Abstract
Background
Increasing evidence suggests that conventional adenomas (CAs) and serrated polyps (SPs) represent two distinct groups of precursor lesions for colorectal cancer (CRC). The influence of common genetic variants on risk of CAs and SPs remain largely unknown.
Methods
Among 27 426 participants within three prospective cohort studies, we created a weighted genetic risk score (GRS) based on 40 CRC-related single nucleotide polymorphisms (SNPs) identified in previous genome-wide association studies; and we examined the association of GRS (per one standard deviation increment) with risk of CAs, SPs and synchronous CAs and SPs, by multivariable logistic regression. We also analysed individual variants in the secondary analysis.
Results
During 18–20 years of follow-up, we documented 2952 CAs, 1585 SPs and 794 synchronous CAs and SPs. Higher GRS was associated with increased risk of CAs [odds ratio (OR) = 1.17, 95% confidence interval (CI): 1.12-1.21] and SPs (OR = 1.09, 95% CI: 1.03-1.14), with a stronger association for CAs than SPs (Pheterogeneity=0.01). An even stronger association was found for patients with synchronous CAs and SPs (OR = 1.32), advanced CAs (OR = 1.22) and multiple CAs (OR = 1.25). Different sets of variants were associated with CAs and SPs, with a Spearman correlation coefficient of 0.02 between the ORs associating the 40 SNPs with the two lesions. After correcting for multiple testing, three variants were associated with CAs (rs3802842, rs6983267 and rs7136702) and two with SPs (rs16892766 and rs4779584).
Conclusions
Common genetic variants play a potential role in the conventional and serrated pathways of CRC. Different sets of variants are identified for the two pathways, further supporting the aetiological heterogeneity of CRC.
Keywords: Colorectal adenomas, polyp, genetics, colorectal cancer
Key Messages
A genetic risk score based on 40 CRC-related SNPs was associated with CAs and SPs, suggesting that genetic factors play a critical role in the conventional and serrated pathways.
Different sets of variants were identified for CAs and SPs, supporting the aetiological heterogeneity of CRC precursors.
Our findings provide a basis for future studies to uncover premalignant biology of colorectum, which may improve early prevention of CRC.
Introduction
Colorectal cancer (CRC) is a heterogeneous disease and may develop through distinct pathways. Approximately 60–80% of CRCs develop through the conventional adenoma (CA)-carcinoma pathway characterized by a series of mutations in oncogenes and tumour suppressor genes.1,2 Although most CAs do not progress to cancer, some grow large, become dysplastic and eventually develop into malignant adenocarcinomas. On the other hand, a recently recognized alternative pathway to CRC, the serrated pathway, is characterized by hypermethylation of CpG islands in gene promoters, an activating point mutation in the oncogene BRAF, and microsatellite instability.3 The serrated pathway contributes to approximately 20–30% of CRC cases, with sessile serrated adenoma/polyps (SSA/Ps) as the major precursor lesion.4 SSA/Ps mainly originate from hyperplastic polyps (HPs), although some SSA/Ps may arise de novo from normal mucosa.5 Traditional serrated adenomas (TSAs) are rare and represent a distinct group of serrated polyps. Because of the flat and subtle endoscopic appearance and the predilection for the proximal colon, serrated polyps (SPs) are believed to contribute disproportionately to the development of ‘interval cancers’ that occur before next screening or surveillance interval after an initially negative colonoscopy, therefore representing a challenge for clinical management.6
Increasing evidence supports an aetiological difference between CAs and SPs. Several lifestyle risk factors for CRC, such as smoking, alcohol intake and obesity, have been more strongly associated with SPs than CAs.7–9 However, data on genetic factors remain sparse and inconsistent. One study created a genetic risk score (GRS) based on 20 CRC-related SNPs and found that the GRS was similarly associated with CAs and HPs,10 wheres another study reported no association between 13 CRC susceptibility single nucleotide polymorphisms (SNPs) and HPs.11 Of note, these studies are limited by the small sample size (n <700 for HPs), lack of pairwise comparison of CAs and SPs, and limited statistical power for individual SNP analysis and subgroup analysis according to histological features of polyps.
Therefore, to extend our knowledge, we performed a comprehensive assessment of 40 established CRC susceptibility variants in relation to CAs and SPs among 27 426 participants from three large prospective cohort studies: the Nurses’ Health Study (NHS), the Nurse’s Health Study 2 (NHS2) and the Health Professionals Follow-up Study (HPFS).
