Abstract
The taxonomy and nomenclature of the genus Aspergillus and its associated sexual (teleomorphic) genera have been greatly stabilised over the last decade. This was in large thanks to the accepted species list published in 2014 and associated metadata such as DNA reference sequences released at the time. It had a great impact on the community and it has never been easier to identify, publish and describe the missing Aspergillus diversity. To further stabilise its taxonomy, it is crucial to not only discover and publish new species but also to capture infraspecies variation in the form of DNA sequences. This data will help to better characterise and distinguish existing species and make future identifications more robust. South Africa has diverse fungal communities but remains largely unexplored in terms of Aspergillus with very few sequences available for local strains. In this paper, we re-identify Aspergillus previously accessioned in the PPRI and MRC culture collections using modern taxonomic approaches. In the process, we re-identify strains to 63 species, describe seven new species and release a large number of new DNA reference sequences.
Key words: Beta-tubulin, DNA barcoding, Calmodulin, GCPSR, Multigene phylogenies, RPB2, Secondary identification markers
Taxonomic novelties: New species: Aspergillus elsenburgensis Visagie, S.M. Romero & Houbraken; Aspergillus heldtiae Visagie; Aspergillus krugeri Visagie; Aspergillus magaliesburgensis Visagie; Aspergillus purpureocrustaceus Visagie; Aspergillus seifertii Visagie & N. Yilmaz; Aspergillus sigurros Visagie
Introduction
Aspergillus is cosmopolitan fungi occurring on a wide range of substrates. Here they fulfil many different functions and have a wide-ranging influence on human and animal life. Even though most species occur as saprophytes living on dead organic material, various species have an (economic) impact on humans (Raper & Fennell 1965).
Human infections caused by Aspergillus are some of the most widely reported for all filamentous fungi (Gianni and Romano, 2004, Balajee et al., 2007). Aspergillus fumigatus, A. flavus and A. terreus attract special interest as human pathogens causing widespread aspergillosis (fungus ball) or bad allergies (Raper and Fennell, 1965, Steinbach et al., 2004, Sugui et al., 2012, de Hoog et al., 2014, Frisvad and Larsen, 2015b), while a much broader spectrum of species is known to cause less invasive and/or superficial infections (Kaur et al., 2000, Zotti and Corte, 2002, Hubka et al., 2012, de Hoog et al., 2014). Aspergillus causes widespread losses for agriculture where they spoil food or grow in agricultural produce, leading to mycotoxin contamination (Perrone et al., 2007, Pitt and Hocking, 2009, Samson et al., 2010, Frisvad and Larsen, 2015a). Aspergillus isolates produce three of the five agriculturally important mycotoxins, including aflatoxins, ochratoxins and fumonisins (Miller, 1995, Frisvad and Larsen, 2015a). The global cost of aflatoxin alone is huge and represents a major problem in developing countries where stunting in children is of major concern (Wu et al., 2008, Pitt et al., 2012, Wu, 2015). Aflatoxin is most commonly produced by A. flavus and A. parasiticus, but many other Aspergilli can produce this devastating mycotoxin. Ochratoxins are commonly produced by Aspergillus species classified in sections Circumdati and Nigri (Frisvad et al., 2004, Frisvad et al., 2011, Davolos et al., 2012, Visagie et al., 2014b), while some sect Nigri species can also produce fumonisins (Frisvad et al., 2011, Frisvad and Larsen, 2015a). On a more positive note, species have industrial applications as producers of enzymes, drugs, organic acids or are used in food fermentations. For example, A. oryzae (the domesticated form of A. flavus) is used in a koji fermentation important for the production of a wide variety of oriental foods (Raper and Fennell, 1965, Varga et al., 2000, Samson et al., 2010, Hong et al., 2013, Kim et al., 2014).
The taxonomy of Aspergillus and its nine associated sexual (or teleomorphic) genera has been greatly stabilised over the last decade. Based on a multigene phylogenetic study, Kocsube et al. (2016) confirmed that Aspergillus is monophyletic and sister to Penicillium as originally shown by Houbraken & Samson (2011). Furthermore, they showed that the genus can be subdivided into six subgenera and several sections, which to a large degree corresponds to associated sexual states. The nomenclatural review and “accepted species list” published by Samson et al. (2014) played a significant role in stabilizing the taxonomy of Aspergillus. It created an “open access” model in the sense that all metadata associated with species names, such as ex-type culture collection accession numbers, sectional classifications, MycoBank numbers and GenBank accession numbers to reference sequences generated from ex-type cultures, were released in the public domain. Calmodulin was proposed as a secondary identification marker to the formal, but rather conserved, ITS DNA barcode (Schoch et al. 2012). All the released data resulted in more reliable species identifications, and new species discovery and its subsequent description are easier and more accurate than ever. Almost anybody with a bit of background knowledge can describe their new species. As a result, the accepted species list grew with more than 100 taxa in the space of 5 years and resulted in the so-called “broad” Aspergillus that has mostly been accepted by the community (Pitt and Taylor, 2014, Pitt and Taylor, 2016, Samson et al., 2014, Kocsube et al., 2016, Samson et al., 2017).
South Africa has great fungal diversity and makes significant contributions to international understanding of a wide range of fungi. Aspergillus is very commonly isolated across South Africa and unlike Penicillium (Schutte 1992), local mycologists were not afraid to attempt identifications down to species level (Cohen, 1950, Swart, 1959, Eicker, 1969, Eicker, 1970a, Eicker, 1970b, Eicker, 1972, Eicker, 1973, Eicker, 1974, Eicker, 1976, Eicker, 1980, van der Merwe et al., 1979, Rabie and Lübben, 1984, Allsop et al., 1987, Watson et al., 1990, Schutte, 1994, Roux and van Warmelo, 1997). These identifications were all based on morphology, meaning that diversity could easily be misidentified due to the known complexities in distinguishing between closely related species without DNA sequence data. Considering the modern methods required to identify species (Samson et al. 2014), we consider Aspergillus to be grossly understudied in South Africa. To our knowledge, the only modern studies reported 23 species isolated from house dust (Visagie et al. 2014a) and seven species from abalone feed collected in the Western Cape (Greeff-Laubscher et al. 2018). The PPRI culture collection housed at the Agricultural Research Council – Plant Health and Protection, Roodeplaat, Pretoria is the biggest repository of Aspergillus in South Africa with close to 500 accessioned strains. The PPRI also houses the old MRC (Medical Research Council) culture collection that contains several Aspergillus. Strains from these collections mostly originate from agricultural sources, but plenty was sourced from environmental collection trips across the country. The aim of this project was to recover as many strains as possible and re-identify them using modern DNA sequencing approaches in order to obtain a baseline knowledge on the diversity of Aspergillus in the country. In this paper, we report on the diversity discovered, formally introduce seven new species and release a large number of valuable DNA reference sequences in the NCBI nucleotide sequence database (GenBank).
Materials & methods
Strains
Strains were recovered from the South African National Collection of Fungi (PPRI) and the Medical Research Council (MRC) collection, both housed at the Agricultural Research Council (ARC; Plant Health and Protection, Roodeplaat). New isolates were obtained during routine identification services provided at PPRI. These originate from a wide range of sources across the country and were deposited into a working collection (CMV) and PPRI. Isolations were made using potato dextrose agar (PDA) or dichloran 18 % glycerol agar (DG18; Oxoid CM0729). Strains and its collection data are summarised in Table 1.
Table 1.
Strains sequenced during the course of this project.
| Species | Strains1 | Section | Location collected / isolated, year | Host | GenBank nr |
|||
|---|---|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | |||||
| Aspergillus chevalieri | PPRI13427 = CMV011F5 | Aspergillus | South Africa, KwaZulu-Natal, Pinetown, 2013 | Soil | – | – | MK451336 | – |
| A. chevalieri | PPRI26000 = CMV003I3 | Aspergillus | South Africa, 2017 | Animal feed | – | MK450979 | MK451332 | – |
| A. chevalieri | PPRI26033 = CMV012H5 | Aspergillus | South Africa, Gauteng, Pretoria, 2018 | Dog food | – | – | MK451338 | – |
| A. chevalieri | PPRI26034 = CMV012H6 | Aspergillus | South Africa, Gauteng, Pretoria, 2018 | Dog food | – | – | MK451339 | – |
| A. chevalieri | PPRI26348 = CMV016E5 | Aspergillus | South Africa, Gauteng, Pretoria, 2019 | Dog food | – | – | MN031422 | – |
| A. chevalieri | PPRI26554 = CMV016D7 | Aspergillus | South Africa, Gauteng, Pretoria, 2019 | Dog food | – | – | MK951911 | – |
| A. chevalieri | PPRI3791 = CMV011B6 | Aspergillus | South Africa, 1986 | – | – | MK451333 | – | |
| A. chevalieri | PPRI4908 = CMV011B7 | Aspergillus | South Africa, Kwazulu Natal, 1993 | Maize kernels (Zea mays) | – | – | MK451334 | – |
| A. chevalieri | PPRI5410 = CMV012B1 | Aspergillus | South Africa, Western Cape, Clanwilliam, 1994 | Rooibos tea (Aspalathus linearis) | – | – | MK451337 | – |
| A. chevalieri | PPRI6331 = CMV011B9 | Aspergillus | South Africa, Gauteng, Pretoria, 1996 | Dried sausage | – | – | MK451335 | – |
| A. montevidensis | CMV012H4 | Aspergillus | South Africa, Gauteng, Pretoria, 2018 | Dog food | – | – | MK451446 | – |
| A. montevidensis | MRC1250 = CMV017A6 | Aspergillus | South Africa, Western Cape, Ceres, 1975 | Apple juice concentrate | – | – | MK951923 | – |
| A. montevidensis | PPRI26035 = CMV012H7 | Aspergillus | South Africa, Gauteng, Pretoria, 2018 | Dog food | – | – | MK451447 | – |
| A. montevidensis | PPRI4851 = CMV011G2 | Aspergillus | South Africa, Gauteng, Johannesburg, 1993 | Air sample | – | – | MK451445 | – |
| A. montevidensis | PPRI6330 = CMV011B8 | Aspergillus | South Africa, Gauteng, Pretoria, 1996 | Dried sausage | – | – | MK451443 | – |
| A. montevidensis | PPRI8674 = CMV011C2 | Aspergillus | South Africa, Gauteng, Johannesburg, 2007 | Wheat (Triticum sp) | – | – | MK451444 | – |
| A. porosus | PPRI3419a = CMV012A8 = CSIR980 | Aspergillus | South Africa, 1988 | – | – | MK451494 | – | |
| A. porosus | PPRI3419b = CMV012A9 = CSIR980 | Aspergillus | South Africa, 1988 | – | – | MK451495 | – | |
| A. proliferans | PPRI6735 = CMV011C1 | Aspergillus | South Africa, Mpumalanga, Piet Retief, 1988 | Bee larvae (Apis mellifera) | – | – | MK451496 | – |
| A. pseudoglaucus | MRC1231 = CMV017A3 | Aspergillus | South Africa, Western Cape, Elgin, 1975 | Apple | – | – | MK951920 | – |
| A. pseudoglaucus | MRC455 = CMV017E9 | Aspergillus | South Africa, unknown | – | – | MN031425 | – | |
| A. pseudoglaucus | MRC462 = CMV017A1 | Aspergillus | South Africa, Pretoria, unknown | – | – | MK951918 | – | |
| A. pseudoglaucus | PPRI26346 = CMV016D9 | Aspergillus | South Africa, Gauteng, Pretoria, 2019 | Dog food | – | – | MK951912 | – |
| A. zutongqii | PPRI3429 = CMV011F7 | Aspergillus | South Africa, Gauteng, Pretoria, 1988 | Lab contaminant | – | – | MK451575 | – |
| A. species | PPRI6060 = CMV004E8 | Candidi | South Africa, Free State, Bloemfontein, 1995 | Dung | MK450633 | MK451000 | MK451330 | – |
| A. tritici | MRC3080 = CMV017B1 | Candidi | South Africa, Mpumalanga, 1982 | Maize (Zea mays) | – | – | MK951927 | – |
| A. tritici | MRC418 = CMV016I7 | Candidi | South Africa, North West Province, Brits, 1971 | Sorghum malt | – | – | MK951916 | – |
| A. ochraceus | PPRI26013 = CMV006D9 | Circumdati | South Africa, Western Cape, 2018 | Wheat (Triticum sp) | – | – | MK451474 | – |
| A. ochraceus | PPRI6335 = CMV007B6 | Circumdati | South Africa, Mpumalanga, Nelspruit, 1997 | Cochecille insects | – | – | MK451476 | – |
| A. ochraceus | PPRI6816 = CMV007B5 | Circumdati | South Africa, North West, Potchefstroom, 1999 | Cowpea (Vigna ungiculata) | – | – | MK451475 | – |
| A. pallidofulvus | CMV012D2 | Circumdati | South Africa, Limpopo, Groblersdal, 2018 | Soil | MK450639 | – | MK451477 | – |
| A. sclerotiorum | PPRI8357 = CMV007B4 | Circumdati | South Africa, 2006 | Rat food | – | – | MK451507 | – |
| A. westerdijkiae | PPRI5061 = CMV007B2 | Circumdati | South Africa, Limpopo, Vaalwater, 1993 | Chrysomelid beetle | – | – | MK451571 | – |
| A. westerdijkiae | PPRI8700 = CMV007B7 | Circumdati | South Africa, Limpopo , Kruger National Park, 2005 | Mopane twigs and leaves (Colophospermum mopane) | – | – | MK451572 | – |
| A. clavatus | PPRI13831 = CMV008F4 | Clavati | South Africa, Gauteng, Bapsfontein, 2014 | Barley seedling (Hordeum vulgare) | – | – | MK451347 | – |
| A. clavatus | PPRI13832 = CMV005I8 | Clavati | South Africa, Gauteng, Bapsfontein, 2014 | Barley seedling (Hordeum vulgare) | – | – | MK451344 | – |
| A. clavatus | PPRI14650 = CMV005I9 | Clavati | South Africa, North Weat, Potchefstroom, 2014 | Animal feed | – | – | MK451345 | – |
| A. clavatus | PPRI17069 = CMV001F9 | Clavati | South Africa, Gauteng, Near Delmas, 2014 | Animal feed, maize kernels | – | – | MK451341 | – |
| A. clavatus | PPRI21896 = CMV006A1 | Clavati | South Africa, Western Cape, Malmesbury, 2016 | Barley sprouted seed (Hordeum vulgare) | – | – | MK451346 | – |
| A. clavatus | PPRI26042 = CMV013A3 | Clavati | South Africa, 2018 | Dragon fruit plant | – | – | MK451349 | – |
| A. clavatus | PPRI26045 = CMV013B4 | Clavati | Swaziland, 2018 | Pig feed | – | – | MK451351 | – |
| A. clavatus | PPRI26493 = CMV013A9 | Clavati | Swaziland, 2018 | Pig feed | – | – | MK951883 | – |
| A. clavatus | PPRI26495 = CMV013B2 | Clavati | Swaziland, 2018 | Pig feed | – | – | MK451350 | – |
| A. clavatus | PPRI4976 = CMV010D7 | Clavati | South Africa, Gauteng, Magaliesburg, 1994 | Soil | – | – | MK451348 | – |
| A. clavatus | PPRI8552 = CMV005I6 | Clavati | South Africa, Mpumalanga, Lydenburg, 2007 | Sunflower seed (Helianthus annuus) | – | – | MK451342 | – |
| A. clavatus | PPRI9818 = CMV005I7 | Clavati | South Africa, Free State, 2008 | Sunflower soil | – | – | MK451343 | – |
| A. giganteus | MRC453 = CMV016I9 | Clavati | South Africa, Pretoria, unknown | – | – | MN031424 | – | |
| A. giganteus | PPRI26019 = CMV008C9 | Clavati | South Africa, Limpopo, 2018 | Chicken feed | MK450637 | MK451147 | MK451418 | – |
| A. seifertii | PPRI26025 = CMV011E3 | Clavati | South Africa, Free State, Golden Gate, unknown | Soil | MK450648 | MK451205 | MK451510 | MK450801 |
| A. seifertii | PPRI3211 = CMV006F5 (ex-type) | Clavati | South Africa, Free State, Golden Gate, 1988 | Grassroots | MK450647 | MK451093 | MK451509 | MK450800 |
| A. dimorphicus | CMV012C9 | Cremei | South Africa, Limpopo, Groblersdal, 2018 | Soil | MK450634 | MK451246 | MK451357 | – |
