Abstract
Dairy cows can experience a negative energy balance (NEB) in early lactation when feed intake is too low to meet the energy requirements for body maintenance and milk production. Metabolic changes occur in mammary gland cells of animals experiencing a negative energy balance. We studied these metabolic changes in milk samples from dairy cows in relation to energy balance status using liquid chromatography–mass spectrometry (QQQ-LC–MS) and nuclear magnetic resonance (1H NMR). NMR and LC–MS techniques are complementary techniques that enabled a comprehensive overview of milk metabolites in our study. Energy balance and milk samples were obtained from 87 dairy cows. A total of 55 milk metabolites were reliably detected, of which 15 metabolites were positively correlated to energy balance and 20 were negatively correlated to energy balance. Cows in NEB produced more milk with increased milk fat yield and higher concentrations of citrate, cis-aconitate, creatinine, glycine, phosphocreatine, galactose-1-phosphate, glucose-1-phosphate, UDP-N-acetyl-galactosamine, UDP-N-acetyl-glucosamine, and phosphocholine but lower concentrations of choline, ethanolamine, fucose, N-acetyl-neuraminic acid, N-acetyl-glucosamine, and N-acetyl-galactosamine. During NEB, we observed an increased leakage of cellular content, increased synthesis of nucleic acids and cell membrane phospholipids, an increase in one-carbon metabolic processes, and an increase in lipid-triglyceride anabolism. Overall, both apoptosis combined with cellular renewal is paramount in the mammary gland in cows in NEB.
Keywords: milk, NMR, LC−MS, metabolomics, integrated analysis, metabolic status, energy balance
Introduction
In early lactation of dairy cows, elevated energy requirements for milk production combined with a relatively low dry matter intake (DMI) can result in an energy deficit or negative energy balance (NEB).1,2 A severe NEB is related to an increased risk of metabolic disorders and diseases, such as fatty liver and ketosis.3,4 In previous studies, it was shown that body reserves were mobilized to meet the nutritional demand of the mammary gland for milk production with concurrent changes in metabolic hormones and plasma metabolites.5,6 The basic metabolic patterns related to the synthesis of milk fat, protein, and lactose are known.7,8 However, modifications of metabolic pathways, especially in relation to energy balance, have not been described yet.
Metabolomics studies aim to detect and quantify small molecules from biofluids through several metabolomics techniques, such as mass spectrometry (MS) and nuclear magnetic resonance (NMR). Integrating the results of different metabolomics techniques can strengthen the interpretation of the metabolic profiles.9−11 Liquid chromatography–mass spectrometry (LC–MS) is a powerful technique with high sensitivity and selectivity.12 High-resolution NMR is a very stable technique with better reproducibility than LC–MS but NMR suffers, relative to LC–MS, from limited sensitivity.13 In recent years, integrated analyses that combine results from LC–MS and NMR have been applied to detect and quantify a wide range of metabolites in biofluids, such as urine, plasma, and milk.14−17 The integration of data from different techniques supports cross-assigning signals from the techniques on the same samples.15 In dairy cows, the integration of MS and NMR data identified biomarkers of heat stress in plasma,16 and the correlation of plasma and milk metabolites was investigated.17 In the past decade, new developments with hydrophilic columns make an analysis of polar metabolites possible using LC–MS. We used a recently introduced pentafluorophenylpropyl (PFPP) column to separate polar metabolites with subsequent identification and quantification using a triple-quadrupole-MS. This combined with high-resolution NMR measurements of the same samples has the potency to detect and quantify more metabolites in milk than hitherto was feasible.
With the combined NMR and LC–MS data sets, we were able to better understand biological pathways affected by NEB and associated alterations in the metabolic status of dairy cows. In our study, 87 dairy cows were studied. In lactation week 2, milk yield, milk composition (fat, protein, and lactose), DMI, and energy balance of individual cows were recorded, as well as milk were collected. From our data, we could obtain a detailed metabolic pattern occurring in cows with severe negative energy balance.
Experimental Section
Animals and Experimental Design
The experimental protocol for the study was approved by the Institutional Animal Care and Use Committee of Wageningen University and conducted at the Dairy Campus research farm (WUR Livestock Research, Lelystad, the Netherlands). The experimental design was described previously.18 Briefly, in total, 91 high-yielding Holstein–Friesian dairy cows averaging 665.6 ± 68.2 kg of body weight (in lactation week 2 after calving) participated in this study. Dairy cows were blocked for parity, expected calving period, and expected milk yield. Within blocks, the cows were randomly assigned to one of two dry period lengths (DPL, 0 day, 2/3 of the cows; or 30 days: 1/3 of the cows) before calving. Prepartum, cows with a 0 day DPL received a lactation ration based on grass silage and corn silage (6.4 MJ net energy for lactation (NE)/kg dry matter (DM)). Cows with a 30 day DPL received a dry cow ration based on grass silage, corn silage, and wheat straw (5.4 MJ NE/kg DM). Postpartum, all cows received the same basal lactation ration as provided to lactating cows prepartum plus additional concentrates. Postpartum, concentrate supply increased stepwise with 0.3 kg/day till 8.5 kg/day on 28 DIM. Bodyweight, milk yield, and feed intake were recorded daily. During lactation, the cows were milked twice daily at ∼0600 and ∼1800 h. In our data set, 75 out of 87 cows experienced a negative energy balance. About one-third of the cows (27 out of 87) were in the 30 day DPL group. The body weight is same (P = 0.93) between dairy cows in the 0 day DPL group (662.5 ± 73.0) and the 30 day DPL group (663.5 ± 65.6) (Table S2). Earlier, we reported the relation of energy balance and metabolites detected in the milk samples of 31 dairy cows in lactation weeks 2 and 7 through LC–MS measurement;19 those 31 cows were not included in the current study. Our previous study has shown that DPL did not affect the milk metabolites profile via a multivariate analysis (principal component analysis), nor did it affect the key metabolites in milk via univariate analysis. Also, in this study, we tested if cows with the 0 day DPL could be separated from cows with the 30 day DPL with an orthogonal partial least squares discriminant analysis for the milk metabolites. The two groups (0 DPL and 30 days DPL) could not be well separated; after cross-validation, the maximum Q2 (the predictability of the model) was less than 0.2, indicating a bad separation of DPL groups using milk metabolites. Here we consider cows with two different DPL’s as one group for further correlation analysis.
