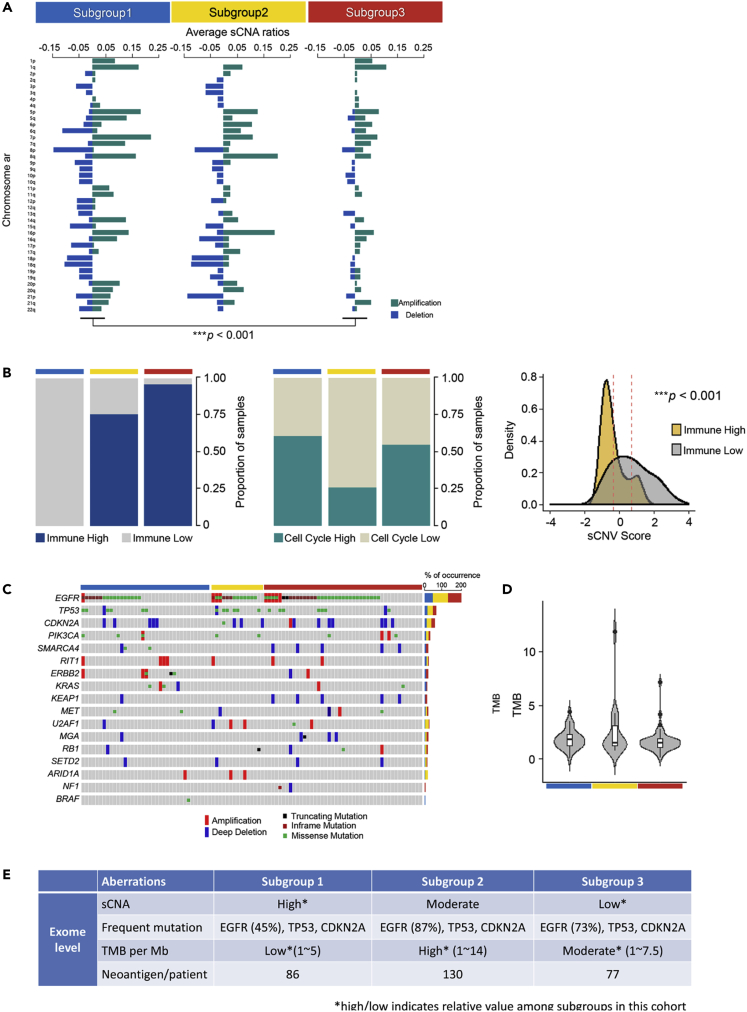
Figure 2.
Exome-Level Subgroup Analysi
(A) Somatic copy number variations. Somatic copy number variations (amplification or deletion) at the level of chromosome arms identified by GISTIC analysis were summarized for each subgroup. Each bar represents average of sCNA ratios across samples. ∗∗∗p<0.001, Student's t test.
(B) Relationship of sCNV score with tumor proliferation and immune evasion. Bar charts show relative proportions between samples with high and low signature scores on two cancer hallmarks (immune and cell cycle signature). Higher- and lower-level samples were based on > 70th and <30th percentiles, respectively. Signature score was determined by the average gene expression level of each signature. Density plot showing distribution of sCNV scores, according to immune score. The x axis is normalized sCNV score across samples. (Immune High: −0.322(±0.127) and Immune Low: 0.704(±0.208)). ∗∗∗p<0.001, Student's t test.
(C) Somatic mutations. From the whole-exome sequencing, 17 genes with most frequent somatic mutations were selected, identifying 5 different gene aberrations: amplification, deep deletion, truncating mutation, inframe mutation, and missense mutation.
(D) Tumor mutation burden (TMB) shown for each subgroup. Every subgroup had TMB at the level of <5 per Mb (mode: ∼2 per Mb). Subgroups 1, 2, and 3 had the least, greatest (up to 14 per Mb), and intermediate (up to 7.5 per Mb) variations, respectively. Most patients, except the outliers with TMB exceeding 5 per Mb, were likely to have immune evasion due to the extremely low probability of immunogenic neoantigen generation. (Subgroup 1: 1.919(±0.146), Subgroup2: 2.644(±0.720), and Subgroup3: 1.700(±0.166).)
(E) Major features at exome level are summarized. Average numbers of neoantigens in each subgroup were calculated as described in Methods (Table S1).