| A. dimorphicus | PPRI26031 = CMV012G4 | Cremei | South Africa, Limpopo, Groblersdal, 2018 | Soil | MK450646 | MK451263 | MK451508 | MK450799 |
| A. wentii | PPRI25999 = CMV003I2 | Cremei | South Africa, 2017 | Animal feed | – | – | MK451569 | – |
| A. wentii | PPRI26048 = CMV013F6 | Cremei | South Africa, Mpumalanga, Barberton, 2018 | Wood in mine | – | – | MK451570 | – |
| A. wentii | PPRI26349 = CMV016E7 | Cremei | South Africa, Mpumalanga, Barberton, 2018 | Wood in mine | – | – | MK951914 | – |
| A. alliaceus | PPRI6826 = CMV007B1 | Flavi | South Africa, Eastern Cape, Port Elizabeth, 1999 | Moth larvae (Cryptophlebia leucotreta) | – | – | MK451307 | – |
| A. flavus | CMV015C6 = 2019-M44 | Flavi | South Africa, 2019 | Wood pallet | – | – | MK951894 | – |
| A. flavus | CMV015C7 = 2019-M44 | Flavi | South Africa, 2019 | Wood pallet | – | – | MK951895 | – |
| A. flavus | CMV015C9 = 2019-M44 | Flavi | South Africa, 2019 | Wood pallet | – | – | MK951896 | – |
| A. flavus | MRC1317 = CMV017A2 | Flavi | South Africa, Western Cape, Somerset West, 1977 | Lemon (Citrus limon) | – | – | MK951919 | – |
| A. flavus | MRC1366 = CMV017A7 | Flavi | South Africa, Western Cape, Ceres, 1978 | Maize (Zea mays), pathotoxicity to sheep | – | – | MK951924 | – |
| A. flavus | MRC1745 = CMV017A8 | Flavi | South Africa, North West Province, Potchefstroom, 1979 | Sorghum malt | – | – | MK951925 | – |
| A. flavus | MRC2526 = CMV017A9 | Flavi | South Africa, unknown | Biltong | – | – | MK951926 | – |
| A. flavus | MRC3732 = CMV017B3 | Flavi | South Africa, Western Cape, Ceres, 1984 | Apple | – | – | MK951929 | – |
| A. flavus | MRC6979 = CMV017B5 | Flavi | South Africa, Mpumalanga, Kruger National Park, unknown | Soil | – | – | MK951931 | – |
| A. flavus | PPRI13141 = CMV002B4 | Flavi | South Africa, Kwazulu Natal, Pietermaritzburg, unknown | Maize (Zea mays) | – | – | MK451376 | – |
| A. flavus | PPRI18143 = CMV001I2 | Flavi | South Africa, 2015 | Rooibos (Aspalathus linearis) | – | – | MK451365 | – |
| A. flavus | PPRI18144 = CMV001I8 | Flavi | South Africa, 2015 | Rooibos (Aspalathus linearis) | – | – | MK451370 | – |
| A. flavus | PPRI18161 = CMV002B3 | Flavi | South Africa, Free State, Bethlehem, 2015 | Wheat (Triticum sp) | – | – | MK451375 | – |
| A. flavus | PPRI18711 = CMV001I4 | Flavi | South Africa, Northwest, Sannieshof, 2015 | Frass of moth (Busseola fusca) feeding inside maize stems | – | – | MK451367 | – |
| A. flavus | PPRI18712 = CMV001I5 | Flavi | South Africa, Northwest, Sannieshof, 2015 | Frass of moth (Busseola fusca) feeding inside maize stems | – | – | MK451368 | – |
| A. flavus | PPRI18713 = CMV001I9 | Flavi | South Africa, Northwest, Coligny, 2015 | Frass of moth (Busseola fusca) feeding inside maize stems | – | – | MK451371 | – |
| A. flavus | PPRI18714 = CMV001I1 | Flavi | South Africa, Northwest, Coligny, 2015 | Frass of moth (Busseola fusca) feeding inside maize stems | – | – | MK451364 | – |
| A. flavus | PPRI18715 = CMV001I6 | Flavi | South Africa, Northwest, Coligny, 2015 | Frass of moth (Busseola fusca) feeding inside maize stems | – | – | MK451369 | – |
| A. flavus | PPRI20581 = CMV002B1 | Flavi | South Africa, Western Cape, Grabouw, 2015 | Insect | – | – | MK451374 | – |
| A. flavus | PPRI22482 = CMV001I3 | Flavi | South Africa, Limpopo, Atlanta, 2016 | Soya beans (Glycine max) | – | – | MK451366 | – |
| A. flavus | PPRI23389 = CMV002A1 | Flavi | South Africa, Western Cape, Stellenbosch, 2016 | Animal feed | – | – | MK451372 | – |
| A. flavus | PPRI25992 = CMV003A4 | Flavi | South Africa, Western Cape, Knysna, 2017 | Hominy chop animal feed | – | – | MK451379 | – |
| A. flavus | PPRI26001 = CMV003I5 | Flavi | South Africa, 2017 | Animal feed | – | – | MK451380 | – |
| A. flavus | PPRI26002 = CMV003I6 | Flavi | South Africa, 2017 | Animal feed | – | – | MK451381 | – |
| A. flavus | PPRI26003 = CMV003I7 | Flavi | South Africa, 2017 | Animal feed | – | – | MK451382 | – |
| A. flavus | PPRI26004 = CMV003I8 | Flavi | South Africa, 2017 | Animal feed | – | – | MK451383 | – |
| A. flavus | PPRI26007 = CMV005E1 | Flavi | South Africa, Gauteng, Sunninghill, 2017 | Groundnut | – | – | MK451384 | – |
| A. flavus | PPRI26022 = CMV008E3 | Flavi | South Africa, Limpopo, 2018 | Chicken feed | – | – | MK451385 | – |
| A. flavus | PPRI26032 = CMV012H1 | Flavi | South Africa, Gauteng, Pretoria, 2018 | Dog food | – | – | MK451387 | – |
| A. flavus | PPRI26036 = CMV012H8 | Flavi | South Africa, Gauteng, Pretoria, 2018 | Dog food | – | – | MK451388 | – |
| A. flavus | PPRI26044 = CMV013B3 | Flavi | Swaziland, 2018 | Pig feed | – | – | MK451389 | – |
| A. flavus | PPRI26345 = CMV016D6 | Flavi | South Africa, Gauteng, Pretoria, 2019 | Dog food | – | – | MK951910 | – |
| A. flavus | PPRI26347 = CMV016E4 | Flavi | South Africa, Gauteng, Pretoria, 2019 | Dog food | – | – | MK951913 | – |
| A. flavus | PPRI26486 = CMV010D4 | Flavi | South Africa, Limpopo, Groblersdal, 2015 | Soil | – | – | MK451386 | – |
| A. flavus | PPRI3274 = CMV002A5 | Flavi | South Africa, Gauteng, Pretoria, 1988 | – | – | MK451373 | – | |
| A. flavus | PPRI7977 = CMV002B5 | Flavi | South Africa, 2005 | – | – | MK451377 | – | |
| A. flavus | PPRI8551 = CMV002B7 | Flavi | South Africa, Mpumalanga, Lydenburg, 2007 | Maize (Zea mays) | – | – | MK451378 | – |
| A. krugeri | PPRI8986 = CMV006G4 (ex-type) | Flavi | South Africa, Limpopo, Kruger National Park, 2005 | Mopane debris (Colophospermum mopane) | MK450655 | MK451098 | MK451517 | MK450808 |
| A. krugeri | PPRI9280 = CMV002C8 | Flavi | South Africa, Limpopo, Kruger National Park, 2005 | Mopane debris (Colophospermum mopane) | MK450654 | MK450928 | MK451516 | MK450807 |
| A. magaliesburgensis | PPRI6165 = CMV007A3 (ex-type) | Flavi | South Africa, Gauteng, Magaliesburg, 1996 | Antlion (Myrmeleontidae) | MK450649 | MK451116 | MK451511 | MK450802 |
| A. nomius | PPRI3753 = CMV002B2 | Flavi | South Africa, Gauteng, Rietondale, 1989 | Termites dead colony | – | MK450926 | MK451473 | – |
| A. parasiticus | PPRI14636 = CMV001H8 | Flavi | South Africa, Gauteng, Bapsfontein, 2014 | Spawnrun on grass | – | – | MK451478 | – |
| A. parasiticus | PPRI14642 = CMV001H9 | Flavi | South Africa, Gauteng, Bapsfontein, 2014 | Spawnrun on grass | – | – | MK451479 | – |
| A. parasiticus | PPRI23021 = CMV002C7 | Flavi | South Africa, Dinaka game reserve, 2016 | Animal feed | – | – | MK451483 | – |
| A. parasiticus | PPRI26046 = CMV013B6 | Flavi | Zambia, Mpangwe, Mpangwe, 2018 | Wheat (Triticum sp) | – | – | MK451489 | – |
| A. parasiticus | PPRI2885 = CMV007A7 | Flavi | South Africa, 1990 | Seed (Watsonin marginata) | – | – | MK451487 | – |
| A. parasiticus | PPRI3754 = CMV007A5 | Flavi | South Africa, Gauteng, Pretoria, 1989 | Termites | – | – | MK451485 | – |
| A. parasiticus | PPRI5183 = CMV007A6 | Flavi | South Africa, Western Cape, Clanwilliam, 1993 | Rooibos tea (Aspalathus linearis) | – | – | MK451486 | – |
| A. parasiticus | PPRI7978 = CMV010B6 | Flavi | South Africa, 2005 | – | – | MK451488 | – | |
| A. parasiticus | PPRI9511 = CMV002B8 | Flavi | South Africa, North West , 2008 | Soil | – | – | MK451480 | – |
| A. parasiticus | PPRI9513 = CMV002G1 | Flavi | South Africa, North West , 2008 | Soil | – | – | MK451484 | – |
| A. parasiticus | PPRI9532 = CMV002C1 | Flavi | South Africa, North West , 2008 | Soil | – | – | MK451481 | – |
| A. parasiticus | PPRI9534 = CMV002C2 | Flavi | South Africa, North West , 2008 | Soil | – | – | MK451482 | – |
| A. pseudonomius | PPRI5063 = CMV002B6 | Flavi | South Africa, Limpopo, Vaalwater, 1992 | Chrysomelid beetle | – | – | MK451505 | – |
| A. tamarii | PPRI26008 = CMV005E2 | Flavi | South Africa, Gauteng, Sunninghill, 2017 | Groundnut | – | – | MK451528 | – |
| A. tamarii | PPRI26010 = CMV005E4 | Flavi | South Africa, 2017 | Soil | – | – | MK451529 | – |
| A. tamarii | PPRI26023 = CMV008E4 | Flavi | South Africa, Limpopo, 2018 | Chicken feed | – | – | MK451531 | – |
| A. tamarii | PPRI2812 = CMV003E1 | Flavi | South Africa, 1991 | Soya beans (Glycine max) | – | – | MK451527 | – |
| A. tamarii | PPRI7392 = CMV007B3 | Flavi | South Africa, 2004 | – | – | MK451530 | – | |
| A. transmontanensis | PPRI14275 = CMV011A5 | Flavi | Zambia, 2013 | Soil | MK450657 | MK451183 | MK451519 | MK450810 |
| A. iizukae | PPRI4965 = CMV007B8 | Flavipedes | South Africa, Gauteng, Pretoria, 1993 | Chrysomelid beetle | – | – | MK451428 | – |
| A. arcoverdensis | PPRI7491 = CMV003C4 | Fumigati | South Africa, 2004 | – | – | MK451311 | – | |
| A. arcoverdensis | PPRI7514 = CMV003C3 | Fumigati | South Africa, 2004 | – | – | MK451310 | – | |
| A. aureolus | PPRI11297 = CMV008A9 | Fumigati | South Africa, Kwazulu Natal, Pinetown, 2011 | Air sample | – | – | MK451321 | – |
| A. aureolus | PPRI3451 = CMV011F8 | Fumigati | South Africa, 1988 | – | – | MK451322 | – | |
| A. elsenburgensis | DTO015G7 | Fumigati | Argentina, La Pampa Province, Chacharramendi | Soil | MT110301 | MT108410 | MT108412 | – |
| A. elsenburgensis | DTO380H5 | Fumigati | Argentina, Catamarca Province | Soil | – | MT108411 | MT108413 | – |
| A. elsenburgensis | DTO381D3 | Fumigati | Argentina, Catamarca Province | Soil | – | – | MT108414 | – |
| A. elsenburgensis | DTO381D8 | Fumigati | Argentina | Soil | MT110302 | – | MT108415 | – |
| A. elsenburgensis | PPRI2994 = CMV011G4 = CSIR1013 (ex-type) | Fumigati | South Africa, Western Cape, Elsenburg, 1986 | Soil | MK450651 | MK451215 | MK451513 | MK450804 |
| A. fischeri | PPRI26026 = CMV011H6 | Fumigati | South Africa, Gauteng, Pretoria, 2018 | Lab contaminant | – | – | MK451359 | – |
| A. fischeri | PPRI3418 = CMV012A7 = CSIR978 | Fumigati | South Africa, 1988 | – | – | MK451363 | – | |
| A. fischeri | PPRI3428 = CMV011I5 = CSIR990 | Fumigati | South Africa, 1988 | – | – | MK451361 | – | |
| A. fischeri | PPRI3488 = CMV012A6 = CSIR1039 | Fumigati | South Africa, 1988 | – | – | MK451362 | – | |
| A. fischeri | PPRI4507 = CMV011I4 = CSIR1094 | Fumigati | South Africa, Eastern Cape, Butterworth, 1986 | Soil | – | – | MK451360 | – |
| A. fumigatiaffinis | PPRI13089 = CMV001G1 | Fumigati | South Africa, Succulent karoo area , unknown | Soil | MK450636 | MK450913 | MK451390 | – |
| A. fumigatiaffinis | PPRI13090 = CMV010I7 | Fumigati | South Africa, Succulent karoo area , unknown | Soil | – | – | MK451392 | – |
| A. fumigatiaffinis | PPRI3210 = CMV004C3 | Fumigati | South Africa, Western Cape, Beaufort West, 1988 | Grass | – | – | MK451391 | – |
| A. fumigatus | CMV015C1 = 2019-M44 | Fumigati | South Africa, 2019 | Wood pallet | – | – | MK951890 | – |
| A. fumigatus | CMV015C2 = 2019-M44 | Fumigati | South Africa, 2019 | Wood pallet | – | – | MK951891 | – |
| A. fumigatus | CMV015C3 = 2019-M44 | Fumigati | South Africa, 2019 | Wood pallet | – | – | MK951892 | – |
| A. fumigatus | CMV015C5 = 2019-M44 | Fumigati | South Africa, 2019 | Wood pallet | – | – | MK951893 | – |
| A. fumigatus | CMV015D8 = 2019-M44 | Fumigati | South Africa, 2019 | Wood pallet | – | – | MK951904 | – |
| A. fumigatus | CMV015D9 = 2019-M44 | Fumigati | South Africa, 2019 | Wood pallet | – | – | MK951905 | – |
| A. fumigatus | MRC435 = CMV016I8 | Fumigati | South Africa, Port Health, 1971 | Rice | – | – | MK951917 | – |
| A. fumigatus | PPRI10161 = CMV002G6 | Fumigati | South Africa, Eastern Cape, 2009 | Silage | – | – | MK451396 | – |
| A. fumigatus | PPRI10162 = CMV002G2 | Fumigati | South Africa, Eastern Cape, 2009 | Silage | – | – | MK451393 | – |
| A. fumigatus | PPRI10498 = CMV003D5 | Fumigati | South Africa, Eastern Cape, Port Elizabeth, 2010 | Maize silage (Zea mays) | – | – | MK451406 | – |
| A. fumigatus | PPRI10499 = CMV002G5 | Fumigati | South Africa, Eastern Cape, Port Elizabeth, 2010 | Maize silage (Zea mays) | – | – | MK451395 | – |
| A. fumigatus | PPRI12665 = CMV002G3 | Fumigati | South Africa, Free State, Luchhof, 2012 | Rye seed (Secale cereale) | – | – | MK451394 | – |
| A. fumigatus | PPRI13084 = CMV002G7 | Fumigati | South Africa, Gauteng, Pretoria, 2013 | Pear | – | – | MK451397 | – |
| A. fumigatus | PPRI13252 = CMV003D6 | Fumigati | South Africa, 2013 | – | – | MK451407 | – | |
| A. fumigatus | PPRI20934 = CMV008B7 | Fumigati | South Africa, 2016 | – | MK451141 | MK451412 | – | |
| A. fumigatus | PPRI25993 = CMV003A5 | Fumigati | South Africa, Western Cape, Knysna, 2017 | Hominy chop animal feed | – | – | MK451398 | – |
| A. fumigatus | PPRI25998 = CMV003H8 | Fumigati | South Africa, 2017 | Animal feed | – | – | MK451409 | – |
| A. fumigatus | PPRI26006 = CMV005D8 | Fumigati | South Africa, Gauteng, Bedfordview, 2018 | Potting soil | – | – | MK451411 | – |
| A. fumigatus | PPRI3283 = CMV003C6 | Fumigati | South Africa, North West, Pella, 1993 | Soil | – | – | MK451399 | – |
| A. fumigatus | PPRI3478 = CMV004C2 | Fumigati | South Africa, Gauteng, Bapsfontein, 1988 | Straw | – | – | MK451410 | – |
| A. fumigatus | PPRI3479 = CMV003C7 | Fumigati | South Africa, Gauteng, Johannesburg, 1988 | Compost | – | – | MK451400 | – |
| A. fumigatus | PPRI3505 = CMV010B9 | Fumigati | South Africa, Gauteng, Denneboom, 1987 | Pine | – | MK451153 | MK451416 | – |
| A. fumigatus | PPRI4975 = CMV003C8 | Fumigati | South Africa, Mpumalanga, Malelane, 1993 | Bagasse | – | – | MK451401 | – |
| A. fumigatus | PPRI5090 = CMV003H1 | Fumigati | South Africa, Mpumalanga, Malelane, 1993 | Decayed mineola | – | MK450976 | MK451408 | – |
| A. fumigatus | PPRI7394 = CMV003C9 | Fumigati | South Africa, 2004 | – | – | MK451402 | – | |
| A. fumigatus | PPRI8522 = CMV003D1 | Fumigati | South Africa, Mpumalanga, Vlakfontein, 2006 | Chickens (Gallus domesticus) | – | – | MK451403 | – |
| A. fumigatus | PPRI8523 = CMV003D2 | Fumigati | South Africa, Mpumalanga, Vlakfontein, 2006 | Chickens (Gallus domesticus) | – | – | MK451404 | – |
| A. fumigatus | PPRI8525 = CMV008F9 | Fumigati | South Africa, Mpumalanga, Vlakfontein, 2006 | Chickens (Gallus domesticus) | – | – | MK451414 | – |
| A. fumigatus | PPRI8527 = CMV008G1 | Fumigati | South Africa, Mpumalanga, Vlakfontein, 2006 | Chickens (Gallus domesticus) | – | – | MK451415 | – |
| A. fumigatus | PPRI8558 = CMV003D3 | Fumigati | South Africa, 2007 | Chickens (Gallus domesticus) | – | – | MK451405 | – |
| A. fumigatus | PPRI8560 = CMV008F8 | Fumigati | South Africa, 2007 | Chickens (Gallus domesticus) | – | – | MK451413 | – |
| A. hiratsukae | PPRI3260 = CMV012G1 = CSIR1064 | Fumigati | South Africa, 1988 | – | – | MK451422 | – | |
| A. hiratsukae | PPRI9172 = CMV008F5 | Fumigati | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | – | MK451421 | – |
| A. hiratsukae | PPRI9185 = CMV004E7 | Fumigati | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | – | MK451420 | – |
| A. hiratsukae | PPRI9190 = CMV004E4 | Fumigati | South Africa, Limpopo, Kruger National Park, 2005 | Mopane twigs and leaves (Colophospermum mopane) | – | – | MK451419 | – |
| A. laciniosus | PPRI3197 = CMV011I6 = CSIR1050 | Fumigati | South Africa, 1988 | – | – | MK451440 | – | |
| A. laciniosus | PPRI3247 = CMV011G5 | Fumigati | South Africa, North West, Pella, 1988 | – | MK451216 | MK451439 | – | |
| A. laciniosus | PPRI3417 = CMV010F8 = CSIR638 | Fumigati | South Africa, 1988 | – | MK451163 | MK451437 | – | |
| A. laciniosus | PPRI3847 = CMV011F6 | Fumigati | South Africa, Kwazulu Natal, Greytown, 1985 | Maize kernels (Zea mays) | – | – | MK451438 | – |