Milk Samples
Milk samples for fat, protein, and lactose percentage (ISO 9622, Qlip, Zutphen, the Netherlands) were collected four times per week (Tuesday afternoon, Wednesday morning, Wednesday afternoon, and Thursday morning). Milk samples were analyzed as a pooled sample per cow per week and used to calculate the average fat, protein, and lactose yields in this week. Milk samples for metabolomics analysis were collected on Wednesday morning in the lactation week 2 and then stored at −20 °C until analysis. Milk production traits were averaged per week. Four milk samples were omitted from the analysis because two cows suffered from clinical mastitis, one dairy cow suffered from metritis, and one cow had locomotion problems in the sampling week. Fat- and protein-corrected milk was calculated as20
Energy Intake and Energy Balance
Roughage and concentrate were supplied separately, daily intakes were recorded per individual cow using roughage intake control troughs (Insentec, Marknesse, the Netherlands). Energy balance was calculated per week according to the Dutch net energy evaluation (VEM) system, as the difference between net energy intake and the estimated net energy requirements for maintenance, and milk yield (1000 VEM = 6.9 MJ of NE).21,22
NMR Measurement and Data Preprocessing
Sample preparation and NMR measurements were performed as described earlier.23,24 Briefly, milk samples were first thawed to room temperature. The fat layer of milk was removed by the addition of deuterated chloroform and centrifugation (12 000 rpm, 15 min, centrifuge 5424, Eppendorf). Subsequently, 175 μL of milk serum was mixed with 175 μL of phosphate buffer (pH = 7.0), and these samples were filtered to remove protein using an Amicon Ultra 0.5 mL 10 kDa cutoff spin filter (Millipore Corp., Billerica, MA) with centrifugation at 12 000 rpm for 15 min. The samples were measured with a 3 mm NMR tube (Bruker matching system) using a Bruker NMR spectrometer Avance III with a 600 MHz/54 mm UltraShielded Plus magnet equipped with a CryoPlatform cryogenic cooling system, a BCU-05 cooling unit and an ATM automatic tuning and matching unit. Measurements were done at 300 K. One-dimensional (1D) nuclear Overhauser enhancement spectroscopy (NOESY) spectra were obtained. Baseline corrections, alignment to the resonance of alanine (1.484 ppm), and intensity calibration to internal maleic acid were done for all spectra. Assignment of metabolites resonances was performed using the published literature, the Human Metabolome Database version 2.0 online library (http://hmdb.ca/) as well as internal standards.
LC–MS Measurements
For quantification of metabolites, a targeted, standardized, and quality controlled metabolic phenotyping was performed based on the LC-QQQ-MS analysis. The sample as prepared for NMR was also used for the analysis with the triple-quadrupole mass spectrometer (Shimadzu LC-QQQ-MS; LC–MS-8040) using the PFPP method as described earlier.25,26 The sample injection volume used was 1 μL, and a single analysis took 25 min. From the LC-QQQ-MS spectra, metabolites were regarded as reliably identified, when more than 60% observations in all samples showed a reliable intensity and peak shape for a metabolite. We selected 60% as a cutoff value because of the large differences in the amounts of metabolites in the different samples. The large variation of energy balance in the cows in our study results in a large variation of concentration of metabolites in the milk samples.
Integrated Analysis and Software
The NMR data sets were aligned, the water region was removed, and the NMR spectra were integrated into 0.01 ppm bins. The intensity of the bins was subsequently correlated to energy balance parameters. Bins, which correlated well to energy balance, were selected and the corresponding NMR resonances (peaks) were specifically integrated by carefully selecting peaks to minimize the overlap in the NMR spectra. The correlation matrix of the selected NMR peaks and LC–MS data sets were subsequently analyzed. In case a metabolite could be identified from both LC–MS and NMR, the intensity of this metabolite was quantified based on its NMR spectrum. The integrated analysis was done as described earlier.15,27 Before correlation analysis, the zero values were replaced with the minimum values for a metabolite. The Pearson correlation coefficient (r) and the corresponding P value were obtained by function “cor.test ()” in R (version 3.4.3).
Results and Discussion
Measurement by LC–MS and NMR and Integrated Analysis
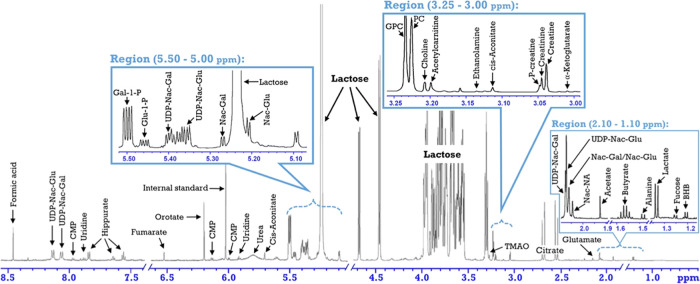
In the LC–MS spectra of milk, 97 metabolites were initially targeted. A LC–MS-based metabolite was regarded as “detected”, when the metabolite in the milk samples of more than 50 of the 87 cows was clearly observed. This resulted in 27 milk metabolites detected by LC–MS (Table S1). In the NMR spectra of milk, lactose dominated the region around 3.52–3.95 ppm, which masked the signal of other metabolites in this region (Figure 1). For instance, glycine is impossible to be detected by NMR in milk samples.28 Nevertheless, many resonances could be clearly observed resulting in 35 NMR-based milk metabolites (Table S1).
Figure 1.
Proton nuclear magnetic resonance spectrum of a milk sample. Abbreviations: BHB, β-hydroxybutyrate; CMP, cytidine monophosphate; Gal-1-P, galactose-1-phosphate; Glu-1-P, glucose-1-phosphate; GPC, glycerophosphocholine; Nac-Gal, N-acetyl-glucosamine; Nac-Glu, N-acetyl-glucosamine; Nac-Na, N-acetyl-neuraminic acid; PC, phosphocholine; P-creatine, phosphocreatine; phosphocholine; TMAO, trimethylamine N-oxide; UDP-Nac-Gal, uridine diphosphate-N-acetyl-galactosamine; and UDP-Nac-Glu, uridine diphosphate-N-acetyl-glucosamine.