Methods
Study participants
The NHS included 121 700 registered female nurses aged 30–55 years at enrolment in 1976; NHS2 included 116 686 registered female nurses aged 25–42 years at enrolment in 1989; and HPFS enrolled 51 529 male health professionals aged 40–75 years at baseline in 1986. Details of the follow-up of the three cohorts have been described previously.12–14
Blood specimens were collected from a subset of the participants of the NHS (n = 32 826) between 1989 and 1990, the NHS2 (n = 29 611) between 1996 and 1999 and the HPFS (n = 18 225) between 1993 and 1995. Detailed procedures for blood collection, handling and storage were described elsewhere.15 Participants who provided blood samples had similar demographic, dietary and lifestyle profiles compared with those who did not.16
In the current study, we included participants who were genotyped in previous genome-wide association studies (GWAS) nested within the NHS (n = 18 498), NHS2 (n = 8274) and HPFS (n = 10 889). These studies were primarily designed to study other outcomes, including breast cancer, pancreatic cancer, glaucoma, endometrial cancer, colorectal cancer, ovarian cancer, glioma, prostate cancer, type 2 diabetes, coronary heart disease, kidney stone, gout, mammographic density, venous thromboembolism and post-traumatic stress disorder.17,18 Because detailed histological information of colorectal polyps was not collected until 1992 for the NHS, 1991 for the NHS2 and 1992 for the HPFS, we used these years as the baseline of the current study. At baseline, we excluded participants who were of non-European origin, had a history of cancer (except non-melanoma skin cancer), colorectal polyp or inflammatory bowel disease, or had no endoscopic examination of lower gastrointestinal tract. Eligible participants were followed for diagnosis of colorectal polyps until 1 June 2012 for the NHS, 1 2011 for the NHS2 and 1 January 2010 for the HPFS. A total of 27 426 participants (NHS: n = 13 101; NHS2: n = 6493; HPFS: n = 7832) were included in the analysis (see flowchart in Supplementary Figure 1, available as Supplementary data at IJE online). The study was approved by the institutional review board at the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health. Written informed consent was obtained from each participant.
Ascertainment of colorectal polyp cases and subtypes
On each biennial questionnaire, participants were asked whether they had undergone a colonoscopy or sigmoidoscopy and whether any colorectal polyp had been diagnosed in the past 2 years. Among all endoscopies, flexible sigmoidoscopies accounted for 27%, with a greater proportion in the earlier years than in the later years (Supplementary Figure 2, available as Supplementary data at IJE online). For those who reported polyp diagnosis, we asked for permission to obtain their endoscopic and pathological records. Investigators blinded to any exposure information reviewed all records, confirmed the diagnosis and extracted relevant clinical and pathological data.
In the current study, CAs included tubular, tubulovillous and villous adenomas and adenomas with high-grade dysplasia; and SPs included hyperplastic polyps and mixed/serrated adenomas. Mixed/serrated adenoma consisted of both mixed polyps (those with both adenomatous and hyperplastic changes in histology) and polyps with any serrated diagnosis (e.g. serrated adenomas, serrated polyps and SSA/Ps). If a participant had both CAs and SPs in an endoscopy, we recorded each type of the polyps separately, and considered the patient as a synchronous SP and CA case.
Genotyping and variant selection
Genotype data were obtained from various GWAS studies nested within the NHS, NHS2 and HPFS cohorts.17,18 Details on genotyping have been described previously17,18 and summarized in the Supplementary materials, available as Supplementary data at IJE online.
A total of 63 CRC susceptibility variants that had been associated with CRC in GWAS were selected as candidate SNPs for this study.19–32 Each group of correlated SNPs (R2 >0.01) was represented by a single SNP that showed the strongest association with CRC in the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) that had no overlap with the current study sample. Finally, a total of 40 variants were included for the analysis (Supplementary Table S1, available as Supplementary data at IJE online).
To examine the cumulative effect of the 40 genetic variants, a weighted GRS was calculated using the equation: GRS, where βi is the regression coefficient for the SNPi in relation to CRC derived from the GECCO datasets using the additive genetic model. Each unit of the GRS represents one risk allele increment. The distribution of GRS in our study samples is presented in Supplementary Figure S3, available as Supplementary data at IJE online, with a higher score indicating greater genetic predisposition to CRC.
Statistical analysis
The current study only included participants who had at least one lower endoscopy during the follow-up. If a participant reported more than one endoscopy during the study period, multiple records from the same participant were included in the analysis. Participants were censored at the diagnosis of the first colorectal polyp or the date of latest endoscopy, whichever occurred first. To account for multiple records per participant and to handle time-varying covariates efficiently, we used an Andersen-Gill data structure with a new record for each 2-year follow-up period during which a participant underwent an endoscopy.
Multivariable logistic regression for clustered data (PROC GENMOD) was used to examine the odds ratio (OR) and 95% confidence interval (CI) of developing CAs, SPs and synchronous SPs and CAs, with participants without any polyp as the reference, according to the quintiles of GRS (cut-offs determined in the controls) and per one standard deviation (SD) increment of GRS. We adjusted for study cohort, time period of endoscopy, number of previous endoscopies, time in years since the most recent endoscopy, age and the top three principal components for population structure. We compared the genetic associations between SPs and CAs through a case-only analysis and calculated the Pheterogeneity.7 In the secondary analysis, we analysed the associations with CAs and SPs for individual SNPs included in the GRS, and corrected for multiple testing using Bonferroni correction (adjusted α = 0.05/40 = 1.3 × 10−3).
Detailed subgroup analysis was performed according to histopathological features of polyps, and major lifestyle factors associated with CRC (Supplementary materials, available as Supplementary data at IJE online). Advanced CAs were defined as at least one CA of ≥10 mm in diameter or with advanced histology (tubulovillous/villous histological features or high-grade/severe dysplasia).33 Given that large (≥10 mm) or proximal SPs have been associated with higher risk for CRC,34,35 we defined SPs located in the proximal colon or with size of ≥10 mm as high-risk SPs. If a participant had more than one polyp in an endoscopy, the size of the largest polyp and the histology of the most advanced lesion were used.