| A. lentulus | PPRI6170 = CMV007I3 | Fumigati | South Africa, Northern Cape, Loffiesdraai, 1996 | Sand | – | MK451134 | MK451442 | – |
| A. lentulus | PPRI7532 = CMV003C5 | Fumigati | South Africa, 2004 | – | MK450952 | MK451441 | – | |
| A. udagawae | PPRI11324 = CMV010I9 | Fumigati | South Africa, Eastern Cape, Port Elizabeth, 2011 | Mealy bug on Citrus | – | MK451179 | MK451543 | – |
| A. udagawae | PPRI26030 = CMV012F7 | Fumigati | South Africa, Limpopo, Groblersdal, 2018 | Soil | – | MK451259 | MK451544 | – |
| A. wyomingensis | PPRI5178 = CMV007I4 | Fumigati | South Africa, Western Cape, Clanwilliam, 1993 | Rooibos tea (Aspalathus linearis) | – | – | MK451574 | – |
| A. wyomingensis | PPRI5573 = CMV007I2 | Fumigati | South Africa, Western Cape, Clanwilliam, 1994 | Rooibos tea (Aspalathus linearis) | – | – | MK451573 | – |
| A. amoenus | PPRI26021 = CMV008E2 | Nidulantes | South Africa, Limpopo, 2018 | Chicken feed | – | – | MK451308 | – |
| A. amoenus | PPRI26047 = CMV013F4 | Nidulantes | South Africa, Mpumalanga, Barberton, 2018 | Wood in mine | – | – | MK451309 | – |
| A. creber | PPRI13168 = CMV002A9 | Nidulantes | South Africa, North West, Mafikeng, 2013 | Chicken house bedding | – | – | MK451352 | – |
| A. creber | PPRI3737 = CMV002G9 | Nidulantes | South Africa, Gauteng, Pretoria, 1989 | Orange (Citrus sinensis) | – | – | MK451353 | – |
| A. creber | PPRI3869 = CMV011F9 | Nidulantes | South Africa, Free State, Bloemfontein, 1990 | Honey flower seed (Melianthus comosus) | – | – | MK451356 | – |
| A. creber | PPRI5081 = CMV002H1 | Nidulantes | South Africa, Mpumalanga, Hazyview, 1993 | Lemon (Citrus limon) | – | – | MK451354 | – |
| A. creber | PPRI9900 = CMV008C2 | Nidulantes | South Africa, Kwazulu Natal, Pinetown, 2008 | – | – | MK451355 | – | |
| A. jensenii | PPRI13238 = CMV001F7 | Nidulantes | South Africa, KwaZulu-Natal, Pinetown, 2013 | Environmental sample | – | – | MK451433 | – |
| A. jensenii | PPRI2806 = CMV011G1 | Nidulantes | South Africa, 1991 | – | – | MK451436 | – | |
| A. jensenii | PPRI5384 = CMV007A9 | Nidulantes | South Africa, 1993 | Flower (Gladiolus corms) | – | – | MK451435 | – |
| A. jensenii | PPRI6329 = CMV003H2 | Nidulantes | South Africa, Kwazulu Natal, 1996 | Contaminant bioproduct | – | MK450977 | MK451434 | – |
| A. nidulans | PPRI20935 = CMV010I1 | Nidulantes | South Africa, 2016 | – | – | MK451456 | – | |
| A. protuberus | PPRI26350 = CMV016F8 | Nidulantes | South Africa, Mpumalanga, Barberton, 2018 | Wood in mine | – | – | MK951915 | – |
| A. protuberus | PPRI5575 = CMV008B2 | Nidulantes | South Africa, 1994 | Diesel fuel filters | – | – | MK451497 | – |
| A. purpureocrustaceus | PPRI3840 = CMV008B3 (ex-type) | Nidulantes | South Africa, Limpopo, 1990 | Plant debris | MK450653 | MK451138 | MK451515 | MK450806 |
| A. purpureocrustaceus | PPRI5548 = CMV008B1 | Nidulantes | South Africa, Western Cape, Cape Town, 1994 | Spider (Palystes castaneus) | MK450652 | MK451137 | MK451514 | MK450805 |
| A. quadrilineatus | PPRI26342 = CMV015B3 | Nidulantes | South Africa, Mpumalanga, Marble Hall, 2019 | – | – | MK951887 | – | |
| A. recurvatus | PPRI3165 = CMV010C4 | Nidulantes | South Africa, 1988 | MK450645 | MK451157 | MK451506 | – | |
| A. rugulosus | MRC3329 = CMV017B2 | Nidulantes | South Africa, Free State, Clocolan, 1983 | Oats | – | – | MK951928 | – |
| A. sydowii | CMV008E1 = 2018-M76/352 | Nidulantes | South Africa, Limpopo, 2018 | Chicken feed | – | – | MK451524 | – |
| A. sydowii | CMV015B9 = 2019-M44 | Nidulantes | South Africa, 2019 | Wood pallet | – | – | MK951889 | – |
| A. sydowii | PPRI12668 = CMV008C6 | Nidulantes | South Africa, KwaZulu-Natal, Pinetown, 2012 | Environmental sample | – | – | MK451523 | – |
| A. sydowii | PPRI13067 = CMV008B4 | Nidulantes | South Africa, KwaZulu-Natal, Pinetown, 2012 | Environmental sample | – | – | MK451521 | – |
| A. sydowii | PPRI13241 = CMV001D6 | Nidulantes | South Africa, KwaZulu-Natal, Pinetown, 2013 | Environmental sample | – | – | MK451520 | – |
| A. sydowii | PPRI3810 = CMV008F1 | Nidulantes | South Africa, Free State, Bloemfontein, 1990 | Honey flower seed (Melianthus comosus) | – | – | MK451525 | – |
| A. sydowii | PPRI3839 = CMV008F2 | Nidulantes | South Africa, 1990 | Watsonia marginata | – | – | MK451526 | – |
| A. sydowii | PPRI6542 = CMV008C5 | Nidulantes | South Africa, Kwazulu Natal, Pinetown, 1997 | Lab shelf | – | – | MK451522 | – |
| A. alabamensis | PPRI25994 = CMV003A6 | Terrei | South Africa, Western Cape, Knysna, 2017 | Hominy chop animal feed | – | MK450947 | MK451300 | MK450758 |
| A. alabamensis | PPRI25996 = CMV003A9 | Terrei | South Africa, Western Cape, Knysna, 2017 | Hominy chop animal feed | – | MK450948 | MK451301 | MK450759 |
| A. alabamensis | PPRI26028 = CMV012E2 | Terrei | South Africa, Limpopo, Groblersdal, 2018 | Soil | – | – | MK451299 | – |
| A. aureoterreus | PPRI13096 = CMV010F6 | Terrei | South Africa, Succulent karoo area , unknown | Soil | – | MK451161 | MK451323 | MK450772 |
| A. carneus | PPRI13094 = CMV010F7 | Terrei | South Africa, Succulent karoo area , unknown | Soil | – | MK451162 | MK451331 | MK450778 |
| A. cf alabamensis | PPRI7492 = CMV004A7 | Terrei | South Africa, 2004 | – | MK450983 | MK451312 | MK450765 | |
| A. cf alabamensis | PPRI8696 = CMV004D7 | Terrei | South Africa, Limpopo , Kruger National Park, 2005 | Soil | – | MK450993 | MK451318 | MK450770 |
| A. cf alabamensis | PPRI8741 = CMV004C9 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | MK450990 | MK451315 | MK450768 |
| A. cf alabamensis | PPRI8747 = CMV004D2 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | MK450991 | MK451316 | MK450769 |
| A. cf alabamensis | PPRI8979 = CMV004D5 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | MK450992 | MK451317 | – |
| A. cf alabamensis | PPRI9150 = CMV004C8 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Mopane debris (Colophospermum mopane) | – | MK450989 | MK451314 | MK450767 |
| A. cf alabamensis | PPRI9184 = CMV004C7 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | MK450988 | MK451313 | MK450766 |
| A. cf alabamensis | PPRI9189 = CMV004E1 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Mopane twigs and leaves (Colophospermum mopane) | – | MK450995 | MK451320 | – |
| A. cf alabamensis | PPRI9206 = CMV004D9 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Mopane twigs and leaves (Colophospermum mopane) | – | MK450994 | MK451319 | MK450771 |
| A. cf allahabadii | PPRI5574 = CMV004C1 | Terrei | South Africa, Western Cape, Clanwilliam, 1994 | Rooibos tea (Aspalathus linearis) | – | MK450987 | MK451302 | MK450760 |
| A. cf allahabadii | PPRI7534 = CMV004E6 | Terrei | South Africa, Limpopo, Kruger National Park, 2003 | Soil | – | MK450999 | MK451306 | MK450764 |
| A. cf allahabadii | PPRI8751 = CMV004E3 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Soil | MK450629 | MK450997 | MK451304 | MK450762 |
| A. cf allahabadii | PPRI8987 = CMV004E2 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Mopane debris (Colophospermum mopane) | MK450628 | MK450996 | MK451303 | MK450761 |
| A. cf allahabadii | PPRI9194 = CMV004E5 | Terrei | South Africa, Limpopo, Kruger National Park, 2005 | Mopane debris (Colophospermum mopane) | – | MK450998 | MK451305 | MK450763 |
| A. citrinoterreus | PPRI7464 = CMV004A6 | Terrei | South Africa, North West, Welwitschia, 2004 | – | – | MK451340 | – | |
| A. heldtiae | PPRI4229 = CMV004A2 (ex-type) | Terrei | South Africa, 1991 | Millet seed | MK450656 | MK450981 | MK451518 | MK450809 |
| A. hortai | PPRI25995 = CMV003A8 | Terrei | South Africa, Western Cape, Knysna, 2017 | Hominy chop animal feed | – | – | MK451423 | – |
| A. hortai | PPRI5864 = CMV004A5 | Terrei | South Africa, Gauteng, Onderstepoort, 1995 | Animal tissue | – | – | MK451424 | – |
| A. hortai | PPRI7533 = CMV004A9 | Terrei | South Africa, Limpopo, Kruger National Park, 2003 | Soil | – | MK450985 | MK451425 | – |
| A. hortai | PPRI8707 = CMV004C5 | Terrei | South Africa, Limpopo , Kruger National Park, 2005 | Mopane (Colophospermum mopane) | – | – | MK451427 | – |
| A. hortai | PPRI9902 = CMV004B1 | Terrei | South Africa, Gauteng, Pretoria, 2008 | Industrial food colourant | – | – | MK451426 | – |
| A. species CBS142751 | PPRI9903 = CMV004A8 | Terrei | South Africa, Gauteng, Pretoria, 2008 | Industrial food colourant | MK450635 | MK450984 | MK451358 | – |
| A. terreus | PPRI10373 = CMV007A8 | Terrei | South Africa, Eastern Cape, Port Elizabeth, 2010 | Maize silage (Zea mays) | – | – | MK451535 | – |
| A. terreus | PPRI13086 = CMV011A4 | Terrei | South Africa, Succulent karoo area , unknown | Soil | – | – | MK451537 | – |
| A. terreus | PPRI20932 = CMV004B3 | Terrei | South Africa, 2016 | – | – | MK451533 | – | |
| A. terreus | PPRI25997 = CMV003H4 | Terrei | South Africa, 2017 | Animal feed | – | – | MK451532 | – |
| A. terreus | PPRI26027 = CMV012E1 | Terrei | South Africa, Limpopo, Groblersdal, 2018 | Soil | – | – | MK451538 | – |
| A. terreus | PPRI26029 = CMV012E3 | Terrei | South Africa, Limpopo, Groblersdal, 2018 | Soil | – | – | MK451539 | – |
| A. terreus | PPRI8282 = CMV010F9 | Terrei | South Africa, 2006 | – | – | MK451536 | – | |
| A. terreus | PPRI8672 = CMV004B9 | Terrei | South Africa, 2007 | Kenaf (Hibiscus cannabinus) | – | – | MK451534 | – |
| A. calidoustus | PPRI15353 = CMV006B3 | Usti | South Africa, KwaZulu-Natal, Pinetown, 2014 | – | – | MK451329 | – | |
| A. insuetus | MRC5597 = CMV017B4 | Usti | South Africa, Western Cape, Cape Town, unknown | Direct scraping off Castle wall | – | – | MK951930 | – |
| A. insuetus | PPRI3456 = CMV006F8 | Usti | South Africa, 1988 | Grass | – | – | MK451429 | – |
| A. pseudodeflectus | PPRI5177a = CMV006F9 | Usti | South Africa, Western Cape, Clanwilliam, 1993 | Rooibos tea (Aspalathus linearis) | MK450644 | MK451096 | MK451503 | – |
| A. pseudodeflectus | PPRI5177b = CMV010G3 | Usti | South Africa, Western Cape, Clanwilliam, 1993 | Rooibos tea (Aspalathus linearis) | – | – | MK451504 | – |
| A. pseudodeflectus | PPRI8971 = CMV005H9 | Usti | South Africa, Limpopo, Kruger National Park, 2005 | Mopane debris (Colophospermum mopane) | MK450642 | MK451064 | MK451498 | – |
| A. pseudodeflectus | PPRI8976 = CMV005I1 | Usti | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | – | MK451499 | – |
| A. pseudodeflectus | PPRI9168 = CMV006A6 | Usti | South Africa, Limpopo, Kruger National Park, 2008 | Mopane twigs and leaves (Colophospermum mopane) | – | – | MK451502 | – |
| A. pseudodeflectus | PPRI9203 = CMV005I2 | Usti | South Africa, Limpopo, Kruger National Park, 2005 | Mopane twigs and leaves (Colophospermum mopane) | MK450643 | MK451065 | MK451500 | – |
| A. pseudodeflectus | PPRI9404 = CMV005I3 | Usti | South Africa, Limpopo, Kruger National Park, 2005 | Soil | – | – | MK451501 | – |
| A. pseudoustus | MRC1233 = CMV017A5 | Usti | South Africa, Western Cape, Drakenstein, 1975 | Apple juice concentrate | – | – | MK951922 | – |
| A. pseudoustus | MRC1234 = CMV017A4 | Usti | South Africa, Western Cape, Drakenstein, 1975 | Apple juice concentrate | – | – | MK951921 | – |
| A. sigurros | PPRI15889 = CMV005I4 (ex-type) | Usti | South Africa, KwaZulu-Natal, Pinetown, 2014 | Environmental sample | MK450650 | MK451066 | MK451512 | MK450803 |
Acronyms of culture collections: PPRI, culture collection of the National Collections of Fungi, housed at the Agricultural Research Council - Plant Health and Protection (ARC), Roodeplaat, South Africa; MRC, culture collection of the Medical Research Council housed at PPRI; CSIR, culture collection of the Council for Scientific and Industrial Research; CMV, working collection housed at the PPRI; DTO, working collection of the Applied and Industrial Mycology group housed the Westerdijk Institute, Utrecht, the Netherlands.
DNA extraction, sequencing, and phylogenetic analysis
DNA was extracted from 7 d old colonies grown on Blakeslee’s (1915) malt extract agar (MEAbl) using the Quick-DNATM Fungal/Bacterial Miniprep Kit (Zymo Research, CA, USA). The 5.8S rDNA internal transcribed spacer regions (ITS), beta-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) genes were amplified in a 25 μl PCR master mix containing 12.5 μl OneTaq® 2X Master Mix with GC Buffer (New England Biolabsinc, MA, USA), 0.5 μl for each primer (10 μM), 10.5 μl milliQ H2O, and 1 μl template DNA. PCR conditions and primers were used as suggested by Samson et al. (2014). Automated sequencing was done at Inqaba Biotechnical Industries (Pty) Ltd (Pretoria, South Africa) using the same primers used for PCR amplification. For RPB2, additional sequencing reactions were performed with internal sequencing primers RPB2-527R (Peterson 2008), RPB2-388F (Peterson 2008), RPB2-F311 (Houbraken & Samson 2011) and RPB2-R310 (Houbraken & Samson 2011).
Contigs were assembled and edited in Geneious Prime v. 2019.2.1 (BioMatters Ltd., Auckland, New Zealand), and new sequences deposited to GenBank (www.ncbi.nlm.nih.gov/genbank/). Accession numbers are listed in Table 1. Sequences were compared to a locally curated reference sequence dataset based on the ex-type sequences published in Samson et al. (2014). Preliminary identifications were made using this dataset in a local BLAST search tool in Geneious. Subsequent reference sequences were selected (Supplementary Table 1) based on these results, with GenBank accession numbers also shown on phylogenetic trees.
All datasets were aligned in MAFFT v. 7.427 (Katoh & Standley 2013) selecting the G-INS-I option, with alignments manually trimmed, adjusted and concatenated in Geneious where needed or appropriate. Aligned datasets were analysed using Maximum Likelihood (ML) and Bayesian tree Inference (BI). For concatenated phylogenies, each gene was treated as separate partitions. ML was performed using IQtree v. 1.6.11 (Nguyen et al. 2015). For each dataset or partition, the most suitable model was calculated using Modelfinder (Kalyaanamoorthy et al. 2017) and ultrafast bootstrapping approximation done using UFBoot2 (Hoang et al. 2018), both integrated into IQtree. BI was performed using MrBayes v. 3.2.7 (Ronquist et al. 2012). The most suitable model for each dataset or partition was selected based on the Akaike information criterion (Akaike 1974) using MrModeltest v. 2.4 (Nylander 2004). Analyses were performed with three sets of four chains (1 cold and three heated) and were stopped at an average standard deviation for split frequencies of 0.01 using the stoprule. Trees were visualised in Figtree v. 1.4.4 (https://github.com/rambaut/figtree/releases) and visually prepared for publication in Affinity Designer v. 1.7.1 (Serif (Europe) Ltd, Nottingham, UK). ML and BI tree topologies did not differ, and thus the former was chosen to present results with both boostrap values and posterior probabilities shown for supported branches.
Several phylogenetic analyses were prepared. Firstly, a total phylogeny based on ITS, BenA, CaM and RPB2 sequence data was calculated which covered all sections detected in this study. Secondly, smaller datasets were prepared based on observed relationships, which allowed for more reliable alignments and more presentable trees. Thirdly, single gene trees were calculated in the case of putative new species to apply the genealogical concordance phylogenetic species recognition concept (GCPSR) (Taylor et al. 2000).