A number of milk metabolites have been reported before using LC–MS or NMR.29−31 We integrated the results from the LC–MS and NMR data sets. Milk metabolites detected both by NMR and LC–MS had a high correlation between the different detection methods LC–MS and NMR, i.e., acetyl-carnitine (r = 0.90), choline (r = 0.92), CMP (r = 0.90), glutamate (r = 0.94), α-ketoglutarate (r = 0.90), uridine (r = 0.93), and valine (r = 0.90). The consistency between the two measurement methods indicated that data obtained by LC–MS were reliable.
Through the integration of LC–MS and NMR, 55 metabolites were detected from milk samples of 87 dairy cows in lactation week 2. Of these 55 milk metabolites, 15 were positively correlated to energy balance (Figure 2) and 20 were negatively correlated to energy balance (Figure 3). The cows in this study have large differences in energy balance (from −500 to 500 kJ/kg 0.75 day). These large differences in energy balance resulted in large differences in the number of metabolites of the milk samples measured as will be shown and discussed below. The most striking correlations to the energy balance of the metabolites measured in the milk samples were cis-aconitate (−0.74), citrate (−0.71), glycine (−0.66), galactose-1-phosphate (−0.64), uridine diphosphate-N-acetyl-galactosamine (−063), choline (0.63), and N-acetyl-neuraminic acid (0.61). A negative correlation between a metabolite and energy balance implies higher amounts of these metabolites when a cow is in negative energy balance and a positive correlation between a metabolite and energy balance indicates lower amounts of these metabolites in cows with negative energy balance. Many metabolites have strong correlations to other metabolites; for instance, choline is strongly correlated to ethanolamine (0.81) and to N-acetyl-neuraminic acid (0.82), or as another example, citrate is strongly correlated to cis-aconitate (0.96) and modestly correlated to glycine (0.61). These correlations between metabolites indicate that in energy balance certain pathways are influenced in mammary gland cells. Based on the changes observed for the metabolites in this study, we think that the major process in the energy balance is cell apoptosis and cell renewal.
Figure 2.
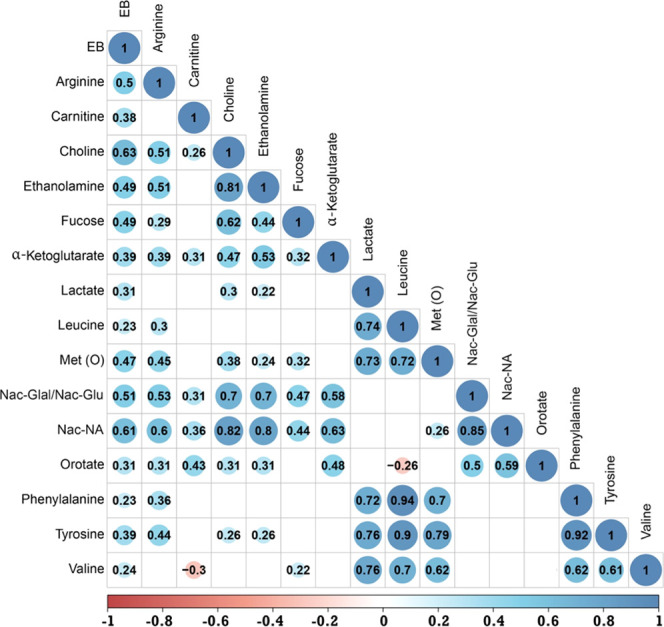
Pearson correlation matrix of metabolites and energy balance (EB) of dairy cows in lactation week 2. A total of 15 milk metabolites are positively correlated to energy balance (P < 0.05). A value of ≤0.35 is considered a low or weak correlation, 0.36–0.67 a modest correlation, and ≥0.68 a strong correlation. Abbreviations: Met (O), methionine sulfoxide; Nac-Gal, N-acetyl-galactosamine; Nac-Glu, N-acetyl-glucosamine; and Nac-NA, N-acetyl-neuraminic acid.
Figure 3.
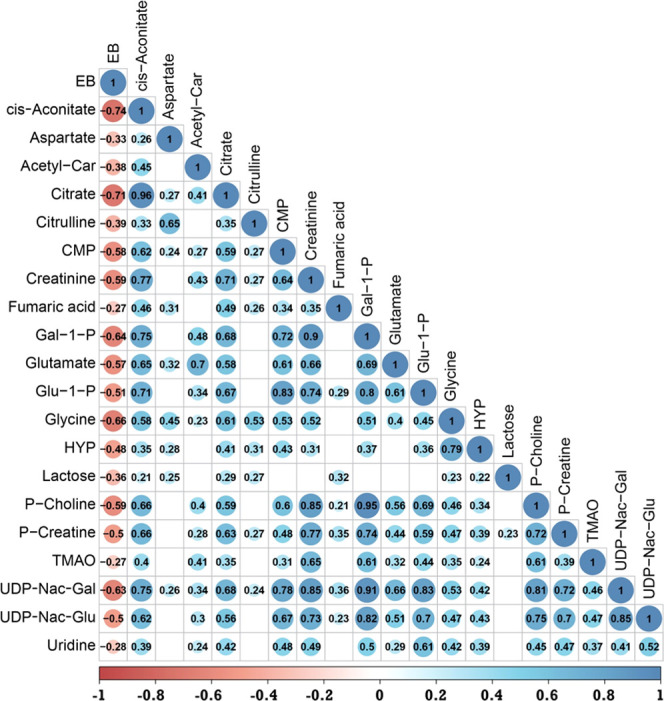
Pearson correlation matrix of metabolites and energy balance (EB) of dairy cows in lactation week 2. A total of 26 milk metabolites are negatively correlated to energy balance (P < 0.05). A value of ≤0.35 is considered a low or weak correlation, 0.36–0.67 a modest correlation, and ≥0.68 a strong correlation. Abbreviations: acetyl-car, acetyl-carnitine; CMP, cytidine monophosphate; Gal-1-P, galactose-1-phosphate; Glu-1-P, glucose-1-phosphate; HYP, hydroxyproline; Nac-Gal, N-acetyl-galactosamine; Nac-Glu, N-acetyl-glucosamine; TMAO, trimethylamine N-oxide; P-choline, phosphocholine; P-creatine, phosphocreatine; UDP-Nac-Gal, uridine diphosphate-N-acetyl-galactosamine; and UDP-Nac-Glu, uridine diphosphate-N-acetyl-glucosamine.
