Abstract
The different developmental stage-associated microbiota of the peach fruit fly, Bactrocera zonata (Diptera: Tephritidae), was characterized using 16S rRNA gene (V3–V4 region) metabarcoding on the Illumina HiSeq platform. Taxonomically, at 97% similarity, there were total 16 bacterial phyla, comprising of 24 classes, 55 orders, 90 families and 134 genera. Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes were the most abundant phyla with Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, Bacteroidia and Bacilli being the most abundant classes. The bacterial genus Enterobacter was dominant in the larval and adult stages and Pseudomonas in the pupal stage. A total of 2645 operational taxonomic units (OTUs) were identified, out of which 151 OTUs (core microbiota) were common among all the developmental stages of B. zonata. The genus Enterobacter, Klebsiella and Pantoea were dominant among the core microbiota. PICURSt analysis predicted that microbiota associated with B. zonata may be involved in membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair processes as well as in cellular processes and signalling. The microbiota that was shared by all the developmental stages of B. zonata in the present study could be targeted and the foundation for research on microbiota-based management of fruit flies.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02381-4) contains supplementary material, which is available to authorized users.
Keywords: Bactrocera zonata, Ontogeny, Microbiota, 16S rRNA gene, Management
Introduction
The dipteran flies of family Tephritidae are commonly known as fruit flies. The peach fruit fly, Bactrocera zonata (Saunders), is a serious insect pest of fruit crops in many parts of the world particularly in the South and South East Asia and in some parts of Africa with more than 50 plant species as host (Kapoor, 1993; Duyck et al. 2004; Choudhary et al. 2012; CABI/EPPO, 2013). Due to huge damage potential, B. zonata is considered as one of the most destructive insect pest species of peach, guava and mango in tropical and subtropical climatic conditions (Choudhary et al. 2012). Most of the studies related to this pest have been limited to its biology, distributions, population genetic structure, host plant association and management through pheromone-based mass trapping as well as with chemical insecticides (Duyck et al. 2004; Choudhary et al. 2012, 2018). However, very few studies have been carried out on the bacterial communities associated with B. zonata, limited to the culture-dependent techniques (Reddy et al. 2014; Naaz et al. 2016), whereas diversity and functional role of bacterial communities has been investigated in many insects groups including fruit flies (Fitt and O’Brien 1985; Lloyd et al. 1986; Prabhakar et al. 2009; Nakajima et al. 2005; Noman et al. 2019).
The bacterial communities in insects mostly live in the gut, hemocoel, mycetomes and within the cells. The bacteria plays a significant role in various metabolic functions of insects, i.e. nutrition, digestion, protection, detoxification of insecticides, extreme thermal tolerance and a source of cues and signals synthesis (semiochemicals) (Dillon and Dillon 2004; Russell and Moran 2006; Oliver et al. 2003; Engel and Moran 2013; Feldhaar, 2011; Douglas, 2015; Hammer and Bowers 2015; Ezenwa et al. 2012; Wingfield et al. 2016; Hosokawa et al. 2017). The insect–bacterial interaction is influenced by various factors like environmental conditions (Parmentier et al. 2016; Hongoh et al. 2006; Zhao et al. 2018; Malacrinò et al. 2018), diet and the host on which insects feed (Ferrari et al. 2007; Hosokawa et al. 2017; Wagner et al. 2015; Medina et al. 2011). Inter- and intraspecific variations have been reported in the bacterial communities of the insects’ species and at different stages of insects’ development (Aharon et al. 2013; Malacrinò et al. 2018).
Recent technology development and availability of cheaper DNA sequencing facilities have enabled the entire metabarcoding profiling and functional role of bacterial communities associated with different stages of insects including fruit flies of genus Bactrocera and Ceratitis (Wang et al. 2013; Wang et al. 2014; Zhao et al. 2018; Yong et al. 2017; Malacrinò et al. 2018; Aharon et al. 2013). However, the profiling of bacterial communities with recent molecular biology culture-independent techniques has not been investigated in the peach fruit fly, Bactrocera zonata.
Based on the current knowledge (Aharon et al. 2013; Andongma et al. 2015; Zhao et al. 2018; Malacrinò et al. 2018), we assumed that the gut microbiota in fruit flies changes with developmental stages (metamorphosis), and this leads to changes in molecular and phenotypic characters that provide the spatio-temporal adaptive advantages to the insect in any environment (Abdelfattah et al. 2017; Malacrinò et al. 2018). Therefore, in the present study, we examined the bacterial community harboured by B. zonata, during ontogeny using metagenomic techniques as well as predicted the functional metabolic activities of the associated microbial communities.
Materials and methods
Ethics statement
The study did not require any permits as it did not involve endangered or protected species.
Sampling for stage-associated microbiota of B. zonata
Fruit flies infested wood apple (Aegle marmelos L.) fruits were collected from Research farm of ICAR Research Complex for Eastern Region, Research Centre (ICAR-RCER, RC), Ranchi, India (23° 45′ N latitude, 85° 30′ E longitude, elevation 620 m AMSL), in May 2018. Four different developmental stages, i.e. first instar larvae, third instar larvae, pupal stage and adult females, were collected. The first instar larvae were directly collected by cutting the infested fruits of wood apple with fruit fly. Different stages of B. zonata were reared on same host fruits in the laboratory conditions according to protocol described by Choudhary et al. (2020) and collected for further experiments. Larvae that emerged from the fruits and moved out to the sand for pupation were collected for the third instar larval stage. A fair number of third instar larvae were left in the soil to pupate in order to obtain pupal stage samples, and other pupae were left for the emergence of adult stage. 5-day-old adult females were collected to study the microbiota associated with the adult stage. All samples (first instar larvae, third instar larvae, pupae and adult females) were placed separately in microtubes and stored in − 80 °C until DNA extraction. Twenty specimens (N = 5 for each developmental stage) were used to compare the microbiota profile of different developmental stages of B. zonata.
DNA extraction, 16S rRNA gene amplification and sequencing
Each sample was separately surface sterilized with 0.1% mercuric chloride for 30 s followed by washing twice with double distilled H2O for 60 s to remove the external contaminations. Then, total DNA was extracted from individual sample using the DNeasy Blood & Tissue Kit (Qiagen, Germany) following the manufacturer’s instructions. The quantity and quality of the extracted DNA were subsequently checked by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Each stage, DNA of five individuals of B. zonata were pooled together to make a single DNA sample for each developmental stage. The pooled DNA of each stage was further used for stage-associated microbiota analysis. The characterization of bacterial community was achieved by targeting the V3-V4 hypervariable regions of 16S rRNA gene with primers 341F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) and 805R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′). Each developmental stage’s library was prepared with a NEBNext Ultra II DNA Library Preparation Kit (New England BioLabs Inc.) according to the manufacturer’s protocol using NEBNextQ5® Hot HiFi PCR Master Mix (NEBNext- New England BioLabs, and the NEBNext oligos kit (2 × 250 bp). Paired-end (PE, 2 × 250 nt) sequencing was performed with an Illumina HiSeq (HiSeq Rapid SBS Kits v2) at AgriGenome Labs, Cochin, Kerala; following the manufacturer’s run protocols (Illumina, Inc., San Diego, CA, USA).