In sensitivity analysis, we examined the associations of GRS with CAs, SPs or synchronous SPs and CAs among participants who were selected as controls in the source GWAS studies of various outcomes. Moreover, to test the robustness of our findings to the secular trend of colonoscopy versus flexible sigmoidoscopy (Supplementary Figure S2, available as Supplementary data at IJE online), we performed a sensitivity analysis by including participants who had undergone colonoscopies only. All statistical analyses were conducted by SAS version 9.4 software (SAS Institute Inc., Cary, NC).
Results
During 18–20 years of follow-up of 27 426 participants in the three cohorts, we documented 2952 CAs, 1585 SPs and 794 synchronous CAs and SPs. As shown in Table 1, participants had a mean age of 63.2 years and 70% were females. Compared with participants without any polyp, those with CAs, SP, or synchronous CAs and SPs were more likely to have a higher GRS and a family history of CRC; they also had a higher body mass index (BMI), smoked more cigarettes, drank more alcohol and were less likely to be physically active or to regularly use aspirin. Moreover, compared with CA cases, SP cases tended to have a higher BMI, smoke more cigarettes and drink more alcohol.
Table 1.
Basic characteristics of study participants in the three cohort studies (NHS, NHS2, HPFS)a
| Overall population | Non-polyp | CA-only | SP-only | Synchronous CA and SP | |
|---|---|---|---|---|---|
| No. of participants | 27 426 | 22 095 | 2952 | 1585 | 794 |
| Age, years | 63.2±10.0 | 63.2±10.1 | 64.1±9.2 | 60.7±9.7 | 64.5±8.5 |
| Female, % | 70 | 71 | 57 | 73 | 58 |
| Genetic risk score (GRS) | 30.5 (3.9) | 30.5 (3.9) | 31.1 (3.9) | 30.8 (3.9) | 31.3 (3.8) |
| Family history of colorectal cancer, % | 22 | 22 | 27 | 26 | 27 |
| Pack-years of smoking | 10.0±16.4 | 9.8±16.2 | 10.5±17.0 | 14.0±20.1 | 16.6±21.1 |
| Never smokers, % | 51 | 51 | 52 | 45 | 40 |
| Past smokers‚ <30 packs/year, % | 35 | 35 | 33 | 33 | 34 |
| Past smokers‚ ≥30 packs/year, % | 9 | 9 | 10 | 14 | 14 |
| Current smokers, <30 packs/year, % | 2 | 2 | 2 | 3 | 3 |
| Current smokers‚ ≥30 packs/year, % | 3 | 3 | 3 | 6 | 9 |
| Body mass index, kg/m2 | 26.4±5.0 | 26.4±5.0 | 26.8±5.0 | 27.2±5.1 | 27.8±5.5 |
| <25, % | 44 | 45 | 41 | 37 | 33 |
| 25.0-29.9, % | 37 | 36 | 38 | 40 | 38 |
| 30.0-34.9, % | 13 | 13 | 14 | 16 | 19 |
| ≥35, % | 6 | 6 | 7 | 8 | 10 |
| Height, cm | 168.7±9.3 | 168.7±9.3 | 168.8±9.1 | 168.5±9.1 | 169.4±8.8 |
| Physical activity, MET-h/weekb | 22.3±19.8 | 22.5±19.9 | 20.9±18.4 | 20.7±17.5 | 19.3±18.0 |
| <7.5, % | 21 | 20 | 22 | 23 | 25 |
| 7.5-14.9, % | 24 | 24 | 25 | 25 | 27 |
| 15-29.9, % | 31 | 31 | 30 | 30 | 30 |
| 30-59.9, % | 20 | 20 | 19 | 18 | 15 |
| ≥60, % | 5 | 5 | 4 | 4 | 3 |
| Alcohol intake, g/day | 7.5±10.3 | 7.4±10.2 | 7.5±10.8 | 8.5±12.0 | 8.5±11.9 |
| Regular aspirin use, %c | 43 | 43 | 41 | 42 | 39 |
NHS, the Nurses’ Health Study; NHS2, the Nurses’ Health Study 2; HPFS, the Health Professionals Follow-up Study; CA, conventional adenoma; SP, serrated polyp; GRS, genetic risk score; MET, metabolic equivalent task.
The presented data are based on repeatedly collected information for each participant up to polyp diagnosis for cases and the end of follow-up period for non-polyps. Cumulative average values across person-endoscopies are presented. Mean±SD is presented for continuous variables and percentage for categorical variables. All variables are adjusted for age and sex except for age and sex themselves.
Physical activity is represented by the product sum of the METS of each specific recreational activity and hours spent on that activity per week.
A standard tablet contains 325 mg aspirin, and regular users were defined as those who used at least two standard tablets per week.
Table 2 shows the associations between GRS and polyp subtypes. Higher GRS was associated with increased risk of CAs (OR per 1-SD increment = 1.17, 95% CI: 1.12–1.21), SPs (OR = 1.09, 95% CI: 1.03–1.14), and synchronous CAs and SPs (OR = 1.24, 95% CI: 1.16–1.32), with a stronger association for CAs than SPs (Pheterogeneity = 0.01). When CAs and SPs were further classified based on their malignant potential, we found that GRS was more strongly associated with advanced CAs (OR = 1.22, −CI: 1.16–1.28) than non-advanced CAs (OR = 1.12, 95% CI: 1.07–1.18) (Pheterogeneity = 0.02), whereas no difference was found for high- and low-risk SPs (OR = 1.10, 95% CI: 1.01–1.19 and OR = 1.08, 95% CI: 1.02–1.15, respectively; Pheterogeneity = 0.82).