Cell Apoptosis and Cell Renewal
The final step in lactose synthesis is a process confined to the Golgi apparatus.32 Lactose concentrations in milk are always very constant because the secretion from the Golgi apparatus is driven by osmotic force related to the lactose concentration in the Golgi. Glucose-1-phosphate (Glu-1-P) and galactose-1-phosphate (Gal-1-P), intermediates in lactose synthesis, were negatively correlated with energy balance, r = −0.51 and −0.64, respectively (Figure 3). The presence of high concentrations of these intermediates in the lactose biosynthesis process indicates that in the mammary gland some cells leak the cellular content into the milk pool due to apoptosis.24 Apoptosis in the mammary gland could be related to or even caused by low plasma IGF-1 concentrations in dairy cows in NEB.33 In mammals, IGF-1 is a cell survival factor and an anti-apoptotic factor.34,35 In dairy cows, the apoptotic index in the mammary gland has been reported to be up to fourfold greater in early lactation than in later lactation.36 After calving, the dramatically increased milk production has been proposed to lead to the removal of apoptotic epithelial cells.37,38 Besides metabolites related to lactose synthesis, a series of intermediates, used for nucleic acids synthesis and present in high concentrations, were observed to have negative correlations with energy balance, i.e., uridine (r = −0.28), CMP (r = −0.58), and glycine (r = −0.66) as shown in Figure 3. Cellular renewal requires extensive DNA and RNA synthesis,39 and a negative correlation between intermediates in nucleic acids synthesis and energy balance could indicate accelerated cell proliferation in dairy cows in NEB. The process of cell proliferation increases the number of mammary epithelial cells critical for milk production. In the mammary gland of dairy cows, the total DNA content increases by 65% around 10 days pre- and postpartum.41 After parturition, mammary epithelial cells continuously increase in number (7), and an increase in the size of the mammary gland could also be promoted by elevated concentrations of growth hormone.40 Cell proliferation is an energy-demanding process42 not only for nucleic acids synthesis but also for cell membrane synthesis.
In a eukaryotic membrane, phosphatidylcholine (PtC) and phosphatidylethanolamine (PtE) account for more than 50% of the total phospholipids.43 The pathway of PtC synthesis from choline and PtE synthesis from ethanolamine using cytidine coenzymes is referred to as the Kennedy pathway.44 In our study, energy balance was positively correlated with choline (r = 0.63) and ethanolamine (r = 0.49). In contrast, phosphocholine was observed to be negatively correlated with energy balance (r = −0.59). The rate-limiting step in the PtC synthesis is the formation of CDP-choline from phosphocholine by cytidine triphosphate (CTP)-phosphocholine cytidylyltransferase (PCT).45 The low amounts of choline and the high amounts of phosphocholine in dairy cows in NEB indicate that PtC biosynthesis is increased. Remarkably, we observed that phosphocholine and choline concentrations were strongly correlated to several metabolites involved in the glycosylation of proteins, N-acetyl-galactosamine (Nac-Gal), N-acetyl-glucosamine (Nac-Glu), N-acetyl-neuraminic acid (Nac-Neu), UDP-Nac-Gal, and UDP-Nac-Glu (Figure 4). In the phospholipid membrane synthesis, proteins present in the membrane need to be glycosylated to obtain cellular stability for signal transduction processes and viral or microbial defense.
Figure 4.
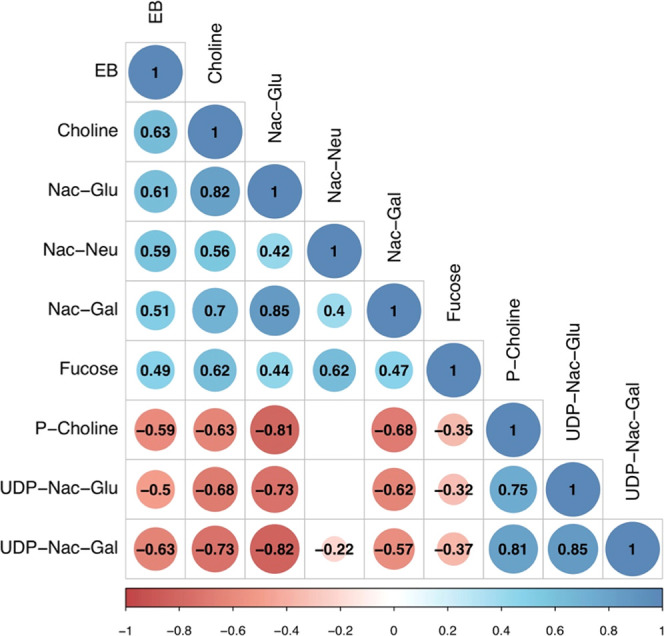
Pearson correlation matrix of metabolites related to glycosylation and lipid synthesis (P < 0.05). A value of ≤0.35 is considered a low or weak correlation, 0.36–0.67 a modest correlation, and ≥0.68 a strong correlation. Abbreviations: Nac-Glu, N-acetyl-glucosamine; Nac-Neu, N-acetyl-neuraminic acid; Nac-Gal, N-acetyl-galactosamine; P-choline, phosphocholine; UDP-Nac-Glu, uridine diphosphate-N-acetyl-glucosamine; and UDP-Nac-Gal, uridine diphosphate-N-acetyl-galactosamine.
In the current study, high amounts of UDP-Nac-Gal and UDP-Nac-Glu and low amounts of N-acetyl-neuraminic acid, Nac-Gal, and Nac-Glu were detected in the milk of cows in NEB. Milk proteins are heavily glycosylated, with N-acetyl-neuraminic acid, Nac-Gal, and Nac-Glu residues as major substituents.46 UDP-N-acetyl-galactosamine and UDP-Nac-Glu are activated substrates used for protein glycosylation. Apparently, the increased amounts of UDP-Nac-Gal and UDP-Nac-Glu in dairy cows in NEB indicates that there is a high demand for protein glycosylation. The observation that these two UDP derivatives used for glycosylation are strongly correlated with choline and phosphocholine suggests that not only the synthesis of membrane phospholipids but also the glycosylation of membrane proteins is of equally high importance for cows in NEB. Glycosylation of milk serum proteins could be related to glycosylation of membrane proteins, as the protein concentration in milk was observed to be strongly correlated with N-acetyl-galactosamine (Nac-Gal), N-acetyl-glucosamine (Nac-Glu), N-acetyl-neuraminic acid, and choline. Possibly, the glycosylation of membrane proteins is the driving force for the glycosylation of cytosolic milk proteins. It has been observed that glycosylation of milk proteins varies depending on the lactation week postpartum,47,48 but differences in glycosylation patterns of cytosolic and membrane proteins of individual cows related to energy status have not been studied in detail.