Sequencing data analysis and statistical analysis
The data from each developmental stage were processed and analysed using the Quantitative Insights Into Microbial Ecology (QIIME, version 1.9.1) pipeline (Caporaso et al. 2010a). The total raw sequence reads were quality checked for different parameters, viz. base quality parameters, base composition distribution and GC distribution. Paired-end reads from HiSeq 2500 sequencing were quality trimmed and joined using FLASH program (Version 1.2.11, < https://ccb.jhu.edu/software/FLASH/ >) (Magoč and Salzberg 2011). The consensus sequences were obtained after removing the low-quality read that did not meet the criteria for further analysis. As a part of pre-processing of sequence reads, chimeras were also removed with de novo (uchime_denovo command) using the UCHIME algorithm implemented in the tool VSEARCH, version1.7.0 (Rognes et al. 2016). Pre-processed reads from all samples were pooled and clustered into OTUs based on their sequence similarity using Uclust programme, version 1.2.22q1 (similarity cut-off = 0.97) (Edgar 2010) and assigned taxonomy using the Silva release 13.5 database as the reference126, with the taxonomy assignment tool PyNAST (Caporaso et al. 2010b). The downstream analyses of the core data were done using Biological Observation Matrix (BIOM) file. The sequences that did not have any alignment against taxonomic database and that were classified as mitochondria or chloroplast, as well as singletons, were filtered out of the data set. Bacterial diversity indices and Heatmap of relative abundance were generated using online Microbiome Analyst platform with the Euclidean distance method (Dhariwal et al. 2017). A Venn diagram to determine the unique and shared OTUs across the developmental stages of B. zonata was drawn using the VennDiagram package in R environment (https://cran.r-project.org/bin/windows/base/). Similarity percentage (SIMPER) analysis was performed to calculate the average dissimilarities in structure of bacterial community between different developmental stages of B. zonata. The difference in structure of bacterial community was tested with χ2 test in the PAST 3.25 software package (Hammer et al. 2001). A phylogenetic tree was constructed with the four most abundant phyla of bacterial community of B. zonata. The longest reads of each bacterial family from four most abundant phyla were selected to compute the maximum likelihood tree with Tamura-Nei as the best fit model with 1000 bootstrap using MEGA 6.0 software (Tamura et al. 2013) and visualized using Interactive Tree Of Life (iTOL version 4.0) (Letunic and Bork 2019). The metabolic functions of the bacterial communities identified from the 16S rRNA sequences were predicted by annotating pathways of OTUs against the KEGG database using PICRUSt v. 1.1.4 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved State) software (Langille et al. 2013).
Results
Description of 16S rRNA gene sequencing reads
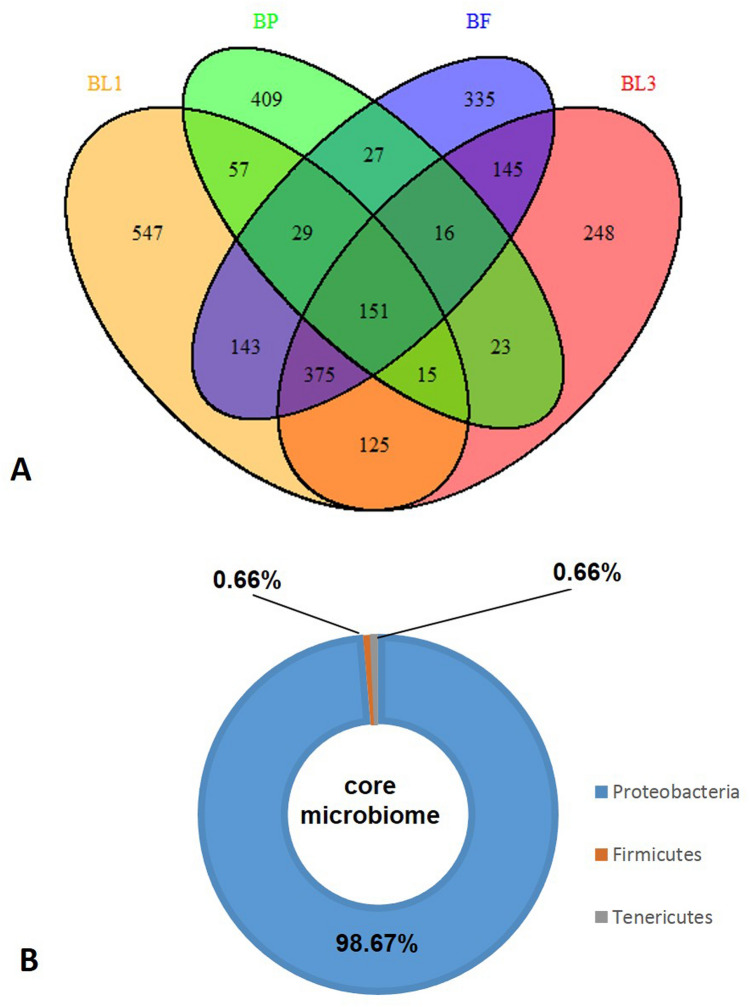
Illumina HiSeq platform was used to sequence the 16S rRNA gene of microbiota associated with different developmental stages of B. zonata. A total of 8,73,093 raw sequence reads of 16S rRNA gene were generated from different developmental stages of B. zonata. Finally, a data set for each stages of B. zonata was prepared after the removal of low quality and chimera from raw sequences (Table 1). Overall, 2645 OUTs were identified from all stages of B. zonata (Table 1). Among them, the lowest number of OTUs (Operational Taxonomic Units) was observed from the pupal stage (BP; 727) and highest number from the first instar larval stage (BL1; 1442) which indicates the complexity of microbial population across the developmental stages (Table 1). The Venn diagram showed that 97% OTUs were shared among different developmental stages of B. zonata (Fig. 1a). A total of 151 OTUs (core microbial community) were common across each stage. Percentage-wise distribution of core microbial community (151 OTUs) reveals that Proteobacteria (> 98%) were the most dominant phylum in each stage, whereas Firmicutes and Tenericutes contributed less than 1% (Fig. 1b). Shannon and Simpson diversity index analysis indicated the highest species richness of bacterial communities in pupal stage of B. zonata (Table 2). The rarefaction curves for the BP (Supplementary Fig. 1) did not attain full plateau, suggesting that bacterial richness in pupal stage of B. zonata was not yet determined completely. Additional sequence sampling is still required to capture the microbiota diversity of B. zonata pupal stage. The taxonomic analyses of all developmental stages are available as supplementary material (Supplementary Table 1) and with clear understanding through sunburst charts of each stage (Supplementary Fig. 2; html links).
Table 1.
Summary of the 16 s rRNA sequencing data
| IDb | No. of bacterial reads | Total consensus sequences | Pre-processed consensus sequences | No. of observed OTUsa | GC content (%) | Avg. length (nt) |
|---|---|---|---|---|---|---|
| BL1 | 250,632 | 143,077 | 118,103 | 1442 | 54.63 | 450 |
| BL3 | 203,871 | 121,364 | 111,049 | 1098 | 50.66 | 450 |
| BP | 216,637 | 110,562 | 66,528 | 727 | 50.58 | 450 |
| BF | 201,953 | 121,261 | 114,815 | 1221 | 50.57 | 450 |
| Total | 2645 |
aOTUs (operational taxonomic units) at the 97% sequence similarity cut-off
bBL1, BL3, BP and BF refer to first instar larvae, third instar larvae, pupa and adult female of Bactrocera zonata
Fig. 1.
OTUs (Operational Taxonomic Units) analysis between different developmental stages of B. zonata at 97% similarity. a Venn diagram showing unique and shared OTUs, of which 151 OTUs shared between all developmental stages. b Percentage distribution of common shared OTUs (151 OTU) between all developmental stages at phylum level. BL1, BL3, BP and BF refers to first instar larvae, third instar larvae, pupa and adult female of Bactrocera zonata
Table 2.
