Abstract
Chitin is one of the most abundant renewable organic materials found on earth. The chitin utilization locus in Flavobacterium johnsoniae, which encodes necessary proteins for complete enzymatic depolymerization of crystalline chitin, has recently been characterized but no detailed structural information on the enzymes was provided. Here we present protein structures of the F. johnsoniae chitobiase (FjGH20) and chitinase B (FjChiB). FjGH20 is a multi-domain enzyme with a helical domain not before observed in other chitobiases and a domain organization reminiscent of GH84 (β-N-acetylglucosaminidase) family members. The structure of FjChiB reveals that the protein lacks loops and regions associated with exo-acting activity in other chitinases and instead has a more solvent accessible substrate binding cleft, which is consistent with its endo-chitinase activity. Additionally, small angle X-ray scattering data were collected for the internal 70 kDa region that connects the N- and C-terminal chitinase domains of the unique 158 kDa multi-domain chitinase A (FjChiA). The resulting model of the molecular envelope supports bioinformatic predictions of the region comprising six domains, each with similarities to either Fn3-like or Ig-like domains. Taken together, the results provide insights into chitin utilization by F. johnsoniae and reveal structural diversity in bacterial chitin metabolism.
Subject terms: SAXS, X-ray crystallography
Introduction
Chitin is as a key component of arthropod exoskeletons and fungal cell walls, and as such one of the most abundant renewable organic materials found on earth. The long chitin polysaccharides, consisting of β(1 → 4)-linked N-acetyl-d-glucosamine (GlcNAc) units, share many properties with cellulose; both polysaccharides are completely insoluble and coalesce into crystalline fibers, and their decomposition is highly challenging. Metabolism of chitin and cellulose proceeds through similar systems, where multiple enzymatic activities are required. Endo-acting enzymes cleave amorphous regions of the crystals, while exo-acting processive enzymes depolymerize the chains from either the reducing- or non-reducing ends. In addition, lytic polysaccharide monooxygenases (LPMOs) introduce chain breaks in crystalline regions, whereas β-glycosidases complete the hydrolysis by converting oligosaccharides into monosaccharides1,2. The chitinolytic machinery of the bacterium Serratia marcescens has served as a model system for enzymatic chitin turnover, and consists of five core enzymes: SmChiA and B (exo-acting processive chitinases), SmChiC (endo-acting non-processive chitinase), CBP21 (LPMO), and a chitobiase converting chitooligosaccharides (CHOs) into GlcNAc2.
Bacteria belonging to the Bacteroidetes phylum are well-known as efficient metabolizers of complex polysaccharides, and many of these species contain so-called polysaccharide utilization loci (PULs) which target distinct glycans3. PULs are discrete gene clusters, comprising all of the proteins needed to bind, transport and enzymatically deconstruct complex polysaccharides, and many PULs targeting both plant- and microbial glycans have been characterized in recent years4–6. No culturable Bacteroidete has yet conclusively been shown to deconstruct cellulose via the actions of a PUL7, but we recently identified and characterized a chitin utilization locus (ChiUL) from the aerobic soil bacterium Flavobacterium johnsoniae8. An atypical feature of the ChiUL compared to other characterized PULs from gut bacteria is that the main chitinase, FjChiA, is not attached to the outer membrane but is instead solubly secreted into the extracellular milieu by the Type IX secretion system8–10. This possibly reflects the fact that the environment of F. johnsoniae is less nutrient dense compared to the gastrointestinal tracts of animals and may require both protein secretion and cellular motility for efficient scavenging for resources11.
The F. johnsoniae ChiUL consists of 11 genes: two SusC/D-like pairs for oligosaccharide capture and transport, an inner membrane transporter, a two-component sensor/regulator pair, and four enzymes. While one of the enzymes of the locus, a predicted glucosamine-6-phosphate deaminase, proved impossible to produce by heterologous expression, the three remaining enzymes directly targeting chitin or CHOs were studied in detail. The large (~ 158 kDa) FjChiA enzyme is essential for efficient chitin depolymerization and growth on chitin8,10, and consists of two catalytic domains, FjChiA_N (N-terminal, exo-acting) and FjChiA_C (C-terminal, endo-acting), both from glycoside hydrolase family 18 (GH18) and sharing only ~ 20 to 30% sequence identity to previously characterized family members. FjChiA_N and FjChiA_C are connected by a ~ 70 kDa segment, FjChiA_M, which lacks significant similarity in primary structure to any known proteins but was shown to bind crystalline polysaccharides and may as such present a novel carbohydrate-binding module (CBM) motif8. The chitin depolymerization activity of the native full-length FjChiA was dramatically better than a combination of the two terminal catalytic domains, indicating the importance for the FjChiA_M domain in this multi-domain enzyme. The remaining GH18 chitinase of the locus, FjChiB (endo-acting), was active on chitin and CHOs, but to a much lesser extent compared to FjChiA. The fourth enzyme, the FjGH20 chitobiase/N-acetylglucosaminidase, was active only on CHOs.