The synthesis of PtC from PC is the rate-limiting step in PtC synthesis, possibly because cytidine triphosphate (CTP) is a rate-limiting metabolite in this process (Figure 5). CTP is used to not only form PtC and PtE in the Kennedy pathway but synthesize nucleic acids. In addition, CTP is synthesized from uridine diphosphate (UTP), which is an intermediate in the synthesis of lactose. Dairy cows in NEB had a greater overall lactose yield (r = −0.59), which indicates that large amounts of UTP were probably used to synthesize lactose perhaps rather than form CTP. Finally, CTP and UTP can be used for DNA and RNA synthesis during cell proliferation,49 creating a high demand for both UTP and CTP for cows in NEB.
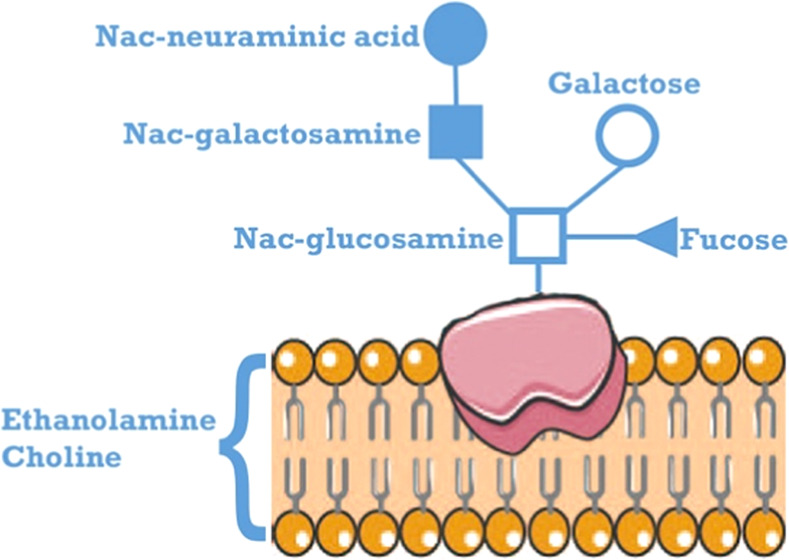
Figure 5.
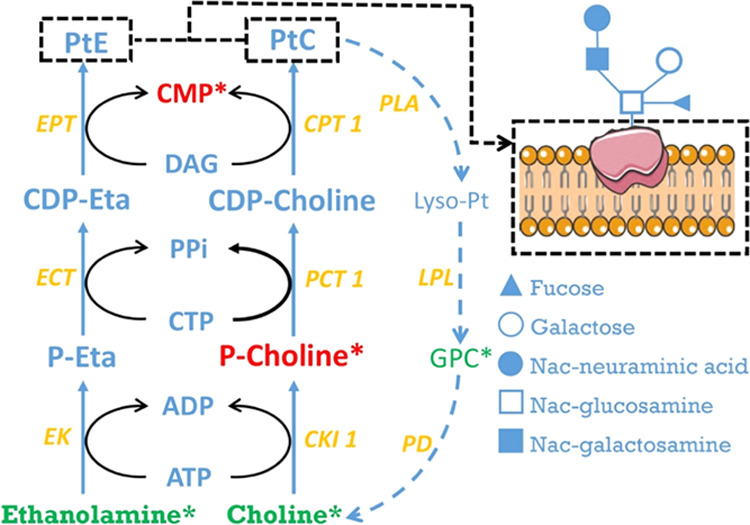
Composition of head groups of a phospholipid bilayer of a cellular membrane. Choline and ethanolamine are two main substrates for the synthesis of phosphatidylcholine (PtC) and phosphatidylethanolamine (PtE), respectively. PtC can be degraded via GPC to choline. Proteins embedded in the cellular membrane are shown to be glycosylated. Abbreviations: ADP, adenosine diphosphate; ATP, adenosine triphosphate; CDP, cytidine diphosphate; CKI 1, choline kinase; CTP, cytidine triphosphate; ECT phosphoethanolamine cytidylyltransferase; EK, ethanolamine kinase; EPT, ethanolaminephosphotransferase; LPL, lysophospholipase; P-Eta, phosphoethanolamine; P-choline, phosphocholine; PD, glycerophosphocholine phosphodiesterase; PLA, phospholipase A2; and PPi, pyrophosphate.
Membrane Biosynthesis
For the synthesis of membrane phospholipids, diglycerides are needed (Figure 4). A diglyceride molecule diacylglycerol (DAG) contains two molecules of fatty acids and one molecule of glycerol. The biosynthesis of free fatty acids is an energy-consuming process, and fatty acids are largely used for milk triglycerides. In cows in NEB, a competition between DAG, which is used for the synthesis of milk triglycerides, and DAG, which is used for the synthesis of phospholipids, can occur, as the amount of triglycerides in the milk of dairy cows in NEB is increased. Glycerophosphocholine (GPC) is a product from the breakdown of PtC (Figure 5). A low GPC/PC ratio was observed for cancer cells in humans.50,51 A low GPC/PC ratio was used to indicate a risk of ketosis in dairy cows.29 In our study, the GPC/PC ratio is positively correlated to the energy balance (r = 0.41), indicating low levels of GPC/PC in NEB. However, this ratio is more the consequence of the high correlation of PC to the energy balance (r = −0.59, indicating high amounts of PC in NEB) than of GPC to energy balance where a very weak relationship was observed (r = 0.18, P = 0.09). The low amounts of GPC, as well as low amounts of N-acetyl-neuramic acid, Nac-Gal, and Nac-Glu, are in our view related to the reprocessing of cellular components through the lysosome, enabling the building blocks to be reprocessed. Therefore, we propose that the correlation of GPC/PC to energy balance is related to the process of cell membrane synthesis during cell proliferation in the mammary gland, leading to high concentrations of PC.
