Abstract
Bacilli can form dormant, highly resistant, and metabolically inactive spores to cope with extreme environmental challenges. In this study, we examined the evolutionary age of Bacillus subtilis sporulation genes using the approach known as genomic phylostratigraphy. We found that B. subtilis sporulation genes cluster in several groups that emerged at distant evolutionary time-points, suggesting that the sporulation process underwent several stages of expansion. Next, we asked whether such evolutionary stratification of the genome could be used to predict involvement in sporulation of presently uncharacterized genes (y-genes). We individually inactivated a representative sample of uncharacterized genes that arose during the same evolutionary periods as the known sporulation genes and tested the resulting strains for sporulation phenotypes. Sporulation was significantly affected in 16 out of 37 (43%) tested strains. In addition to expanding the knowledge base on B. subtilis sporulation, our findings suggest that evolutionary age could be used to help with genome mining.
Keywords: sporulation, genomic phylostratigraphy, genome mining, Bacillus subtilis
Introduction
Certain bacteria, such as Bacilli and Clostridia, are known to develop spores. These are dormant, highly resistant, and metabolically inactive life forms that are capable of surviving extreme conditions (Hilbert and Piggot 2004). Spores can germinate into fully functional vegetative cells once they encounter favorable conditions. In Bacillus subtilis, arguably the best characterized sporulation model organism, spore development lasts for several hours and requires spatial and temporal coordination in expression of hundreds of genes (Hilbert and Piggot 2004). Sporulation genes are structurally and functionally diverse (signal transduction systems, sigma factors and transcriptional regulators, metabolic enzymes, and structural proteins of the spore coating), because spore development requires a complete reconfiguration of the cellular envelope and metabolism. Understanding of B. subtilis sporulation is quite extensive, but not complete, and new sporulation genes are still being identified (Traag et al. 2013; Meeske et al. 2016).
Despite their structural and functional diversities, sporulation genes all contribute to a common developmental program. In that sense, sporulation is similar to developmental processes in multicellular organisms, where expression of thousands of functionally diverse genes needs to be spatially and temporally regulated (Hilbert and Piggot 2004; Yanai 2018). It has been previously shown that the evolutionary history of a gene is strongly associated with expression constraints and biological functions, given that it affects the level of gene integration into the cellular environment (Domazet-Lošo and Tautz 2010; Capra et al. 2013). Recent advances in computational biology made it possible to explore potential connections between gene evolution and functional evolution. With the method of genomic phylostratigraphy, gene age can be inferred by considering an orthologous group of genes to represent descendent lineages of the deepest speciation node, or to be the result of divergence between the two most distant homologs (Domazet-Loso et al. 2007). Therefore, a phylostratigraphy analysis of any given genome classifies its genes into phylostrata (PS), each populated with those genes whose protein families emerged at a specific node (time-point) in the tree of life. Then, the age of a gene is defined as the most recent common ancestor of the species found in that phylostratum. Based on such phylostratigraphy analyses, several authors stipulated that if a new biological function emerged at a given time-point in evolution, then the genes related to that function should be enriched in the corresponding phylostratum. This finding has been confirmed in cases such as, for example, cancer development in Metazoa (Trigos et al. 2017) and development of sensory systems in vertebrates (Sestak et al. 2013).
In this study, we asked whether B. subtilis sporulation genes, by analogy to developmental genes of Metazoa, might also cluster in terms of evolutionary age. By examining the B. subtilis whole genome phylostratigraphy map (Ravikumar et al. 2018), we found that B. subtilis sporulation genes arose at several time-points in evolution (significantly enriched in four different PS), suggesting that the sporulation process underwent several stages of expansion. We then hypothesized that genes without an assigned function (y-genes) that belong to sporulation-enriched PS might have a higher than average probability to be involved in sporulation. To test this, we inactivated a representative set of 37 genes of unknown function from sporulation-enriched PS and established that 16 (43%) of these gene knockouts lead to a sporulation phenotype. Various types of phenotypes were observed, with either losses or gains in terms of numbers of produced spores and their structural integrity/functionality. Some of the more striking phenotypes were related to spore coat assembly, regulation of sporulation timing, and a regulatory switch for prophage induction during sporulation.
Results and Discussion
Bacillus subtilis Sporulation Genes Are Enriched in PS2 and PS8–10
Bacillus subtilis genes that are known or suspected to be involved in sporulation are annotated in two major databases: SubtiWiki (Zhu and Stülke 2018) and SporeWeb (Eijlander et al. 2014). Most of the annotations in these databases are based on functional characterization data, that is, there is experimental evidence that gene inactivation or overexpression leads to a measurable sporulation phenotype. However, in some cases, the annotations are based only on gene transcription data (Nicolas et al. 2012), that is, there is a record of differential transcription of the gene at some stage of sporulation or germination. In such cases, the gene is merely suspected to be involved in sporulation, but there is no direct evidence for gene mutation leading to a sporulation phenotype. The current entries SubtiWiki (Zhu and Stülke 2018) and SporeWeb (Eijlander et al. 2014) are not fully overlapping, and by consequence, there is no consensus list of sporulation genes. Hence, our starting point for this study was to establish a consensus list of known sporulation genes. As our experimental setup relies on phenotype analysis, we adopted the more stringent definition of a sporulation gene: gene mutation should lead to a measurable sporulation phenotype. Based on this criterion, a manual literature search of phenotype data for all entries in the two sporulation databases yielded 361 known sporulation genes (supplementary table S1, Supplementary Material online). Next, we examined the distribution of these sporulation genes in the available B. subtilis phylostratigraphy map (Ravikumar et al. 2018). Known sporulation genes account for 8.6% of the B. subtilis gene set, therefore, we considered any PS with a percentage higher than that as sporulation gene-enriched (fig. 1A). It should be noted that the B. subtilis genome follows a well-established trend: most genes belong to the old PS, whereas younger PS contain progressively fewer genes (Ravikumar et al. 2018). About 195 sporulation genes were found in B. subtilis PS1. However, they account for only 7% of the total, so PS1 was not considered as sporulation-enriched. Sporulation genes are enriched in PS2 (30%): the time period corresponding to separation of Bacteria from Archaea and Eukarya, and PS8–10 (20%, 19%, and 15%, respectively): the time period corresponding to successive branching points of Bacilli, Bacilliales, and Bacilliaceae (fig. 1A). An enrichment of sporulation genes can also be observed in PS13 (11%). However, this PS contains only nine genes in total, of which one is a known sporulation gene and the rest are prophage genes. We therefore disregarded it in our further analyses. Sporulation gene content in all other PS was below the genome average.
Fig. 1.