In the carbohydrate-active enzymes database (CAZy; https://www.cazy.org12), GH18 is a large family comprising over 20,000 members with close to 500 having been biochemically characterized to date. Of these, 93 enzymes are currently listed as structurally determined (April 2020). While this is a small subset compared to the family size, the number of solved structures provides deep insight into the enzymes’ structure–function relationships. The catalytic domains of GH18 enzymes consist of (β/α)8 barrels which are often appended by CBMs or other chitin-binding structures that are important for overall enzyme efficiency13–15. A notable feature present in some GH18 members is a region/domain inserted between the β-strand 7 and α-helix 7 of the barrel, referred to as the chitinase insertion domain (CID), which is proposed to aid in defining the processivity of the enzymes by forming a (partial) tunnel over a bound polysaccharide chain. Structural information on GH20 enzymes is sparser, with 22 structures currently deposited to the protein data bank (PDB). While several GH20 enzymes have been shown to be multi-modular and the family as a whole contains a range of substrate specificities, the enzymes all comprise a core catalytic domain consisting of a (β/α)8 barrel that carries conserved catalytic residues16–20.
In our previous work, we solved the structures of the surface-tethered SusD-like proteins CusDI and CusDII, however the structural basis for the activity of the enzymes of the ChiUL remained unresolved8. Here we present the crystal structures of the FjChiB chitinase and the FjGH20 chitobiase, as well as a small-angle X-ray scattering (SAXS) envelope of the novel FjChiA_M domain. The protein structures reveal features consistent with their enzymatic activities and reveal a novel domain in FjGH20. The ab initio modelling of the FjChiA_M SAXS envelope indicated an elongated protein which, supported by bioinformatic analyses, is suggested to be comprised of distinct modules. Collectively, the results add to our understanding of the structure–function relationships of these important enzymes that are involved in the metabolism of the abundant polysaccharide chitin.
Results and discussion
Structure of FjGH20
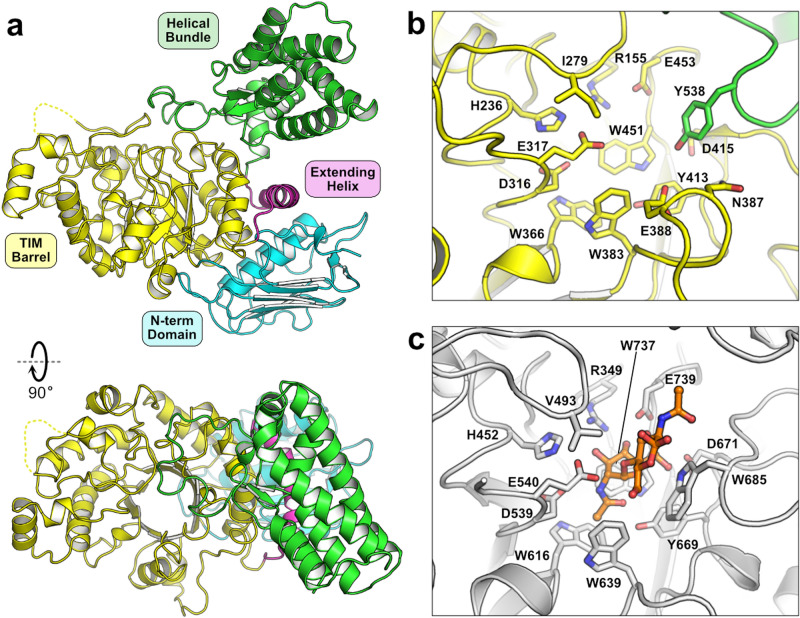
GH20 family members are multi-domain proteins with a TIM-barrel domain containing the catalytic site. The predicted catalytic domain of FjGH20 shares 25 to 38% sequence identity with previously characterized GH20 family members, but the overall amino acid sequence identity is as low as 15% compared to certain studied enzymes. Based on these data, we hypothesized that FjGH20 might also contain structural features or domains previously not seen in other family members. Thus, to illuminate these features we pursued structural determination of the FjGH20 by X-ray crystallography and were able to solve the protein structure from diffraction data extending to 1.70 Å resolution. The crystallographic asymmetric unit contained two protein molecules whose interaction gives rise to a buried surface area of ~ 11%, as determined by PISA21. Analysis of the protein in solution by size exclusion chromatography was consistent with a monomer unit being the prominent species (≥ 80%) and thus the dimer observed in the crystal structure is likely not of biological relevance. Each protomer consists of three domains: a central domain (residues 145–502) composed of a (β/α)8 TIM-barrel, an N-terminal domain (residues 1–144) comprising a six-stranded β-sheet and two α-helices where the helices are sandwiched between the β-sheet and the central domain, and a C-terminal domain (residues 504–673) consisting of an eight-helix bundle with two short β-strands (Fig. 1). An α-helix (residues 481–503) extends from and across α-helix 8 of the TIM-barrel to connect the central domain to the C-terminal helical bundle. Most of the polypeptide chains are well defined in the electron density except for residues 243–261 and 421–429 in both protomers, which are not visible and are poorly defined, respectively, suggesting a greater degree of flexibility of these regions.