Citrate detected from NMR spectra was negatively correlated with energy balance (r = −0.71). Citrate is an important metabolite involved in cellular energy metabolism (Figure 5). In mitochondria, citrate is an intermediate in the tricarboxylic acid cycle where citrate can be isomerized into cis-aconitate. cis-Aconitate is also very well correlated with energy balance (r = −0.74). However, fumarate, which is easily measured by NMR, was observed not to be correlated to NEB. These observations indicate that citrate is mainly used extramitochondrial to form acetyl-CoA to be used for fatty acids synthesis.52 In the current study, energy balance was negatively correlated with milk fat yield (r = −0.78) and milk fat yield was positively correlated to citrate levels (r = 0.53), indicating that citrate is used for milk fat synthesis in cows in NEB. We propose to use citrate levels as an indicator of energy status.53,54 The concentration of milk citrate has a wide variation throughout lactation.55 Dairy cows have a greater concentration of milk citrate in early lactation than in mid lactation,54 which could be explained by the improved energy balance in mid lactation.
In earlier studies, it was observed that cows in negative energy balance had lower plasma glucose, lower insulin, and lower insulin-growth-factor-1 (IGF-1) but higher BHB and free fatty acid concentrations. Cows in negative energy balance have more metabolic disorders as ketosis and fatty liver. Cows in positive energy balance have higher levels of insulin, glucose, and IGF-1.33 An increased level of IGF-1 could have anabolic effects on glucose metabolism, so the higher levels of IGF-1 in cows with positive energy balance result in a more anabolic condition. Glucose is in dairy cows is not only used as an energy source but also a precursor to synthesize lactose in the mammary gland cells. Low insulin levels will have an effect on glucose uptake by insulin-responsive tissue. Insulin is important in the growth hormone to IGF-1 relationship in cows. Under normal physiological conditions, growth hormone induces IGF-1 synthesis and IGF-1 negatively regulates growth hormone (GH) production in a feedback loop. In cows in negative energy balance, this regulation of IGF-1 and growth hormone is uncoupled in the liver and results in reduced IGF-1 concentrations, although growth hormone concentrations are elevated. How this uncoupling of growth hormone to IGF-1 is regulated is to be studied in more detail in the future.
Conclusions
In this study, 55 metabolites were detected and reliably quantified from milk serum of dairy cows in lactation week 2 using NMR and LC–MS through integrated analysis. A large number of metabolites (20) were negatively and 15 metabolites were positively related to the energy balance of cows. Based on these data, we concluded that apoptosis and cellular proliferation occurs in cows with NEB with increases in the synthesis of nucleic acids, cell membrane phospholipids, protein glycosylation, one-carbon metabolism, and lipid metabolism.
Acknowledgments
The authors thank the Chinese Scholarship Council (CSC, China) for financial support of the PhD study of the first author. The authors thank the staff of the Dairy Campus (Lelystad, the Netherlands) and Gerrit Remmelink for their assistance during the experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00706.
Milk metabolites detected either by liquid chromatography–mass spectrometry (LC–MS), nuclear magnetic resonance (NMR), or both (Table S1); and the number, milk yield, body weight, and energy balance (EB) of dairy cows with two dry period length (DPL, 0 day, or 30 day) (Table S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- de Vries M. J.; Veerkamp R. F. Energy balance of dairy cattle in relation to milk production variables and fertility. J. Dairy Sci. 2000, 83, 62–69. 10.3168/jds.S0022-0302(00)74856-9. [DOI] [PubMed] [Google Scholar]
- Bell A. W. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J. Anim. Sci. 1995, 73, 2804–2819. 10.2527/1995.7392804x. [DOI] [PubMed] [Google Scholar]
- Ingvartsen K. L.; Dewhurst R. J.; Friggens N. C. On the relationship between lactational performance and health: is it yield or metabolic imbalance that cause production diseases in dairy cattle? A position paper. Livest. Prod. Sci. 2003, 83, 277–308. 10.1016/S0301-6226(03)00110-6. [DOI] [Google Scholar]
- Friggens N. C.; Andersen J. B.; Larsen T.; Aaes O.; Dewhurst R. J. Priming the dairy cow for lactation: a review of dry cow feeding strategies. Anim. Res. 2004, 53, 453–473. 10.1051/animres:2004037. [DOI] [Google Scholar]
- Bauman D. In Regulation of Nutrient Partitioning During Lactation: Homeostasis and Homeorhesis Revisited, Ruminant Physiology: Digestion, Metabolism, Growth and Reproduction; 2000; pp 311–328.