Morphology
Morphological characterisation and species descriptions were made using standardised protocols published in Samson et al. (2014). Colony characters were captured on Czapek yeast autolysate agar (CYA), CYA with 5 % NaCl (CYAS), DG18, MEAbl (Oxoid LP0039 malt extract, Oxoid LP0034 peptone), MEA (Samson et al. 2010), oatmeal agar (OA), yeast extract sucrose agar (YES) and creatine sucrose agar (CREA). Strains were three-point inoculated on these media in 90 mm Petri dishes. Plates were incubated in darkness for 7 d at 25 °C, with additional CYA plates incubated at 30 and 37 °C. Colour names and codes used in descriptions follow Kornerup & Wanscher (1967). Microscopic observations were made using a Zeiss AXIO Imager.M2 compound and Zeiss AXIO Zoom.V16 microscopes equipped with AxioCaM MRc5 and 512 cameras driven by Zen Blue v. 2.3 software (Carl Zeiss CMP GmbH, Göttingen, Germany). Colonies were captured with a Sony NEX-5N camera. Extended Depth of Field analysis and stacking of colony texture micrographs were performed in Helicon Focus v. 7.5.4 (HeliconSoft, Kharkiv, Ukraine). Plates were prepared in Affinity Photo v. 1.7.1 (Serif (Europe) Ltd, Nottingham, UK). For aesthetic purposes, micrographs were adjusted using the "inpainting brush tool" without altering areas of scientific significance.
Results
Strains
Of the ±320 PPRI strains selected for this study, ±250 were viable with 218 selected for sequencing. Eighteen MRC strains were sequenced. New isolations resulted in 65 strains, of which 51 were deposited in PPRI. DNA reference sequences (350 total: 24 ITS, 52 BenA, 250 CaM, 28 RPB2) were generated and submitted to GenBank during this study. Identified strains belonged to 63 species, representing 11 sections of Aspergillus. Seven of the species were found to be novel species and are described below in the Taxonomy section.
Phylogeny
For a general overview of results, a total phylogeny was calculated including all sequences generated during this study and reference sequences summarised in Supplementary Table 1. Results were summarised as a circular tree (Supplementary Fig. 1) and subsequently used as a baseline to calculate more focused phylogenies used to confirm final identifications and show relationships of the novel species.
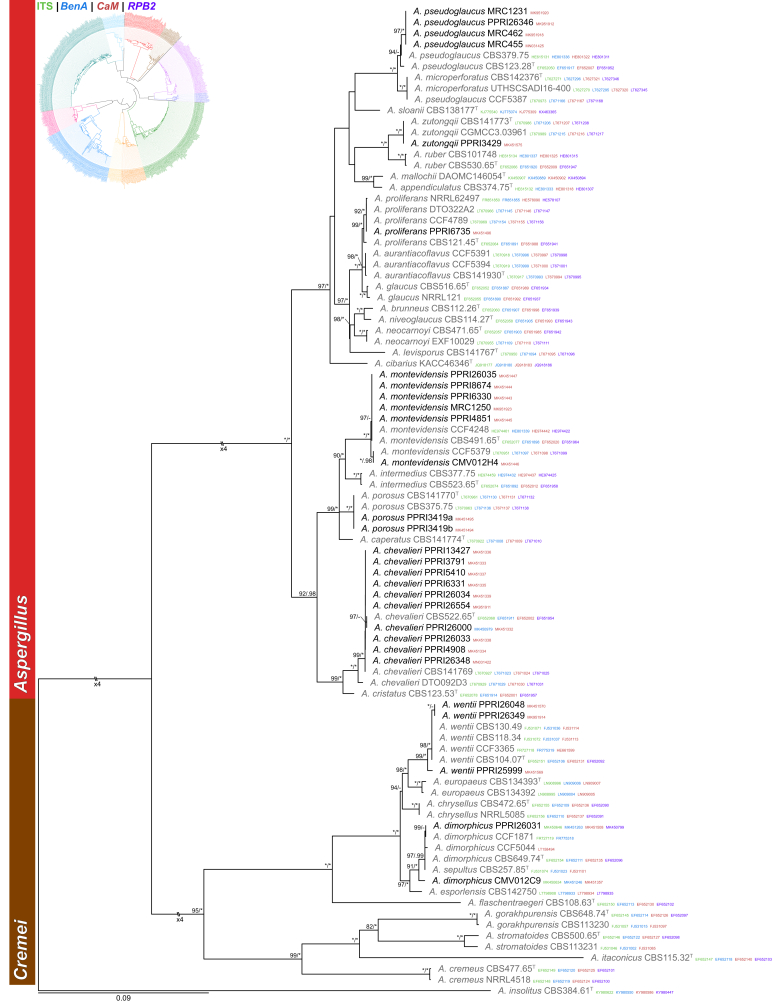
Sections Aspergillus and Cremei (Fig. 1) — We identified six section Aspergillus species including A. chevalieri, A. montevidensis, A. porosus, A. proliferans, A. pseudoglaucus and A. zutongqii. This section was reviewed recently and two recently described species A. porosus and A. zutongqii are detected here (Chen et al. 2017). From section Cremei, we identified A. wentii and A. dimorphicus. Aspergillus dimorphicus and A. sepultus are phylogenetically identical. Since A. dimorphicus (Mehrotra & Prasad 1969) is the older name, A. sepultus (Tuthill & Christensen 1986) is synonymised with the former.
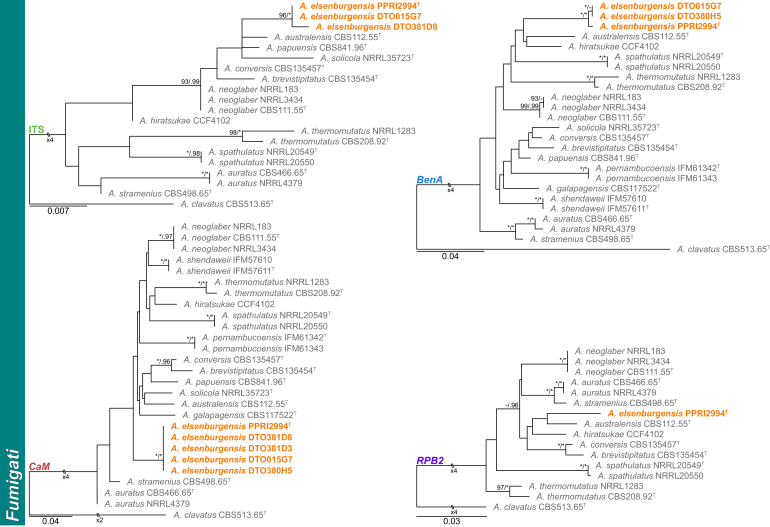
Fig. 1.
Multigene phylogeny of Aspergillus sect Aspergillus and Cremei based on a combined ITS, BenA, CaM and RPB2 dataset. Strains identified during this study are shown in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
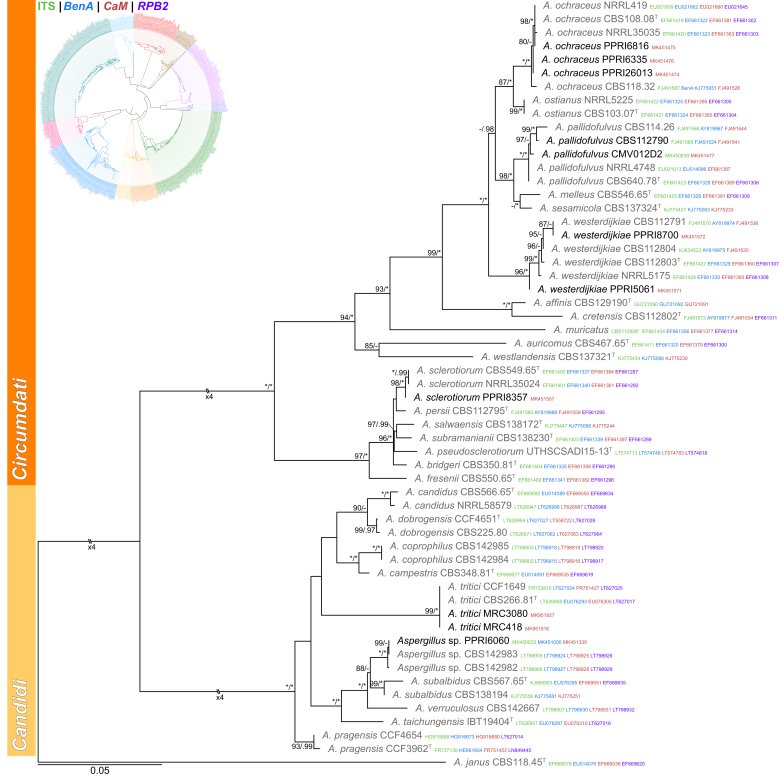
Sections Candidi and Circumdati (Fig. 2) — In section Candidi, only two species were identified. One strain represented A. tritici, while PPRI6060 resolved in a unique clade closely related to A. subalbidus, which represents a new species that will be described in a different paper. Section Circumdati typically contains ochratoxin A producing species (Visagie et al., 2014b, Frisvad and Larsen, 2015a). Our study respectively identified strains as A. ochraceus, A. pallidofulvus, A. sclerotiorum and A. westerdijkiae.
Fig. 2.
Multigene phylogeny of Aspergillus sect Circumdati and Candidi based on a combined ITS, BenA, CaM and RPB2 dataset. Strains identified during this study are shown in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
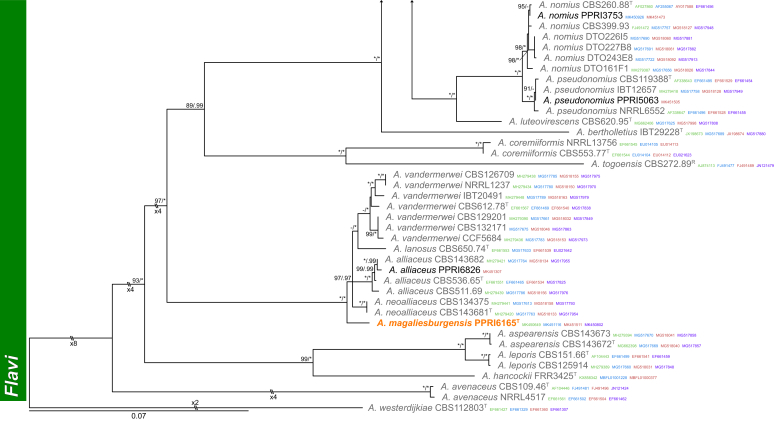
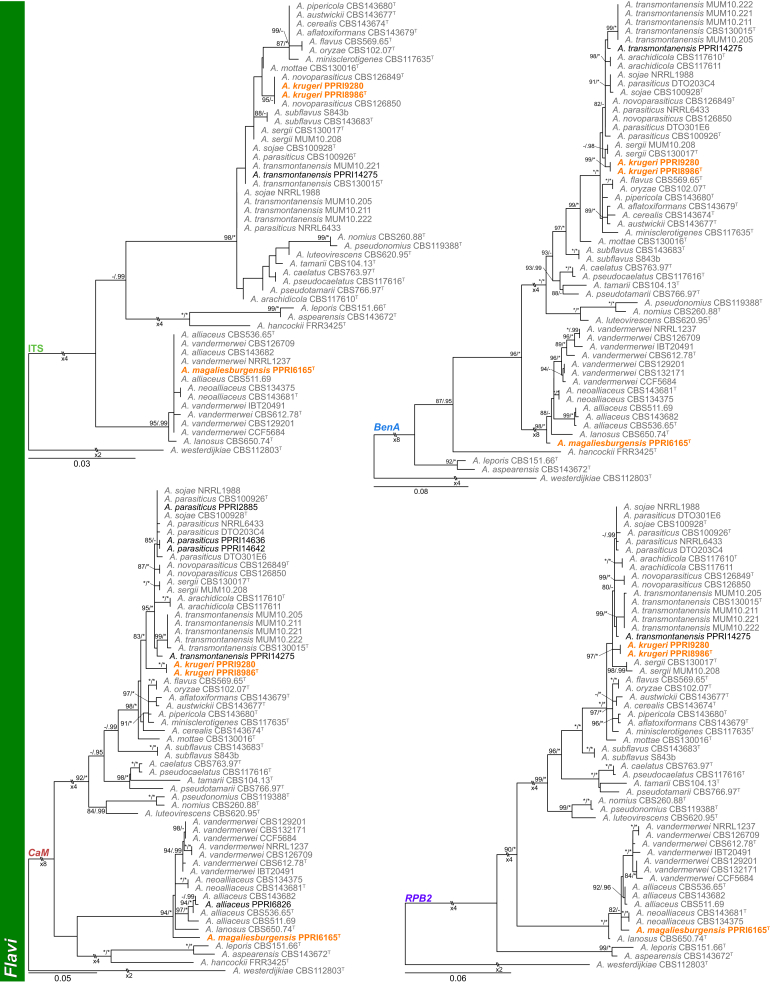
Section Flavi (Fig. 3) — Among strains identified during this study, section Flavi was well represented. Seven known (A. alliaceus, A. flavus, A. nomius, A. parasiticus, A. pseudonomius, A. tamarii and A. transmontanensis) and two new species were detected. PPRI14275 consistently grouped basal to the A. transmontanensis clade. This single strain was morphologically identical to latter and we, therefore, identified it as A. transmontanensis. PPRI8986 and PPRI9280 formed a well-supported clade basal to the A. parasiticus clade and is described below as A. krugeri. PPRI6165 represented a unique lineage in the A. vandermerweii, A. lanosus, A. alliaceus and A. neoalliaceus clade, and is described as A. magaliesburgensis below.
Fig. 3.
Multigene phylogeny of Aspergillus sect Flavi based on a combined ITS, BenA, CaM and RPB2 dataset. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
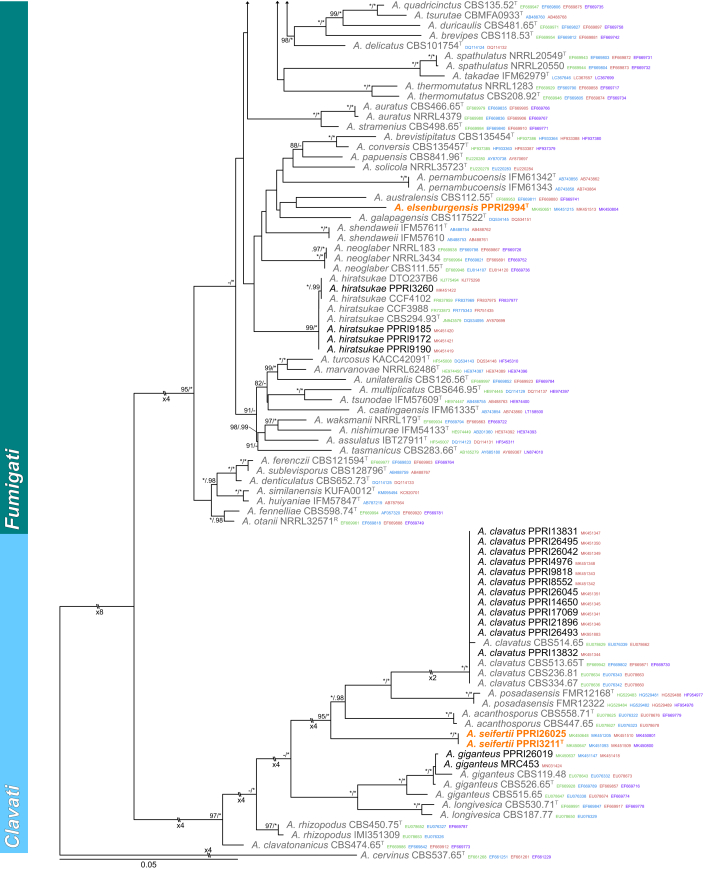
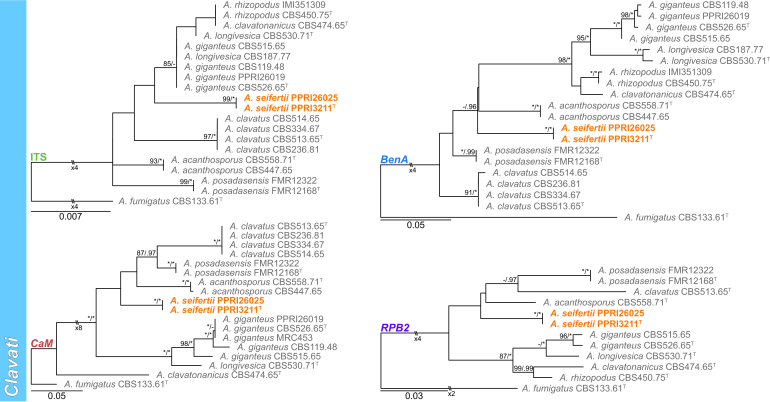
Sections Fumigati and Clavati (Fig. 4) — Section Fumigati was well represented amongst strains identified during this study. Strains were identified into 11 known (A. arcoverdensis, A. aureolus, A. fischeri, A. fumigatiaffinis, A. fumigatus, A. hiratsukae, A. laciniosus, A. lentulus, A. udagawae and A. wyomingensis) and one new species described below as A. elsenburgensis. The multigene phylogeny resolved this strain as sister species to A. australensis. Strains previously identified as A. laciniosus resolved in two distinct clades. One clade containing the ex-type (CBS 117721T) for A. laciniosus and the other the ex-type (CBM-FA884T) for A. takakii. Three species were identified in section Clavati, including A. clavatus, A. giganteus and a new species described below as A. seifertii.
Fig. 4.
