Abstract
To prescribe effective antibiotics to patients with bacterial infections in a timely manner and to avoid the misuse of antibiotics, a rapid antimicrobial susceptibility test (AST) is essential. However, conventional AST methods require more than 16 h to provide results; thus, we developed an electrical AST (e-AST) system, which provides results within 6 h. The proposed e-AST is based on an array of 60 aptamer-functionalized capacitance sensors that are comparable to currently available AST panels and a pattern-matching algorithm. The performance of the e-AST was evaluated in comparison with that of broth microdilution as the reference test for clinical strains isolated from septic patients. A total of 4,554 tests using e-AST showed a categorical agreement of 97% with a minor error of 2.2%, major error of 0.38%, and very major error of 0.38%. We expect that the proposed e-AST could potentially aid antimicrobial stewardship efforts and lead to improved patient outcomes.
Subject terms: Biological techniques, Biotechnology
Introduction
Bacterial infection-related complications such as sepsis are serious diseases with mortality rates ranging from 30 to 50%1–3. If appropriate treatment is delayed, survival chances decrease drastically. Thus, rapid antimicrobial susceptibility testing (AST) is required to prescribe the patient with effective antibiotics and avoid their misuse4. However, conventional phenotype-based AST methods, including broth dilution and disk diffusion methods, require more than 3 d to show AST results5,6. Genotypic methods, such as PCR, enable the rapid identification of strains carrying specific antibiotic resistance genes7. However, they have several disadvantages, such as reliance on high bacterial concentrations to provide sufficient DNA and manual sample handling steps, such as lysing bacterial cells to extract nucleic acids. Further, the extracted DNA contains DNA from both living and dead cells, leading to a high false positive rate. Hence, phenotype methods remain the “gold standard” for clinical AST.
To reduce AST time, various phenotype AST techniques based on optical imaging8–14, microfluidic channels15–19, and other biosensors4,20 have been pursued. For example, several microscopy-based methods have been developed to measure bacterial growth every 10 min as clonal aggregates multiply in Mueller–Hinton media, to determine antibiotic-induced morphological changes in single bacterial cells10, and to measure the number of bacteria in a microfluidic channel19. In addition, rapid AST using biochemical, optical, and isothermal measurements has been reported21,22.
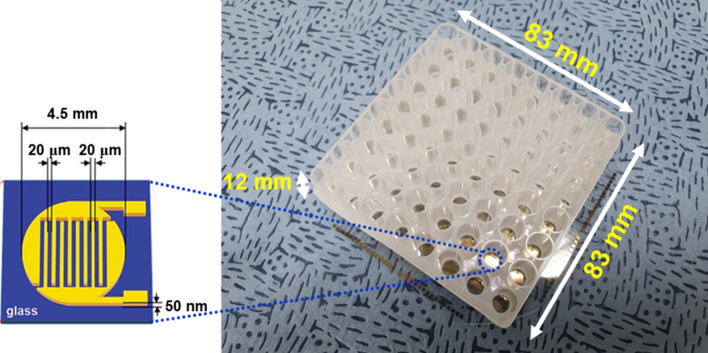
Here, we report electrical AST (e-AST) chips composed of 60 aptamer-functionalized capacitance sensors (Fig. 1). Bacteria act as dielectric particles; thus, if bacteria are bound to the sensor surface via the aptamer, the capacitance changes. As a result, bacterial activities, such as growth and death, can be monitored in real-time by measuring the capacitance change, allowing more rapid AST23,24. For AST of clinical samples, it is necessary to simultaneously test various antimicrobial agents at different concentrations; therefore, we fabricated sensor arrays consisting of 60 sensors. Among the 60 sensors, two and three sensors were used for negative and positive controls, respectively, and the other 55 sensors were used to measure the antibiotic susceptibility to 11 antibiotics at five different concentrations (see Supplementary Table S1). To evaluate the performance of the e-AST system, 30 clinical strains, most of which are frequently found in sepsis, were tested (Table 1). The categorical agreement between the e-AST system and gold standard broth microdilution (BMD) test was estimated to be 97%, meeting the requirements of the United States Food and Drug Administration (FDA).
Figure 1.
Photograph of capacitance sensor array consisting of 60 capacitance sensors. The inset shows a schematic of capacitance sensor.
Table 1.
Representative microorganism from sepsis in clinical strains.
| Bacteria | Number of strains | Clinical strains |
|---|---|---|
| Gram-negative | ||
| E. coli | 5 | U433, U556a, B12327, U5307, U6267a |
| A. baumannii | 5 | P762, T800, R4197, R4299, R4356 |
| P. aeruginoa | 5 | PAE1, PAE2, PAE3, PAE4, PAE5 |
| K. pneumoniae | 5 | KP1a, KP2a, KP3, KP4a, KP5a |
| Gram-positive | ||
| S. aureus | 5 | P101, T82, C970, R4308, R4319 |
| E. faecalis | 5 | R1238, U554, U5179, U4879, U5064 |
aExtended-spectrum β-lactamase (ESBL) positive strains.