Diversity of bacterial community in different developmental stages of Bactrocera zonata
| IDa | Shannon index | Simpson index | Chao 1 | Ace | Fischer |
|---|---|---|---|---|---|
| BL1 | 1.89 | 0.55 | 716.66 | 756.11 | 104.18 |
| BL3 | 1.19 | 0.32 | 757.42 | 705.14 | 97.46 |
| BP | 2.44 | 0.75 | 315.63 | 310.75 | 44.95 |
| BF | 1.29 | 0.32 | 726.24 | 720.56 | 100.28 |
aBL1, BL3, BP and BF refer to first instar larvae, third instar larvae, pupa and adult female of Bactrocera zonata
The taxonomic composition of microbiota associated with different developmental stages
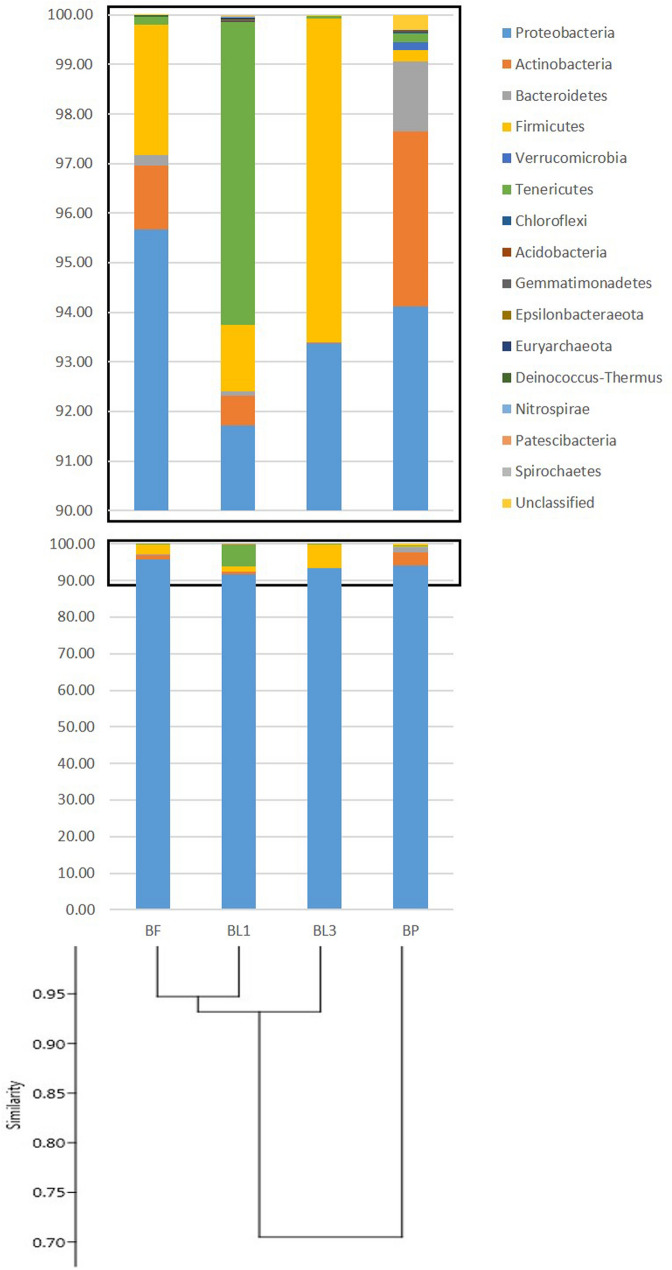
Microbiota associated with B. zonata in the present study were categorized into 16 bacterial phyla including an unassigned bacterial phylum, comprising of 24 classes, 55 orders, 90 families and 134 genera. Among bacterial phyla, Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes were the most abundant across the developmental stages with varying percentages (Table 3 and Fig. 2). Proteobacteria were consistently predominant phylum in all developmental stages (Fig. 2). Proteobacteria dominated with an average of 93.7% (91.72% in BL1 to 95.67% in BF) is followed by Firmicutes with an average 2.6% (0.22% in BP to 6.52% in BL3), Actinobacteria, 1.3% (0.02% in BL3 to 3.52% in BP), and Bacteroidetes, 0.43% (0% in BL3 to 1.41% in BP) of the total reads (Fig. 2 and Supplementary Table 1). However, other 12 phyla, viz. Acidobacteria, Tenericutes, Verrucomicrobia, Epsilonbacteraeota, Deinococcus-Thermus, Gemmatimonadetes, Epsilonbacteraeota, Nitrospirae, Patescibacteria, Chloroflexi, Spirochaetes, Euryarchaeota and other unclassified phylum, were found at very low relative abundance. The exception was Tenericutes in first instar larvae (BL1) which contributed 6.11% of the total reads in this stage. SIMPER analysis revealed that average dissimilarities in the bacterial community structure across developmental stages were primarily due to the sharing of phyla Proteobacteria and Actinobacteria between stages (Table 3).
Table 3.
Average dissimilarity in bacterial community and Chi-square test values between developmental stages
| Taxon | Average dissimilarity (%) | |||||
|---|---|---|---|---|---|---|
| BL1-BL3 | BL1-BP | BL1-BF | BL3-BP | BL3-BF | BP-BFa | |
| Proteobacteria | 2.92 | 4.47 | 2.31 | 7.39 | 0.61 | 6.78 |
| Actinobacteria | 0.97 | 2.33 | 0.45 | 3.30 | 0.52 | 2.78 |
| Tenericutes | 1.09 | 0.84 | 0.89 | 0.25 | 0.20 | 0.06 |
| Firmicutes | 0.10 | 0.27 | 0.07 | 0.17 | 0.17 | 0.34 |
| Bacteroidetes | 0.49 | 1.10 | 0.40 | 1.58 | 0.08 | 1.50 |
| Euryarchaeota | 0.10 | 0.10 | 0.10 | 0.00 | 0.00 | 0.00 |
| Acidobacteria | 0.10 | 0.03 | 0.10 | 0.14 | 0.00 | 0.14 |
| Nitrospirae | 0.03 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 |
| Gemmatimonadetes | 0.03 | 0.10 | 0.03 | 0.14 | 0.00 | 0.14 |
| Chloroflexi | 0.03 | 0.24 | 0.03 | 0.28 | 0.00 | 0.28 |
| Patescibacteria | 0.03 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 |
| Spirochaetes | 0.03 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 |
| Deinococcus-Thermus | 0.02 | 0.07 | 0.03 | 0.05 | 0.00 | 0.04 |
| Epsilonbacteraeota | 0.01 | 0.03 | 0.03 | 0.02 | 0.05 | 0.07 |
| Verrucomicrobia | 0.00 | 0.34 | 0.00 | 0.34 | 0.00 | 0.34 |
| Unknown | 0.08 | 1.13 | 0.05 | 1.22 | 0.03 | 1.18 |
| overall | 6.07 | 11.17 | 4.62 | 14.87 | 1.66 | 13.67 |
| df = | 11 | 14 | 10 | 12 | 9 | 12 |
| chi square test | 10.53 | 3.29 | 14.70 | 6.70 | 25.94* | 6.70 |
| p < 0.5 | NS | p < 0.10 | NS | p < 0.01 | NS | |
aBL1, BL3, BP and BF refer to first instar larvae, third instar larvae, pupa and adult female of Bactrocera zonata
Fig. 2.