Multigene phylogeny of Aspergillus sect Fumigati and Clavati based on a combined ITS, BenA, CaM and RPB2 dataset. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
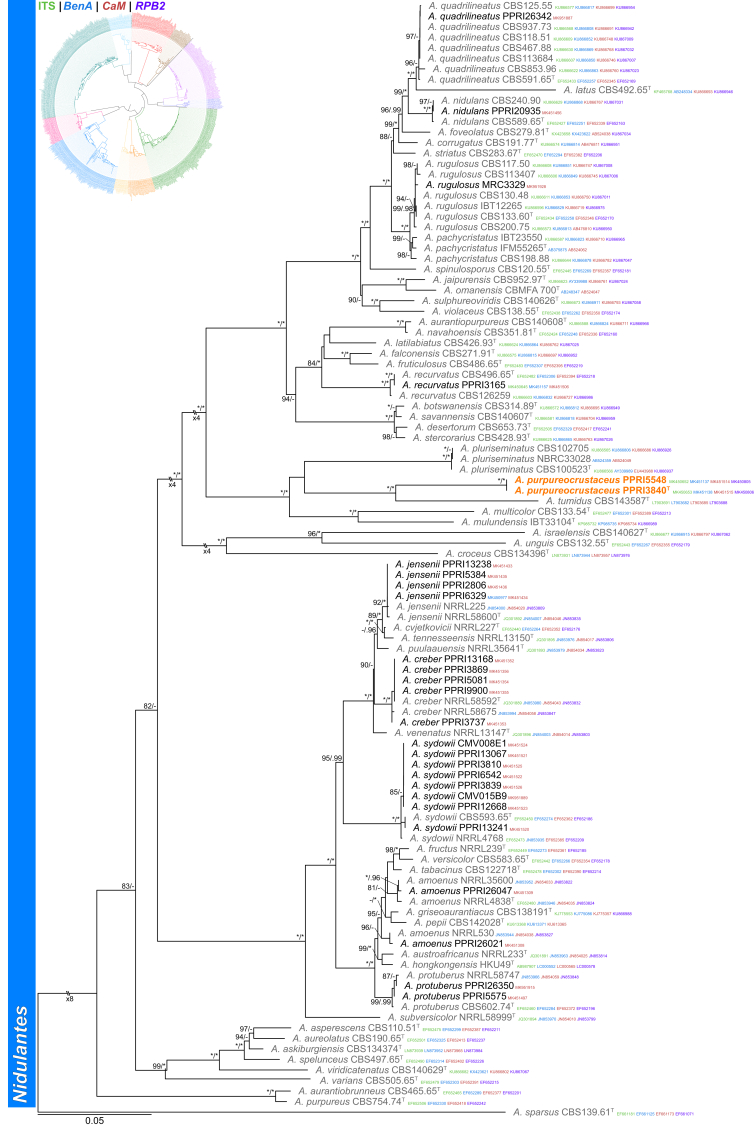
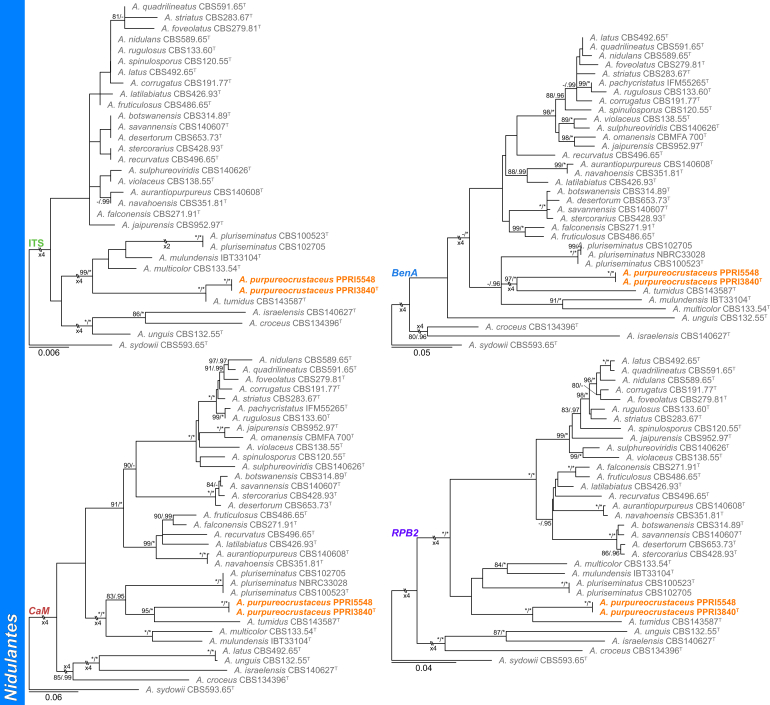
Section Nidulantes (Fig. 5) — Ten section Nidulantes species were identified during this study: A. amoenus, A. creber, A. jensenii, A. nidulans, A. protuberus, A. quadrilineatus, A. recurvatus, A. rugulosus, A. sydowii, and one new species described below as A. purpureocrustaceus. The new species resolved as a close relative of A. tumidus.
Fig. 5.
Multigene phylogeny of Aspergillus sect Nidulantes based on a combined ITS, BenA, CaM and RPB2 dataset. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
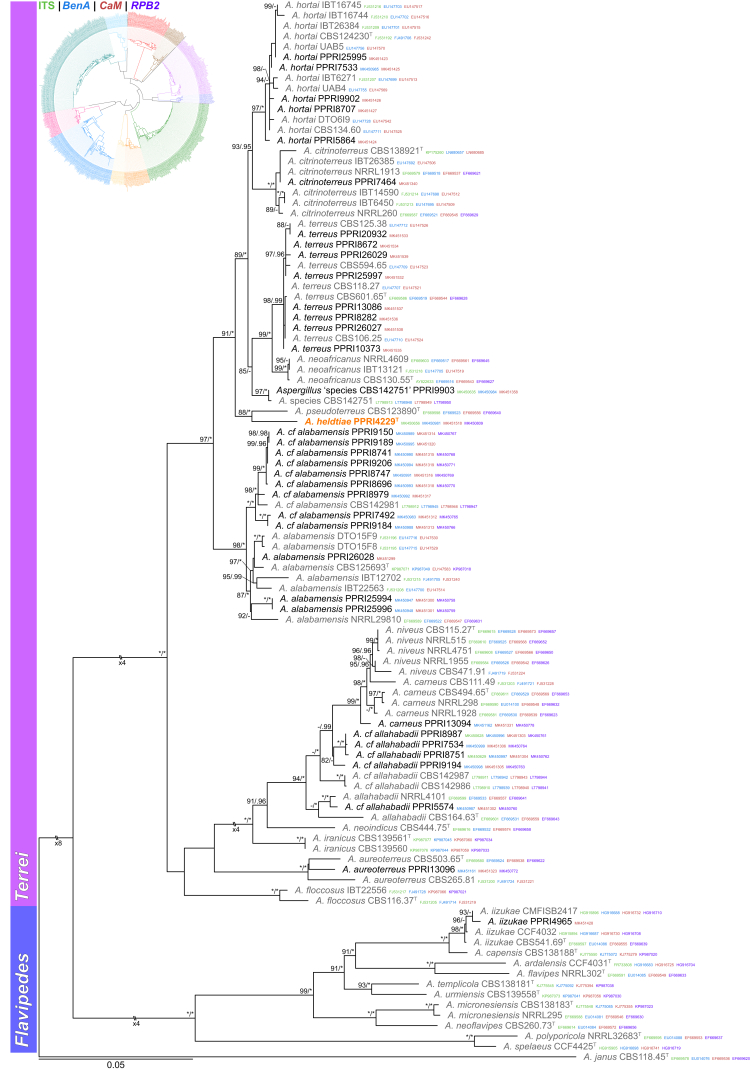
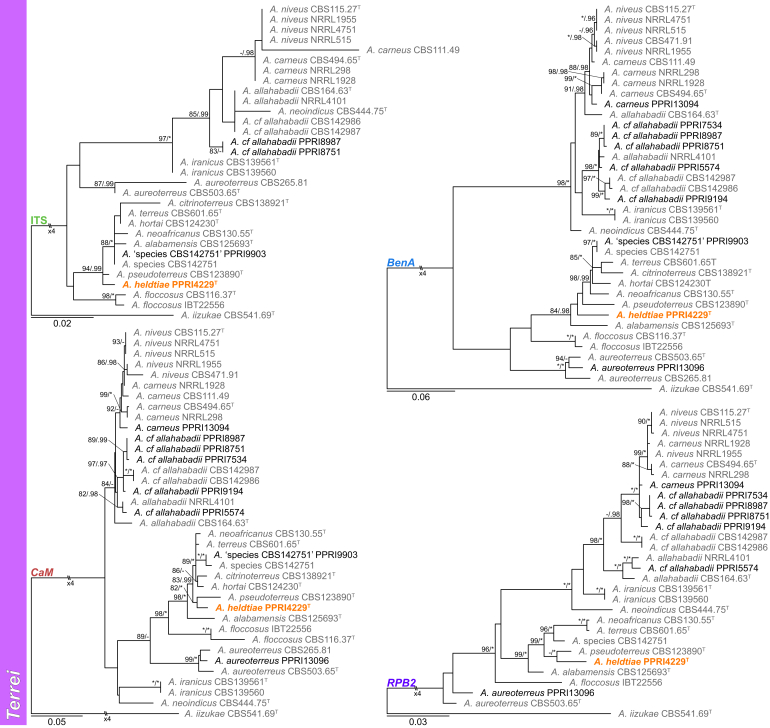
Sections Terrei and Flavipedes (Fig. 6) — Ten section Terrei species were identified during this study: A. alabamensis, A. aureoterreus, A. carneus, A. citrinoterreus, A. hortai, A. terreus and four new species. One of these new species is described below as A. heldtiae, which consistently resolved as a sister species to A. pseudoterreus. The remaining three species or clades were temporarily named A. cf. alabamensis, A. cf. allahabadii and Aspergillus sp. CBS 142751 as they will be described in a separate paper. Aspergillus iizukae was the only species identified from section Flavipedes.
Fig. 6.
Multigene phylogeny of Aspergillus sect Terrei and Flavipedes based on a combined ITS, BenA, CaM and RPB2 dataset. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
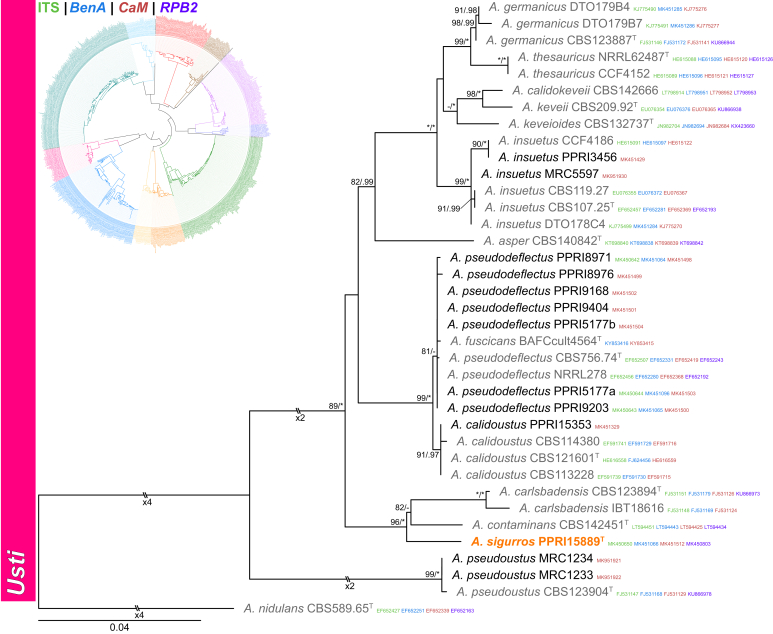
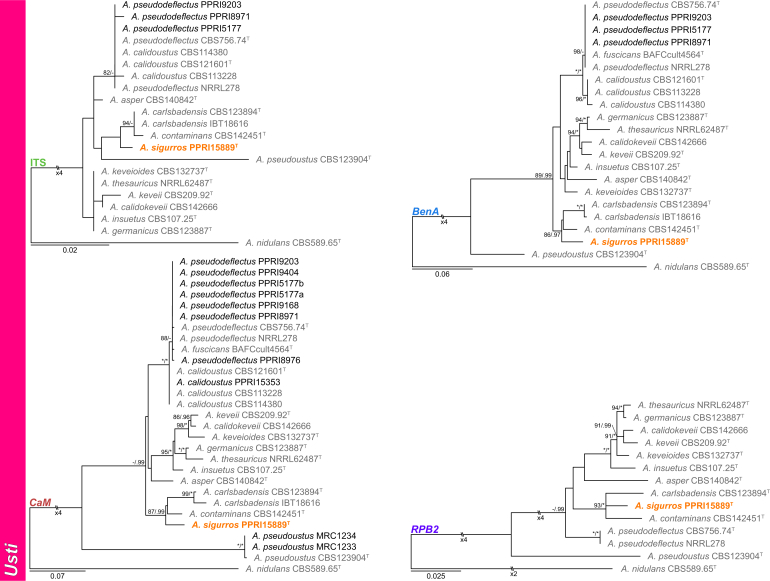
Section Usti (Fig. 7) — Five section Usti species were identified during this study and included A. calidoustus, A. insuetus, A. pseudodeflectus and A. pseudoustus, while one new species is described below as A. sigurros. The new species resolved in a clade with A. carlsbadensis and A. contaminans. Based on phylogenetic results, the more recently described A. fuscicans (Romero et al. 2018) should be considered a synonym of the older A. pseudodeflectus (Samson & Mouchacca 1975).
Fig. 7.
Multigene phylogeny of Aspergillus sect Usti based on a combined ITS, BenA, CaM and RPB2 dataset. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. GenBank accession numbers for ITS (green), BenA (blue), CaM (maroon) and RPB2 (purple) are given behind strain numbers. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
Section Nigri — The PPRI collection contained a large number of black Asperillus strains classified in section Nigri. Full results will be published elsewhere. Strains were identified into nine species as A. aculeatus, A. brasiliensis, A. brunneoviolaceus, A. japonicus, A. neoniger, A. niger, A. piperis, A. tubingensis and A. welwitschiae.
Morphology
We introduce seven new species in the Taxonomy section below. These species belong to sections Clavati, Flavi, Fumigati, Nidulantes, Terrei and Usti based on the phylogenetic analyses. Strains conformed to the general morphological characters previously observed for species accepted in these sections. All of the new species were compared with respective close relatives, with notes provided on distinguishing characters after each species description in the Taxonomy section.
Taxonomy
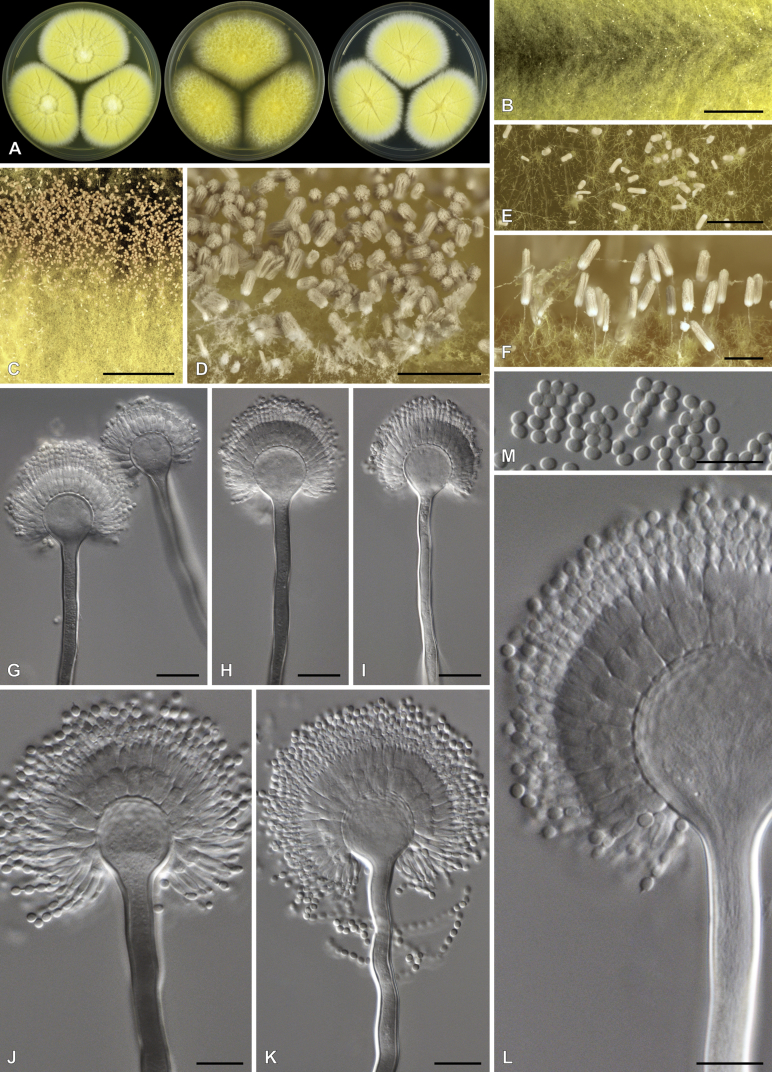
Aspergillus elsenburgensis Visagie, S.M. Romero & Houbraken, sp. nov. MycoBank MB834199. Fig. 14.
Fig. 14.
Aspergillus elsenburgensis. A. Colonies, from left to right, CYA, MEAbl, DG18. B–E. Close-up of colonies on DG18 (B, C), CYA (D) and OA (E). F. Ascoma. G, H. Asci and ascospores. I–L. Conidiophores. M. Conidia. Scale bars: B, D, E = 1 mm; C = 0.2 mm; F, G = 20 μm; H–M = 10 μm.
Etymology: Latin, elsenburgensis, named after Elsenburg, the town the ex-type was collected from.
Classification: Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Fumigati.
Diagnosis: Colonies showing faster growth at 37 °C than at 25 °C, white and floccose, ascomata produced in aerial hyphae after prolonged incubation, white to cream colored, sporulation very sparse, conidiophores with short stipes (10–70 μm) and small globose conidia (1.5–2 μm).
Typus: South Africa, Western Cape, Elsenburg, soil, June 1986, (holotype PREM 62313, culture ex-type PPRI 2994 = CMV 011G4 = CSIR1013).
ITS Barcode: MK450651 (alternative identification markers: BenA = MK451215; CaM = MK451513; RPB2 = MK450804).
Colony diam (7 d, in mm): CYA 35–40; CYA 30 °C 50–53; CYA 37 °C 50–60; CYAS 8–12; MEAbl 55–60; MEA 40–45; DG18 20–25; YES 45–50; OA 45–50; CREA 35–36.