Table 2.
Association between genetic risk score (GRS) and risk of colorectal polyp subtypes in the three cohort studies (NHS, NHS2, HPFS)a
| Polyp subtype | Quintiles of GRS |
Per 1-SD increment of GRS | Ptrend | Pheterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| Lowest | Second | Middle | Fourth | Highest | ||||
| Conventional adenoma (CA)-only | ||||||||
| n | 466 | 555 | 573 | 614 | 744 | 2952 | ||
| Mean of GRS | 25.2 | 28.3 | 30.5 | 32.5 | 36.1 | 31.1 | ||
| OR (95% CI) | 1(Ref) | 1.21(1.06-1.37) | 1.25(1.10-1.41) | 1.33(1.18-1.51) | 1.63(1.44-1.83) | 1.17(1.12-1.21) | <.001 | Ref 1 |
| Non-advanced | ||||||||
| n | 282 | 310 | 301 | 338 | 397 | 1628 | ||
| Mean of GRS | 25.2 | 28.3 | 30.5 | 32.5 | 36.0 | 30.9 | ||
| OR (95% CI) | 1(Ref) | 1.12(0.95-1.32) | 1.09(0.92-1.28) | 1.22(1.04-1.43) | 1.44(1.23-1.68) | 1.12(1.07-1.18) | <.001 | Ref 2 |
| Advancedb | ||||||||
| n | 184 | 245 | 272 | 276 | 347 | 1324 | ||
| Mean of GRS | 25.2 | 28.4 | 30.5 | 32.5 | 36.1 | 31.2 | ||
| OR (95% CI) | 1(Ref) | 1.34(1.10-1.63) | 1.49(1.23-1.80) | 1.50(1.24-1.82) | 1.91(1.59-2.29) | 1.22(1.16-1.28) | <.001 | 0.02(vs. Ref 2) |
| Serrated polyp (SP) onlyc | ||||||||
| n | 293 | 278 | 303 | 344 | 367 | 1585 | ||
| Mean of GRS | 25.1 | 28.4 | 30.5 | 32.5 | 36.0 | 30.8 | ||
| OR (95% CI) | 1(Ref) | 0.94(0.80-1.12) | 1.03(0.88-1.22) | 1.18(1.00-1.38) | 1.24(1.06-1.45) | 1.09(1.03-1.14) | <.001 | 0.01(vs. Ref 1) |
| Low-risk | ||||||||
| n | 181 | 171 | 185 | 219 | 231 | 987 | ||
| Mean of GRS | 25.1 | 28.3 | 30.5 | 32.5 | 35.8 | 30.8 | ||
| OR (95% CI) | 1(Ref) | 0.94(0.76-1.16) | 1.02(0.83-1.25) | 1.21(0.99-1.47) | 1.25(1.03-1.53) | 1.08(1.02-1.15) | 0.01 | Ref 3 |
| High-risk | ||||||||
| n | 99 | 97 | 103 | 112 | 122 | 533 | ||
| Mean of GRS | 25.1 | 28.4 | 30.4 | 32.5 | 36.3 | 30.9 | ||
| OR (95% CI) | 1(Ref) | 0.98(0.74-1.30) | 1.04(0.79-1.38) | 1.14(0.87-1.50) | 1.23(0.94-1.60) | 1.10(1.01-1.19) | 0.04 | 0.82(vs. Ref 3) |
| Synchronous CA and SP | ||||||||
| n | 104 | 151 | 163 | 177 | 199 | 794 | ||
| Mean of GRS | 25.2 | 28.4 | 30.5 | 32.6 | 36.3 | 31.3 | ||
| OR (95% CI) | 1(Ref) | 1.48(1.15-1.90) | 1.60(1.25-2.05) | 1.72(1.35-2.20) | 1.96(1.54-2.49) | 1.24(1.16-1.32) | <.001 | 0.13(vs. Ref 1) |
NHS, the Nurses’ Health Study; NHS2, the Nurses’ Health Study 2; HPFS, the Health Professionals Follow-up Study; CA, conventional adenoma; SP, serrated polyp; GRS, genetic risk score; OR, odds ratio; CI, confidence interval; Ref, reference.£
Multivariable logistic regression model adjusted for study cohort (NHS, NHS2, HPFS), time period of endoscopy (in 2-year intervals), number of previous endoscopies (continuous), time in years since the most recent endoscopy (continuous), age (continuous), and top three principal components for population structure. The Pheterogeneity was calculated by comparing genetic associations between subgroups through a case-only analysis.
Advanced group denotes at least one CA of ≥10 mm in diameter or with advanced histology (tubulovillous/villous histological features or high-grade/severe dysplasia) or ≥3 CAs regardless of histology or size.
High-risk group denotes SPs located in proximal colon or with size of ≥10 mm.
When stratified by histopathological features of polyps (Table 3), the GRS association was stronger for multiple CAs (OR = 1.25, 95% CI: 1.17–1.34) than a single CA (OR = 1.13, 95% CI: 1.08–1.18) (Pheterogeneity = 0.01), and for CAs with advanced histology (tubulovillous, villous or high-grade dysplasia) (OR = 1.26, 95% CI: 1.18–1.35) than tubular CAs (OR = 1.14, 95% CI: 1.09-1.20) (Pheterogeneity = 0.02).