UPGMA clustering of B. zonata samples at different developmental stages according to community composition and structure with relative abundance of bacterial community at the phylum level. High-quality sequences obtained from different developmental stages were clustered in operational taxonomic units (OTUs) according to the open-reference method at a 97% of similarity. BL1, BL3, BP and BF refer to first instar larvae, third instar larvae, Pupa and Adult female of Bactrocera zonata
Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, Bacteroidia and Bacilli were the predominant classes of bacterial communities found in the present study. These comprised more than 93% of total reads of all the developmental stages. The most abundant class was Gammaproteobacteria, accounting more than 90% individually in all developmental stages except in the pupal stage (BP) (74.93%) followed by class Alphaproteobacteria (19.09%) (Supplementary Table 1). Across all the developmental stages except pupal stage, the most abundant family was Enterobacteriaceae ranged from 90.10% in first instar (BL1) to 95.35% in adult fly (BF). The pupal stage was only accounted for 22.71% in Enterobacteriaceae because 53.15% of Proteobacteria reads were not classified up to family level. Therefore, the most abundant family in the pupal stage was Pseudomonadaceae (28.62%). When OTUs assigned to the genus level, genus Enterobacter was the most dominant genus found across the developmental stages of B. zonata, which comprised of 64.69%, 83.48% and 82.63% in BL1, BL3 and BF stage, respectively. The pupal stage harboured Pseudomonas (28.62%) as the most abundant genus followed by Enterobacter (15.47%). However, the first instar larval stage consisted of Candidatus-Bacilloplasma lineage which comprised of 5.81%. (Supplementary Table 1).
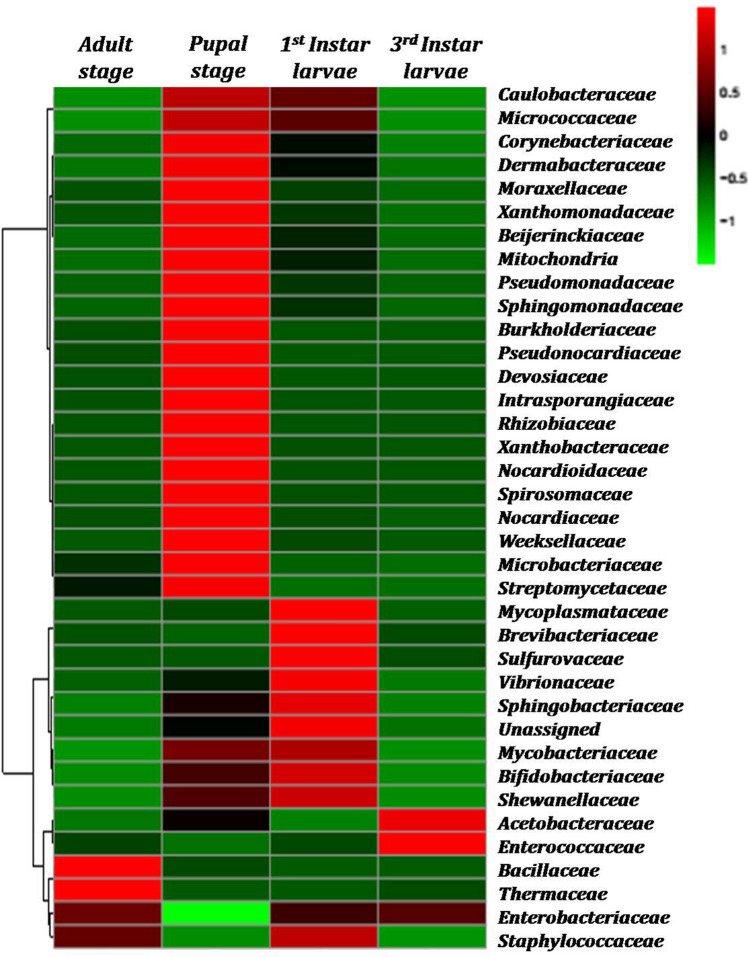
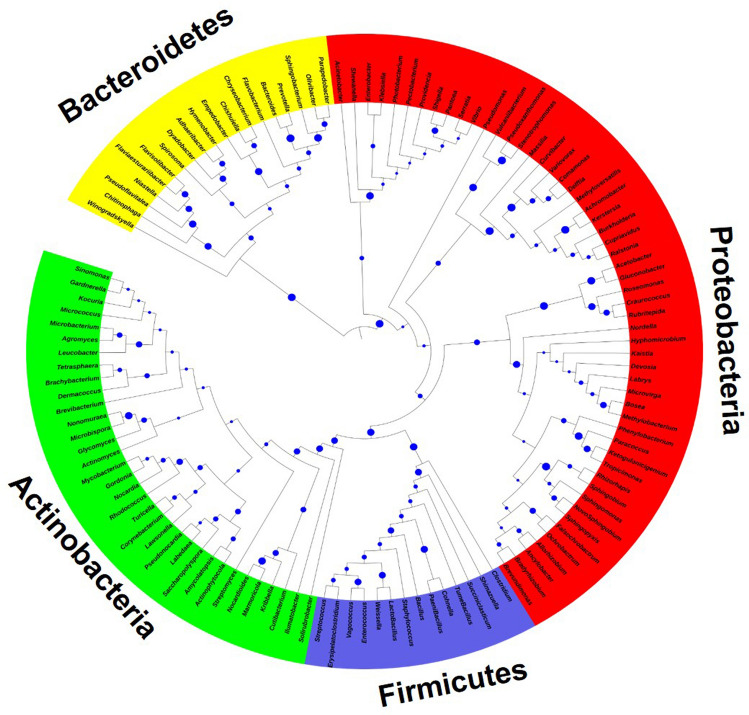
The different developmental stages of B. zonata were tested, representing top picked 37 different families from different phylum. Only a few bacterial OTUs displayed high relative abundance, as shown in the heat map of the OTUs (Fig. 3). UPGMA clustering of OTUs across developmental stages of B. zonata revealed that changes in microbiota composition were not associated with the development of successive stages (Fig. 2). Microbiota profile of third instar larvae and adult were the most similar, whereas microbiota associated with pupae was least shared with those associated with other developmental stages. Phylogenetically, bacterial families of phylum Actinobacteria, Firmicutes and Bacteroidetes were placed in the respective single clade and Proteobacteria were further grouped into classes Alphaproteobacteria, Deltaproteobacteria and Gammaproteobacteria (Fig. 4).
Fig. 3.
Heat maps showing the relative abundances of 16S rRNA gene OTUs between different developmental stages, at family level. The colours indicate relative abundances ranging from green (lower abundances) to red (higher abundances) (indicated in a scale of − 1 to 1)
Fig. 4.