Colony characters (25 °C, 7 d): CYA colonies surface floccose, mycelial areas white, ascomata present after prolonged incubation, produced in aerial hyphae, sparse sporulation present after >2 wk incubation, greenish, soluble pigment absent, exudate clear, reverse pigmentation yellowish white to pale yellow (3A2–3A3). MEAbl colonies surface floccose, mycelial areas white, ascomata present after prolonged incubation, produced in aerial hyphae, sporulation absent, sparse sporulation present after >2 wk incubation, greenish, soluble pigment absent, exudate clear, reverse pigmentation yellowish white to pale yellow (3A2–3A3). YES colonies surface floccose, mycelial areas white, sporulation absent, soluble pigment absent, exudate clear, reverse pigmentation yellowish white to pale yellow (3A2–3A3). DG18 colonies surface floccose, mycelial areas white, sporulation sparse, white but becomes greenish with age, soluble pigment absent, exudate clear, reverse pigmentation yellowish white to pale yellow (3A2–3A3). CREA colonies weak growth, acid not produced.
Micromorphology: Conidial heads radiate. Conidiophores uniseriate. Stipes hyaline, smooth, 10–70 × 2.5–4(–4.5) μm. Vesicles subclavate, phialides cover 50 % of head, 5–8 μm wide. Phialides ampulliform, 4.5–6 × 2–3 μm. Conidia globose, smooth, 1.5–2.5 × 1.5–2.5 μm, (2.06 ± 0.15 × 2 ± 0.16, n = 56) μm, length/width 1.03 ± 0.05. Ascomata neosartorya-like, white to cream, abundant on OA, 70–270 μm. Asci 8-spored, 9–15 μm. Ascospores smooth, with 2 prominent equatorial furrow, globose to subglobose from the top, 4–5 × 3.5–5 μm (4.5 ± 0.2 × 4.2 ± 0.3, n = 43) μm, length/width 1.08 ± 0.07.
Notes: The multigene phylogeny resolves A. elsenburgensis as a close relative of A. australensis and A. galapagensis in section Fumigati (Fig. 4, Fig. 8). The new species grows faster on MEAbl and have somewhat longer stipes than A. australensis (55–60 vs 40–45 mm; up to 70 μm vs up to 30 μm (Samson et al. 2007)). Compared to A. galapagensis, A. elsenburgensis shows slightly faster growth on most media, while it also produces smaller conidia (1.5–2 vs 2.5–3 μm (Samson et al. 2007)).
Fig. 8.
Single gene phylogenies of Aspergillus sect Fumigati based on ITS, BenA, CaM and RPB2. Strains from new species are shown in orange text and reference strains in grey text. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
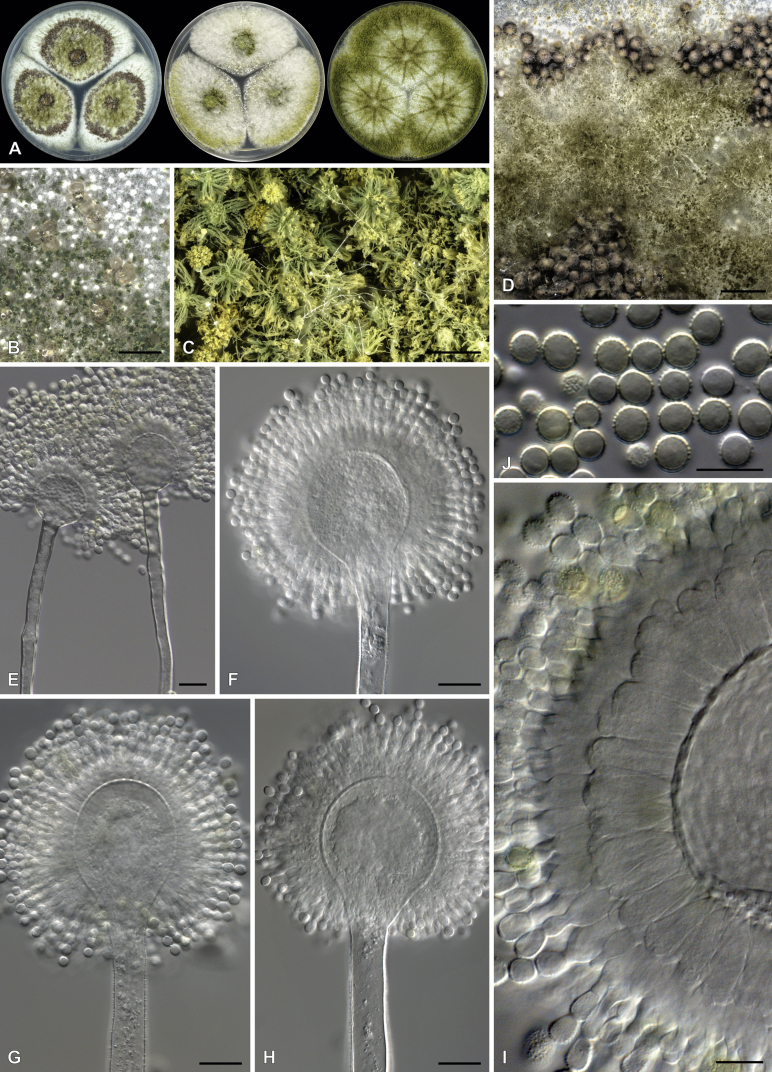
Aspergillus heldtiae Visagie, sp. nov. MycoBank MB834200. Fig. 15.
Fig. 15.
Aspergillus heldtiaeA. Colonies, from left to right, CYA, MEAbl, DG18. B–F. Close-up of colonies on CYA (B), DG18 (C, D) and MEAbl (E, F). G–L. Conidiophores. M. Conidia. Scale bars: B–C = 2 mm; D, E = 0.5 mm; F = 0.2 mm; G–I = 20 μm; J–M = 10 μm.
Etymology: Latin, heltdiae, named after Margaret Vinci Heldt, the creator of the beehive hairstyle that was popular during the 1960s and famously Marge Simpson’s choice of hairstyle. This species resembles the beehive when observed through a dissection microscope.
Classification: Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Terrei.
Diagnosis: Colonies showing rapid growth, bright yellow mycelial areas, cinnamon sporulation, conidiophores biseriate, vesicle 17–28 μm, stipes hyaline with a small proportion darkened, conidia smooth, globose to subglobose, 2–2.5 μm.
Typus: South Africa, unknown, Millet seed, June 1991, (holotype PREM 50864, culture ex-type PPRI 4229 = CMV 004A2).
ITS Barcode: MK450656 (alternative identification markers: BenA = MK450981; CaM = MK451518; RPB2 = MK450809).
Colony diam (7 d, in mm): CYA 54–58; CYA 30 °C 53–56; CYA 37 °C 60–65; CYAS 50–55; MEAbl 55–60; MEA 36–38; DG18 55–65; YES > 70; OA 33–35; CREA 30–32.
Colony characters (25 °C, 7 d): CYA colonies surface floccose, mycelial areas greenish yellow (1A8), sporulation sparse, cinnamon colored, soluble pigment absent, exudate absent, reverse pigmentation olive brown (4B8), light yellow (3A5). MEAbl colonies surface floccose, mycelial areas yellow (2A7), sporulation sparse, cinnamon colored, soluble pigment absent, exudate absent, reverse pigmentation olive brown (4B8), light yellow (3A5). YES colonies surface floccose, mycelial areas greenish yellow (1A8), sporulation sparse, cinnamon colored, soluble pigment absent, exudate absent, reverse pigmentation olive brown (4B8), light yellow (3A5). DG18 colonies surface floccose, mycelial areas greenish yellow (1A8), sporulation sparse, cinnamon colored, soluble pigment absent, exudate absent, reverse pigmentation olive brown (4B8), light yellow (3A5). CREA colonies strong growth, weak acid production.
Micromorphology: Conidial heads columnar. Conidiophores biseriate. Stipes hyaline, small proportion darkened, smooth, 140–330 × 5–8 μm. Vesicles globose, metulae cover 100 % of head, 17–28 μm wide. Metulae 6.5–8.5 × 3–4 μm. Phialides ampulliform, 5.5–7.5 × 2–2.5 μm. Conidia globose to subglobose, smooth, 2–2.5 × 2–2.5 μm, (2.4 ± 0.1 × 2.1 ± 0.1, n = 52) μm, length/width 1.15 ± 0.07. Ascomata not observed.
Notes: Phylogenies resolve A. heldtiae as a close relative of A. pseudoterreus in section Terrei (Fig. 6, Fig. 9). Both species produce bright yellow colonies with cinnamon colored sporulation. However, A. pseudoterreus produce conidiophores in distinctive loosely bundled synnema (Samson et al. 2011a), which is absent in the new species. Aspergillus heldtiae produces a minor proportion of darkened stipes, which are not reported for A. pseudoterreus.
Fig. 9.
Single gene phylogenies of Aspergillus sect Terrei based on ITS, BenA, CaM and RPB2. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
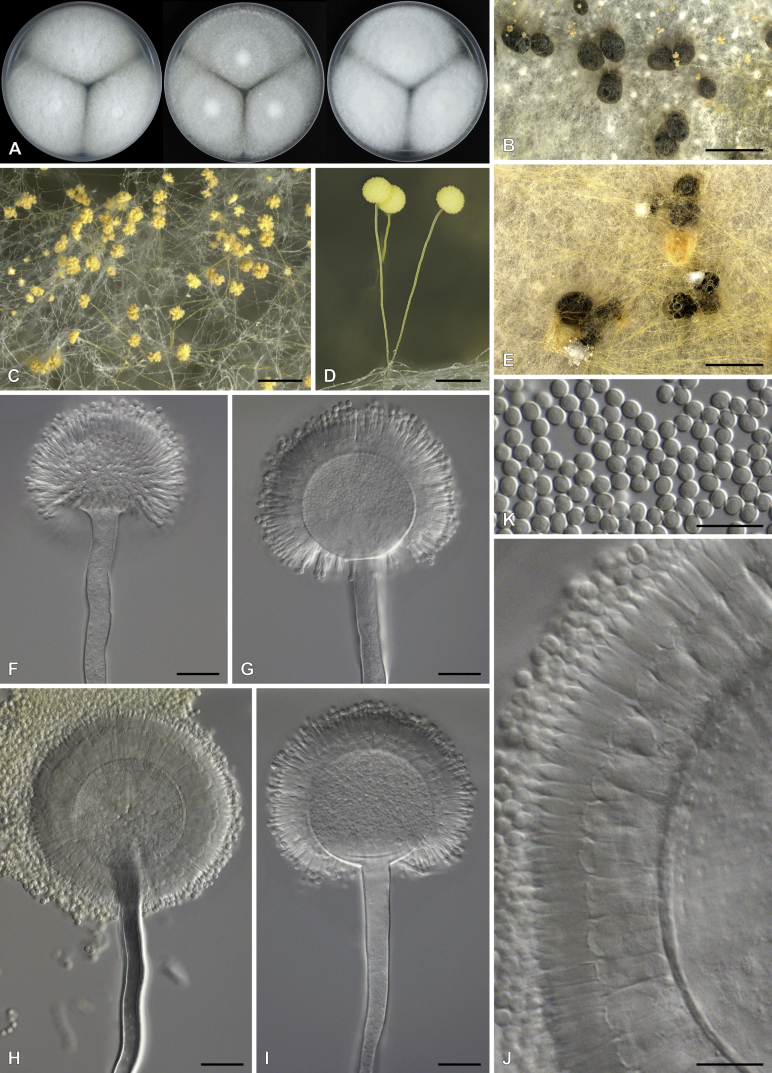
Aspergillus krugeri Visagie, sp. nov. MycoBank MB834203. Fig. 16.
Fig. 16.
Aspergillus krugeri. A. Colonies, from left to right, CYA, MEAbl, DG18. B–D. Close-up of colonies on MEAbl (B), DG18 (C) and CYA (D). E–I. Conidiophores. J. Conidia. Scale bars: B, D = 2 mm; C = 0.5 mm; F, G = 20 μm; H–M = 10 μm.
Etymology: Latin, krugeri, named after the Kruger National Park, the National Park where the ex-type was collected from.
Classification: Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Flavi.
Diagnosis: Colonies on CYA showing rapid growth at 25 °C and moderate growth at 37 °C, dense sporulation, greyish to dark green, dark brown sclerotia abundant, conidial heads radiate, splitting into 3 or more columns, conidiophores uni- to biseriate, stipes rough, vesicle 40–80 μm wide, conidia broadly ellipsoid, rough, 4–7 × 3.5–6.5 μm.
Typus: South Africa, Kruger National Park, Mopane tree debris (Colophospermum mopane), October 2005, collected by E.J. vd Linde (holotype PREM 62309, culture ex-type PPRI 8986 = CMV 006G4).
ITS Barcode. MK450655 (alternative identification markers: BenA = MK451098; CaM = MK451517; RPB2 = MK450808).
Colony diam (7 d, in mm): CYA 60–70; CYA 30 °C 65–70; CYA 37 °C 40–47; CYAS 55–58; MEAbl > 70; MEA > 70; DG18 > 70; YES > 70; OA 52–56; CREA 33–36.
Colony characters (25 °C, 7 d): CYA colonies surface granular and velutinous, mycelial areas white, sporulation moderately dense to dense, greyish green to dark green (29E7–F7) colored, sclerotia abundant, white when young becoming brown to almost purplish, soluble pigment absent, exudate clear, reverse pigmentation pale yellow to dull yellow (3A3–B3), olive brown (4D4) below sclerotia. MEAbl colonies surface granular and velutinous, mycelial areas white, sporulation moderately dense to dense, greyish green to dark green (29E7–F7) colored, sclerotia abundant, white when young becoming brown to almost purplish, soluble pigment absent, exudate clear, reverse pigmentation pale yellow to dull yellow (3A3–B3). YES colonies surface velutinous, granular and floccose, mycelial areas white, sporulation dense, greyish green (29E7–30E7) colored, covering white to brown to almost purplish sclerotia, soluble pigment absent, exudate clear, reverse pigmentation greyish yellow (4B5), pale yellow to light yellow (4A3–5). DG18 colonies surface velutinous, mycelial areas white, sporulation dense, greyish green (29E7–30E7) colored, covering white to brown sclerotia, soluble pigment absent, exudate absent, reverse pigmentation pale yellow to dull yellow (3A3–B3). CREA colonies weak growth, weak acid production.
Micromorphology: Conidial heads radiate, splitting into 3 or more columns. Conidiophores uniseriate to biseriate with an equal ratio. Stipes hyaline, rough, 350–1000(–1300) × 10–18(–21) μm. Vesicles globose to spathulate, metulae/phialides cover 100 % of head, 40–80 μm wide. Metulae 11–22 × 5–10 μm. Phialides ampulliform, 10–15 × 4.5–7 μm. Conidia broadly ellipsoid, rough, 4–7 × 3.5–6.5 μm, (5.5 ± 0.7 × 5.1 ± 0.6, n = 72) μm, length/width 1.08 ± 0.04. Ascomata not observed. Sclerotia white when young, becoming dark brown with age, 370–850 μm.
Notes: Aspergillus krugeri belongs to the A. flavus-clade (Frisvad et al. 2019) and is closely related to A. arachidicola, A. parasiticus, A. novoparasiticus, A. sergii and A. transmontanensis (Fig. 3, Fig. 10). These species are morphologically similar, but colony growth rates can distinguish between them. Aspergillus krugeri grows faster than A. parasiticus (40–60 mm), A. sergii (<55 mm) and A. transmontanensis (55–57 mm) on CYA (Soares et al. 2012). On CYA at 37 °C, Aspergillus krugeri grows more restricted than A. arachidicola (60–70 mm), A. novoparasiticus (58–63 mm), A. sergii (<60 mm) and A. transmontanensis (55–57 mm) (Pildain et al., 2008, Gonçalves et al., 2012).
Fig. 10.
Phylogenies of Aspergillus sect Flavi based on ITS, BenA, CaM and RPB2. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
Aspergillus magaliesburgensis Visagie, sp. nov. MycoBank MB834204. Fig. 17.
Fig. 17.
Aspergillus magaliesburgensis. A. Colonies, from left to right, CYA, MEAbl, DG18. B–E. Close-up of colonies after prolonged incubation on CYA (B–D) and MEAbl (E). F–J. Conidiophores. K. Conidia. Scale bars: B, E = 2 mm; C = 0.2 mm; D = 0.5 mm; F–L = 20 μm; J, K = 10 μm.
Etymology: Latin, magaliesburgensis, named after Magaliesburg, the town the ex-type was collected from.
Classification: Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Flavi.
Diagnosis: Colonies pale, sparse intense yellow sporulation becoming cinnamon with age. Stipes yellow, conidia smooth, globose to subglobose, 2.5–3.5 × 2.5–3.5 μm. Sclerotia present.
Typus: South Africa, Gauteng, Magaliesburg, from an Antlion (Myrmeleontidae), April 1996, collected by J. Pieterse (holotype PREM 62314, culture ex-type PPRI 6165 = CMV 007A3).
ITS Barcode: MK450649 (alternative identification markers: BenA = MK451116; CaM = MK451511; RPB2 = MK450802).
Colony diam (7 d, in mm): CYA 65–70; CYA 30 °C 60–65; CYA 37 °C 50–55; CYAS 65-70; MEAbl > 70; MEA 53–56; DG18 > 70; YES > 70; OA 55–60; CREA 55–60.
Colony characters (25 °C, 7 d): CYA colonies surface floccose, mycelial areas white, some yellow aerial mycelia present, sporulation absent after 7 d, intense yellow when present, with age cinnamon, sclerotia present, black when mature, soluble pigment absent, exudate absent, reverse pigmentation yellowish white to pale yellow (2A2–3). MEAbl colonies surface floccose, mycelial areas white, sporulation absent after 7 d, intense yellow when present, with age cinnamon, sclerotia present, black when mature, soluble pigment absent, exudate absent, reverse pigmentation pale yellow to dull yellow (3A3–B3). YES colonies surface floccose, mycelial areas white, sporulation very sparse, bright yellow, black when mature, soluble pigment absent, exudate absent, reverse pigmentation pale yellow to greyish yellow (4A3–B3). DG18 colonies surface floccose, mycelial areas white, some yellow aerial mycelia present, sporulation absent, soluble pigment absent, exudate absent, reverse pigmentation yellowish white to pale yellow (2A2–3). CREA colonies weak growth, acid not produced.