Identification of new sporulation genes by phylostratigraphy. (A) The distribution of Bacillus subtilis-known sporulation genes (see Materials and Methods for definition of known sporulation genes) in PS1–15. Whole genome average of sporulation genes is shown with a dotted line. Total number of genes and number of known sporulation genes are indicated above each PS. PS with enriched known sporulation genes is highlighted in yellow. Deviations from the expected frequencies were tested by χ2 test, and indicated by **P < 0.05 or ***P < 0.01. (B) Summary of the gene candidates selected for assessing sporulation phenotypes, from sporulation-enriched PS2 and PS8–10. Letters next to gene names represent transcription profiles published by Nicolas et al. (2012): N (white) stands for no differential transcription during sporulation, S (blue) stands for strong, and W (light blue) for weak differential transcription during sporulation, respectively. Protein detected at 4 h after sporulation initiation using mass spectrometry are marked with red squares and those detected by protein–GFP fusions with green squares. Corresponding gene size is shown with gray bars. (C) Transcriptional profiles of the selected candidate genes during growth in LB (LB), sporulation (SPO), and germination (G). Candidates were classified into three groups: no differential transcription during sporulation (left panel), strong differential transcription during sporulation (middle panel), and weak differential transcription during sporulation (right panel), as shown in B individual transcriptional profiles of candidate genes are shown in gray, and the average for the whole group is indicated in red.
Interestingly, many of the major sporulation regulators whose genes are present in PS1, such as Spo0A and SigK (Hilbert and Piggot 2004) are known to functionally interact with more recent proteins. Such regulatory proteins may have existed prior to the emergence of sporulation and could have been repurposed for a new function when sporulation emerged, via new protein–protein interactions. For example, the gene for Sda, a protein that mediates the phosphorylation status of Spo0A (Hilbert and Piggot 2004) is found in PS9. The gene for BofC, a protein that controls maturation of SigK (Hilbert and Piggot 2004), is found in PS8. Spo0E, a very important negative regulator of sporulation (Hilbert and Piggot 2004), is encoded by a gene from PS10. These examples indicate that regulation of sporulation continued evolving long after its emergence. There are several well-established cases of complex developmental processes where most of the related genes emerged in a single phylostratum (Trigos et al. 2017). Sporulation seems to present an exception to this rule, with many of the B. subtilis sporulation genes being very old (PS1 and PS2), and a significant amount of them emerging later in evolution, clustering in PS8–10. Sporulation is by no means the only exception to this rule. For example, genes involved in biofilm and competence development in B. subtilis display a very similar pattern. Most biofilm genes are found in PS1, but genes for some important biofilm regulators are found in more recent PS. For example, BslA, an inhibitor of KinA autophosphorylation, is encoded by a gene in PS8 (Cairns et al. 2014). SlrA (negative regulator of SlrR), SinI (negative regulator of SinR) (Cairns et al. 2014), and DegQ (stimulator of DegU-dependent phosphorylation by DegS) (Do et al. 2011) are all encoded by genes in PS10. Similar distribution is found for competence-related genes, with most of them being in PS2, but the master regulator ComK (Schultz et al. 2009) is encoded by a gene in PS8. It seems that, in bacteria, the emergence time-points of genes involved in the same process may differ.
Selection of Uncharacterized Genes from Sporulation-Enriched PS for Phenotype Testing
Sequenced bacterial genomes are functionally annotated to a large extent, but roughly 20–25% of bacterial genes remain uncharacterized. Therefore, new genome-mining approaches are still needed to tap into this unexploited potential. Several studies based on phylostratigraphy suggested that genes related to any given developmental process are enriched in the PS where the process originated (Trigos et al. 2017). We therefore asked whether this enrichment could be used to infer functions of presently uncharacterized genes. In B. subtilis, the bulk of sporulation genes originated in PS1, but the sporulation process in its present form critically depends on the sporulation genes that appeared in PS2 and PS8–10. We hypothesized that these sporulation-enriched PS might be a good place to look for new sporulation genes. To test this hypothesis, we tested whether the currently uncharacterized genes from PS2 and PS8–10 might be involved in sporulation. Bacillus subtilis PS2 and PS8–10 contain 249 uncharacterized genes. However, not all the corresponding proteins can be automatically assumed to be present in the cell during sporulation. If a protein could not be detected during sporulation, it is unlikely to be involved in the process. Hence, in order to narrow down our selection, we decided to work with a subset of genes whose proteins could be experimentally detected during sporulation, using either mass spectrometry (MS) proteomics or N-terminal GFP fusions (fig. 1B and supplementary fig. S1A, Supplementary Material online). This expression check was used as a “yes–no” cut-off criterion for selecting candidates for further testing, and it should not be confused with the notion of differential expression. This procedure yielded a set of 37 candidate genes (fig. 1B and C). In conclusion, all of the selected candidates satisfy the following criteria: 1) they are uncharacterized, and specifically, no experimental evidence exists in the literature about their mutation leading to a measurable sporulation phenotype, and 2) their proteins can be detected in a sporulating B. subtilis culture.
Some authors have argued that genes with similar transcription profiles tend to be involved in similar cellular processes, and by consequence, have suggested that gene transcription profiles can be used to predict gene function (Wu et al. 2002; Kim et al. 2016). This principle has also been used for some annotations of sporulation genes in SubtiWiki (Zhu and Stülke 2018) and SporeWeb (Eijlander et al. 2014), based on a large transcriptional study performed on B. subtilis. Among our 37 candidate genes, there are approximately equal subsets of genes with and without differential transcription during sporulation, according to Nicolas et al. (2012). About 18 of the selected candidates showed no differential transcription during sporulation (fig. 1C left panel) and 19 showed a more- or less-pronounced differential transcription. Of the latter, nine exhibited strong differential transcription during sporulation (fig. 1C middle panel) and ten weak differential transcriptions (fig. 1C right panel). It should be noted that among the 37 candidate genes, only 2 are in operons containing known sporulation genes: yvcB and yjaU (supplementary fig. S1B, Supplementary Material online).