Materials and methods
Fabrication of e-AST chip
The e-AST chip, consisting of 60 sensors, was fabricated on a glass substrate (Fig. 1). Each sensor had interdigitated Au electrodes with a width of 20 µm and spacing of 20 µm, which were patterned by conventional photolithography and lift-off techniques. For bacterial culture, a 300 μl-acrylic well (Nobel Biosciences Inc., Hwaseong-Si, South Korea) was attached to the array sensor using 3 M VHBTM tape. Next, the bacteria-aptamers were immobilized on the sensor surface.
Bacterial strains and their growth condition
Thirty clinical isolates comprising four Gram-negative strains (Escherichia coli, Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa) and two Gram-positive strains (Staphylococcus aureus and Enterococcus faecalis) were used for the study (Table 1). Two extended-spectrum β-lactamase (ESBL)-producing E. coli and four ESBL-producing K. pneumoniae were included. Sepsis-causing clinical bacterial strains from various specimens (blood, pus, and urine) were collected from the laboratories of Yonsei University Severance Hospital (Seoul, South Korea). They were subcultured on blood agar plates (KOMED Life Science Co., Ltd., Seoul, South Korea) for 20 to 24 h before the test and their antimicrobial resistance was performed with VITEK 2 systems (bioMerieux with three AST cards). Mueller Hinton Broth (cation-adjusted) (BD Biosciences, San Jose, CA, USA) was used as the growth medium. This study was approved by the Institutional Review Board of Severance Hospital, Yonsei University Health System (4-2017-1179).
Aptamer immobilization on the sensor surface
The sequences of the DNA aptamers, specific to each bacterial species, were as follows: E. coli, 5′-GCA ATG GTA CGG TAC TTC CCC ATG AGT GTT GTG AAA TGT TGG GAC ACT AGG TGG CAT AGA GCC GCA AAA GTG CAC GCT ACT TTG CTA A-3′25; A. baumannii, 5′-TAC ATG GTC AAC CAA ATT CTT GCA AAT TCT GCA TTC CTA CTG T-3′26; K. pneumoniae, 5′-GCA ATG GTA CGG TAC TTC C(N45)-CAA AAG TGC ACG CTA CTT TGC TAA-3′27; P. aeruginosa, 5′-CCC CCG TTG CTT TCG CTT TTC CTT TCG CTT TTG TTC GTT TCG TCC CTG CTT CCT TTC TTG-(CH2)3′-SH-3′28; S. aureus, 5′-GCA ATG GTA CGG TAC TTC CTC GGC ACG TTC TCA GTA GCG CTC GCT GGT CAT CCC ACA GCT ACG TCA AAA GTG CAC GCT ACT TTG CTA A-3′29; and E. faecalis, 5′-ATC CAG AGT GAC GCA GCA CGA CAC GTT AGG TTG GTT AGG TTG GTT AGT TTC TTG TGG ACA CGG TGG CTT A-3′30. All aptamers were custom synthesized by Genotech Inc. (Daejeon, South Korea) and dissolved at 10 μM in distilled water for further use.
To detect various bacteria, mixtures of various bacterial aptamers were immobilized on the sensor surface, as previously reported23. First, the sensors were treated with O2 plasma for 1 min to form OH groups on the glass substrate. After the O2 plasma treatment, the sensors were treated with 10% (3-aminopropyl) triethoxysilane (Sigma, St. Louis, MO, USA) for 2 h at room temperature, followed by washing with 10 mM sulfuric acid for 15 min and then with distilled water. Subsequently, the sensors were treated with 10 mM 3,3′-dithiopropionic acid for 2 h at room temperature, followed by washing with distilled water. To activate the carboxylic groups, the sensors were treated with 12.9 mM N-3-dimethylaminopropyl (Sigma) and 8.7 mM N-hydroxysulfosuccinimide (Sigma) for 1 h, and then washed with ethanol. Finally, the sensors were incubated in a solution of 3 μM amine-modified aptamer for 6 h and washed thoroughly with distilled water.
Antibiotic coating on the sensor surface
For each antimicrobial agent, a stock solution of 1 mg/ml was prepared in ultra-pure water, and diluted in the growth medium. Ten microliters of the antimicrobial solutions were added to the wells of the Gram-negative and Gram-positive chips. They were then dried using a freeze-dryer.