- Clark C. E. F.; Fulkerson W. J.; Nandra K. S.; Barchia I.; Macmillan K. L. The use of indicators to assess the degree of mobilisation of body reserves in dairy cows in early lactation on a pasture-based diet. Livest. Prod. Sci. 2005, 94, 199–211. 10.1016/j.livprodsci.2004.11.038. [DOI] [Google Scholar]
- Rezaei R.; Wu Z.; Hou Y.; Bazer F. W.; Wu G. Amino acids and mammary gland development: nutritional implications for milk production and neonatal growth. J. Anim. Sci. Biotechnol. 2016, 7, 20 10.1186/s40104-016-0078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D. R.; Kensinger R. S.; Harrell R. J.; Bauman D. E. Nutrient uptake and endocrine regulation of milk synthesis by mammary tissue of lactating sows. J. Anim. Sci. 1995, 73, 36–56. 10.2527/1995.73suppl_236x. [DOI] [Google Scholar]
- Nicholson J. K.; Lindon J. C. Systems biology: metabonomics. Nature 2008, 455, 1054–1056. 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- Fernie A. R.; Trethewey R. N.; Krotzky A. J.; Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- Saccenti E.; Tenori L.; Verbruggen P.; Timmerman M. E.; Bouwman J.; van der Greef J.; Luchinat C.; Smilde A. K. Of monkeys and men: A metabolomic analysis of static and dynamic urinary metabolic phenotypes in two species. PLoS One 2014, 9, e106077 10.1371/journal.pone.0106077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R.; Hageman J. A.; van Eeuwijk F.; Kooke R.; Flood P. J.; Wijnker E.; Keurentjes J. J. B.; Lommen A.; van Eekelen H. D. L. M.; Hall R. D.; Mumm R.; de Vos R. C. H. Improved batch correction in untargeted MS-based metabolomics. Metabolomics 2016, 12, 88 10.1007/s11306-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S.; Vervoort J.; Moco S.; Bino R. J.; De Vos R. C. H.; Bino R. Metabolomics technologies and metabolite identification. TrAC, Trends Anal. Chem. 2007, 26, 855–866. 10.1016/j.trac.2007.08.003. [DOI] [Google Scholar]
- Zhang S.; Gowda G. A. N.; Ye T.; Raftery D. Advances in NMR-based biofluid analysis and metabolite profiling. Analyst 2010, 135, 1490–1498. 10.1039/c000091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockford D. J.; Holmes E.; Lindon J. C.; Plumb R. S.; Zirah S.; Bruce S. J.; Rainville P.; Stumpf C. L.; Nicholson J. K. Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: application in metabonomic toxicology studies. Anal. Chem. 2006, 78, 363–371. 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- Tian H.; Zheng N.; Wang W.; Cheng J.; Li S.; Zhang Y.; Wang J. Integrated Metabolomics Study of the Milk of Heat-stressed Lactating Dairy Cows. Sci. Rep. 2016, 6, 24208 10.1038/srep24208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M. S.; Almstetter M. F.; Nürnberger N.; Sigl G.; Gronwald W.; Wiedemann S.; Dettmer K.; Oefner P. J. Correlations between milk and plasma levels of amino and carboxylic acids in dairy cows. J. Proteome Res. 2013, 12, 5223–5232. 10.1021/pr4006537. [DOI] [PubMed] [Google Scholar]
- van Hoeij R. J.; Dijkstra J.; Bruckmaier R. M.; Gross J. J.; Lam T. J. G. M.; Remmelink G. J.; Kemp B.; van Knegsel A. T. M. The effect of dry period length and postpartum level of concentrate on milk production, energy balance, and plasma metabolites of dairy cows across the dry period and in early lactation. J. Dairy Sci. 2017, 100, 5863–5879. 10.3168/jds.2016-11703. [DOI] [PubMed] [Google Scholar]
- Xu W.; Vervoort J.; Saccenti E.; van Hoeij R.; Kemp B.; van Knegsel A. Milk Metabolomics Data Reveal the Energy Balance of Individual Dairy Cows in Early Lactation. Sci. Rep. 2018, 8, 15828 10.1038/s41598-018-34190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CVB. Chemische samenstellingen en nutritionele waarden van voedermiddelen (in Dutch). Productschap Diervoeder; CBV (Centraal Veevoeder Bureau): The Hague, the Netherlands, 2011.
- CVB. Veevoedertabel. Gegevens over chemische samenstelling, verteerbaarheid en voederwaarde vanvoedermiddelen; CVB: Lelystad, the Netherlands, 2005.
- Van Es A. J. H. Feed evaluation for dairy cows. Livest. Prod. Sci. 1975, 2, 95–107. 10.1016/0301-6226(75)90029-9. [DOI] [Google Scholar]
- Lu J.; Boeren S.; van Hooijdonk T.; Vervoort J.; Hettinga K. Effect of the DGAT1 K232A genotype of dairy cows on the milk metabolome and proteome. J. Dairy Sci. 2015, 98, 3460–3469. 10.3168/jds.2014-8872. [DOI] [PubMed] [Google Scholar]
- Lu J.; Antunes Fernandes E.; Paez Cano A. E.; Vinitwatanakhun J.; Boeren S.; van Hooijdonk T.; van Knegsel A.; Vervoort J.; Hettinga K. A. Changes in milk proteome and metabolome associated with dry period length, energy balance, and lactation stage in postparturient dairy cows. J. Proteome Res. 2013, 12, 3288–3296. 10.1021/pr4001306. [DOI] [PubMed] [Google Scholar]
- Suzuki M.; Nishiumi S.; Kobayashi T.; Azuma T.; Yoshida M. LC–MS/MS-based metabolome analysis detected changes in the metabolic profiles of small and large intestinal adenomatous polyps in Apc Min/+ mice. Metabolomics 2016, 12, 68 10.1007/s11306-016-0988-7. [DOI] [Google Scholar]
- Matsubara A.; Izumi Y.; Nishiumi S.; Suzuki M.; Azuma T.; Fukusaki E.; Bamba T.; Yoshida M. Supercritical fluid extraction as a preparation method for mass spectrometry of dried blood spots. J. Chromatogr. B 2014, 969, 199–204. 10.1016/j.jchromb.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Maher A. D.; Hayes B.; Cocks B.; Marett L.; Wales W. J.; Rochfort S. J. Latent biochemical relationships in the blood-milk metabolic axis of dairy cows revealed by statistical integration of 1H NMR spectroscopic data. J. Proteome Res. 2013, 12, 1428–1435. 10.1021/pr301056q. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Tzur D.; Knox C.; Eisner R.; Guo A. C.; Young N.; Cheng D.; Jewell K.; Arndt D.; Sawhney S.; Fung C.; Nikolai L.; Lewis M.; Coutouly M.-A.; Forsythe I.; Tang P.; Shrivastava S.; Jeroncic K.; Stothard P.; Amegbey G.; Block D.; Hau D. D.; Wagner J.; Miniaci J.; Clements M.; Gebremedhin M.; Guo N.; Zhang Y.; Duggan G. E.; Macinnis G. D.; Weljie A. M.; Dowlatabadi R.; Bamforth F.; Clive D.; Greiner R.; Li L.; Marrie T.; Sykes B. D.; Vogel H. J.; Querengesser L. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M. S.; Buttchereit N.; Miemczyk S. P.; Immervoll A.-K.; Louis C.; Wiedemann S.; Junge W.; Thaller G.; Oefner P. J.; Gronwald W. NMR metabolomic analysis of dairy cows reveals milk glycerophosphocholine to phosphocholine ratio as prognostic biomarker for risk of ketosis. J. Proteome Res. 2012, 11, 1373–1381. 10.1021/pr201017n. [DOI] [PubMed] [Google Scholar]
