmer JD, Downie SR.. 1997. The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet. 31(5):419–429. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshetkar AM, Datar MN, Tamhankar S, Li P, Choudhary RK.. 2019. Understanding evolution in Poales: insights from Eriocaulaceae plastome. PLoS One 14(8):e0221423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzentas N. 2010. Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26(20):2620–2621. [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I.. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28(7):2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Wing RA, Gruissem W.. 1989. The chloroplast genome exists in multimeric forms. Proc Natl Acad Sci U S A. 86(11):4156–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Nascimento Vieira L, et al. 2016. The plastome sequence of the endemic Amazonian conifer, Retrophyllum piresii (Silba) C.N.Page, reveals different recombination events and plastome isoforms. Tree Genet Genomes 12:929. [Google Scholar]
- Doyle J, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ.. 1992. Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc Natl Acad Sci U S A. 89(16):7722–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas DVet al. . 2015. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions and accelerated rate of evolution in clpP Sci Rep. 5(1): 10.1038/srep16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erixon P, Oxelman B.. 2008. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS One 3(1):e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo D, et al. 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet Genomes 9(2):489–498. [Google Scholar]
- Fu C-N, et al. 2019. Prevalence of isomeric plastomes and effectiveness of plastome super-barcodes in yews (Taxus) worldwide. Sci Rep. 9(1):2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray BN, Ahner BA, Hanson MR.. 2009. Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res. 18(4):559–572. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK.. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 70(2):149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK.. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28(1):583–600. [DOI] [PubMed] [Google Scholar]
- Guo W, et al. 2014. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol Evol. 6(3):580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon C, Maliga P.. 2014. Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Res. 21(4):417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK.. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 66(4):350–361. [DOI] [PubMed] [Google Scholar]
- Harris ME, Meyer G, Vandergon T, Vandergon VO.. 2013. Loss of the acetyl-CoA carboxylase (accD) gene in Poales. Plant Mol Biol Rep. 31(1):21–31. [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K.. 2008. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 8(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka J, et al. 1989. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 217(2–3):185–194. [DOI] [PubMed] [Google Scholar]
- Hsu C-Y, Wu C-S, Chaw S-M.. 2016. Birth of four chimeric plastid gene clusters in Japanese umbrella pine. Genome Biol Evol. 8(6):1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104(49):19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, et al. 2015. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One 10(11):e0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-J, Lee H-L.. 2005. Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cells 19(1):104–113. [PubMed] [Google Scholar]
- Knox EB. 2014. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc Natl Acad Sci U S A. 111(30):11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R, Tewari KK.. 1979. Inverted repeats in chloroplast DNA from higher plants. Proc Natl Acad Sci U S A. 76(1):41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr DJG, Katz LA.. 2009. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. BioTechniques 47(4):857–866. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-L, Jansen RK, Chumley TW, Kim K-J.. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 24(5):1161–1180. [DOI] [PubMed] [Google Scholar]
- Lilly JW, Havey MJ, Jackson SA, Jiang J.. 2001. Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell 13(2):245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, et al. 2013. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene 528(2):120–131. [DOI] [PubMed] [Google Scholar]
- Magee AM, et al. 2010. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20(12):1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, Igloi GL, Kössel H.. 1995. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 251(5):614–628. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N.. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186(2):299–317. [DOI] [PubMed] [Google Scholar]
- Martínez-Alberola F, et al. 2013. Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PLoS One 8(11):e79685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelangeli FA, Davis JI, Stevenson DW.. 2003. Phylogenetic relationships among Poaceae and related families as inferred from morphology, inversions in the plastid genome, and sequence data from the mitochondrial and plastid genomes. Am J Bot. 90(1):93–106. [DOI] [PubMed] [Google Scholar]
- Morris LM, Duvall MR.. 2010. The chloroplast genome of Anomochloa marantoidea (Anomochlooideae; Poaceae) comprises a mixture of grass-like and unique features. Am J Bot. 97(4):620–627. [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ.. 2004. Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol. 335(4):953–970. [DOI] [PubMed] [Google Scholar]
- Orton LM, Burke SV, Duvall MR.. 2019. Plastome phylogenomics and characterization of rare genomic changes as taxonomic markers in plastome groups 1 and 2 Poeae (Pooideae; Poaceae). PeerJ 7:e6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. 1983. Chloroplast DNA exists in two orientations. Nature 301(5895):92–93. [Google Scholar]
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19(1):325–354. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF.. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29(2):537–550. [DOI] [PubMed] [Google Scholar]