Maximum likelihood tree constructed for phylogenetic analysis of most dominant taxa of bacterial community (on the basis of > 1% abundance of phylum in at least two developmental stages and present across all the developmental stage using 16S rRNA gene sequences) associated with the developmental stages of B. zonata. Bootstrap values were obtained from a search with 1000 replicates
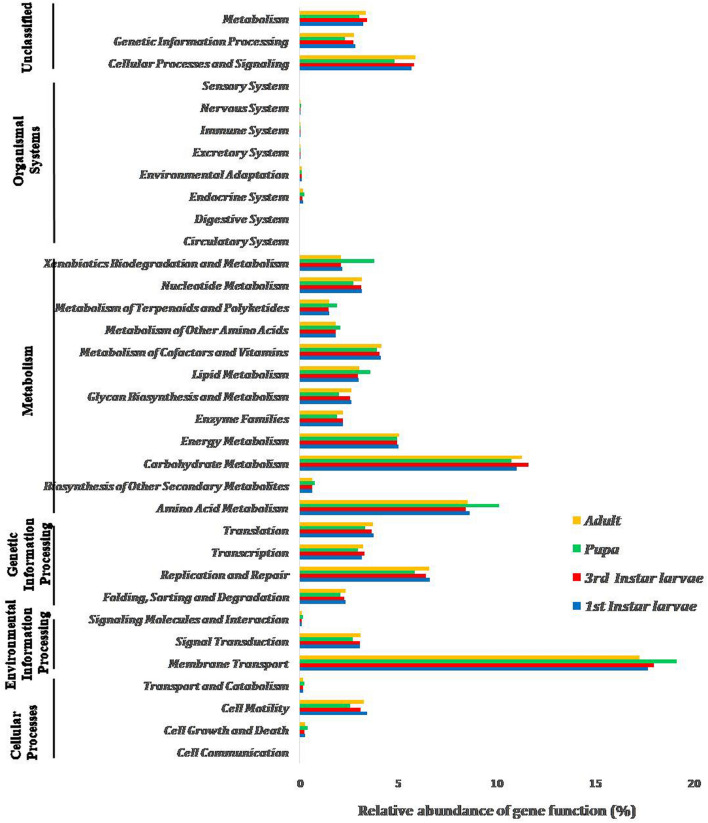
Functional prediction of microbiota across developmental stages
The functional potentials of microbiota associated with developmental stages of B. zonata were predicted using PICRUSt analysis. A total of 6538 KEGG Orthology (KO) groups were observed in the predicted metagenomes involving in membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair processes and cellular processes and signalling (Fig. 5). The highest number of functional potential of carbohydrate metabolism, replication and repair processes, cellular processes and signalling and signal transduction of bacterial communities were predicted in the pupal stage compared to other stages. Based on the OTU abundance, some predicted pathways are likely to be more abundant in adult stage compared with the pupal stage (e.g. carbohydrate metabolism, replication and repair processes, energy metabolism as well as cell motility).
Fig. 5.
Predicted metabolic functions of bacterial communities associated with the different developmental stages of B. zonata. All of the predicted KEGG metabolic pathways are shown at the second hierarchical level and grouped by major functional categories
Genes involved in the membrane transport system are assumed to be associated with the MFS (Major Facilitator Superfamily) transporters. MFS are capable of transporting small solutes in response to ion gradients, ATP-Binding Cassette (ABC) transporters transport a variety of substrates ranging from small ions to macromolecules coupled with the ATP hydrolysis. PTS (phosphoenolpyruvate-dependent phosphotransferase) system can also be involved in the uptake of carbohydrates and convert them into their respective phosphoesters during transportation (Supplementary Table 2). During ontogeny, the abundant OUT-predicted genes were present for the enzymes transketolase and phosphoglycerate mutase that were involved in two different carbohydrate metabolism pathways, viz. pentose-phosphate pathway and glycolysis, respectively. A large number of OTUs were annotated for the “antibiotic transport system ATP-binding proteins” and the “antibiotic transport system permease proteins” in all developmental stages. The high numbers of OTUs were also recorded with proteases that were involved in the protein metabolism pathways (Supplementary Table 2).
Discussion
This is the first ever report using Illumina HiSeq sequencing that revealed the microbiota associated with B. zonata during its ontogeny. Previously, only the gut microbiota of B. zonata had been studied with culture-based and conventional molecular methods (Naaz et al. 2016; Reddy et al. 2014). These methods allow the identification of less than 1% of the insect’s associated microbiota (Hill et al. 2000). Metabarcoding approaches on the Illumina HiSeq platform have proven better to resolve the complexity of the associated microbial community during the ontogeny in fruit flies (Yong et al. 2017; Zhao et al. 2018; Aharon et al. 2013; Malacrinò et al. 2018).
Diversity and predicted functions of microbiota associated with B. zonata
The present study showed that Proteobacteria was dominant phylum associated across the developmental stages of B. zonata. This is consistent with the previous reports on microbiota associated with Bactrocera species, i.e. B. dorsalis, B. carambolae, B. minax, B. tryoni, B. neohumeralis, B. jarvasi and B. cacuminata (Wang et al. 2014; Yong et al. 2017; Morrow et al. 2015; Deutscher et al. 2018; Zhao et al. 2018; Augustinos et al. 2019). In contrast to this, Firmicutes was the dominant phylum in the adult stage of B. dorsalis (Andongma et al. 2015). In this study, the phylum Actinobacteria was the most predominant bacterial community in the pupal stage of B. zonata in comparison with immature (larvae) and adult stages. Recently, association of phylum Actinobacteria with pupal stage of B. dorsalis has been reported with their possible specific role in the pupal stage of the fruit flies (Zhao et al. 2018), as both the Bactrocera species are very similar in their geographical distribution, climatic requirements, biology as well as their host plant association.
The host polyphagy and adaptation to a wide variety of environment by the insects is made possible by the presence of a variety of digestive enzymes and the association with metabolically capable symbiotic bacteria (Berasategui et al. 2016). Our result showed that members of Enterobacteriaceae were the most dominant family during larval and adult stage, whereas members of Pseudomonadaceae were the predominant in the pupal stage of B. zonata. The dominance of the members of Enterobacteriaceae during larval and adult stage of B. dorsalis may have their role in sugar metabolism (Zhao et al. 2018). The results suggest that the process of digestion may differ with the developmental stage, resulting in changes in the microbiota composition. Similarly, host diet is known to influence gut microbiota composition in fruit flies (Malacrinò et al. 2018). The change in gut bacterial diversity was observed when adults of B. dorsalis were fed with a full diet versus a sugar diet (Wang et al. 2011). The family Enterobacteriaceae, known to be associated with most of the fruit fly species, also play a vital role in courtship and reproduction (Ben Ami et al. 2010).
Fruit flies mostly feed on low-nitrogen containing fruits and vegetables and cannot be able to synthesize some of the essential amino acids. Enterobacteriaceae (diazotrophic bacteria) are known to play a role in atmospheric nitrogen fixation (Behar et al. 2005) and may also contribute to host fitness by limiting the proliferation of pathogenic bacteria found in the medfly, Ceratitis capitata (Behar et al. 2008). In the present study, members of Enterobacteriaceae were dominant (> 90%) among bacterial communities across all the developmental stages of B. zonata except pupa (< 23%). This suggests that Enterobacteriaceae may play a valuable role for the development of host fly, B. zonata. In the pupal stage, Pseudomonas was dominant followed by Enterobacter and Achromobacter (> 10% relative abundance) which is in line with the report on B. carambolae (Yong et al. 2017). Pseudomonas spp. is also reported as an entomopathogen in diamondback moth (Indiragandhi et al. 2007), and it can synthesize a toxic molecule with potential antiparasitic activity in mosquitoes (Azambuja et al. 2005) and low fitness in Ceratitis capitata (Behar et al. 2008). We report the association of genus, Candidatus-Bacilloplasma for the first time with B. zonata and found abundantly in the first instar (5.81%). Candidatus-Bacilloplasma is a lineage of Tenericutes which was first time described in Porcellio scaber (Kostanjsek et al. 2007). The presence of coevolved symbiotic bacteria has been reported in other fruit flies such as Bactrocera oleae which harbours “Candidatus Erwinia dacicola” (Capuzzo et al. 2005; Estes et al. 2012). So, the genus Candidatus may have specific role in the development of B. zonata as symbiotic bacteria. The variations in the bacterial communities across developmental stages of B. zonata may reflect changes in the habitat during its life stages.