Micromorphology: Conidial heads radiate. Conidiophores biseriate. Stipes yellow, smooth, (350–)900–1150 × (6–)8–12 μm. Vesicles globose, metulae cover 100 % of head, 40–85 μm wide. Metulae 8–12(–16) × 3.5–5.5 μm. Phialides ampulliform, 7.5–10 × 2–3 μm. Conidia globose to subglobose, smooth, 2.5–3.5 × 2.5–3.5 μm, (3.1 ± 0.2 × 2.9 ± 0.3, n = 52) μm, length/width 1.08 ± 0.12. Ascomata not observed. Sclerotia black when mature, 550–1500 μm.
Notes: Phylogenies resolve A. magaliesburgensis in section Flavi in the A. alliaceus clade (Frisvad et al. 2019), containing A. alliaceus, A. lanosus, A. neoalliaceus and A. vandermerwei (Fig. 3, Fig. 10). BenA, CaM and RPB2 can be used to identify the new species. Aspergillus magaliesburgensis produces sclerotia, and these structures are absent in A. vandermerwei, while A. lanosus typically produces bright yellow colonies. The new species is distinct from A. neoalliaceus based on the subglobose to ellipsoid conidia of the latter. Morphologically, A. magaliesburgensis and A. alliaceus could not be distinguished from each other. We do note that “faintly yellow conidiophores” were previously observed for A. alliaceus (Raper & Fennell 1965), while A. magaliesburgensis produce conidiophores with distinctly yellow stipes.
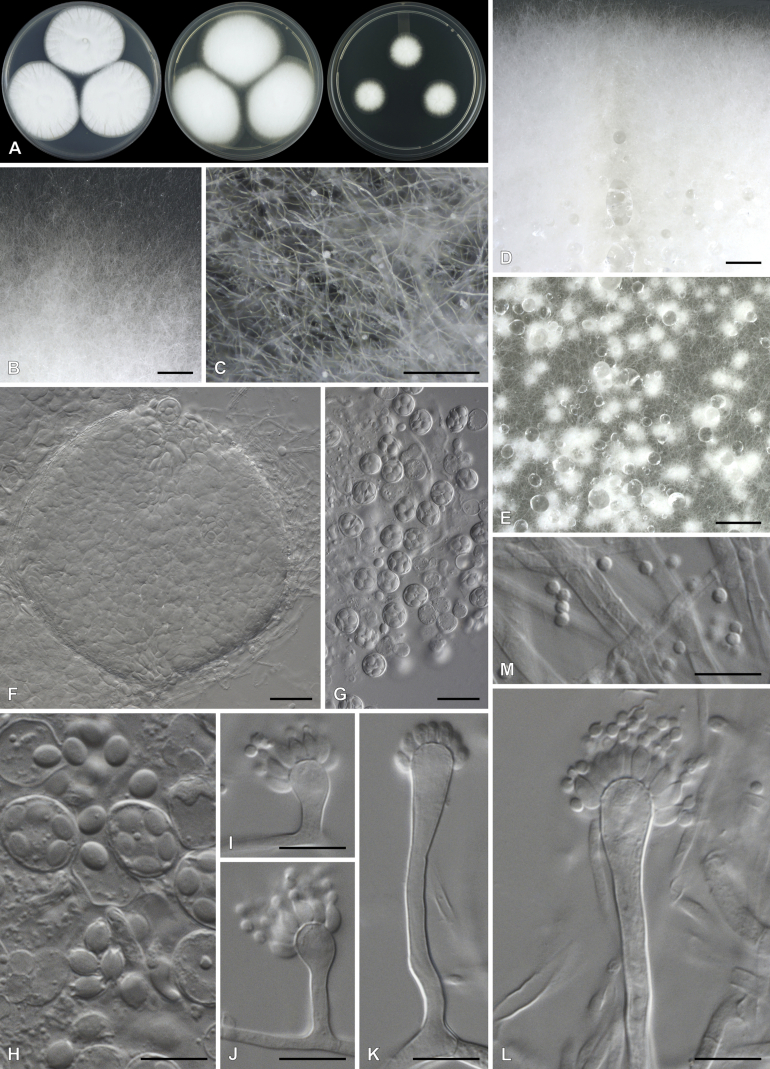
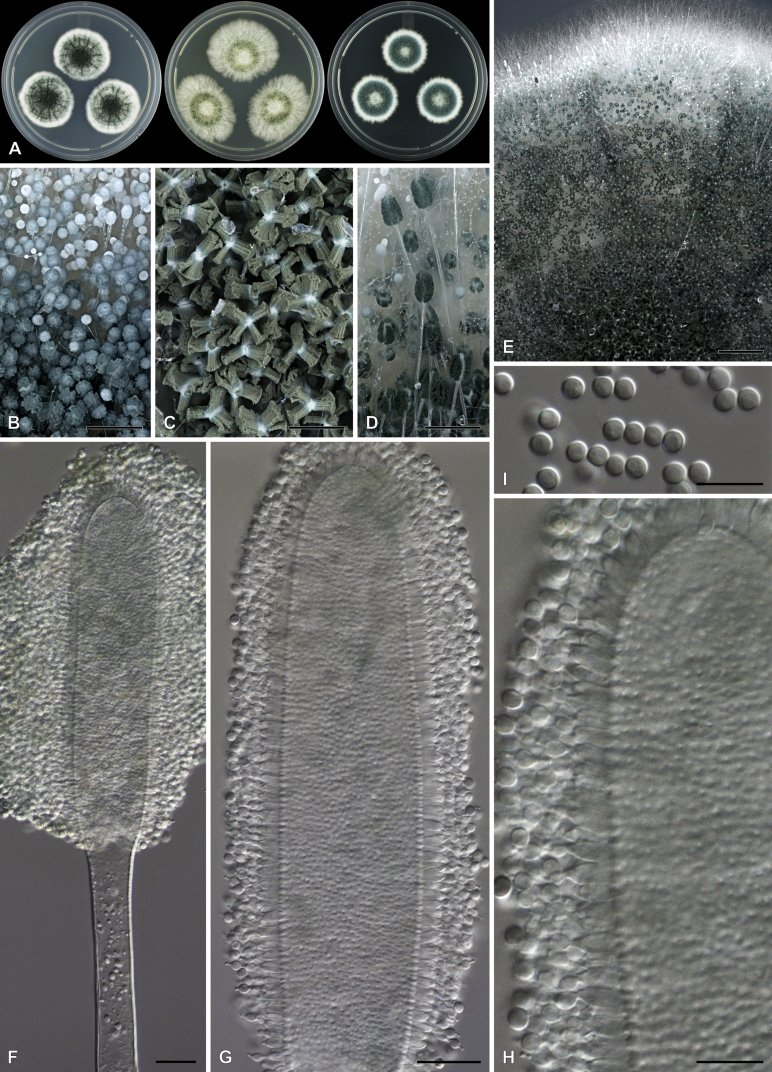
Aspergillus purpureocrustaceus Visagie, sp. nov. MycoBank MB834205. Fig. 18.
Fig. 18.
Aspergillus purpureocrustaceus. A. Colonies, from left to right, CYA, MEAbl, DG18. B–E. Close-up of colonies on MEAbl (B), DG18 (C, E) and CYA (D). F. Hülle cells. G. Potential immature asci. H–K. Conidiophores. L. Conidia. Scale bars: B, C, D = 2 mm; E = 0.5 mm; F = 20 μm; G–K = 10 μm.
Etymology: Latin, named purpureocrustaceus, meaning purple and crust, in reference to the colonies on CYA and MEAbl that turn purple and crust-like with age.
Classification: Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Nidulantes.
Diagnosis: Colonies crust-like and very hard due to abundant Hülle cells produced on surface, having a reddish brown to purple color, sporulation sparse to absent, conidiophores biseriate, stipes 130–310 μm, conidia globose to subglobose, rough, 3.5–4.5(–5) × 3–4.5 μm.
Typus: South Africa, Limpopo, plant debris, January 1990, (holotype PREM 62264, culture ex-type PPRI 3840 = CMV 008B3).
Additional material examined: South Africa, Western Cape, Cape Town, Huntsman spider (Palystes castaneus), January 1994, collected by N. Larsen & H. Robertson PPRI 5548 = CMV 008B1.
ITS Barcode: MK450653 (alternative identification markers: BenA = MK451138; CaM = MK451515; RPB2 = MK450806).
Colony diam (7 d, in mm): CYA 40–41 (25–26); CYA 30 C 10–15; CYA 37 °C no growth; CYAS 25–28; MEAbl 45–47 (33–35); MEA 38–41; DG18 35–40; YES 53–60; OA 25–30; CREA 27–30.
Colony characters (25 °C, 7 d): CYA colonies surface floccose, mycelial areas yellow to grey, sporulation absent, Hülle cells abundant, reddish brown, becoming purple and crust-lie with age, soluble pigment absent, exudate reddish brown and clear, reverse pigmentation dark brown (6F8), yellowish brown (5D4–5). MEAbl colonies surface floccose, mycelial areas yellow to grey, sporulation absent, Hülle cells abundant, reddish brown, becoming purple and crust-lie with age, soluble pigment absent, exudate reddish brown and clear, reverse pigmentation dark brown (6F8), yellowish brown (5D4–5). YES colonies surface floccose, mycelial areas yellow to grey, sporulation absent, Hülle cells abundant, reddish brown, soluble pigment absent, exudate reddish brown and clear, reverse pigmentation olive brown (4F8), pale yellow (4A2). DG18 colonies surface floccose, mycelial areas yellow to grey, sporulation sparse, greyish green (28D6), Hülle cells abundant, reddish brown, soluble pigment absent, exudate reddish brown and clear, reverse pigmentation dark brown (6F8), yellowish brown (5D4–5). CREA colonies weak growth, acid not produced.
Micromorphology: Conidial heads radiate. Conidiophores biseriate. Stipes hyaline, smooth, 130–310 × 5–7.5 μm. Vesicles subclavate, metulae cover 75–100 % of head, 10–20 μm wide. Metulae 6–11.5 × 3–5.5 μm. Phialides ampulliform, 7–10 × 3–4 μm. Conidia globose, rough, 3.5–4.5(–5) × 3–4.5 μm, (4.1 ± 0.4 × 3.9 ± 0.4, n = 28) μm, length/width 1.05 ± 0.04. Hülle cells globose to subglobose, occurring in hard crusts with reddish purple color, 13–25 μm. Ascomata not observed.
Notes: Phylogenies resolve A. purpureocrustaceus in a clade of section Nidulantes with A. multicolor, A. mulundensis, A. pluriseminatus and A. tumidus (Fig. 5, Fig. 11). This group of species typically produce abundant Hülle cells, often giving the colony a reddish to purple color with age (Roy et al., 1987, Stchigel and Guarro, 1997, Chen et al., 2016, Crous et al., 2018). Comparing these species, only A. multicolor and A. mulundensis are capable of growth on CYA at 37 °C. Aspergillus pluriseminatus can be distinguished from the other species in this clade by the presence of a sexual state and absence of asexual state. Compared to the new species, A. tumidus grows more restricted on MEA (38–41 vs 22–23 mm), grows more rapidly on CYA at 30 °C (10–15 vs 32–34 mm), with its colony appearance dominated by good sporulation.
Fig. 11.
Single gene phylogenies of Aspergillus sect Nidulantes based on ITS, BenA, CaM and RPB2. Strains from new species are shown in orange text and reference strains in grey text. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
Aspergillus seifertii Visagie & N. Yilmaz, sp. nov. MycoBank MB834206. Fig. 19.
Fig. 19.
Aspergillus seifertii. A. Colonies, from left to right, CYA, MEAbl, DG18. B–E. Close-up of colonies on DG18 (B) and CYA (C–E). F–H. I. Conidiophores. I. Conidia. Scale bars: B–D = 0.5 mm; E = 2 mm; F, G = 20 μm; H–I = 10 μm.
Etymology: Latin, seifertii, named after Dr. Keith A. Seifert, a prominent Canadian mycologist specialised on mycotoxigenic genera and other hyphomycetes.
Classification – Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Clavati.
Diagnosis — Colonies greyish to dark green, producing large conidiophores with clavate heads, stipes up to 6 mm long, vesicles 26–60 μm wide, up to 210 μm long.
Typus: South Africa, Free State, Golden Gate National Park, Grassroots, January 1988, collected by R. Anelich (holotype PREM 49066, culture ex-type PPRI 3211 = CMV 006F5).
Additional material examined: South Africa, Free State, Golden Gate National Park, Soil, 2018, collected by R. Jacobs, PPRI 26025 = CMV 011E3; CMV 011E4.
ITS Barcode: MK450647 (alternative identification markers: BenA = MK451093; CaM = MK451509; RPB2 = MK450800).
Colony diam (7 d, in mm): CYA 33–35; CYA 30 °C 35–38; CYA 37 °C 2–3; CYAS 10–12; MEAbl 40–45; MEA 38–40; DG18 25–28; YES 45–50; OA 28–35; CREA 20–25.
Colony characters (25 °C, 7 d): CYA colonies surface floccose, mycelial areas white, sporulation moderately dense, greyish green to dark green (25D5–F5), soluble pigment absent, exudate clear, reverse pigmentation pale green to pale yellow (30A3–1A3–2A3). MEAbl colonies surface floccose, mycelial areas white, sporulation sparse, dark green (25F5), soluble pigment absent, exudate clear, reverse pigmentation pale green to pale yellow (30A3–1A3–2A3). YES colonies surface floccose, mycelial areas white, sporulation dense, greyish green to dark green (25D5–F5), soluble pigment absent, exudate clear, reverse pigmentation yellowish white to yellow (3A2–6). DG18 colonies surface floccose, mycelial areas white, sporulation moderately dense, dull green to dark green (25D4–F5), soluble pigment absent, exudate absent, reverse pigmentation pale green to pale yellow to light yellow (30A3–1A3–2A3–3A4). CREA colonies weak growth, acid not produced.
Micromorphology: Conidial heads clavate, with age splitting into 3–4 divergent columns. Conidiophores uniseriate. Stipes hyaline, smooth, up to 6 mm × 17–24 μm. Vesicles clavate, phialides cover 100 % of head, 26–60 μm wide, up to 210 μm long. Phialides ampulliform, 7–9.5 × 2.5–3.5 μm. Conidia globose, smooth, 3–4 × 3–4 μm, 3.4 ± 0.2 × 3.3 ± 0.19, n = 59) μm, length/width 1.03 ± 0.05. Ascomata not observed.
Notes: Phylogenies resolves Aspergillus seifertii as a unique lineage in section Clavati (Fig. 4, Fig. 12). Generally, species from this section produce blue-green conidia and clavate conidiophores (except for A. posadasensis for which only a sexual reproductive state was reported (Marin-Felix et al. 2014)). The stipe and vesicle length are generally good characters to distinguish between these species (Varga et al. 2007). Aspergillus clavatus and A. seifertii produce conidiophores with stipes of up to 3 and 6 mm, respectively, while A. giganteus and A. longivesica can grow several cm in length. The remaining section Clavati species have stipes shorter than 1 mm. Vesicle length is also a useful character. Aspergillus clavati, A. seifertii, A. giganteus and A. longivesica have vesicles up to 200, 210, 600 and 3200 μm, respectively.
Fig. 12.
Single gene phylogenies of Aspergillus sect Terrei based on ITS, BenA, CaM and RPB2. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
Aspergillus sigurros Visagie, sp. nov. MycoBank MB834207. Fig. 20.
Fig. 20.
Aspergillus sigurros. A. Colonies, from left to right, CYA, MEAbl, DG18. B–E. Close-up of colonies on CYA (B), DG18 (C, D) and MEAbl (E). F. Hülle cells. G–K. I. Conidiophores. I. Conidia. Scale bars: B, C, E = 2 mm; D = 0.5 mm; F–H = 20 μm; I–K = 10 μm.
Etymology: Latin, sigurros, named after Sigurrós, one of Keith A. Seifert’s favourite music groups.
Classification: Eurotiomycetes, Eurotiales, Aspergillaceae, Aspergillus section Usti.
Diagnosis: Colonies with grey to brownish moderately dense sporulation, growth on CYA at 30 °C 10–14 mm; conidiophores with brown stipes, vesicles 11–25 μm wide, conidia spiny, globose 3–4 × 3–4 μm.
Typus: South Africa, KwaZulu-Natal, Pinetown, unknown environmental sample, April 2014, collected by M. Truter (holotype PREM 62308, culture ex-type PPRI 15889 = CMV 005I4 = 2014-M62/147).
ITS Barcode: MK450650 (alternative identification markers: BenA = MK451066; CaM = MK451512; RPB2 = MK450803).
Colony diam (7 d, in mm): CYA 34–35; CYA 30 °C 10–15; CYA 37 °C no growth; CYAS 27–29; MEAbl 29–31; MEA 28–30; DG18 28–32; YES 39–42; OA 32–35; CREA 25–26.
Colony characters (25 °C, 7 d): CYA colonies surface floccose, mycelial areas white, sporulation moderately dense, brownish grey to brown (5E2–5–6E5–2), soluble pigment absent, exudate clear, minute droplets, reverse pigmentation olive (2D4), yellowish white (3A2). MEAbl colonies surface floccose, mycelial areas white, sporulation moderately dense, grey (5E1–6E1) to brown (6E6), soluble pigment absent, exudate absent, reverse pigmentation olive (2D4), yellowish white (3A2). YES colonies surface floccose, mycelial areas white, sporulation sparse to moderately dense, greyish brown (5D3–6D3), soluble pigment absent, exudate absent, reverse pigmentation brownish orange (5C5), yellowish white (3A2). DG18 colonies surface floccose, mycelial areas white, sporulation moderately dense, brownish grey to brown (5E2–5–6E5–2), soluble pigment absent, exudate clear, minute droplets, reverse pigmentation brown (5E5), olive (2D4), yellowish white (3A2). CREA colonies strong growth, acid not produced.
Micromorphology: Conidial heads radiate. Conidiophores biseriate. Stipes brown, smooth, (85–)120–360 × 2.5–6.5 μm. Vesicles globose, metulae cover 50–75 % of head, 11–25 μm wide. Metulae 6–10 × 3–4 μm. Phialides ampulliform, 6–8.5 × 2.5–3.5 μm. Conidia globose, spiny to somewhat wart-like, some covered in sheath, 3–4 × 3–4 μm, (3.2 ± 0.2 × 3.2 ± 0.2, n = 53) μm, length/width 1.03 ± 0.04. Hülle cells irregularly elongated, in scattered groups, 22–60 × 11–20 μm. Ascomata not observed.