Table 3.
Association between genetic risk score (GRS) (per 1-SD increment) and risk of colorectal polyp subtypes according to polyp features in the three cohort studies (NHS, NHS2, HPFS)a
| Polyp feature | Conventional adenoma only |
Serrated polyp only |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean of GRS | OR (95% CI) | P | P heterogeneity | n | Mean of GRS | OR (95% CI) | P | P heterogeneity | |
| Subsite | ||||||||||
| Proximal colon | 1468 | 31.1 | 1.18 (1.12–1.24) | <0.001 | Ref | 488 | 30.8 | 1.09 (0.99–1.19) | 0.07 | Ref |
| Distal colon | 1442 | 31.1 | 1.18 (1.12–1.24) | <0.001 | 0.83 | 758 | 31.0 | 1.15 (1.07–1.24) | <0.001 | 0.21 |
| Rectum | 527 | 31.2 | 1.19 (1.09–1.29) | <0.001 | 0.95 | 582 | 30.8 | 1.06 (0.98–1.15) | 0.12 | 0.83 |
| Size | ||||||||||
| <10 mm | 1763 | 31.0 | 1.14 (1.09–1.19) | <0.001 | Ref | 1401 | 30.8 | 1.08 (1.02–1.13) | 0.01 | Ref |
| ≥10 mm | 1067 | 31.2 | 1.21 (1.14–1.28) | <0.001 | 0.09 | 149 | 30.9 | 1.12 (0.94–1.33) | 0.20 | 0.61 |
| Multiplicity | ||||||||||
| Single | 2069 | 31.0 | 1.13 (1.08–1.18) | <0.001 | Ref | 922 | 30.8 | 1.08 (1.02–1.15) | 0.02 | Ref |
| Multiple | 873 | 31.3 | 1.25 (1.17–1.34) | <0.001 | 0.01 | 663 | 30.8 | 1.09 (1.01–1.18) | 0.02 | 0.77 |
| Histology | ||||||||||
| Tubular | 1727 | 31.0 | 1.14 (1.09–1.20) | <0.001 | Ref | |||||
| Tubulovillous, villous or high–grade dysplasia | 764 | 31.4 | 1.26 (1.18–1.35) | <0.001 | 0.02 | |||||
NHS, the Nurses’ Health Study; NHS2, the Nurses’ Health Study 2; HPFS, the Health Professionals Follow-up Study; GRS, genetic risk score; OR, odds ratio; CI, confidence interval; Ref, reference.
Multivariable logistic regression model adjusted for study cohort (NHS, NHS2, HPFS), time period of endoscopy (in 2-year intervals), number of previous endoscopies (continuous), time in years since the most recent endoscopy (continuous), age (continuous) and top three principal components for population structure. The Pheterogeneity was calculated by comparing genetic associations between subgroups through a case-only analysis.
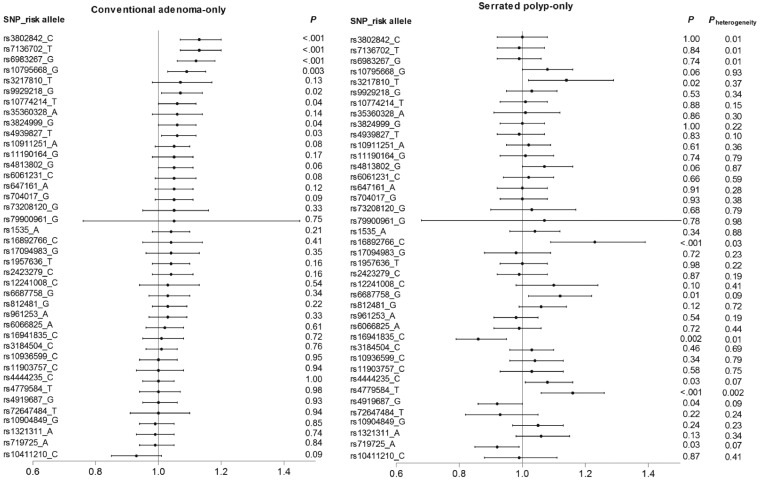
We further analysed the association of individual SNPs with CAs and SPs. In general, the catalogue of SNPs associated with CAs appeared to be different from that associated with SPs (Figure 1; Supplementary Table S3, available as Supplementary data at IJE online). No correlation was found for the magnitudes of the associations between each of the 40 SNPs and risk of CAs and SPs (Spearman correlation coefficient r = 0.02). The heterogeneity test showed that the associations for six SNPs (rs16892766, rs16941835, rs3802842, rs4779584, rs6983267 and rs7136702) were statistically different between CAs and SPs (Pheterogeneity <0.05). After Bonferroni correction for multiple testing, three SNPs were associated with risk of CAs in the same direction as reported previously for CRC: rs3802842, rs6983267 and rs7136702; whereas two SNPs showed an association with risk of SPs in the same direction as reported previously for CRC: rs16892766 and rs4779584; the OR per each risk allele ranged from 1.12 to 1.23. We also summarize the major functional evidence for these SNPs in relation to CRC in Table 4.
Figure 1.
Association between 40 colorectal cancer susceptibility variants and risk of conventional adenoma-only and serrated polyp-only in the three cohort studies (NHS, NHS2, HPFS). Pheterogeneity was calculated to test the difference in the association of each of the SNPs with risk of conventional adenoma and serrated polyp through case-control analysis.