In the present study, 151 OTUs were common among all the developmental stages of B. zonata and suggest the possibility of vertical transmission of these bacterial communities (Fig. 1A). Vertical transition is common for some of the bacterial communities in tephritid flies (Andongma et al. 2015; Lauzon et al. 2009). The core microbiota of phylum Proteobacteria was comprised of predominant Enterobacter followed by Klebsiella and Pantoea. Some other Proteobacteria were also present in B. zonata, viz. Pectobacterium, Providencia, Serratia and Shigella. These bacterial communities have also been found in the gut of other tephritids (Wang et al. 2011; Liu et al. 2016, 2018; Yong et al. 2017; Morrow et al. 2015; Deutscher et al. 2018; Zhao et al. 2018; Augustinos et al. 2019). Our data are in support of previous studies, at least regarding the “key” players, mainly few genera belonging to Gammaproteobacteria. The genus Stenotrophomonas was found at low levels in the pupa (≤ 0.5%) and in the first instar larvae (< 0.01%) which was earlier recorded in the gut of B. zonata adult flies (Reddy et al. 2014) and also in the adult and larval stage of B. dorsalis (Thaochan et al. 2013; Yong et al. 2017). The diversity and abundance of the bacterial community in insect gut may be influenced by metamorphosis, pH, from acidic to extremely alkaline conditions in the gut compartments of various insects as well as the partial oxygen pressure from the outside environment (Moll et al. 2001; Dillon and Dillon 2004; Engel and Moran 2013).
The present study also provides predictions of the metabolic functions of the microbiota associated with developmental stages of B. zonata. PICRUSt analysis predicted the high abundance of genes involved in membrane transport (especially those connected with ABC transporters) (Fig. 5) which might be related to antibiotic resistance. This is because of ATP-binding cassette (ABC)-type multidrug transporters use a free energy of ATP hydrolysis to pump drugs out of the cell (Putman et al. 2000). The amino acid metabolism could be a vital metabolic function of microbiota for insects feeding on diets with low nitrogen content (Ben-Yosef et al. 2014). However, carbohydrates metabolism, especially pectin hydrolysis, could be an important in larval stage, when fruits have low protein content (Prabhakar et al. 2009). The predicted glutathione S-transferase gene with high abundance might have the ability to detoxify insecticides and xenobiotics (Hu et al. 2014). However, these in silico predicted functions need to be validated in vitro in future research.
The association of symbiotic bacteria in fruit flies is very important for their nutrition, defence, development and fitness to environmental changes. In the present study, information could be used to develop a target symbioticide and manipulation of particular bacterial community harboured during ontogenesis process for the development of Integrated Pest Management programmes. Such pest management programmes have been explored earlier by researchers, i.e. Wolbachia-induced cytoplasmic incompatibility in the medfly (Zabalou et al. 2004), use of symbioticide for fruit fly management (Belcari and Bobbio 1999; Sood and Prabhakar 2009), attracting odours (Naaz et al. 2016), enhancing the success of sterile insect technique (Niyazi et al. 2004), declining the pesticide resistance (Cheng et al. 2017), mass rearing of parasitoids (Leroy et al. 2011) and so on (Prabhakar et al. 2008; Noman et al. 2019). In addition, the symbionts of insect are being exploited as promising sources for novel bioactive metabolites (Dettner 2011). Chemical defence mechanisms provided by microbial symbionts to their host against pathogens, parasites, parasitoids and predators have been studied in several insects, including planthoppers (Fredenhagen et al. 1987), beetles (Kellner 2002), psyllids (Nakabachi et al. 2013) and solitary wasps (Kaltenpoth and Engl, 2014). Actinobacteria are known as important insect’s symbiont for providing defence for their host by producing antimicrobial activity compounds (Kaltenpoth 2009). The moderate abundance of Actinobacteria in the pupal stage of B. zonata as compared to other developmental stages supports the idea that Actinobacteria plays a defensive role in the pupal stage of B. zonata. This is consistent with the previous observation that B. dorsalis has abundant Actinobacteria in the pupae. The presence of Dermabacteraceae and Streptomycetaceae in pupae, in our study, may indicate a defensive function. Members of genus Streptomyces are well known as the main source of antibiotics with diverse biological activities (Arasu et al. 2008). The genus Brachybacterium of the family Dermabacteraceae were also identified and known to express strong antimicrobial activity (Liu et al. 2011). Therefore, pupae of B. zonata could be utilized as a source of bacteria with antimicrobial activity.
In conclusion, this study is the first report of the microbiota diversity and abundance associated during ontogenesis of B. zonata using next-generation sequencing. Distinct microbial flora association with specific developmental stages of B. zonata might suggest a specific role of microbial community to fulfil the developmental needs of particular stages in this insect. The results of the present study also supplement information to the available literature and provide a better understanding of the microbiota associated with B. zonata and other tephritids. Future research is needed to validate the in silico predicted functions of each taxa of microbiota in the in vitro conditions, identifying their capability as source of novel bioactive compounds as a bio-control agent, and the possibilities for targeted manipulation in future management programmes of fruit flies are an exciting research challenge.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the anonymous reviewers and executive editor-in-chief for their valuable comments and suggestion on the earlier version of this paper.
Author contributions
NN, JSC and BD designed the study; NN and JSC carried out the experiments; NN and JSC analysed the data; and NN, JSC, AC, AD and BD shared in scoping the study, data interpretation and writing the manuscript. All the authors have read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Accession numbers
The final Sequence Read Achieves (SRAs) of each developmental stage were deposited in the NCBI SRA database with the SRA Accession Numbers: SRX6875946—SRX6875949 under the Bio-Project PRJNA570100.
References
- Abdelfattah A, Malacrinò A, Wisniewski M, Cacciola SO, Schena L. Metabarcoding: a powerful tool to investigate microbial communities and shape future plant protection strategies. Biol Control. 2017;120:1–10. doi: 10.1016/j.biocontrol.2017.07.009. [DOI] [Google Scholar]
- Aharon Y, Pasternak Z, Ben Yosef M, Behar A, Lauzon C, Yuval B, et al. Phylogenetic, metabolic, and taxonomic diversities shape Mediterranean fruit fly microbiotas during ontogeny. Appl Environ Microb. 2013;79(1):303–313. doi: 10.1128/AEM.02761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andongma AA, Wan L, Dong Y-C, Li P, Desneux N, White JA, Niu CY. Pyrosequencing reveals a shift in symbiotic bacteria populations across life stages of Bactrocera dorsalis. Sci Rep. 2015;5:9470. doi: 10.1038/srep09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasu MV, Duraipandiyan V, Agastian P, Ignacimuthu S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. J Med Mycol. 2008;18:147–153. [Google Scholar]