Real-time capacitance measurements
Subcultured bacterial colonies were released to growth media and the suspension was diluted to a concentration of 105 CFU/ml for the AST experiments. 200 μl of bacterial solution was added to each well of the e-AST chip. Then, the e-AST chip was mounted on a capacitance impedance analyzer (Proteomtech Inc., Seoul, South Korea) capable of simultaneously measuring the capacitance of 60 sensors (see Supplementary Fig. S1). It was then placed inside an incubator maintained at 37 °C. The capacitance was measured with a peak-to-peak alternating current signal of 10 mV at a frequency of 1,000 Hz. The capacitance impedance analyzer inside the incubator was linked via Bluetooth to the computer, and the data from each sensor were collected every 16.5 min.
Broth microdilution test
For the BMD tests, antimicrobial solutions were prepared from the stock solution. Antimicrobial agents at the appropriate concentration were pipetted into the wells of BD Falcon™ 96 MicroWell plates (BD Biosciences, San Jose, CA, USA) at a volume of 200 μl, and the wells were then inoculated with 10 μl bacterial stock solution, each at a final concentration of 5 × 105 CFU/ml31. After 16 h of incubation at 37 °C, the MIC values of the microdilution wells were read as the concentration, at which more than 80% reduction in growth was observed, as compared to that in the control by unaided visual inspection5,32.
Estimation of similarity measure
A similarity measure was estimated using a pattern-matching algorithm33. Briefly, the complexity of the (ΔC/C0) × d(ΔC/C0)/dt curves was reduced using principal component analysis, and the feature of each curve was extracted via linear discriminant analysis. Then, the similarity measure was determined by a/(a + b), where a and b are the Euclidean distance to the features of cell-free media curve and the positive control curves, respectively. For cell-free media and the positive control, the similarity measure was expected to be 0 and 1, respectively. The program was coded using MATLAB.
Statistical analysis
The 95% confidence intervals for the proportion of CA, including mE, ME, and VME between the e-AST, VITEK 2 Systems, and gold standard BMD method, were also calculated.
Results and discussion
Relationship between real-time capacitance and bacterial growth
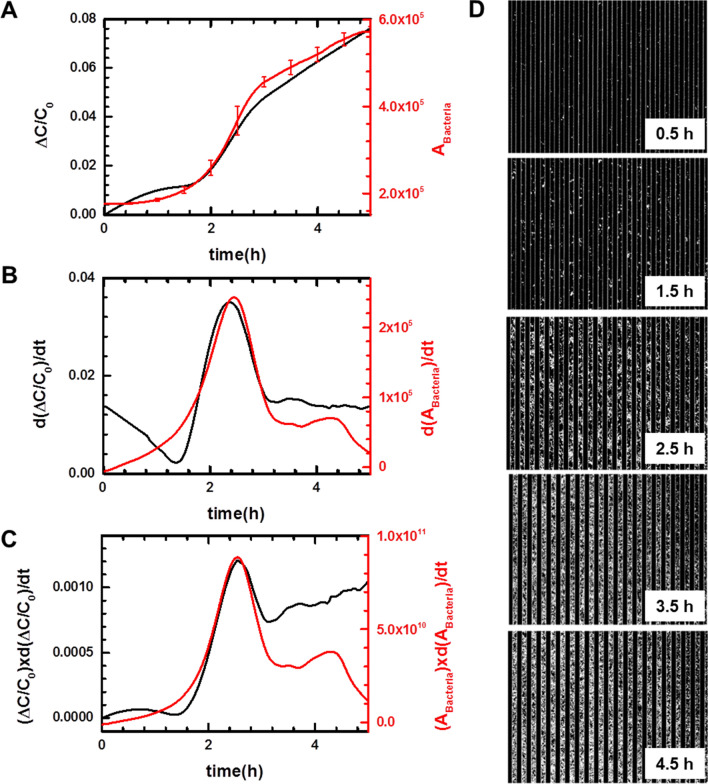
Prior to the AST test, we measured the real-time capacitance when E. coli U433 of 105 CFU/ml was cultured in media (black curve, Fig. 2A), where C0 is the initial capacitance value and ΔC = C − C0. The capacitance increased slowly from 0 to 2 h, and rapidly from 2 to 2.9 h; thereafter, it increased steadily. Accordingly, the capacitance increase rate, calculated numerically by differentiating the capacitance with respect to time, d(ΔC/C0)/dt, shows a peak at 2.4 h (black curve, Fig. 2B). To investigate the relationship between the real-time capacitance and the number of bacteria bound to the sensor surface via aptamers, phase contrast optical images were acquired when E. coli U433 at 105 CFU/ml was cultured in a Chamlide chamber (Live Cell Instrument, Inc., Seoul, South Korea) maintained at 37 °C (Fig. 2D).
Figure 2.
(A) Time dependence of normalized capacitance change (ΔC/C0, black curve) and the area occupied by bacteria (ABacteria, red curve) when E. coli U433 was cultured. (B) d(ΔC/C0)/dt (black curve) and d(ABacteria)/dt (red curve) numerically calculated from the data shown in (A). (C) (ΔC/C0) × d(ΔC/C0)/dt (black curve) and (ABacteria) × d(ABacteria)/dt (red curve). (D) Time-lapse phase contrast optical images acquired when E. coli U433 was cultured in a Chamlide chamber.