About 43% of Candidates from Sporulation-Enriched PS Exhibit a Sporulation Phenotype
All candidate genes (fig. 1B) were inactivated using a seamless gene inactivation procedure (minimizing polar effects). Growth profiles of the resulting mutant strains indicated no major effects on vegetative growth (supplementary fig. S2, Supplementary Material online). The only exception was ΔykzB, which grew at a normal rate but reached stationary phase already at OD600 of 1.2, whereas the other mutants and the wild type reached stationary phase at OD600 ∼1.7. The time-point when exponential growth ceases in the sporulation medium is considered as the sporulation initiation point (T0). T0 was not affected in any of the mutant strains. Spores become visible by phase contrast microscopy at T5 (5 h after T0) and start appearing as mature at T7 (7 h after T0) (Hilbert and Piggot 2004). Thus, to capture not only the capability of spore development but also the kinetics, we examined the fraction of spore forming cells by phase contrast microscopy at time-points T5, T7, and T9 (fig. 2A). Wild type was used as a reference, and a knockout strain of a known sporulation inhibitor, Δspo0E (Hilbert and Piggot 2004), was used as a positive control. As shown in figure 2A, 12 mutants exhibited a statistically significant sporulation phenotype in this assay (P < 0.05), with 9 mutants developing more and 3 mutants fewer spores than the wild type. In addition to the number of visible forespores, the ability of mutants to develop functional heat-resistant spores was examined at T20 (fig. 2B). In this assay, 12 mutants exhibited a statistically significant phenotype (P < 0.05), with 3 strains developing more and 9 strains fewer heat-resistant spores than the wild type. Interestingly, only six mutants had a similar phenotype pattern in both assays. ΔyscB, ΔygaB, and ΔykqC had more visible forespores and heat-resistant spores than the wild type, and we classified them as mutant category I (fig. 2C). ΔygzC, ΔyqkB, and ΔyizD developed fewer visible forespores and heat-resistant spores than the wild type, and were classified as mutant category II (fig. 2C). For the remaining mutants, the total forespore and heat-resistant spore counts were not in agreement (categories III and IV). Category III mutants (fig. 2C) produced more visible forespores than the wild type but exhibited only wild type or inferior levels of heat-resistant spores. Category IV mutants (fig. 2C) had wild-type levels of visible spores, but subwild-type levels of heat-resistant spores. In categories III and IV, the subpopulation of cells that are committed to sporulation at T5–9 is either larger than the population of forespores that successfully complete sporulation by T20, or their sporulation process results in structurally/functionally compromised spores. As structural defects were suspected for categories III and IV, we examined spores made by those mutants under a scanning electron microscope (SEM) (supplementary fig. S2C, Supplementary Material online). This revealed extensive spore morphology defects, such as distortions and cavities for ΔyizC, ΔyvcB, ΔycjQ, ΔykzB, ΔypmP, ΔyueH, ΔyrzL, ΔyphF and ΔyybN, and leakage of spore content from spores of ΔyusN. Overall, out of 37 experimentally tested candidates from sporulation-enriched PS, 16 (43%) exhibited a clear sporulation phenotype in either of the assays. As previously mentioned, roughly one-half of the tested candidates exhibited differential transcription during sporulation (fig. 1B and C), whereas the others were not differentially transcribed. The percentage of verified sporulation phenotypes was very similar in both categories. Among the 18 candidates with no differential transcription during sporulation, 8 (44%) had a sporulation phenotype, whereas 8 out of 19 (42%) candidates with differential transcription tested positive. The success rate was identical for nondifferentially and differentially transcribed genes. Therefore, our results do not support the notion that transcriptional profiling is sufficient to define a sporulation phenotype.
Fig. 2.
Sporulation phenotypes of strains with individual gene knockouts. (A) Total forespore counts observed by phase contrast microscopy 5, 7, and 9 h after initiation of sporulation. Counts are expressed as the log2 ratio of cells with visible spores/total cells for each strain, normalized with respect to the wild type. (B) Heat-resistant spore counts are shown for each strain, expressed as the log2 ratio of the heat-resistant spores/total cells, normalized with respect to the wild type. In (A) and (B), ΔsigF was used as negative control (zero spores, not shown) and Δspo0E as positive control (hypersporulating strain). The t-test result is indicated by **P < 0.05 or ***P < 0.01. (C) Summary of sporulation phenotypes for all examined strains. Phenotype of more forespores/heat-resistant spores compared with the wild type is indicated in red, and fewer forespores/heat-resistant spores are indicated in green. No significant phenotype is indicated with gray. Roman numerals I–IV indicate four classes of mutant phenotypes, described in the main text.
Sporulation Phenotypes in Regulation of Timing, Coat Assembly, and Prophage Switch
We performed some follow-up experiments in order to assess which types of phenotypes appear in different categories of our mutants, although a full characterization of molecular mechanisms by which the candidate genes influence sporulation is outside the scope of this study. The following genes were subjected to further investigation: ΔygaB (category I), ΔyizD (category II), ΔykzB (category III), and ΔyphF (category IV). The selected genes had the most pronounced phenotypes in their respective categories. ΔygaB produced the highest level of visible and heat-resistant spores in category I; ΔyizD produced the second lowest count of visible and heat-resistant spores in category II, but with the strongest defect in terms of heat-resistant spores (difference between the level of visible and heat-resistant spores); ΔykzB mutant produced the highest counts of visible spores in category III; and ΔyphF was the strain with the lowest counts of heat-resistant spores in category IV. All of these grew at a similar rate as the wild type, all except ΔykzB entered sporulation at a similar cell density, and all initiated sporulation at the same time as the wild type (supplementary fig. S2A, Supplementary Material online). As sporulation depends on a complex transcriptional regulation, we performed a transcriptome analysis of these mutants grown in the sporulation medium, at T0 and T3 (supplementary table S2, Supplementary Material online and fig. 3A). Principal component analysis revealed that ΔykzB was the only strain with a global perturbation of the transcriptome at both time-points of T0 and T3 (supplementary fig. S3, Supplementary Material online). ΔygaB global transcription was somewhat affected at T0 but not T3, and the two remaining mutants clustered with the wild type at both time-points (supplementary fig. S3, Supplementary Material online). The transcriptome data cover only sporulation events up to T3, because we found that it difficult to isolate good mRNA quality samples from later stage spores. To follow the transcription of key sporulation genes from T4 onward, we focused on later stage sporulation sigma factors, SigG and SigK through promoters responsive to these sigma factors, sspB and gerE, respectively (expression monitored using promoter–YFP/CFP fusions, fig. 3B).
Fig. 3.

Regulation of sporulation genes in ΔygaB, ΔyizD, ΔykzB, and ΔyphF strains. (A) Differentially expressed genes involved in sporulation in ΔygaB, ΔyizD, ΔykzB, and ΔyphF compared with wild type at sporulation T0 and T3. The list of genes was derived from SubtiWiki (Category: 4. Lifestyles-Sporulation). The fraction of differentially expressed genes in each category is presented in red for up-regulated genes and in green for down-regulated genes. (B) Genetic context of reporter gene fusions and fluorescence microscopy of wild-type Bacillus subtilis cells expressing YFP from a SigG-dependent sspB promoter and CFP from a SigK-dependent gerE promoter. The entire cells are visible in bright field images, the cell membranes are visualized by red fluorescence (FM4-64 staining), and yellow and cyan fluorescence represent reporter gene activity. (C) Left panel: the activity of SigG and SigK in mutant strains, expressed as the fraction of cells with yellow (sspB promoter) or cyan (gerE promoter) fluorescence/total cells, normalized with respect to the wild type. The t-test result of protein activity for each strain is indicated by **P < 0.05 or ***P < 0.01, individually. Central and right panel: the transcription of sigG, sigK, sspB, and gerE, in mutant strains during sporulation at T4 and T5, detected by quantitative reverse-transcriptional PCR. The change of gene expression in mutants versus wild type is expressed as log2 ratio. The t-test result of gene transcription is indicated by ***P < 0.01. The P values of gene transcription for four different genes in four different mutants all fall into the same category, and are thus indicated only once for each mutant.