Table 4.
Association between individual variants and risk of colorectal polyp subtypes in the three cohort studies (NHS, NHS2, HPFS)a
| SNP | Locus/gene | Risk/other allele | Reported OR for CRC (reference) | Risk allele frequency | OR (95% CI) b | P c | Functional evidence (supplementary reference) |
|---|---|---|---|---|---|---|---|
| Conventional adenoma only | |||||||
| rs6983267 | 8q24.21/MYC | G/T | 1.21 (32) | 0.51 | 1.12(1.06-1.18) | <0.1 × 10−3 | Affecting the binding of TCG4 and interaction with the oncogene MYC4 |
| rs3802842 | 11q23.1/ COLCA1, COLCA2 | C/A | 1.11 (30) | 0.29 | 1.13(1.07-1.20) | <0.1 × 10−3 | Decreasing the expression of potential tumour suppressor gene COLCA1 and COLCA25 |
| rs7136702 | 12q13.13/LARP4, DIP2B | T/C | 1.06 (28) | 0.35 | 1.13(1.07-1.20) | <0.1 × 10−3 | Increasing the expression of DIP2B which may modulate DNA methylation in CRC6 |
| Serrated polyp only | |||||||
| rs16892766 | 8q23.3/EIF3H | C/A | 1.25 (31) | 0.08 | 1.23(1.09-1.39) | 0.7 × 10−3 | Increasing the expression of the oncogene EIF3H8 |
| rs4779584 | 15q13.3/GREM1, SCG5 | T/C | 1.15 (27) | 0.19 | 1.16(1.06-1.26) | 0.9 × 10−3 | NA |
NHS, the Nurses’ Health Study; NHS2, the Nurses’ Health Study 2; HPFS, the Health Professionals Follow-up Study; CRC, colorectal cancer; OR, odds ratio; CI, confidence interval; NA, not available.
Multivariable logistic regression model adjusted for study cohort (NHS, NHS2, HPFS), time period of endoscopy (in 2-year intervals), number of previous endoscopies (continuous), time in years since the most recent endoscopy (continuous), age (continuous) and top three principal components for population structure.
Comparing conventional adenoma or serrated polyp with non-polyp.
Bonferroni correction with P<1.3 × 10−3 (0.05/40) was considered statistically significant.
In the stratified analysis, no interaction was noted between demographic or lifestyle factors and GRS (Supplementary Table S4, available as Supplementary data at IJE online). In the sensitivity analysis, we observed similar associations of the GRS with CAs and SPs when restricting to participants who were selected as controls in previous GWAS studies (Supplementary Table S5, available as Supplementary data at IJE online) and who had undergone colonoscopies only (Supplementary Table S6, available as Supplementary data at IJE online).
Discussion
In this large study of 27 426 participants from three prospective cohort studies, a GRS comprising 40 CRC-related variants was associated with higher risk of CRC precursors, with a stronger association observed for CAs than SPs, particular for advanced CAs. The analysis of individual SNPs revealed that different sets of variants were associated with CAs and SPs. Our findings provide novel genetic evidence for the aetiological difference of conventional and serrated pathways in CRC and have implications for developing tailored screening and surveillance strategies for CRC prevention.
Although the genetic architecture of CRC has been investigated extensively, the role of common genetic variants in CRC precursors remains poorly understood. Compared with previous studies that investigated only a limited number of SNPs for CAs,10,11,36 we systematically evaluated 40 CRC-related SNPs identified in eqrlier GWAS in relation to both CAs and SPs. Given the particular importance of SPs in the development of ‘interval cancer’, our study represents the first step to examine the potential of integrating genetic susceptibility information for tailored colonoscopy screening and surveillance of polyps for better prevention of CRC.36
The observation that the GRS for CRC susceptibility had a stronger association with CAs than SPs is not surprising, because CRC arising from the serrated pathway accounts for a relatively small fraction of all CRC cases (20–30%) and is under- represented in the existing GWASs that were used for creation of the GRS. Therefore, further GWAS specifically for SP-related CRC are needed. Nevertheless, among the 40 variants that have been identified thus far for CRC, we found that different sets of SNPs were associated with the risk of CAs and SPs, suggesting that different inherited factors drive the conventional and serrated pathways to CRC. Previous studies have demonstrated the different alterations at somatic and epigenetic levels between CA- and SP-related CRCs.37 In contrast to the conventional pathway involving chromosomal instability, the serrated pathway is characterized by high incidence of activating BRAF mutations and epigenetic silencing of the DNA mismatch repair system.37 In the current study, we provide novel evidence for different germline mutations underlying these two pathways, further supporting the molecular heterogeneity of CAs and SPs.
The strength of the association between GRS and CAs differed by adenoma characteristics. In particular, GRS was more strongly related to the risk of multiple and advanced CAs that have a higher likelihood of progressing into cancer. Similar findings have been reported in previous studies.10,11,36 Moreover, individuals with a family history of CRC have been found more likely to develop multiple CAs.38 These findings together support that genetic susceptibility to sporadic CRC may be, at least partly, mediated by predisposition to CA multiplicity and advancement.