The area occupied by bacteria (ABacteria) was estimated using MATLAB built-in function, which is used to detect the edge line of cells, as a function of time (red curve, Fig. 2A). As for the capacitance, ABacteria exhibited three regions depending on the increasing rate. When ABacteria was differentiated with respect to time, d(ABacteria)/dt exhibited a peak at the same time as d(ΔC/C0)/dt (Fig. 2B), indicating that bacterial growth is closely related to real-time capacitance. In particular, when d(ΔC/C0)/dt and d(ABacteria)/dt were multiplied by ΔC/C0 and ABacteria, respectively, similar behaviors were observed before 3.5 h (Fig. 2C). Therefore, the capacitance data were analyzed in terms of (ΔC/C0) × d(ΔC/C0)/dt.
Determination of antimicrobial susceptibility using real-time capacitance measurements
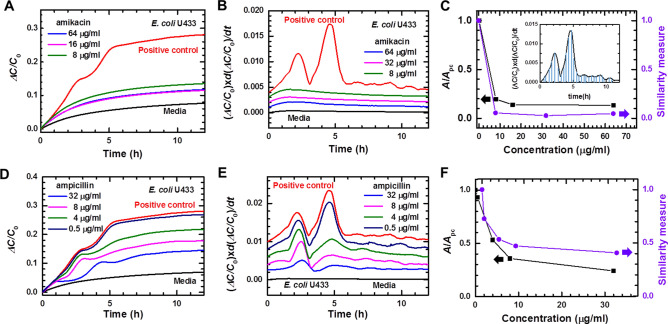
To investigate whether real-time capacitance measurements can be employed to perform ASTs, we measured the real-time capacitance for E. coli U433 treated with different concentrations of amikacin (Fig. 3A) and ampicillin (Fig. 3D), and then calculated (ΔC/C0) × d(ΔC/C0)/dt (Fig. 3B, E). For amikacin, (ΔC/C0) × d(ΔC/C0)/dt showed no peaks at all concentrations, indicating that E. coli U433 is susceptible to amikacin. In contrast, peaks were observed at all concentrations of ampicillin, indicating that U433 is resistant to ampicillin. To determine the antimicrobial susceptibility more quantitatively, we calculated the area enclosed by the curve of (ΔC/C0) × d(ΔC/C0)/dt and the x-axis (inset of Fig. 3C). For all concentrations of amikacin, A/Apc was smaller than 0.2, where Apc is the area estimated for the positive control (black symbols, Fig. 3C). In contrast, A/Apc was higher than 0.2 for all concentrations of ampicillin (black symbols, Fig. 3F). In addition to A/Apc, we estimated a similarity measure using a pattern-matching algorithm, which shows the closeness of (ΔC/C0) × d(ΔC/C0)/dt curve obtained in the presence of antimicrobial agents to that of positive control (purple symbols, Fig. 3C, F). The similarity measure was lower than 0.1 for amikacin and higher than 0.4 for ampicillin. The comparison with the results obtained from the gold standard BMD tests and VITEK 2 systems (see Supplementary Table S2) suggests that the cutoff values between bacterial growth and inhibition are approximately 0.2 and 0.4 for A/Apc and the similarity measure, respectively. A/Apc and the similarity measure exhibited similar behaviors; however, in comparison with the A/Apc, the similarity measure provides a clearer distinction between bacterial growth and inhibition. Therefore, the similarity measure is adopted for the proposed e-AST chips.
Figure 3.
Time dependence of ΔC/C0 measured for E. coli U433 treated with different concentrations of (A) amikacin and (D) ampicillin. Positive control represents the data obtained from E. coli U 433 without antibiotics, and media represents the data of cell-free media. (ΔC/C0) × d(ΔC/C0)/dt numerically calculated for E. coli U433, which was treated with different concentrations of (B) amikacin and (E) ampicillin. The (ΔC/C0) × d(ΔC/C0)/dt curves are shifted to facilitate viewing. The area enclosed by the (ΔC/C0) × d(ΔC/C0)/dt curve and the X-axis, which is denoted by the shaded region in the inset of (C), normalized by the area of positive control (Apc) (A/Apc, black symbols) and similarity measures (purple symbols) estimated using the pattern matching algorithm for E. coli U433, which was treated with different concentrations of (C) amikacin and (F) ampicillin.