Class I mutant ΔygaB produced a progressively increasing proportion of forespores at T5–T9 and more heat-resistant spores at T20 compared with the wild type (fig. 2). Overexpression of ygaB led to an opposite effect (fig. 4), suggesting that YgaB antagonizes sporulation. At T0, the expression of genes in all sporulation regulons was perturbed in ΔygaB (fig. 3A). About 55% of genes involved in the sporulation phosphorelay were up-regulated, along with most of the genes in regulons of KipR, RghR, and Spo0A (supplementary fig. S4A, Supplementary Material online). Sporulation master regulator Spo0A and sigma factors SigF and SigG were also overexpressed (supplementary fig. S4B, Supplementary Material online). This global perturbation in expression of sporulation genes disappeared to a large extent by T3 (fig. 3A). However, at later stages of sporulation (T4–T5), the ΔygaB strain still exhibited a slightly higher proportion of cells with active SigG and SigK (expression of YFP/CFP from sspB and gerE promoters) (fig. 3B and C), accompanied by increased levels of SigG and SigK expression (qRT-PCR) (fig. 3C). This probably accounts for the gradual increase of visible spore counts between T5–T9 in ΔygaB (fig. 2). Our findings suggest a strongly elevated commitment of ΔygaB cells to sporulation at T0 (timing of entry into sporulation not affected), leading to an increased number of functional spores at the end of the sporulation process. It is presently not clear how YgaB antagonizes sporulation. YgaB does not have any discernable DNA-binding domain, so presumably, the effect on transcription is indirect. One possible explanation comes from the protein–protein interaction profile of YgaB (Marchadier et al. 2011). It has been reported that YgaB physically interacts, among others, with two proteins involved in spore development: SpoVR (Beall and Moran 1994) and CtpB (Hilbert and Piggot 2004). It is not immediately apparent whether SpoVR interaction may contribute to the observed ΔygaB phenotype. CtpB is known to cleave SpoIVFA, alleviating inhibition on SpoIVFB, and leading to the production of functional mature SigK, which was found to be excessively activated in ΔygaB. Interestingly, SpoVR is a mother cell protein (Beall and Moran 1994), and CtpB acts in forespore and intracellular space between the mother cell and the forespore (Mastny et al. 2013). Our localization studies with GFP-YgaB indicated that the protein is uniformly distributed in the mother cell and the forespore (fig. 5A).
Fig. 4.
The effect of strains with overexpressed ygaB, yizD, ykzB, and yphF on spore development. (A) Total forespore counts observed by phase contrast microscopy at 5, 7, and 9 h after initiation of sporulation. Counts are expressed as the log2 ratio of cells with visible spores/total cells for each strain, normalized with respect to the wild type. (B) Heat-resistant spore counts are shown for each strain. Counts are expressed as the log2 ratio of the spores/total cells, normalized with respect to the wild type. The t-test result for overexpression strains is indicated by **P < 0.05 or ***P < 0.01.
Fig. 5.
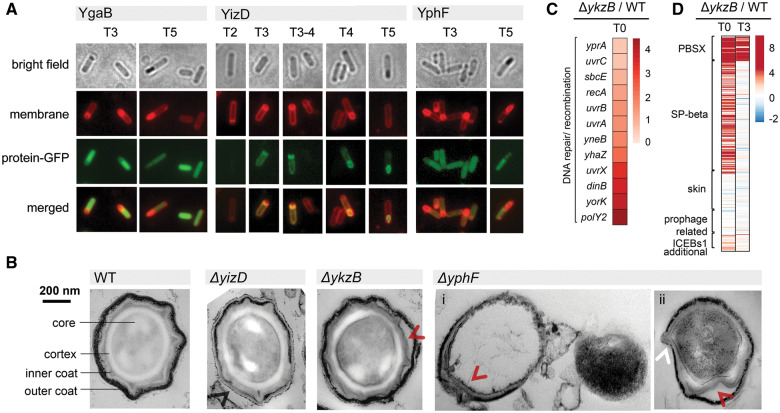
Function of ygaB, yizD, ykzB, and yphF during sporulation. (A) Localization of YgaB, YizD, and YphF during sporulation: for YgaB and YphF at T3 and T5 and for YizD between T2 and T5. The entire cells are visible in bright field images, the cell membranes are visualized by red fluorescence (FM4-64 staining), and green fluorescence represent the localization of protein-GFP. (B) Thin section transmission electron microscopy of spores developed by ΔyizD, ΔykzB, and ΔyphF strains. Spores were isolated by lysozyme treatment for ΔyizD, ΔykzB, and ΔyphF (i) and ΔyphF (ii) was isolated without lysozyme treatment. Spore core, cortex, inner coat, and outer coat are indicated in a control image from the wild type. Abnormalities in spore architecture are indicated by arrows: white arrow indicates thinner outer coat, red arrow indicates space between outer and inner coat, and black arrow indicates the leakage of spore content from outer coat. (C) Heatmap of differential expression of genes involved in DNA quality control in ΔykzB strain compared with wild type at T0. Strain name is indicated under each column. (D) Heatmap of differential expression of prophage-related genes in ΔykzB strain compared with wild type at T0 and T3, respectively.
Class II mutant ΔyizD produced fewer functional spores than the wild type, and its spores exhibited visible structural defects under SEM (supplementary fig. S2C, Supplementary Material online). Overexpression of yizD led to a moderate increase in numbers of visible forespores and heat-resistant spores compared with wild type (fig. 4), without any effect on vegetative growth (supplementary fig. S2B, Supplementary Material online), confirming that YizD contributes positively to the sporulation process. Localization assays with GFP-YizD suggested that this protein is expressed in the mother cell (supplementary fig. S1A, Supplementary Material online). This is consistent with a previous study by Arrieta-Ortiz et al. (2015), which identified yizD as a novel SigE-dependent gene. However, YizD possesses two predicted transmembrane helices (supplementary fig. S5, Supplementary Material online), and the N-terminal fusion with GFP that was used in our initial localization screen could have compromised the correct insertion of the protein in the membrane. We therefore expressed full-length YizD fused to the GFP via its C-terminus and observed that YizD-GFP is localized in the mother cell, with a propensity to accumulate in the outer membrane of the forespore from T3 to T5 (fig. 5A). Concomitant with the start of YizD accumulation in the outer membrane of the forsepore at T3, in ΔyizD, we observed strong down-regulation of genes belonging to the SigE, SigF, SigG, SigK, SpoIIID, SpoVT, and GerE regulons (Hilbert and Piggot 2004) (supplementary fig. S4A, Supplementary Material online). Interestingly, the expression of the corresponding sigma factors and master regulators was not significantly affected in the mutant (supplementary fig. S4B, Supplementary Material online). At T5, the proportion of cells with active SigK was significantly lower in ΔyizD (fig. 3C). Accordingly, the levels of sigK transcripts, and gerE transcripts known to be under SigK control, were also lower (fig. 3C). SigK is known to control the expression of a large fraction of spore coat proteins (Hilbert and Piggot 2004). As ΔyizD spores exhibited visible defects under SEM (supplementary fig. S2C, Supplementary Material online), we examined their structure further, using transmission electron microscopy (TEM). This revealed that ΔyizD spores have a considerably less-dense outer spore coat compared with the wild type, and exhibit loose adhesion of inner and outer coats, resulting in a visible interspace (fig. 5B). Overall, these results suggest that the SigE-dependent transmembrane protein YizD plays a role in the regulation of synthesis and/or assembly of the spore coat, acting from the outer membrane of the forespore on the mother cell side.