To our knowledge, only two studies have evaluated the association between CRC susceptibility variants and SPs.10,11 Consistent with our findings, a case-control study of 642 HPs reported a positive association between a GRS based on 20 CRC-related SNPs and HP risk.10 By classifying SPs into high- and low-risk subgroups according to size and subsite, we found a similar association with GRS. Also, no heterogeneity was observed according to multiplicity of SPs. Therefore, it is possible that the CRC susceptibility variants might primarily affect the initiation rather than progression of SPs. In contrast with the conventional pathway, transition to high-risk SPs might be driven by environmental factors that cause a series of somatic events. Indeed, a much stronger association with the risk of SPs than CAs has been noted for three major risk factors for CRC, including smoking, alcohol consumption and obesity,8,9,39 which all have strong capability to elicit somatic mutations or epigenetic alterations.40,41 The hypothesis is further supported by the fact that SPs-associated molecular features, such as BRAF mutations, CpG island methylation and microsatellite instability, are more often acquired through somatic events.42 We also identified several individual SNPs that were associated with CAs and SPs, and have discussed their implications in CRC in the Supplementary materials, available as Supplementary data at IJE online.
The current study has several strengths, including the large sample size, systematic selection of SNPs, prospective assessment of endoscopic use and polyp diagnosis within three well-established cohorts, and collection of detailed histopathological information of polyps based on pathology reports. Moreover, diagnostic documentation of both CAs and SPs allowed us to compare their associations with common genetic variants, thus providing critical insight into the aetiological heterogeneity.
Our study also has some limitations. First, given the evolving nature and lack of consensus regarding the diagnostic criteria for specific subtypes of SPs, we were unable to separate HPs, SSA/Ps and TSAs based on the review of pathology records. However, since large size and proximal subsite have both been established as strong predictors for the likelihood of SPs progressing into advanced neoplasia, the consistent findings in the stratified analysis according to size and subsite suggest that our observed associations reflect to a large degree the effect of common genetic variants on the serrated pathways for colorectal carcinogenesis. Second, our study included Caucasians only, and the findings may not be generalizable to non-Caucasian populations. Third, despite the large sample size, the statistical power for individual SNP analysis remains limited, especially for SNPs with low allele frequencies. Finally, given the functional heterogeneity of the SNPs, further studies are needed to examine the potential genetic and environmental interactions in the development of CAs and SPs at the individual variant level.
In summary, a GRS based on 40 CRC susceptibility variants was positively associated with risk of CAs and SPs, with a stronger association for CAs than SPs. Different sets of variants were associated with CAs and SPs. These data support the aetiological heterogeneity of CAs and SPs. Further studies are needed to confirm our findings and examine the potential of genetic variants for tailored colonoscopy screening and surveillance for better prevention of CRC.
Funding
This work was supported by the American Cancer Society Mentored Research Scholar Grant (MRSG-17–220-01 - NEC to MS); by the U.S. National Institutes of Health grants (P01 CA87969; UM1 CA186107; P01 CA55075; UM1 CA167552; U01 CA167552; P50 CA127003; K24 DK098311, R01 CA137178, R01 CA202704, R01 CA176726 to ATC; R01 CA151993, R35 CA197735 to SO; K99 CA215314 R00 CA215314 to MS; K01 DK110267 to ADJ); and by grants from GECCO: National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088; U01 CA164930), National Key R&D Program of China (2017YFC0908300 to DH), the National Natural Science Foundation of China (81502873 to DH), the American Institute for Cancer Research (KW), the Project P Fund for Colorectal Cancer Research, the Friends of the Dana-Farber Cancer Institute, Bennett Family Fund and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. ATC is a Stuart and Suzanne Steele MGH Research Scholar. The funders had no role in design or conduct of the study; collection, management, analysis or interpretation of the data; or preparation, review or approval of the manuscript.
Supplementary Material
Acknowledgements
We would like to thank the participants and staff of the Nurses’ Health Study, the Nurses’ Health Study 2 and the Health Professionals Follow-up Study for their valuable contributions, as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Author Contributions
DH and ADJ performed statistical analysis and drafted the manuscript. XH, ATC, MJ, MKG, SO, PK, CT, UP, SAB, YL, ZH, and HS were involved in the acquisition, analysis, and interpretations of data. MS, KW, and ELG were responsible for study design. All authors critically assessed, edited, and approved the final manuscript.
Conflict of interest: None declared.