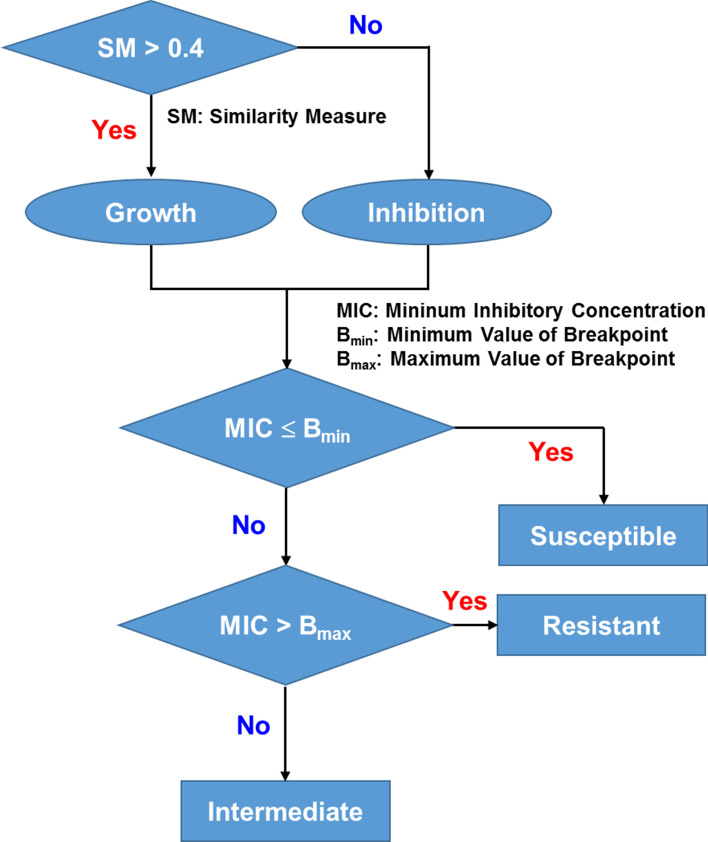
The antimicrobial susceptibility was determined, as shown in Fig. 4. Assuming that the cutoff of similarity measure between bacterial growth and inhibition is 0.4, the MIC was estimated to be less than 8 μg/ml for amikacin. It is lower than the breakpoint of 8 – 16 μg/ml suggested by CLSI; therefore, E. coli U433 was determined to be susceptible to amikacin. For ampicillin, the MIC was estimated to be greater than 32 μg/ml, higher than the breakpoint of 2 – 8 μg/ml, indicating that E. coli U433 is resistant to ampicillin. The gold standard BMD tests and VITEK 2 systems also showed that E. coli U433 is susceptible to amikacin and resistant to ampicillin, indicating that antimicrobial susceptibility can be determined using the e-AST system.
Figure 4.
Data interpretation and susceptibility determination. SM denotes similarity measure.
Performance of e-AST system in clinical samples
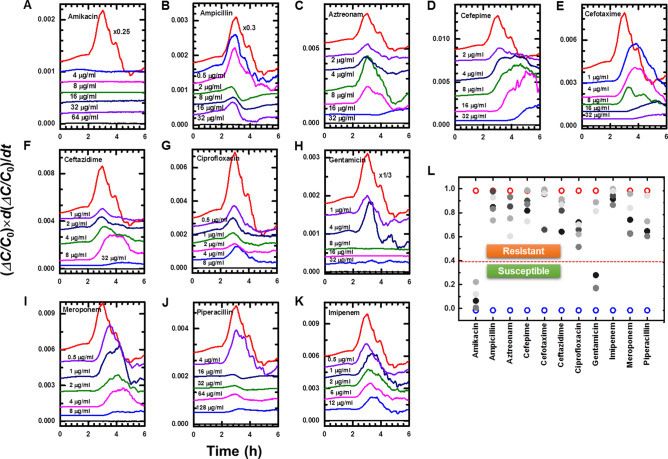
For the AST tests, Gram-negative and Gram-positive chips were prepared. The list of antibiotics and their concentrations is presented in Table S1, which was selected from VITEK 2 kits used at Yonsei University Severance Hospital. To evaluate the performance of the e-AST system, five bacterial isolates per strain were tested for four Gram-negative strains (E. coli, A. baumannii, P. aeruginosa, and K. pneumoniae) and two Gram-positive strains (S. aureus and E. faecalis) (Table 1). Figure 5A – K show the typical results measured for A. baumannii R4197 using the Gram-negative e-AST chip. The data were monitored for 6 h because the time dependence data (Fig. 3) revealed that antimicrobial susceptibility could be determined within 6 h.
Figure 5.
(A–K) Time dependence of (ΔC/C0) × d(ΔC/C0)/dt obtained using Gram-negative e-AST chip for A. baumannii R4197. The (ΔC/C0) × d(ΔC/C0)/dt curves are shifted to facilitate viewing. Positive control (red line) represents the data obtained from A. baumannii R4197 without antibiotics, and media (black line) represents the data of cell-free media. (L) Similarity measures estimated using the pattern matching algorithm for A. baumannii R4197. Higher concentration is indicated by the darker color. Blue and red circles indicate the similarity measure of cell-free media and A. baumannii R4197 without antibiotics, respectively.