Notes: Aspergillus sigurros resolves as a close relative of P. carlsbadensis and P. contaminans in section Usti (Fig. 7, Fig. 13). Compared to A. carlsbadensis, the new species produces conidiophores with broader vesicles (11–25 vs 10–14 μm), larger conidia (3–4 vs 2.5–3 μm) and grows more restricted on CYA at 30 °C (10–15 vs 28–32 mm) (Samson et al. 2011b). Microscopically A. contaminans and A. sigurros are very similar. However, the new species grows faster on CYA at 30 °C (4–5 vs 10–14 mm) (Crous et al. 2017).
Fig. 13.
Single gene phylogenies of Aspergillus sect Usti based on ITS, BenA, CaM and RPB2. Strains from new species are shown in orange text, strains identified during this study in black text and reference strains in grey text. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; ∗ = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp).
Discussion
With this project, we aimed to re-identify strains previously lodged in the PPRI and MRC culture collections as Aspergillus or its old associated sexual state genera (e.g. Eurotium, Emericella etc.). Unfortunately, a large proportion of strains in PPRI were either badly contaminated or not viable (±35 %). As a result, only 250 strains were included in this particular study, with 354 new DNA reference sequences (ITS 24; BenA 52; CaM 250; RPB2 28) generated and published on GenBank. South African Aspergillus was found to be relatively diverse with 63 species identified belonging to 11 sections (sections Aspergillus, Candidi, Circumdati, Clavati, Cremei, Flavi, Flavipedes, Fumigati, Nidulantes, Terrei and Usti). This does not include the 11 Aspergillus sect Nigri species that will be published elsewhere. Among the 63 species, seven were found to be new and are described in the Taxonomy section above. One problem experienced during this project was that for the new species, very few strains were available, e.g. four new species were represented by only one strain, while the remaining three new species had only two strains. This situation is frequent when sequencing smaller collections around the world. For A. elsenburgensis we were fortunate that CBS had several additional strains sequenced. Even though not ideal, comparisons based on morphology, multigene phylogenies and single gene trees applying genealogical concordance phylogenetic species recognition (Taylor et al. 2000), leaves little doubt about the novelty of the new species introduced here.
Sequence based identifications of PPRI and MRC strains was relatively straight forward thanks to the secondary identification marker CaM and associated database (Samson et al. 2014). Throughout the genus and between different sections, the primer pairs cmd5&cmd6 performed (Hong et al. 2005) well. For only a minor proportion of strains, additional ITS, BenA and/or RPB2 sequences were needed to confirm the CaM based identifications. ITS and BenA posed no problems in terms of amplification using proposed methods of (Samson et al. 2014), but RPB2 was difficult to amplify using either 5F&7CR (Liu et al. 1999) or 5FEur&7CREur (Houbraken et al. 2012). Both primer sets provided intermitted hits and misses, with the internal sequencing primers F310, R310, 388F and 527R (Houbraken & Samson 2011) at the end needed to obtain high quality sequences contigs.
Several Aspergillus strains belonging to sect Terrei were tentatively identified during this study. Both A. allahabadii and A. alabamensis appears to contain a large degree of infraspecies variation and potentially contain a large number of new species. Even though the multigene phylogeny (Fig. 6) appears to indicate that several new species may exist, we did not feel comfortable introducing new species in a difficult clade without having more data from other regions of the world. Similarly, PPRI 14275 potentially represents a new species closely related to A. transmontanensis from section Flavi. However, since no consistent morphological differences were observed in this strain, we decided to not introduce a phylogenetic species for this single strain.
One of the big challenges we face in Aspergillus is to discover the missing biodiversity. This can either be in the form of new species discovery and/or isolation of additional strains of already known species. Phylogenetic approaches and their incorporation into our species concepts resulted in rather aggressive approaches. It is not ideal to introduce new species based on one or two strains, but as is obvious from this study, often one is left with that as the only option. Monotypic species are frequent in Aspergillus with 118 of the 415 accepted species represented by a single strain (the ex-type), while 80 species are represented by two strains. Within a modern taxonomy like that employed in Aspergillus, this creates problems on several levels, but most pressing is infraspecies variation for species that are often not captured. This is true from a morphological and DNA sequence perspective, but especially concerning the latter, it creates difficulties with identifications. It is not uncommon to find strains that show a few nucleotide differences from the ex-type sequence. Trying to identify such a sequence becomes very complicated amongst monotypic species as one will often not know if the strain belongs to a new species or if they found infraspecies variation within a known species. Studies that generate a lot of additional reference sequences are thus of great importance, not only to discover new species but also to discover infraspecies variation which ultimately makes future species delineations and thus identifications easier. For taxonomic revisions it is crucial to have as much data as possible available, as recently illustrated for the A. viridinutans species complex, where it was found that A. parafelis and A. pseudofelis should be considered synonyms of the genetically diverse A. felis (Hubka et al. 2018). Expanded efforts to isolate and identify fungi should thus remain a priority in important genera such as Aspergillus.
Acknowledgements
We dedicate this paper to Dr Keith A. Seifert on the occasion of his retirement from Agriculture and Agri-Food Canada (Ottawa Research and Development Centre). Apart from his valuable contributions to the international mycological community, he has been a role model, mentor, colleague and friend to the authors on this paper. We thank Stella Romero for providing additional strains and data used for the description of A. elsenburgensis. CMV would like to acknowledge the Foundational Biodiversity Information Programme (FBIP) of the National Research Foundation of South Africa for financial support provided under grant nr 110441 (reference FBIS170406226088). The authors would like to thank Konstanze Bensch and Shaun Pennycook who provided Latin assistance.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2020.02.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Akaike H. A new look at the statistical model identification. IEEE Transations on Automatic Control. 1974;19:716–723. [Google Scholar]
- Allsop N., Olivier D.L., Mitchell D.T. Fungal populations associated with root systems of proteaceous seedlings at a lowland fynbos site in South Africa. South African Journal of Botany. 1987;53:365–369. [Google Scholar]
- Balajee S.A., Houbraken J., Verweij P.E., et al. Aspergillus species identification in the clinical setting. Studies in Mycology. 2007;59:39–46. doi: 10.3114/sim.2007.59.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee A.F. Lindner's roll tube method of separation cultures. Phytopathology. 1915;5:68–69. [Google Scholar]
- Chen A.J., Frisvad J.C., Sun B.D., et al. Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Studies in Mycology. 2016;84:1–118. doi: 10.1016/j.simyco.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.J., Hubka V., Frisvad J.C., et al. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology. 2017;88:37–135. doi: 10.1016/j.simyco.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C. The occurrence of fungi in the soil, after different burning and grazing treatments of the veld in the Transvaal. South African Journal of Science. 1950;46:26–264. [Google Scholar]
- Crous P.W., Wingfield M.J., Burgess T.I., et al. Fungal planet description sheets: 625–715. Persoonia. 2017;39:270–467. doi: 10.3767/persoonia.2017.39.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Burgess T.I., et al. Fungal planet description sheets: 716–784. Persoonia. 2018;40:239–392. doi: 10.3767/persoonia.2018.40.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davolos D., Persiani A.M., Pietrangeli B., et al. Aspergillus affinis sp. nov., a novel ochratoxin A-producing Aspergillus species (section Circumdati) isolated from decomposing leaves. International Journal of Systematic and Evolutionary Microbiology. 2012;62:1007–1015. doi: 10.1099/ijs.0.034785-0. [DOI] [PubMed] [Google Scholar]
- de Hoog G.S., Guarro J., Gene J., et al. 2014. Atlas of clinical fungi; p. 1126. [Google Scholar]
- Eicker A. Microfungi from surface soil of forest communities in Zululand. Transactions of the British Mycological Society. 1969;53:381–392. [Google Scholar]
- Eicker A. Ecological observations on soil fungi. South African Journal of Science. 1970;66:327–334. [Google Scholar]
- Eicker A. Vertical distribution of fungi in zululand soils. Transactions of the British Mycological Society. 1970;55:45–57. [Google Scholar]
- Eicker A. The occurrence and isolation of South African thermophilic fungi. South African Journal of Science. 1972;68:150–155. [Google Scholar]
- Eicker A. The mycoflora of Eucalyptus maculata leaf litter. Soil Biology and Biochemistry. 1973;5:441–448. [Google Scholar]
- Eicker A. The mycoflora of an alkaline soil of the open-savannah of the Transvaal. Transactions of the British Mycological Society. 1974;63:281–288. [Google Scholar]
- Eicker A. Non parasitic mycoflora of the phylloplane and litter of Panicum coloratum. Transactions of the British Mycological Society. 1976;67:275–281. [Google Scholar]
- Eicker A. Mesophilic fungi associated with cultivation of Agaricus brunnescens. Transactions of the British Mycological Society. 1980;74:465–470. [Google Scholar]
- Frisvad J.C., Frank J.M., Joubraken J., et al. New ochratoxin A producing species of Aspergillus section Circumdati. Studies in Mycology. 2004;50:23–43. [Google Scholar]
- Frisvad J.C., Hubka V., Ezekiel C.N., et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology. 2019;93:1–63. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Larsen T.O. Chemodiversity in the genus Aspergillus. Applied Microbiology and Biotechnology. 2015;99:7859–7877. doi: 10.1007/s00253-015-6839-z. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Larsen T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Frontiers in Microbiology. 2015;6:e1485. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Larsen T.O., Thrane U., et al. Fumonisin and ochratoxin production in industrial Aspergillus niger strains. Plos One. 2011;6 doi: 10.1371/journal.pone.0023496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni C., Romano C. Clinical and histological aspects of toenail onychomycosis caused by Aspergillus spp.: 34 cases treated with weekly intermittent terbinafine. Dermatology. 2004;209:104–110. doi: 10.1159/000079593. [DOI] [PubMed] [Google Scholar]
- Gonçalves S.S., Stchigel A.M., Cano J.F., et al. Aspergillus novoparasiticus: a new clinical species of the section Flavi. Medical Mycology. 2012;50:152–160. doi: 10.3109/13693786.2011.593564. [DOI] [PubMed] [Google Scholar]
- Greeff-Laubscher M.R., Beukes I., Marais G.J., et al. The occurrence of mycotoxigenic fungi in abalone feed in South Africa. African Journal of Marine Science. 2018;40:383–394. [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., et al. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Go S., Shin H., et al. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97:1316–1329. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- Hong S.-B., Lee M., Kim D.-H., et al. Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. Plos One. 2013;8 doi: 10.1371/journal.pone.0063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Spierenburg H., Frisvad J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie van Leeuwenhoek. 2012;101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Barrs V., Dudová Z., et al. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia - Molecular Phylogeny and Evolution of Fungi. 2018;41:142–174. doi: 10.3767/persoonia.2018.41.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Kubátová A., Mallátová N., et al. Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Medical Mycology. 2012;50:601–610. doi: 10.3109/13693786.2012.667578. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Mittal N., Kakkar M., et al. Otomycosis: a clinicomycologic study. Ear Nose Throat Journal. 2000;79:606–609. [PubMed] [Google Scholar]
- Kim D.-H., Kim S.-H., Kwon S.-W., et al. Aspergillus cumulatus sp. nov., from rice straw and air for meju fermentation. Journal of Microbiology and Biotechnology. 2014;24:334–336. doi: 10.4014/jmb.1312.12006. [DOI] [PubMed] [Google Scholar]
- Kocsube S., Perrone G., Magista D., et al. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology. 2016;85:199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A., Wanscher J.H. 2nd edn. Methuen & Co Ltd; London, England: 1967. Methuen handbook of colour. [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Marin-Felix Y., Cano-Lira J.F., Guarro J., et al. Leiothecium cristatum sp. nov. and Aspergillus posadasensis sp. nov., two species of Eurotiales from rainforest soils in South America. International Journal of Systematic and Evolutionary Microbiology. 2014;64:2871–2877. doi: 10.1099/ijs.0.063164-0. [DOI] [PubMed] [Google Scholar]
- Mehrotra B.S., Prasad R. Aspergillus dimorphicus and Emericella cleistominuta spp.nov. from Indian Soils. Transactions of the British Mycological Society. 1969;52:331–349. [Google Scholar]
- Miller J.D. Fungi and mycotoxins in grain - implications for stored-product research. Journal of Stored Products Research. 1995;31:1–16. [Google Scholar]
- Nguyen L.T., Schmidt H.A., Von Haeseler A., et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander A.J.J. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Perrone G., Susca A., Cozzi G., et al. Biodiversity of Aspergillus species in some important agricultural products. Studies in Mycology. 2007;59:53–66. doi: 10.3114/sim.2007.59.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Pildain M.B., Frisvad J.C., Vaamonde G., et al. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. International Journal of Systematic and Evolutionary Microbiology. 2008;58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Hocking A.D. 2009. Fungi and food spoilage. [Google Scholar]
- Pitt J.I., Taylor J.W. Aspergillus, its sexual states and the new international code of nomenclature. Mycologia. 2014;106:1051–1062. doi: 10.3852/14-060. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Taylor J.W. (2441) Proposal to conserve the name Aspergillus (Fungi: Eurotiales: Trichocomaceae) with a conserved type to maintain also the name Eurotium. Taxon. 2016;65:631–632. [Google Scholar]
- Pitt J.I., Wild C.P., Baan R.A., et al. Vol. 158. 2012. pp. 119–129. (Economics of mycotoxins: evaluating costs to society and cost-effectiveness of interventions). [PubMed] [Google Scholar]
- Rabie C.J., Lübben A. The mycoflora of sorghum malt. South African Journal of Botany. 1984;3:251–255. [Google Scholar]
- Raper K.B., Fennell D.I. 2nd edn. Williams & Wilkins Company; Baltimore, USA: 1965. The genus Aspergillus. [Google Scholar]
- Romero S.M., Comerio R.M., Barrera V.A., et al. Aspergillus fuscicans (Aspergillaceae, Eurotiales), a new species in section Usti from Argentinean semi-arid soil. Phytotaxa. 2018;343:67–74. [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., et al. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C., van Warmelo K.T. A survey of the mycobiota of a natural Karoo pasture. Bothalia. 1997;27:167–183. [Google Scholar]
- Roy K., Mukhopadhyay T., Reddy G., et al. Mulundocandin, a new lipopeptide antibiotic I. Taxonomy, fermentation, isolation and characterization. The Journal of Antibiotics. 1987;40:275–280. doi: 10.7164/antibiotics.40.275. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Hong S., Peterson S.W., et al. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Studies in Mycology. 2007;59:147–203. doi: 10.3114/sim.2007.59.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Houbraken J., Thrane U., et al. 2010. Food and indoor fungi; p. 390. [Google Scholar]
- Samson R.A., Hubka V., Varga J., et al. Response to Pitt & Taylor 2016: conservation of Aspergillus with A. niger as the conserved type is unnecessary and potentially disruptive. Taxon. 2017;66:1439–1446. [Google Scholar]
- Samson R.A., Mouchacca J. Additional notes on species of Aspergillus, Eurotium and Emericella from Egyptian desert soil. Antonie van Leeuwenhoek. 1975;41:343–351. doi: 10.1007/BF02565069. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Peterson S.W., Frisvad J.C., et al. New species in Aspergillus section Terrei. Studies in Mycology. 2011;69:39–55. doi: 10.3114/sim.2011.69.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Varga J., Meijer M., et al. New taxa in Aspergillus section Usti. Studies in Mycology. 2011;69:81–97. doi: 10.3114/sim.2011.69.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J., et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S., et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte A.L. An overview of Penicillium (Hyphomycetes) and associated teleomorphs in southern Africa. Bothalia. 1992;22:77–91. [Google Scholar]
- Schutte A.L. An overview of Aspergillus (Hyphomycetes) and associated teleomorphs in southern Africa. Bothalia. 1994;24:171–185. [Google Scholar]
- Soares C., Rodrigues P., Peterson S.W., et al. Three new species of Aspergillus section Flavi isolated from almonds and maize in Portugal. Mycologia. 2012;104:682–697. doi: 10.3852/11-088. [DOI] [PubMed] [Google Scholar]
- Stchigel A.M., Guarro J. A new species of Emericella from Indian soil. Mycologia. 1997;89:937–941. [Google Scholar]
- Steinbach W.J., Benjamin D.K., Kontoyiannis D.P., et al. Infections due to Aspergillus terreus: a multicenter retrospective analysis of 83 cases. Clinical Infectious Diseases. 2004;39:192–198. doi: 10.1086/421950. [DOI] [PubMed] [Google Scholar]
- Sugui J.A., Peterson S.W., Clark L.P., et al. Aspergillus tanneri sp. nov., a new pathogen that causes invasive disease refractory to antifungal therapy. Journal of Clinical Microbiology. 2012;50:3309–3317. doi: 10.1128/JCM.01509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart H.J. Observations on fungus cultures from the collection in the Botany Department of the University of the Witwatersrand 2. Some ascosporic species of Aspergillus. South African Journal of Science. 1959;55:51–53. [Google Scholar]
- Taylor J.W., Jacobson D.J., Kroken S., et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Tuthill D.E., Christensen M. Aspergillus sepultus, a new species in the Aspergillus ochraceus group. Mycologia. 1986;78:475–477. [Google Scholar]
- van der Merwe W.J.J., Eicker A., Marasas W.F., et al. Aerospora of an Eragrostis curvula pasture in South Africa. Onderstepoort Journal of Veterinary Research. 1979;46:19–25. [PubMed] [Google Scholar]
- Varga J., Due M., Frisvad J.C., et al. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Studies in Mycology. 2007;59:89–106. doi: 10.3114/sim.2007.59.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Kevei F., Hamari Z., et al. 2000. Genotypic and phenotypic variability among black aspergilli; pp. 397–411. [Google Scholar]
- Visagie C.M., Hirooka Y., Tanney J.B., et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology. 2014;78:63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Varga J., Houbraken J., et al. Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati) Studies in Mycology. 2014;78:1–61. doi: 10.1016/j.simyco.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R.T., Anelich R., Schutte A.L. Fungi in the gut contents of Namib Desert dune Lepismatidae (Thysanura: Insecta) Madoqua. 1990;17:53–54. [Google Scholar]
- Wu F. Global impacts of aflatoxin in maize: trade and human health. World Mycotoxin Journal. 2015;8:137–142. [Google Scholar]
- Wu F., Liu Y., Bhatnagar D. Cost-effectiveness of aflatoxin control methods: economic incentives. Toxin Reviews. 2008;27:203–225. [Google Scholar]
- Zotti M., Corte A.M. Aspergillus persii: a new species in Circumdati section. Mycotaxon. 2002;83:269–278. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.