Class III mutant ΔykzB produced an increased number of visible forespores, but only a fraction of them resulted in heat-resistant spores, meaning that the overall numbers of heat-resistant spores corresponded roughly to the wild-type levels (fig. 2). Consistently, overexpression of ykzB was found to inhibit sporulation, without any effect on vegetative growth (fig. 4 and supplementary fig. S2B, Supplementary Material online). A microscopy examination of GFP-YkzB indicated that the protein was weakly expressed and exhibited no specific localization during sporulation (supplementary fig. S1A, Supplementary Material online). Some smaller morphological defects in ΔykzB spores were observable by SEM (supplementary fig. S2C, Supplementary Material online) and TEM (fig. 5B). The spores presented fairly normal architecture and thickness of the inner and outer coats, with sporadic loose adhesion of the coats observable in limited areas. In terms of the global transcriptome, ΔykzB was most severely affected of all the examined strains. Although expression of genes related to sporulation in ΔykzB was not affected at T0, it was strongly perturbed at T3 (fig. 3A). Genes related to late sporulation, including spore coating and small acid-soluble spore proteins belonging to SpoVT and GerE regulons were up-regulated, whereas those related to spore initiation in RghR and Spo0A regulons were down-regulated (supplementary fig. S4A, Supplementary Material online). A corresponding pattern was observed in the transcriptional profiles of the mentioned regulators themselves (supplementary fig. S4B, Supplementary Material online). Overall, at T3, the transcription profile of ΔykzB corresponded to changes expected to occur in the wild type at T5, indicating a timely start but accelerated progress of sporulation. At T4 and T5, ΔykzB strain exhibited a higher proportion of cells with active SigG and SigK, accompanied by increased levels of transcription and activation of SigK and SigG (fig. 3C). The accelerated process of sporulation in this strain resulted in more visible forespores, but these do not all develop into fully heat-resistant spores, indicating that acceleration compromises the proper maturation of spores. Accordingly, ΔykzB also exhibited an upshift in expression of genes which are known to participate in quality control of DNA packaged into spores (fig. 5C), such as, for example, recA and uvrC (Setlow B and Setlow P 1996). Interestingly, inactivation of ykzB exhibited another feature not observed in other mutants: it induced overexpression of genes of prophage PBSX, SP-β, and mobile genetic elements ICEBs1 at T0 (fig. 5D). It is known that the excision of the SP-β prophage reconstitutes the gene spsM, which is essential for spore envelope maturation (Abe et al. 2014). It has been argued that prophage induction and sporulation are connected in evolutionary and regulatory terms (Lewis et al. 1998). YkzB seems to be important for coordinating these two processes, and in its absence, both prophage induction and sporulation get accelerated.
Class IV mutant ΔyphF produced a similar number of visible forespores as the wild type, but its spores were not heat resistant and exhibited severe structural defects (fig. 2). Overexpression of yphF resulted in a moderate but statistically significant increase in numbers of visible forespores and heat-resistant spores (fig. 4). Similar to ΔyizD, ΔyphF presented very few changes in the overall expression of sporulation genes at T0 and T3 (fig. 3A and supplementary fig. S4, Supplementary Material online). Those changes concerned mostly the genes encoding spore coat proteins at T3: spoIVA, cotW, yisJ, spoVIF, cotS, ytrI, cotZ were down-regulated, and spoVS and spoVM were up-regulated (supplementary fig. S6, Supplementary Material online). SpoIVA is the primary factor in attaching the spore coat to the outer forespore membrane, acting via SpoVM (McKenney et al. 2013). All the other affected genes also contribute to spore coating at different levels, from assembly to anchoring (Driks 1999; Kuwana et al. 2003; McKenney et al. 2013). Deregulation of the spore coat assembly implied abnormal coating of ΔyphF spores, which was confirmed by TEM observations (fig. 5B). ΔyphF spores lost the normal coating architecture; they exhibited an incomplete outer coat and large-scale deformations of the inner coat. Moreover, ΔyphF spores were sensitive to lysozyme (fig. 5B), which is consistent with the known phenotype of ΔspoVIF (Kuwana et al. 2003). The genes found to be differentially expressed in ΔyphF: ytrI and spoIVA are SigE regulated genes, spoVIF and cotS are SigK regulated genes, and cotW and cotZ are regulated by both SigE and SigK (Eichenberger et al. 2003; Steil et al. 2005). As the expression of sigE was not affected in ΔyphF at T3, we checked the sigK expression and activity at T4 and T5 (fig. 3C). Although the proportion of cells with active SigK and SigG was same as in the wild type at T4 and slightly reduced at T5, the expression of sigG, sigK, SigG-dependent sspB and SigK-dependent gerE was reduced in the mutant strain at both T4 and T5. Unlike YizD, YphF-GFP did not exhibit a spore-specific localization. At time-points T3–T5, YphF–GFP fusion localized to the cytosol, and was present both in the mother cell and in the forespore at T3, gradually disappearing from the forespore toward T5 (fig. 5A). YphF is known to be a part of the AbrB regulon (Chumsakul et al. 2011), and therefore likely to peak out early in sporulation, when AbrB synthesis is repressed by Spo0A. ΔyphF phenotype shares some similarities with ΔyizD (SigK deregulation, spore coating affected), with a few significant differences: most of the deregulated genes code for spore coat proteins and the coating perturbation is more severe than in all other examined mutants (lysozyme sensitivity, larger structural defects under TEM). It is presently unclear how YphF affects the expression of coating genes.
Conclusion
Overall, 43% of the tested candidate genes were found to have a sporulation phenotype in this study. Our results suggest that PS enriched for known sporulation genes are a good place to look for new sporulation genes. This suggests that genomic phylostratigraphy might offer a viable route for predicting functions of uncharacterized genes and thus contribute to genome mining. The approach is complementary to the currently employed procedures for assigning functions to uncharacterized genes, which typically depend on homology searches based on primary (Traag et al. 2013) or tertiary structures (Roy et al. 2010), experimental functional screens (Meeske et al. 2016), and in some cases, on transcriptional profiling (Wu et al. 2002; Kim et al. 2016). In our study, differential transcription during sporulation did not correlate strongly with sporulation phenotypes. The prediction success rate was in fact identical for differentially and nondifferentially transcribed genes. Regarding operon colocalization as a prediction criterion used for bacterial genomes, one candidate gene (yvcB) localized in the same operon with a known sporulation gene tested positive, and the other one (yjaU) tested negative. This could be explained by the fact that yjaU is separated from the known sporulation gene by an alternative terminator and yvcB is not. In searchers based only on transcriptional profiling and operon colocalization, roughly 50% of the candidates detected in our study would have been missed. Equally, it should be noted that 20 of the candidate genes were shorter than 300 bp (fig. 1B), and therefore also likely to be missed by classic experimental screening methods based on random gene interruption. Therefore, we would like to argue that phylostratigraphy showed a complementary value with regards to other types of profiling.