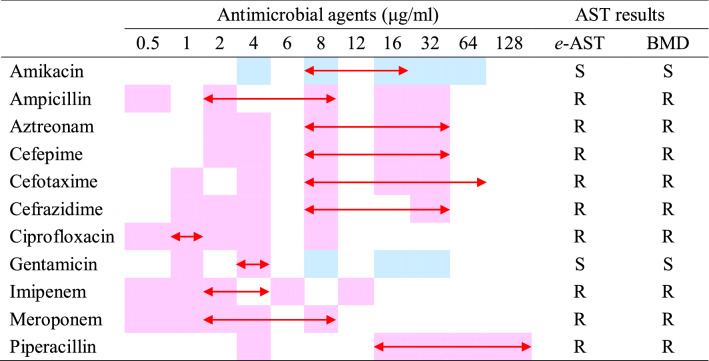
From the capacitance measured as a function of time, (ΔC/C0) × d(ΔC/C0)/dt was numerically calculated, and the similarity measures were estimated using a pattern-matching algorithm (Fig. 5L). The antimicrobial susceptibilities determined by the flow chart in Fig. 4 are summarized in Table 2 for various antibiotics. A. baumannii R4197 was determined to be susceptible to amikacin and gentamicin, and resistant to other antibiotics. These results are consistent with the BMD results, supporting the application possibility of e-AST chips to perform ASTs.
Table 2.
The e-AST results for A. baumannii R4197 and the comparison with gold standard BMD.
For the e-AST and BMD test: SM (similarity measure) < 0.4, blue color; SM > 0.4, pink color; red arrow, MIC interpretive criterion (μg/mL); S, susceptible; R, resistant.
To estimate the error rates of the e-AST system, a total of 4,554 tests were performed and discrepancies were determined by comparing the results of the e-AST chips with those of the BMD test as the reference (Table 3 and see Supplementary Table S3). According to the guidance from the FDA, the discrepancies were classified as minor error, major error, very major error, and category agreement (CA). Minor errors (mEs) represent the strains that were interpreted as “intermediate” using the e-AST chip but were determined to be either sensitive or resistant by the reference test. Major errors (MEs) represent the strains that were interpreted as “resistant” using the e-AST chip but were determined to be sensitive by the reference test. Very major errors (VMEs) represent the strains that were interpreted as “sensitive” using the e-AST chip but were determined to be resistant by the reference test. CA is the same category of rates (S, I, R) provided by the e-AST chip and the reference test. For all strains, CA was higher than 95%, and mEs, MEs, and VMEs rates were lower than 5%, 1%, and 1.8%, respectively. For P. aeruginosa, 187 tests were conducted and there were 4 mEs, 1 ME, and 1 VME. The ME and VME occurred in ceftazidime and imipenem, respectively (see Supplementary Table S3).
Table 3.
Discrepancy rate using clinical isolates.
| Bacteria | No. Test | Discrepancy number | Discrepancy rate (%) | |||||
|---|---|---|---|---|---|---|---|---|
| mEs | MEs | VMEs | mEs | MEs | VMEs | CA | ||
| Gram negative | ||||||||
| E. coli | 187 | 3 | 2 | 0 | 1.60 | 1.07 | 0 | 97.33 |
| A. baumannii | 165 | 4 | 0 | 0 | 2.42 | 0 | 0 | 97.58 |
| P. aeruginoa | 187 | 4 | 1 | 1 | 2.14 | 0.53 | 0.53 | 96.79 |
| K. pneumoniae | 176 | 3 | 1 | 0 | 1.70 | 0.57 | 0 | 97.73 |
| 715 | 14 | 4 | 1 | 1.96 | 0.56 | 0.14 | 97.34 | |
| Gram positive | ||||||||
| S. aureus | 165 | 0 | 0 | 3 | 0 | 0 | 1.82 | 98.18 |
| E. faecalis | 165 | 9 | 0 | 0 | 5.46 | 0 | 0 | 94.55 |
| 330 | 9 | 0 | 3 | 2.73 | 0 | 0.91 | 96.37 | |
| Total | 1,045 | 23 | 4 | 4 | 2.20 | 0.38 | 0.38 | 97.03 |
The e-AST results were compared with the BMD results to calculate the discrepancy rates. For e-AST: mE minor error, ME major error, VME very major error, CA categorical agreement.
In the case of S. aureus, there were 3 VMEs among 165 tests. The 3 VMEs were from S. aureus (R4319) treated with clindamycin (see Supplementary Table S3). In the case focusing on the six β-lactam resistant strains composed of ESBL, corresponding to 220 combinations of antibiotic-bacteria tests, we observed 216 (98.18%) CA, 2 (0.96%) mEs, 2 (0.96%) MEs, and 0 (0%) VME (Table 4). Overall, the total CA, mEs, MEs, and VMEs rates were 97.03%, 2.20%, 0.38%, and 0.38%, respectively. These values satisfy the CLSI recommendations, in which the acceptable inter-method VMEs, MEs, and mEs rates are 1.5%, 3%, and 10%, respectively34.