- Park S, An B, Park S.. 2018. Reconfiguration of the plastid genome in Lamprocapnos spectabilis: IR boundary shifting, inversion, and intraspecific variation. Sci Rep. 8(1):13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, et al. 2017. Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of Geranium. Genome Biol Evol. 9(6):1766–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peredo EL, King UM, Les DH.. 2013. The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLoS One 8(7):e68591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poczai P, Hyvönen J.. 2017. The complete chloroplast genome sequence of the CAM epiphyte Spanish moss (Tillandsia usneoides, Bromeliaceae) and its comparative analysis. PLoS One 12(11):e0187199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X-J, Wu C-S, Chaw S-M, Yi T-S.. 2017. Insights into the existence of isomeric plastomes in Cupressoideae (Cupressaceae). Genome Biol Evol. 9(4):1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley F, Weil JH.. 1985. Organization and sequence of five tRNA genes and of an unidentified reading frame in the wheat chloroplast genome: evidence for gene rearrangements during the evolution of chloroplast genomes. Curr Genet. 9(6):495–503. [DOI] [PubMed] [Google Scholar]
- Rabah SO, et al. 2019. Passiflora plastome sequencing reveals widespread genomic rearrangements. J Syst Evol. 57(1):1–14. [Google Scholar]
- Redwan RM, Saidin A, Kumar SV.. 2015. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 15(1):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R.. 2006. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34(16):4537–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman TA, Jansen RK.. 2014. The plastid genomes of flowering plants In: Maliga P, editor. Chloroplast biotechnology: methods and protocols. Vol. 1132 London: Springer Science and Business Media, LLC. [Google Scholar]
- Ruhlman TA, Jansen RK.. 2018. Chapter eight—aberration or analogy? The atypical plastomes of Geraniaceae In: Chaw S-M, Jansen RK, editors. Advances in botanical research. Vol. 85. Plastid genome evolution. London: Academic Press; p. 223–262. [Google Scholar]
- Ruhlman TA, Zhang J, Blazier JC, Sabir JSM, Jansen RK.. 2017. Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am J Bot. 104(4):559–572. [DOI] [PubMed] [Google Scholar]
- Ruhlman TA, et al. 2015. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir J, et al. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12(6):743–754. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, et al. 2015. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am J Bot. 102(7):1115–1127. [DOI] [PubMed] [Google Scholar]
- Scharff LB, Koop H-U.. 2006. Linear molecules of tobacco ptDNA end at known replication origins and additional loci. Plant Mol Biol. 62(4–5):611–621. [DOI] [PubMed] [Google Scholar]
- Schwarz EN, et al. 2015. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J Syst Evol. 53(5):458–468. [Google Scholar]
- Shaver JM, Oldenburg DJ, Bendich AJ.. 2006. Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224(1):72–82. [DOI] [PubMed] [Google Scholar]
- Shrestha B, et al. 2019. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol Phylogenet Evol. 138:53–64. [DOI] [PubMed] [Google Scholar]
- Sinn BT, Sedmak DD, Kelly LM, Freudenstein JV.. 2018. Total duplication of the small single copy region in the angiosperm plastome: rearrangement and inverted repeat instability in Asarum. Am J Bot. 105(1):71–14. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR.. 2012. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol Evol. 4(3):294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solórzano S, et al. 2019. De novo assembly discovered novel structures in genome of plastids and revealed divergent inverted repeats in Mammillaria (Cactaceae, Caryophyllales). Plants 8(10):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DB, Palmer JD, Thompson WF.. 1986. Structural evolution and flip-flop recombination of chloroplast DNA in the fern genus Osmunda. Curr Genet. 10(11):835–841. [Google Scholar]
- Stevens PF. 2001. onwards. Angiosperm phylogeny website. Version 14, July 2017 (and more or less continuously updated since). Available from: http://www.mobot.org/MOBOT/research/APweb.
- Sun Y-X, et al. 2013. Complete plastid genome sequencing of Trochodendraceae reveals a significant expansion of the inverted repeat and suggests a Paleogene divergence between the two extant species. PLoS One 8(4):e60429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, et al. 2017. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Kuehl JV, Boore JL, Jansen RK.. 2007. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot. 94(3):302–312. [DOI] [PubMed] [Google Scholar]
- Tsumura Y, Suyama Y, Yoshimura K.. 2000. Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga. Mol Biol Evol. 17(9):1302–1312. [DOI] [PubMed] [Google Scholar]
- Untergasser A, et al. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M-L, Blazier JC, Govindu M, Jansen RK.. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31(3):645–659. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Ruhlman TA, Jansen RK.. 2017. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214(2):842–851. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, depamphilis CW, Müller KF, Quandt D.. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-S, Lin C-P, Hsu C-Y, Wang R-J, Chaw S-M.. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F-H, et al. 2010. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 10(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki WP, Burke SV, Swingley WD, Duvall MR.. 2016. The first complete plastid genome from Joinvilleaceae (J. ascendens; Poales) shows unique and unpredicted rearrangements. PLoS One 11(9):e0163218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. 2016. Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biol Evol. 8(3):622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-J, Ma P-F, Li D-Z.. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One 6(5):e20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP.. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209(4):1747–1756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.