To gain an overview of different types of phenotypes related to sporulation genes discovered by phylostratigraphy, we performed some follow-up studies with genes from four difference categories of mutants: ygaB, yizD, ykzB, and yphF. The four genes seem to be involved in sporulation in very different ways. Phenotyping of the category I mutant ΔygaB indicated that this protein acts as a dampener of sporulation, and in its absence many sporulation phosphorelay genes are overexpressed at T0. However, the effect seems to be transient, with near wild-type expression levels at T3 and onward. Category II mutant ΔyizD produced fewer functional spores than the wild type, and its spores exhibited structural defects. YizD seems to be involved in modulating the levels of SigK, which could account for its spore coat phenotype (Hilbert and Piggot 2004). At present, an additional, more direct functional role cannot be excluded. In ΔykzB strain (category III mutant), sporulation was accelerated but resulted in fewer functional spores, possibly due to a miss-regulation of the SP-β prophage switch. YphF (category IV) seems to be important for the expression of spore coat proteins, and thus contributes to spore architecture. One overarching motif in all observed phenotypes is that they involve a rather high level of regulation. The candidate proteins most probably do not have a direct enzymatic or structural role in sporulation, nor are they likely to regulate expression of sporulation genes by binding to DNA regulatory elements directly (DNA-binding motifs absent in all cases). Their phenotypes are moderate, and in no case is sporulation completely abolished. Rather, these proteins seem to act through protein–protein interactions or other presently unknown pathways, to tweak and modulate the expression or activity of key sporulation regulators. This, in addition to most of them being short genes and thus not amenable for knockouts by insertion, probably accounts for the fact that they were not picked up as sporulation genes in previous functional screenings.
Our study demonstrated that phylostratigraphy can contribute to identifying new sporulation genes in B. subtilis. To get a clearer picture of how useful this method can be for genome mining, it would be interesting to examine whether the function-predictive power of phylostratigraphy might extend to other types of developmental phenomena and other species of sequenced microorganisms. If so, it could become an important tool in genome mining and annotation pipelines. As for the results of the current study, further characterization of the identified sporulation genes will be needed to gain full mechanistic insight in their role in sporulation.
Materials and Methods
Phylostratigraphy Analysis of B. subtilis Sporulation Genes
For all sporulation genes listed in either SubtiWiki (Zhu and Stülke 2018) or SporeWeb (Eijlander et al. 2014), primary literature was checked for documented sporulation phenotypes. Our requirement for a sporulation phenotype was that a gene knockout or gene overexpression has been shown to lead to a measurable change in the sporulation process, including but not limited to changes in total spore counts, percentage of resistant spores, structural integrity of spores or sporulation timing. Sole evidence of differential transcription of a gene during sporulation was not considered as sufficient to assign a sporulation phenotype. By this criterion, we performed a manual literature search of phenotype data for all entries in the two sporulation databases. Distribution of resulted genes in the previously published B. subtilis phylostratigraphy map (Ravikumar et al. 2018) was analyzed.
Bacterial Strains and Growth Conditions
Cells were routinely grown in LB, containing chloramphenicol 5 µg/ml, phleomycin 2 µg/ml, neomycin 5 µg/ml for B. subtilis, or ampicillin 100 µg/ml for Escherichia coli, as appropriate. Escherichia coli DH5α was used for plasmid construction. Bacillus subtilis strains BS33 (TF8a λPr-neo::ΔaupP), in which the neomycin-resistance gene is under the control of the Lambda Pr promoter (Bidnenko et al. 2013), were used for construction of seamless knockout mutants.
Genetic Manipulation of B. subtilis
All PCR primers used in this study are listed in supplementary table S3, Supplementary Material online. Construction of the seamless knockout mutants in B. subtilis was performed using the modified mutation delivery method described previously (Bidnenko et al. 2013). Briefly, the DNA fragments flanking the target gene were amplified from the primer pairs gene_1/gene_2 and gene_3/gene_4, and fused to both ends of the phleomycin-resistance cassette with primers gene_1 and gene_4 through joining PCR. The resulting fragment was used to transform BS33 cells, and transformants were selected for phleomycin resistance and neomycin sensitivity. Counter-selection for phleomycin sensitivity and neomycin resistance was applied to select clones which had lost the insertion cassette from the chromosome via recombination between the flanking direct repeats. The obtained seamless knockout mutants were verified by PCR using the primer pairs gene_5 and gene_6.
To examine the translation of the candidate genes, whose proteins were not identified by MS, the native promoters of these genes with the initial 30–60 bp of their corresponding reading frames were fused to a gfp gene without the start codon. The gfp was amplified from pSG1729 (Lewis and Marston 1999) using primer pairs GFP_1/GFP_2, and inserted into pBS1C (Radeck et al. 2013) (using XbaI and PstI restriction sites) to generate the vector pBS1C-GFP. The DNA fragments harboring the promoter and the first 30–60 bp of the candidate genes were amplified with the primer pair gene_GFP_1/gene_GFP_2, and inserted into the pBS1C-GFP (using NotI and XbaI restriction sites).
To examine the activity of sigma factors SigG and SigK, we constructed the fusion of the sspB promoter with the CFP (cyan fluorescent protein)-encoding gene, and the gerE promoter with the YFP (yellow fluorescent protein)-encoding gene. Primer pairs sspB_1/sspB_2 and gerE_1/gerE_2 were used to amplify the indicated promoters, and the PCR products were inserted into pICFP/pIYFP (Veening et al. 2004) (using ApaI and EcoRI restriction sited). Primer pairs sspB_3/sspB_4 and gerE_3/gerE_4 were used to amplify the fusion of sspB promoter-cfp and gerE promoter-yfp obtained in the previous step. The obtained DNA fragments were then inserted into pBS1C (using XbaI and PstI restriction sites). The resulting plasmids were used to transform BS33 cells, and transformants were selected for chloramphenicol resistance and amyE deficiency.
Sporulation Assay
Sporulation was induced as described previously (Bidnenko et al. 2013). Briefly, cells grown overnight on solid DSM medium were inoculated in preheated liquid DSM at OD600 0.05 and incubated at 37 °C with shaking of 200 rpm until end of exponential growth, which time-point was considered as sporulation initiation (T0). Sporulation assays were performed as described previously (Bidnenko et al. 2013). For examination of heat-resistant spores, sporulation-induced samples were kept growing for 20 h after T0. Half of the culture was plated directly in dilutions on LB plates, and the other half was treated at 80 °C for 10 min before plating. Colonies were counted after 24 h of incubation at 37 °C. The capability to develop heat-resistant spores was calculated as the ratio of colonies forming units in heat-treated versus untreated samples, normalized with respect to the wild type. Three independent experiments were performed. For the examination of visible forespores, cells were taken from sporulation-induced samples at time-points T5, T7, and T9 (5, 7, and 9 h after T0), and examined by the Leica DMI4000 B microscope using phase contrast. The capability to develop visible forespores was calculated as the ratio of cells with visible forespores to total cells, normalized with respect to the wild type. Three independent experiments were performed for each strain, and a minimum of 500 cells were examined for each sample.