- Augustinos AA, Tsiamis G, Cáceres C, Abd-Alla AMM, Bourtzis K. Taxonomy, diet, and developmental stage contribute to the structuring of gut-associated bacterial communities in tephritid pest species. Front Microbiol. 2019;10:2004. doi: 10.3389/fmicb.2019.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azambuja P, Garcia ES, Ratcliffe NA. Gut microbiota and parasite transmission by insect vectors. Trends Parasitol. 2005;21:568–572. doi: 10.1016/j.pt.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Behar A, Yuval B, Jurkevitch E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol. 2005;14(9):2637–2643. doi: 10.1111/j.1365-294X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- Behar A, Yuval B, Jurkevitch E. Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J Insect Physiol. 2008;54:1377–1383. doi: 10.1016/j.jinsphys.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Belcari A, Bobbio E. The use of copper in control of the olive fly - Bactrocera oleae. Inf Fitopatol. 1999;49:52–55. [Google Scholar]
- Ben Ami E, Yuval B, Jurkevitch E. Manipulation of the microbiota of mass-reared mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J. 2010;4:28–37. doi: 10.1038/ismej.2009.82. [DOI] [PubMed] [Google Scholar]
- Ben-Yosef M, Pasternak Z, Jurkevitch E, Yuval B. Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J Evol Biol. 2014;27:2695–2705. doi: 10.1111/jeb.12527. [DOI] [PubMed] [Google Scholar]
- Berasategui A, Shukla S, Salem H, Kaltenpoth M. Potential applications of insect symbionts in biotechnology. Appl Microbiol Biotechnol. 2016;100:1567–1577. doi: 10.1007/s00253-015-7186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CABI/EPPO (2013) Bactrocera zonata. [Distribution map]. Distribution Maps of Plant Pests, No.December, 3rd revision, CABI, Wallingford, UK, Map 125.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, De Santis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuzzo C, Firrao G, Mazzon L, Squartini A, Girolami V. “Candidatus Erwinia dacicola”, a co-evolved symbiotic bacterium of the olive fly, Bactrocera oleae (Gmelin) Int J Syst Evol Microbiol. 2005;55:1641–1647. doi: 10.1099/ijs.0.63653-0. [DOI] [PubMed] [Google Scholar]
- Cheng D, Guo Z, Riegler M, Xi Z, Liang G, Xu Y. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel) Microbiome. 2017;5:13. doi: 10.1186/s40168-017-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary JS, Kumari A, Das B, Maurya S, Kumar S. Diversity and population dynamic of fruit flies species in methyl eugenol based parapheromone traps in Jharkhand region of India. Ecoscan. 2012;1:57–60. [Google Scholar]
- Choudhary JS, Naaz N, Lemtur M, Das B, Singh AK, Bhagwati BP, Prabhakar CS. Genetic analysis of Bactrocera zonata (Diptera: Tephritidae) populations from India based on cox1 and nad1 gene sequences. Mitochondrial DNA Part A. 2018;29(5):727–736. doi: 10.1080/24701394.2017.1350952. [DOI] [PubMed] [Google Scholar]
- Choudhary JS, Mali SS, Naaz N, Mukherjee D, Maonaro DB, Singh AK, Rao MS, Bhatt BP. Predicting the population growth potential of Bactrocera zonata (Saunders) (Diptera: Tephritidae) using temperature development growth models and their validation in fluctuating temperature condition. Phytoparasitica. 2020;48:1–13. doi: 10.1007/s12600-019-00777-4. [DOI] [Google Scholar]
- Dettner K (2011) Potential pharmaceuticals from insects and their co-occurring microorganisms. In Insect Biotechnology: A. Vilcinskas (ed), Biologically-Inspired Systems, Springer, Dordrecht, Vol. 2.
- Deutscher AT, Burke CM, Darling AE, Riegler M, Reynolds OL, Chapman TA. Near full-length 16S rRNA gene next-generation sequencing revealed Asaia as a common midgut bacterium of wild and domesticated Queensland fruit fly larvae. Microbiome. 2018;6:85. doi: 10.1186/s40168-018-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King I, Agellon LB, Xia J. MicrobiomeAnalyst—a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:180–188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon R, Dillon V. The gut bacteria of insects: non-pathogenic interactions. Annu Rev Entomol. 2004;49(1):71–79. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Multiorganismal Insects: diversity and function of resident microorganisms. Annu Rev Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyck PF, Sterlin JF, Quilici S. Survival and development of different life stages of Bactrocera zonata (Diptera: Tephritidae) reared at five constant temperatures compared to other fruit fly species. Bull Entomol Res. 2004;94:89–93. doi: 10.1079/ber2003285. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Engel P, Moran NA. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- Estes AM, Hearn DJ, Burrack HJ, Rempoulakis P, Pierson EA. Prevalence of Candidatus Erwinia dacicola in wild and laboratory olive fruit fly populations and across developmental stages. Environ Entomol. 2012;41(2):265–274. doi: 10.1603/EN11245. [DOI] [PubMed] [Google Scholar]
- Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Microbiology: animal behaviour and the microbiome. Science. 2012;338:198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- Feldhaar H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol. 2011;36(5):533–543. doi: 10.1111/j.1365-311.2011.01318.x. [DOI] [Google Scholar]
- Ferrari J, Scarborough CL, Godfray HCJ. Genetic variation in the effect of a facultative symbiont on host-plant use by pea aphids. Oecologia. 2007;153(2):323–329. doi: 10.1007/s00442-007-0730-2. [DOI] [PubMed] [Google Scholar]
- Fitt GP, O’Brien RW. Bacteria associated with four species of Dacus (Diptera: Tephritidae) and their role in the nutrition of the larvae. Oecol (Berl) 1985;67:447–454. doi: 10.1007/BF00384954. [DOI] [PubMed] [Google Scholar]
- Fredenhagen A, Tamura SY, Kenny PTM, Komura H, Naya Y, Nakanishi K, et al. Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J Am Chem Soc. 1987;109:4409–4411. doi: 10.1021/ja00248a055. [DOI] [Google Scholar]
- Hammer Ø, Harper D, Ryan P. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- Hammer TJ, Bowers MD. Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia. 2015;179(1):1–14. doi: 10.1007/s00442-015-3327-1. [DOI] [PubMed] [Google Scholar]
- Hill GT, Mitkowski NA, Aldrich-Wolfe L, Emele LR, Jurkonie DD, Ficke A, et al. Methods for assessing the composition and diversity of soil microbial communities. Appl Soil Ecol. 2000;15(1):25–36. doi: 10.1016/S0929-1393(00)00069-X. [DOI] [Google Scholar]
- Hongoh Y, Ekpornprasit L, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, et al. Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol Ecol. 2006;15(2):505–516. doi: 10.1111/j.1365-294X.2005.02795.x. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. Obligate symbiont involved in pest status of host insect. Proc R Soc Lond [Biol] 2017;274:1979–1984. doi: 10.1098/rspb.2007.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Dou W, Wang JJ, Jia FX, Wang JJ. Multiple glutathione S-transferase genes: identification and expression in oriental fruit fly, Bactrocera dorsalis. Pest Manag Sci. 2014;70:295–303. doi: 10.1002/ps.3558. [DOI] [PubMed] [Google Scholar]
- Indiragandhi P, Anandham R, Madhaiyan M, Poonguzhali S, Kim GH, Saravanan VS, et al. Cultivable bacteria associated with larval gut of prothiofos-resistant, prothiofos-susceptible and field-caught populations of diamondback moth, Plutella xylostella and their potential for antagonism towards entomopathogenic fungi and host insect nutrition. J Appl Microbio. 2007;103:2664–2675. doi: 10.1111/j.1365-2672.2007.03506.x. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M. Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol. 2009;17:529–535. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, Engl T. Defensive microbial symbionts in Hymenoptera. Funct Ecol. 2014;28:315–327. doi: 10.1111/1365-2435.12089. [DOI] [Google Scholar]
- Kapoor VC. Indian fruit flies (Insecta: Diptera: Tephritidae) New York: International Sciences Publisher; 1993. p. 228. [Google Scholar]
- Kellner RL. Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera: Staphylinidae) Insect Biochem Mol Biol. 2002;32:389–395. doi: 10.1016/S0965-1748(01)00115-1. [DOI] [PubMed] [Google Scholar]