- Klein M. S.; Almstetter M. F.; Schlamberger G.; Nürnberger N.; Dettmer K.; Oefner P. J.; Meyer H. H. D.; Wiedemann S.; Gronwald W. Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J. Dairy Sci. 2010, 93, 1539–1550. 10.3168/jds.2009-2563. [DOI] [PubMed] [Google Scholar]
- Sundekilde U. K.; Poulsen N. A.; Larsen L. B.; Bertram H. C. Nuclear magnetic resonance metabonomics reveals strong association between milk metabolites and somatic cell count in bovine milk. J. Dairy Sci. 2013, 96, 290–299. 10.3168/jds.2012-5819. [DOI] [PubMed] [Google Scholar]
- Kuhn N. J.; Carrick D. T.; Wilde C. J. Lactose synthesis: the possibilities of regulation. J. Dairy Sci. 1980, 63, 328–336. 10.3168/jds.S0022-0302(80)82934-1. [DOI] [PubMed] [Google Scholar]
- van Hoeij R. J.; Kok A.; Bruckmaier R. M.; Haskell M. J.; Kemp B.; van Knegsel A. T. M. Relationship between metabolic status and behavior in dairy cows in week 4 of lactation. Animal 2019, 13, 640–648. 10.1017/S1751731118001842. [DOI] [PubMed] [Google Scholar]
- Capuco A. V.; Ellis S. E.; Hale S. A.; Long E.; Erdman R. A.; Zhao X.; Paape M. J. Lactation persistency: Insights from mammary cell proliferation studies. J. Anim. Sci. 2003, 81, 18–31. 10.2527/2003.81suppl_318x. [DOI] [PubMed] [Google Scholar]
- Flint D. J.; Knight C. H. Interactions of Prolactin and Growth Hormone (GH) in the Regulation of Mammary Gland Function and Epithelial Cell Survival. J. Mammary Gland Biol. Neoplasia 1997, 2, 41–48. 10.1023/A:1026321529451. [DOI] [PubMed] [Google Scholar]
- Capuco A. V.; Wood D. L.; Baldwin R.; Mcleod K.; Paape M. J. Mammary cell number, proliferation, and apoptosis during a bovine lactation: relation to milk production and effect of bST1. J. Dairy Sci. 2001, 84, 2177–2187. 10.3168/jds.S0022-0302(01)74664-4. [DOI] [PubMed] [Google Scholar]
- Monks J.; Smith-Steinhart C.; Kruk E. R.; Fadok V. A.; Henson P. M. Epithelial cells remove apoptotic epithelial cells during post-lactation involution of the mouse mammary gland. Biol. Reprod. 2008, 78, 586–594. 10.1095/biolreprod.107.065045. [DOI] [PubMed] [Google Scholar]
- Miller N.; Delbecchi L.; Petitclerc D.; Wagner G. F.; Talbot B. G.; Lacasse P. Effect of stage of lactation and parity on mammary gland cell renewal. J. Dairy Sci. 2006, 89, 4669–4677. 10.3168/jds.S0022-0302(06)72517-6. [DOI] [PubMed] [Google Scholar]
- Huberman J. A.; Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 1968, 32, 327–341. 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Kleinberg D. L. Early mammary development: growth hormone and IGF-1. J. Mammary Gland Biol. Neoplasia 1997, 2, 49–57. 10.1023/A:1026373513521. [DOI] [PubMed] [Google Scholar]
- Akers R. M.Lactation and the Mammary Gland; Blackwell Publishing Company, 2002. [Google Scholar]
- Salazar-Roa M.; Malumbres M. Fueling the cell division cycle. Trends Cell Biol. 2017, 27, 69–81. 10.1016/j.tcb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Gibellini F.; Smith T. K. The Kennedy pathway—de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010, 62, 414–428. 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- Kennedy E. P.; Weiss S. B. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 1956, 222, 193–214. [PubMed] [Google Scholar]
- Li Z.; Vance D. E. Thematic review series: glycerolipids. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008, 49, 1187–1194. 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- O’Riordan N.; Kane M.; Joshi L.; Hickey R. M. Structural and functional characteristics of bovine milk protein glycosylation. Glycobiology 2014, 24, 220–236. 10.1093/glycob/cwt162. [DOI] [PubMed] [Google Scholar]
- Bonfatti V.; Chiarot G.; Carnier P. Glycosylation of κ-casein: Genetic and nongenetic variation and effects on rennet coagulation properties of milk. J. Dairy Sci. 2014, 97, 1961–1969. 10.3168/jds.2013-7418. [DOI] [PubMed] [Google Scholar]
- Robitaille G.; Ng-Kwai-Hang K.-F.; Monardes H. G. Variation in the N-acetyl neuraminic acid content of bovine κ-casein. J. Dairy Res. 1991, 58, 107–114. 10.1017/S0022029900033550. [DOI] [Google Scholar]
- Kates J. R.; McAuslan B. R. Poxvirus DNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1967, 58, 134. 10.1073/pnas.58.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio E.; Mezzanzanica D.; Alberti P.; Spadaro F.; Ramoni C.; D’Ascenzo S.; Millimaggi D.; Pavan A.; Dolo V.; Canevari S.; Podo F. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005, 65, 9369–9376. 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- Aboagye E. O.; Bhujwalla Z. M. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999, 59, 80–84. [PubMed] [Google Scholar]
- Morrish F.; Noonan J.; Perez-Olsen C.; Gafken P. R.; Fitzgibbon M.; Kelleher J.; VanGilst M.; Hockenbery D. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J. Biol. Chem. 2010, 285, 36267–36274. 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baticz O.; Tömösközi S.; Vida L.; et al. Relationship between concentration of citrate and ketone bodies in cow’s milk. Acta Vet. Hung. 2002, 50, 253–261. 10.1556/avet.50.2002.3.1. [DOI] [PubMed] [Google Scholar]
- Garnsworthy P. C.; Masson L. L.; Lock A. L.; Mottram T. T. Variation of Milk Citrate with Stage of Lactation and De Novo Fatty Acid Synthesis in Dairy Cows. J. Dairy Sci. 2006, 89, 1604–1612. 10.3168/jds.S0022-0302(06)72227-5. [DOI] [PubMed] [Google Scholar]
- Banks W.; Clapperton J. L.; Girdler A. K.; Steele W. Effect of inclusion of different forms of dietary fatty acid on the yield and composition of cow’s milk. J. Dairy Res. 1984, 51, 387–395. 10.1017/S0022029900023682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.