Scanning Electron Microscopy
Spores were purified by lysozyme treatment and salt and detergent washes as described previously (Nicholson and Setlow 1990). Briefly, sporulation-induced cells were harvested 24 h after T0, and incubated at 37 °C for 1 h in 50 mM Tris–Cl, pH 7.2 with 50 µg/ml lysozyme. Cells were then subjected to five washes in 1 M NaCl, deionized water, 0.05% SDS, 50 mM Tris–Cl, pH 7.2. Spores were fixed overnight with 3% glutaraldehyde. The fixed spores were dehydrated with a series of washes with increasing ethanol concentration (40%, 50%, 60%, 70%, 80%, 90%, and 100%) for 10 min each. Thin films were prepared by using dehydrated spores on cover glass and dried for 2 h at room temperature. The dried samples were sputter coated with gold (5 nm) before imaging. SEM imaging was performed with the Supra 60 VP microscope (Carl Zeiss AG).
Transmission Electron Microscopy
TEM was performed as described previously (Korch and Doi 1971). In brief, pure spores isolated by lysozyme from ΔyizD, ΔykzB, and ΔyphF, and sporulation culture of ΔyphF fixed in modified Karnovsky fixative (2.5% glutaraldehyde, 2% formaldehyde, 0.02% sodium azide in 0.05 M Na-cacodylate buffer). Fixation was microwave-assisted using Leica AMW. The samples were then incubated in 1.5% potassium ferrocyanide/2% osmium tetroxide in cacodylate buffer, dehydrated, and infiltrated with Durcupan resin in Leica EM AMW. The samples were finally polymerized in BEEM capsules. All the samples were handled identically. Ultrathin sections (70 nm) were imaged with a LEO 912 OMEGA TEM.
Fluorescence Microscopy
Cells were examined during vegetative growth, and at time-points T3 and T5 during sporulation. Samples were observed through a LeicaCTR4000 inverted microscope. For examination of the activity of SigG, cyan fluorescence was detected, and for SigK, yellow fluorescence was detected. Cells were stained with FM4-64 (Molecular Probes) to visualize the cell membrane. Three independent experiments were performed for each strain, and a minimum of 500 cells were examined for each sample.
Mass Spectrometry Analyses
Sporulation-induced samples were harvested at T4 and resuspended in 100 mM Tris, pH 8.0 with 10 mM EDTA and 4% (w/v) SDS. Cell lysates were produced by mechanical glass bead disruption and proteins were precipitated from the crude extract using chloroform and methanol. The protein pellet was dissolved in 6 M urea, 2 M thiourea in 10 mM Tris–HCl pH 8.0. About 100 µg of the purified protein extract was digested either in-solution or in-gel. Samples taken for in-solution digestion (Ravikumar et al. 2014) were reduced for 1 h at room temperature with 1 mM dithiothreitol and alkylated for 1 h in the dark with 5.5 mM iodoacetamide (IAA). Proteins were next predigested with LysC (Wako Chemicals GmbH) for 3 h followed by overnight digestion with Trypsin (Promega), at room temperature. Peptides obtained as a result were fractionated based on their differences in their isoelectric point using the 3100 Offgel Fractionator (Agilent). Samples taken for in-gel digestion were separated on a NuPage Bis–Tris 4–12% gradient gel (Invitrogen) based on the manufacturer’s instructions. Fractions were processed and stage-tipped as described earlier (Ravikumar et al. 2014), prior to being loaded onto the mass spectrometer.
The MS analyses were performed on an QExactive HF mass spectrometer interfaced with Easy-nLC1200 liquid chromatography system (Thermo Fisher Scientific). Peptides were trapped on an Acclaim Pepmap 100 C18 trap column (100 μm×2 cm, particle size 5 μm, Thermo Fischer Scientific) and separated on an in-house constructed C18 analytical column (300×0.075 mm I.D., 3 μm, Reprosil-Pur C18, Dr Maisch) using the gradient from 6% to 38% acetonitrile in 0.2% formic acid over 45 min followed by an increase to 80% acetonitrile in 0.2% formic acid for 5 min at a flow of 300 nl/min. The instrument was operated in data-dependent mode, where the precursor ion mass spectra were acquired at a resolution of 60,000, and the ten most intense ions were isolated in a 1.2-Da isolation window and fragmented using collision energy HCD settings at 28. MS2 spectra were recorded at a resolution of 30,000, charge states 2–4 were selected for fragmentation, and dynamic exclusion was set to 20 s. An inclusion list with a tolerance of 10 ppm including theoretical tryptic peptides of the proteins of interest was additionally used.
Acquired MS spectra were processed with MaxQuant v 1.5.1.0 (Cox and Mann 2008). Database search was performed against a target-decoy database of B. subtilis 168 from UniProt (taxonomy ID 224308). Endoprotease Trypsin/P was fixed as the protease with a maximum missed cleavage of two. A false discovery rate of 1% was applied at the peptide and protein level.
RNA Sequencing
Cells were harvested from sporulation-induced samples at time-points T0 and T3. Total RNA extraction was performed through RNeasy Mini Kit (Qiagen), and the quality examination was performed by Bioanalyzer (Agilent) through Agilent RNA 6000 Nano Kit. After removing rRNA from total RNA samples through Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina), library was prepared through TruSeq mRNA stranded Bacteria HT (Illumina). RNAseq was performed by paired end reads with 75 bp from each end, and >100 times coverage per samples, at Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark.
RNAseq Data Processing
Pair-ended Illumina RNA sequences were mapped on the reference genome of Bacillus subtilis subsp. subtilis str. 168 (NCBI Assembly accession: ASM904v1; GCF_000009045.1) using BBMap V37.66. After mapping, a quality check was performed using FastQC V0.11.7. In total, 269,675,258 pair-ended 75-bp sequences were mapped on the reference genome. Each mutant/wild type was represented with sequences from three technical replicates, respectively. Samples were represented between 6,514,042 and 12,829,840 reads. The SAMtools package V1.6 (Li et al. 2009) was used to generate, sort, and index BAM files for downstream data analysis. Subsequent RNAseq data processing was performed in R V3.4.2. (R Core Team 2017) using a custom-made scripts, utilizing the Bioconductor package DESeq2 V1.18.1 (Love et al. 2014). The count matrix, containing the number of reads overlapping each gene in the samples, was generated using the “Union” mode of the summarizeOverlaps function in R package GenomicAlignments V1.14.2 (Lawrence et al. 2013).
PCA Analysis
The principal component analysis was performed in R with the Bioconductor package DESeq2 V1.18.1(Love et al. 2014) and plotPCA function using raw expression counts previously transformed to the log2 scale with the rlog function of the same package. PCA plot was made in R package ggplot2 V2.2.1 (Wickham 2016) with the function ggplot.