Table 4.
Discrepancy rate of two representative ESBL-positive isolates from sepsis patients.
| ESBL-bacteria | No. strains | No. test | Discrepancy number | Discrepancy rate (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| mEs | MEs | VMEs | mEs | MEs | VMEs | CA | |||
| E. coli | 2 | 77 | 1 | 0 | 0 | 1.29 | 0 | 0 | 98.70 |
| K. pneumoniae | 4 | 143 | 1 | 2 | 0 | 2.09 | 0.69 | 0 | 97.20 |
Extended-spectrum β-lactamase (ESBL) positive strains.
Conclusion
We developed e-AST systems based on an array of 60 aptamer-functionalized capacitance sensors and a pattern-matching algorithm for rapid AST. Bacterial behaviors, such as growth and inhibition, were monitored in real-time by measuring the capacitance change, thereby allowing the determination of antimicrobial susceptibility within 6 h, which is shorter in comparison with that by the conventional methods (~ 16 h). To validate the clinical usefulness of e-AST, the e-AST results were compared with the results of gold standard BMD for six clinical strains from septic patients, including ESBL-positive E. coli and K. pneumoniae as antibiotic-resistant pathogens. The discrepancies between e-AST and BMD tests were estimated to be 2.20% mE, 0.38% ME, and 0.38%, which are lower than the FDA requirements (mE ≤ 10%, ME ≤ 3%, and VME ≤ 1.5%). These results demonstrate that e-AST can be applied for rapid AST. Diagnosis of sepsis by e-AST may cost more than gold standard method; however, it is expected that e-AST system could increase survival rate of sepsis patients.
Supplementary information
Acknowledgements
This work was supported by the Nano-Convergence Foundation (Grant No. R201801311) and Advanced Technology Center + Program (ATC+, Grant No. 20008864) funded by the Ministry of Trade, Industry and Energy (MOTIE, KOREA) and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, Future Planning and the Korean government (MEST) (Grant Nos. 2017R1D1A1B03028551, 2019R1H1A2080077, 2019R1H1A2079842, 2020R1A202011942, 2020R1A2C1006092, and 2019R1A6A1A03032869) and Brain Korea 21 PLUS Project for Medical Science.
Author contributions
Conceived and designed the experiments: K.S.L., S.M.L., J.S.S., and K.H.Y. Performed the experiments: K.S.L., S.M.L., J.O., I.H.P., and M.H. Analyzed the data: K.S.L., S.M.L., D.Y., K.J.L., J.S.S., and K.H.Y. Wrote the paper: S.M.L., J.S.S., and K.H.Y.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kyo-Seok Lee and Sun-Mi Lee.
Contributor Information
Sun-Mi Lee, Email: sunmilee@yonsei.ac.kr.
Jeon-Soo Shin, Email: jsshin6203@yuhs.ac.
Kyung-Hwa Yoo, Email: khyoo@yonsei.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-020-70459-3.
References
- 1.Theisen M, et al. The glutamate-rich protein (GLURP) of plasmodium falciparum is a target for antibody-dependent monocyte-mediated inhibition of parasite growth in vitro. Infect. Immun. 1998;66:11–17. doi: 10.1128/IAI.66.1.11-17.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, et al. Sepsis and septic shock. Nat. Rev. Dis. Primers. 2016;2:16045. doi: 10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent JL, et al. Sepsis in European intensive care units: results of the SOAP study. Crit. Care Med. 2006;34:344–353. doi: 10.1097/01.CCM.0000194725.48928.3A. [DOI] [PubMed] [Google Scholar]
- 4.Behera B, et al. Emerging technologies for antibiotic susceptibility testing. Biosens. Bioelectron. 2019;142:111552. doi: 10.1016/j.bios.2019.111552. [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen JH, Ferraro MJ, Reller LB, Weistein M. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 2009;49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 6.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substance. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 7.Rolain JM, Mallet MN, Fournier PE, Raoult D. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 2004;54:538–541. doi: 10.1093/jac/dkh324. [DOI] [PubMed] [Google Scholar]
- 8.Metzger S, Frobel R, Dunne A, Dunne WM., Jr Rapid simultaneous identification and quantitation of Staphylococcus aureus and Pseudomonas aeruginosa directly from bronchoalveolar lavage specimens using automated microscopy. Diagn. Microbiol. Infect. Dis. 2014;79:160–165. doi: 10.1016/j.diagmicrobio.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 9.Choi J, et al. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip. 2013;13:280–287. doi: 10.1039/C2LC41055A. [DOI] [PubMed] [Google Scholar]
- 10.Choi J, et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014;6:174. doi: 10.1126/scitranslmed.3009650. [DOI] [PubMed] [Google Scholar]