- Kostanjsek R, Strus J, Avgustin G. “Candidatus Bacilloplasma”, a novel lineage of mollicutes associated with the hindgut wall of the terrestrial isopod porcellio scaber (Crustacea: Isopoda) Appl Environ Microbiol. 2007;73(17):5566–5573. doi: 10.1128/AEM.02468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, et al. Predictive functional profling of microbial communities using 16S rRNA marker gene sequence. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzon C, McCombs S, Potter S, Peabody N. Establishment and vertical passage of Enterobacter (Pantoea) agglomerans and Klebsiella pneumonia through all life stages of the Mediterranean fruit fly (Diptera: Tephritidae) Ann Entomol Soc Am. 2009;102:85–95. [Google Scholar]
- Leroy PD, Sabri A, Heuskin S, Thonart P, Lognay G, et al. Microorganisms from aphid honeydew attract and enhance the efficacy of natural enemies. Nat Comm. 2011;2:348. doi: 10.1038/ncomms1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:256–259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZX, Huang K, Xiao HD, Chen QH, He JW, Chen YG. Screening and preliminary identification of the antimicrobial strains associated with Anthopleura xanthogrammica. Chin J Antibiot. 2011;36:416–420. [Google Scholar]
- Liu LJ, Martinez-Sañudo I, Mazzon L, Prabhakar CS, Girolami V, Deng YL, Dai Y, Li ZH. Bacterial communities associated with invasive populations of Bactrocera dorsalis (Diptera: Tephritidae) in China. Bull Entomol Res. 2016;106:718–728. doi: 10.1017/S0007485316000390. [DOI] [PubMed] [Google Scholar]
- Liu S, Chen Y, Li W, Tang G, Yang Y, Jiang HB, Dou W, Wang JJ. Diversity of bacterial communities in the intestinal tracts of two geographically distant populations of Bactrocera dorsalis (Diptera: Tephritidae) J Econ Entomol. 2018;111(6):2861–2868. doi: 10.1093/jee/toy231. [DOI] [PubMed] [Google Scholar]
- Lloyd AC, Drew RAI, Teakle DS, Hayward AC. Bacteria associated with some Dacus species (Diptera: Tephritidae) and their host fruit in Queensland. Aust J Biol Sci. 1986;39:361–368. [Google Scholar]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malacrinò A, Campolo O, Medina RF, Palmeri V. Instar- and host-associated differentiation of bacterial communities in the Mediterranean fruit fly Ceratitis capitata. PLoS One. 2018;13(3):e0194131. doi: 10.1371/journal.pone.0194131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RF, Nachappa P, Tamborindeguy C. Differences in bacterial diversity of host-associated populations of Phylloxer anotabilis Pergande (Hemiptera: Phylloxeridae) in pecan and water hickory. J Evol Biol. 2011;24(4):761–71. doi: 10.1111/j.1420-9101.2010.02215.x. [DOI] [PubMed] [Google Scholar]
- Moll RM, Romoser WS, Modrzakowski MC, Moncayo AC, Lerdthusnee K. Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J Med Entomol. 2001;38:29–32. doi: 10.1603/0022-2585-38.1.29. [DOI] [PubMed] [Google Scholar]
- Morrow JL, Frommer M, Shearman FC, Riegler M. The microbiome of field-caught and laboratory-adapted Australian Tephritid fruit fly species with different host plant use and specialisation. Microb Ecol. 2015;70:498–508. doi: 10.1007/s00248-015-0571-1. [DOI] [PubMed] [Google Scholar]
- Naaz N, Choudhary JS, Prabhakar CS, Moanaro MS. Identification and evaluation of cultivable gut bacteria associated with peach fruit fly, Bactrocera zonata (Diptera: Tephritidae) Phytoparasitica. 2016;44:165–176. [Google Scholar]
- Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 2013;23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Hongoh Y, Usami R, Kudo T, Ohkuma M. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol Ecol. 2005;54:247–255. doi: 10.1016/j.femsec.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Niyazi N, Lauzon CR, Shelly TE. Effect of probiotic adult diets on fitness components of Sterile male Mediterranean fruit flies (Diptera: Tephritidae) under laboratory and field cage conditions. J Econ Entomol. 2004;97(5):1570–1580. doi: 10.1603/0022-0493-97.5.1570. [DOI] [PubMed] [Google Scholar]
- Noman MS, Liu L, Bai Z, Li Z. Tephritidae bacterial symbionts: potentials for pest management. Bull Entomol Res. 2019;1:14. doi: 10.1017/S0007485319000403. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci. 2003;100(4):1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier L, Meeus I, Mosallanejad H, de Graaf DC, Smagghe G. Plasticity in the gut microbial community and uptake of Enterobacteriaceae (Gammaproteobacteria) in Bombus terrestris bumblebees’ nests when reared indoors and moved to an outdoor environment. Apidologie. 2016;47(2):237–250. doi: 10.1007/s13592-015-0393-7. [DOI] [Google Scholar]
- Prabhakar CS, Sood P, Mehta PK. Protein hydrolyzation and pesticide tolerance by gut bacteria of Bactrocera tau (Walker) Pest Manage Econ Zoo. 2008;16:123–129. [Google Scholar]
- Prabhakar C, Sood P, Kapoor V, Kanwar S, Mehta P, Sharma P. Molecular and biochemical characterization of three bacterial symbionts of fruit fly, Bactrocera tau (Tephritidae: Diptera) J Gen Appl Microbiol. 2009;55:479–487. doi: 10.2323/jgam.55.479. [DOI] [PubMed] [Google Scholar]
- Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64:672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K, Sharma K, Singh S. Attractancy potential of culturable bacteria from the gut of peach fruit fly, Bactrocera zonata (Saunders) Phytoparasitica. 2014 doi: 10.1007/s12600-014-0410-9. [DOI] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. Peer J. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Moran NA. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc Lond [Biol] 2006;273(1586):603–610. doi: 10.1098/rspb.2005.3348PMID:16537132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Prabhakar CS. Molecular diversity and antibiotic sensitivity of gut bacterial symbionts of fruit fly Bactrocera tau Walker. J Biol Control. 2009;23(3):213–220. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaochan N, Sittichaya W, Sausa-ard W, Chinajariyawong A. Incidence of Enterobacteriaceae in the larvae of the Polyphagous Insect Bactrocera papayae Drew & Hancock (Diptera: Tephritidae) infesting different host fruits. Philipp Agric Sci. 2013;96:384–391. [Google Scholar]
- Wagner SM, Martinez AJ, Ruan Y-M, Kim KL, Lenhart PA, Dehnel AC, et al. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol. 2015;29(11):1402–1410. doi: 10.1111/1365-2435.12459. [DOI] [Google Scholar]
- Wang A, Yao Z, Zheng W, Zhang H. Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) based on 454 pyrosequencing. PLoS ONE. 2014;9:e106988. doi: 10.1371/journal.pone.0106988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jin L, Zhang H. Comparison of the diversity of the bacterial communities in the intestinal tract of adult Bactrocera dorsalis from three different populations. J Appl Microbiol. 2011;110:1390–1401. doi: 10.1111/j.1365-2672.2011.05001.x. [DOI] [PubMed] [Google Scholar]
- Wingfield MJ, Garnas JR, Hajek A, Hurley BP, de Beer ZW, Taerum SJ. Novel and co-evolved associations between insects and microorganisms as drivers of forest pestilence. Biol Invasions. 2016;18:1045–1056. [Google Scholar]
- Yong HS, Song SL, Chua KO, Lim PE. High diversity of bacterial communities in developmental stages of Bactrocera carambolae (Insecta: Tephritidae) revealed by IlluminaMiSeq sequencing of 16S rRNA gene. Curr Microbiol. 2017;74:1076–1082. doi: 10.1007/s00284-017-1287-x. [DOI] [PubMed] [Google Scholar]
- Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, Bourtzis K. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci USA. 2004;101(42):15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zhang X, Chen Z, Wang Z, Lu Y, Cheng D. The divergence in bacterial components associated with Bactrocera dorsalis across developmental stages. Front Microbiol. 2018;9:114. doi: 10.3389/fmicb.2018.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.