References
- 1. Fearon ER, Vogelstein B.. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 2. Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007;50:113–30. [DOI] [PubMed] [Google Scholar]
- 3. Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol 2011;42:1–10. [DOI] [PubMed] [Google Scholar]
- 4. Snover DC, Ahnen DJ, Burt RW. et al. Serrated polyps of the colon and rectum and serrated polyposis In: Bosman FT, Carneiro F, Hruban RH. et al. (eds). WHO Classification of Tumours of the Digestive System. 4th edn Lyon, France: IARC Press, 2010. [Google Scholar]
- 5. Rashtak S, Rego R, Sweetser SR, Sinicrope FA.. Sessile serrated polyps and colon cancer prevention. Cancer Prev Res 2017;10:270–78. [DOI] [PubMed] [Google Scholar]
- 6. Sweetser S, Smyrk TC, Sinicrope FA.. Serrated colon polyps as precursors to colorectal cancer. Clin Gastroenterol Hepatol 2013;11:760–67; quiz e54–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M.. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology 2018;155:355–73.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davenport JR, Su T, Zhao Z. et al. Modifiable lifestyle factors associated with risk of sessile serrated polyps, conventional adenomas and hyperplastic polyps. Gut 2018;67:456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burnett-Hartman AN, Passarelli MN, Adams SV. et al. Differences in epidemiologic risk factors for colorectal adenomas and serrated polyps by lesion severity and anatomical site. Am J Epidemiol 2013;177:625–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang B, Shrubsole MJ, Li G. et al. Association of genetic variants for colorectal cancer differs by subtypes of polyps in the colorectum. Carcinogenesis 2012;33:2417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burnett-Hartman AN, Newcomb PA, Hutter CM. et al. Variation in the association between colorectal cancer susceptibility loci and colorectal polyps by polyp type. Am J Epidemiol 2014;180:223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giovannucci E, Ascherio A, Rimm EB, Colditz GA, Stampfer MJ, Willett WC.. Physical activity, obesity, and risk for colon cancer and adenoma in men. Ann Intern Med 1995;122:327–34. [DOI] [PubMed] [Google Scholar]
- 13. Grodstein F, Martinez ME, Platz EA. et al. Postmenopausal hormone use and risk for colorectal cancer and adenoma. Ann Intern Med 1998;128:705–12. [DOI] [PubMed] [Google Scholar]
- 14. Nimptsch K, Malik VS, Fung TT. et al. Dietary patterns during high school and risk of colorectal adenoma in a cohort of middle-aged women. Int J Cancer 2014;134:2458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bertrand KA, Giovannucci E, Liu Y. et al. Determinants of plasma 25-hydroxyvitamin D and development of prediction models in three US cohorts. Br J Nutr 2012;108:1889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunter DJ, Hankinson SE, Hough H. et al. A prospective study of NAT2 acetylation genotype, cigarette smoking, and risk of breast cancer. Carcinogenesis 1997;18:2127–32. [DOI] [PubMed] [Google Scholar]
- 17. Lindstrom S, Loomis S, Turman C. et al. A comprehensive survey of genetic variation in 20 691 subjects from four large cohorts. PLoS One 2017;12:e0173997.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joshi AD, Andersson C, Buch S. et al. Four susceptibility loci for gallstone disease identified in a meta-analysis of genome-wide association studies. Gastroenterology 2016;151:351–63.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng C, Matsuda K, Jia WH. et al. Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology 2016;150:1633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schumacher FR, Schmit SL, Jiao S. et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun 2015;6:7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Al-Tassan NA, Whiffin N, Hosking FJ. et al. A new GWAS and meta-analysis with 1000Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep 2015;5:10442.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H, Burnett T, Kono S. et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat Commun 2014;5:4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang B, Jia WH, Matsuda K. et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet 2014;46:533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peters U, Jiao S, Schumacher FR. et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology 2013;144:799–807.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jia WH, Zhang B, Matsuo K. et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet 2013;45:191–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dunlop MG, Dobbins SE, Farrington SM. et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet 2012;44:770–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomlinson IP, Carvajal-Carmona LG, Dobbins SE. et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet 2011;7:e1002105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houlston RS, Cheadle J, Dobbins SE. et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet 2010;42:973–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Houlston RS, Webb E, Broderick P. et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 2008;40:1426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tenesa A, Farrington SM, Prendergast JG. et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 2008;40:631–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tomlinson IP, Webb E, Carvajal-Carmona L. et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 2008;40:623–30. [DOI] [PubMed] [Google Scholar]
- 32. Tomlinson I, Webb E, Carvajal-Carmona L. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007;39:984.. [DOI] [PubMed] [Google Scholar]
- 33. Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR.. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012;143:844–57. [DOI] [PubMed] [Google Scholar]
- 34. Schreiner MA, Weiss DG, Lieberman DA.. Proximal and large hyperplastic and nondysplastic serrated polyps detected by colonoscopy are associated with neoplasia. Gastroenterology 2010;139:1497–502. [DOI] [PubMed] [Google Scholar]
- 35. Gao Q, Tsoi KK, Hirai HW. et al. Serrated polyps and the risk of synchronous colorectal advanced neoplasia: a systematic review and meta-analysis. Am J Gastroenterol 2015;110:501–09; quiz 10. [DOI] [PubMed] [Google Scholar]
- 36. Weigl K, Thomsen H, Balavarca Y, Hellwege JN, Shrubsole MJ, Brenner H.. Genetic risk score is associated with prevalence of advanced neoplasms in a colorectal cancer screening population. Gastroenterology 2018;155:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strum WB. Colorectal adenomas. N Engl J Med 2016;374:1065–75. [DOI] [PubMed] [Google Scholar]
- 38. Wark PA, Wu K, van 't Veer P, Fuchs CF, Giovannucci EL.. Family history of colorectal cancer: a determinant of advanced adenoma stage or adenoma multiplicity? Int J Cancer 2009;125:413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M.. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology 2018;155:355–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Supek F, Lehner B.. Clustered mutation signatures reveal that error-prone DNA repair targets mutations to active genes. Cell 2017;170:534–47.e23. [DOI] [PubMed] [Google Scholar]
- 41. Weisenberger DJ, Levine AJ, Long TI. et al. Association of the colorectal CpG island methylator phenotype with molecular features, risk factors, and family history. Cancer Epidemiol Biomarkers Prev 2015;24:512–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Je IJ, Vermeulen L, Meijer GA, Dekker E.. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol 2015;12:401–09. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.