- 11.Choi J, et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci. Rep. 2017;7:1148. doi: 10.1038/s41598-017-01278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredborg M, et al. Real-time optical antimicrobial susceptibility testing. J. Clin. Microbiol. 2013;51:2047–2053. doi: 10.1128/JCM.00440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredborg M, et al. Rapid antimicrobial susceptibility testing of clinical isolates by digital time-lapse microscopy. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:2385–2394. doi: 10.1007/s10096-015-2492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chantell C. Multiplexed automated digital microscopy for rapid identification and antimicrobial susceptibility testing of bacteria and yeast directly from clinical samples. Clin. Microbiol. Newsl. 2015;37:161–167. doi: 10.1016/j.clinmicnews.2015.10.001. [DOI] [Google Scholar]
- 15.Kim SC, Cestello-Blanco S, Inoue K, Zare RN. Miniaturized antimicrobial susceptibility test by combining concentration gradient generation and rapid cell culturing. Antibiotics (Basel). 2015;4:455–466. doi: 10.3390/antibiotics4040455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Zhen L, Liu J, Wu J. Rapid antibiotic susceptibility testing in a microfluidic pH sensor. Anal. Chem. 2013;85:2787–2794. doi: 10.1021/ac303282j. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Lee S, Kim JK, Chung HJ, Jeon JS. Microfluidic-based observation of local bacterial density under antimicrobial concentration gradient for rapid antibiotic susceptibility testing. Biomicrofluidics. 2019;13:014108. doi: 10.1063/1.5066558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, et al. Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal. Chem. 2013;85:3971–3976. doi: 10.1021/ac4004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan R, et al. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens. Bioelectron. 2013;49:118–125. doi: 10.1016/j.bios.2013.04.046. [DOI] [PubMed] [Google Scholar]
- 20.Syal K, et al. Current and emerging techniques for antibiotic susceptibility test. Theranostics. 2017;7:1795–1805. doi: 10.7150/thno.19217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puttaswamy S, Gupta SK, Regunath H, Smith LP, Sengupta S. A comprehensive review of the present and future antibiotic susceptibility testing (AST) systems. Arch. Clin. Microbiol. 2018;9:1–9. doi: 10.4172/1989-8436.100083. [DOI] [Google Scholar]
- 22.Idelevich EA, Becker K. How to accelerate antimicrobial susceptibility testing. Clin. Micorbiol. Infect. 2019;25:1347–1365. doi: 10.1016/j.cmi.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Jo N, et al. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growth and antibiotic susceptibility. Biosens. Bioelectron. 2018;102:164–170. doi: 10.1016/j.bios.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Song SH, et al. Vertical capacitance aptasensors for real-time monitoring of bacterial growth and antibiotic susceptibility in blood. Biosens. Bioelectron. 2019;143:111623. doi: 10.1016/j.bios.2019.111623. [DOI] [PubMed] [Google Scholar]
- 25.Kim YS, Song MY, Jumg J, Kim BC. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell-systematic evolution of ligands by exponential enrichment approach. Anal. Biochem. 2013;436:22–28. doi: 10.1016/j.ab.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Rasoulinejad S, Gargari SLM. Aptamer-nanobody based ELASA for specific detection of Acinetobacter baumannii isolates. J. Biotechnol. 2016;231:46–54. doi: 10.1016/j.jbiotec.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 27.Kim, B. C., Kim, Y. S., Chung, J. Y., Jurng, J. S. & Song, M. Y. Single-stranded nucleic acid aptamers specifically binding to klebsiella pneumoniae and method for detecting k. pneumonia using the same.” US Patent 9 284 550. Accessed 20 Nov 2013.
- 28.Yoo SM, Kim DK, Lee SY. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta. 2015;132:112–117. doi: 10.1016/j.talanta.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Lian Y, He F, Wang H, Tong F. A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus. Biosens. Bioelectron. 2015;65:314–319. doi: 10.1016/j.bios.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Turek D, Van SD, Johnson J, Ocsoy I, Tan W. Molecular recognition of live methicillin-resistant Staphylococcus aureus cells using DNA aptamers. World J. Transl. Med. 2013;2:67–74. doi: 10.5528/wjtm.v2.i3.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wikler MA, et al. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically - Seventh Edition: Approved Standard M7–A10. Wayne, PA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 32.Patel JB, et al. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically – Tenth Edition: Approved Standard M7–A10. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 33.Murty MN, Devi VS. Pattern Recognition: An Algorithmic Approach. 1. New York: Springer; 2011. [Google Scholar]
- 34.Powell M, et al. Development of In Vitro Susceptibility Testing Criteria and Quality Control Parameters: Fourth Edition: Approved Standard M23–A4. Wayne, PA: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.