Supplementary Material
Acknowledgments
We are grateful to Sara Koska for her help with transcriptome data analysis. We acknowledge the Centre for Cellular Imaging at the University of Gothenburg, Sweden and the National Microscopy Infrastructure, NMI (VR-RFI 2016-00968) for helping with TEM. The Novo Nordisk Foundation Center for Biosustainability, Next Generation Sequencing laboratory, is acknowledged for assistance with RNA sequencing. This research was supported by grants from the Swedish Research Council (V.R., 2015-05319), the Danish Research council (DFF, 9040-00075B), and the Novo Nordisk Foundation (NNF10CC1016517) to I.M.
Raw data of RNA sequencing are available at ArrayExpress with accession number E-MTAB-8124.
References
- Abe K, Kawano Y, Iwamoto K, Arai K, Maruyama Y, Eichenberger P, Sato T.. 2014. Developmentally-regulated excision of the SPβ prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS Genet. 10(10):e1004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu T, Greenfield A, Shuster B, Barry SN, Gallitto M, Liu B, Kacmarczyk T, et al. 2015. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol. 11:839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Moran CP Jr.. 1994. Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J Bacteriol. 176(7):2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidnenko V, Shi L, Kobir A, Ventroux M, Pigeonneau N, Henry C, Trubuil A, Noirot-Gros MF, Mijakovic I.. 2013. Bacillus subtilis serine/threonine protein kinase YabT is involved in spore development via phosphorylation of a bacterial recombinase. Mol Microbiol. 88(5):921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns LS, Hobley L, Stanley-Wall NR.. 2014. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol. 93(4):587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Stolzer M, Durand D, Pollard KS.. 2013. How old is my gene? Trends Genet. 29(11):659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S.. 2011. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 39(2):414–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- Do TH, Suzuki Y, Abe N, Kaneko J, Itoh Y, Kimura K.. 2011. Mutations suppressing the loss of DegQ function in Bacillus subtilis (natto) poly-γ-glutamate synthesis. Appl Environ Microbiol. 77(23):8249–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Brajković J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23:533–539. [DOI] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D.. 2010. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468(7325):815–818. [DOI] [PubMed] [Google Scholar]
- Driks A. 1999. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 63(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, González-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, et al. 2003. The sigma E regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol. 327(5):945–972. [DOI] [PubMed] [Google Scholar]
- Eijlander RT, de Jong A, Krawczyk AO, Holsappel S, Kuipers P.. 2014. SporeWeb: an interactive journey through the complete sporulation cycle of Bacillus subtilis. Nucleic Acids Res. 42(D1):D685–D691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ.. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 68(2):234–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Suo B, Emmons SW.. 2016. Gene function prediction based on developmental transcriptomes of the two sexes in C. elegans Cell Rep.17:917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch CT, Doi RH.. 1971. Electron microscopy of the altered spore morphology of a ribonucleic acid polymerase mutant of Bacillus subtilis. J Bacteriol. 105(3):1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana R, Yamamura S, Ikejiri H, Kobayashi K, Ogasawara N, Asai K, Sadaie Y, Takamatsu H, Watabe K.. 2003. Bacillus subtilis spoVIF (yjcC) gene, involved in coat assembly and spore resistance. Microbiology 149(10):3011–3021. [DOI] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ.. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol. 9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Marston AL.. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227(1):101–110. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Brannigan JA, Offen WA, Smith I, Wilkinson AJ.. 1998. An evolutionary link between sporulation and prophage induction in the structure of a repressor: anti-repressor complex. J Mol Biol. 283(5):907–912. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchadier E, Carballido-López R, Brinster S, Fabret C, Mervelet P, Bessières P, Noirot-Gros MF, Fromion V, Noirot P.. 2011. An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: exploration by an integrative approach. Proteomics 11(15):2981–2991. [DOI] [PubMed] [Google Scholar]
- Mastny M, Heuck A, Kurzbauer R, Heiduk A, Boisguerin P, Volkmer R, Ehrmann M, Rodrigues CD, Rudner DZ, Clausen T.. 2013. CtpB assembles a gated protease tunnel regulating cell-cell signaling during spore formation in Bacillus subtilis. Cell 155(3):647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eichenberger P.. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 11(1):33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Rodrigues CD, Brady JH, Lim C, Bernhardt TG, Rudner DZ.. 2016. High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol. 14(1):e1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL, Setlow P.. 1990. Sporulation, germination and outgrowth In: Harwood CL, Cutting SM, editors. Molecular biology methods for Bacillus. Chichester: John Wiley & Son; p. 432–450. [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335(6072):1103–1106. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2017. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: https://www.R-project.org/. [Google Scholar]
- Radeck J, Kraft K, Bartels J, Cikovic T, Dürr F, Emenegger J, Kelterborn S, Sauer C, Fritz G, Gebhard S, et al. 2013. The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J Biol Eng. 7(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar V, Nalpas NC, Anselm V, Krug K, Lenuzzi M, Šestak MS, Domazet-Lošo T, Mijakovic I, Macek B.. 2018. In-depth analysis of Bacillus subtilis proteome identifies new ORFs and traces the evolutionary history of modified proteins. Sci Rep. 8(1):17246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar V, Shi L, Krug K, Derouiche A, Jers C, Cousin C, Kobir A, Mijakovic I, Macek B.. 2014. Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC. Mol Cell Proteomics. 13(8):1965–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y.. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 5(4):725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN.. 2009. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc Natl Acad Sci U S A. 106(50):21027–21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestak MS, Božičević V, Bakarić R, Dunjko V, Domazet-Lošo T.. 2013. Phylostratigraphic profiles reveal a deep evolutionary history of the vertebrate head sensory systems. Front Zool. 10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow B, Setlow P.. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J Bacteriol. 178(12):3486–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steil L, Serrano M, Henriques AO, Völker U.. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151(2):399–420. [DOI] [PubMed] [Google Scholar]
- Traag BA, Pugliese A, Eisen JA, Losick R.. 2013. Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. J Bacteriol. 195(2):253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigos AS, Pearson RB, Papenfuss AT, Goode DL.. 2017. Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc Natl Acad Sci U S A. 114(24):6406–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J-W, Smits WK, Hamoen LW, Jongbloed JD, Kuipers OP.. 2004. Visualization of differential gene expression by improved cyan fluorescent protein and yellow fluorescent protein production in Bacillus subtilis. Appl Environ Microbiol. 70(11):6809–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Wu LF, Hughes TR, Davierwala AP, Robinson MD, Stoughton R, Altschuler SJ.. 2002. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet. 31(3):255–265. [DOI] [PubMed] [Google Scholar]
- Yanai I. 2018. Development and evolution through the lens of global gene regulation. Trends Genet. 34(1):11–20. [DOI] [PubMed] [Google Scholar]
- Zhu B, Stülke J.. 2018. SubtiWiki in 2018: from genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 46(D1):D743–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.