Figure 1.
Structure of FjGH20. The overall structure of the enzyme is shown with the individual domains annotated by color (a). The active site architectures of FjGH20 (b) and SmGH20 (c; PDB accession 1qbb) are shown with key residues lining the substrate-binding pocket. The structure of SmGH2020 was determined in complex with chitobiose (orange carbons). A notable difference between the two enzymes is the presence of a tyrosine (Tyr538) from a loop of the helical bundle domain in FjGH20 filling the void left by the absence of a conserved tryptophan (Trp685 in SmGH20), which is involved in substrate binding by ring stacking with a GlcNAc residue in chitobiose.
Structural features of FjGH20 and comparison to homologous structures
The FjGH20 N-terminal and central domains, although only sharing up to 35% sequence identity over the domains, are closely related in structure to several GH20 members, such as the β-hexosaminidases from Bacteroides thetaiotaomicron (BT0459; PDB accession 6q63) and Homo sapiens (PDB accession 1o7a; root mean square deviation of Cα atoms ~ 2.5 Å, as determined by DALI22). The N-terminal domain of GH20 family members is ubiquitous and bears structural resemblance to certain CBMs, but a defined biological role for this domain remains undetermined. The catalytic site for the N-acetylhexosaminidase activity in these enzymes is found in the cleft of the TIM-barrel. Several of the active site residues shown to be important for substrate binding and catalysis in GH20 members are conserved in FjGH20, including the H-x-G-G-D-E motif where a glutamic acid (Glu317) is proposed to act as a general acid/base in the reaction. The aspartic acid in this motif (Asp316), along with a conserved tyrosine residue (Tyr413), are proposed to position and polarize the N-acetyl group for the substrate-assisted catalytic mechanism20,23,24. The FjGH20 substrate-binding pocket is distinct in one significant way compared to the archetypical GH20 from S. marcescens (SmGH20)20,25 in that FjGH20 lacks the extended loop that is located between β-strand 7 and α-helix 7 in SmGH20 (Fig. 1). This alteration of the FjGH20 TIM-barrel leads to loss of a conserved active site tryptophan residue (Trp685), which is involved in sugar binding in the + 1 subsite26 of SmGH20. Interestingly, a portion of a loop extending from the FjGH20 helical bundle domain wraps around, and closes off, one side of the active site cleft in the TIM barrel with a tyrosine residue (Tyr538) that projects its sidechain into a position similar to the Trp685 residue in SmGH20. A similar arrangement occurs in the homologous human β-hexosaminidase, which also has a short loop between β-strand 7 and α-helix 7 of the TIM-barrel and has a tyrosine residue in a position similar to Tyr538 in FjGH20. Interestingly, in the human protein the tyrosine residue originates from a loop in a different protomer27. It seems unlikely that this different feature of the substrate-binding sites significantly affects overall activity since FjGH20, human β-hexosaminidase, and a GH20 enzyme from B. thetaiotaomicron, which lacks an analogous aromatic residue, maintain enzymatic activity toward their target substrates8,28.
The FjGH20 C-terminal helical bundle is a distinct domain amongst structurally determined GH20 family members. The GH20 family is distantly related to GH84, which also contains enzymes with N‐acetyl‐β‐hexosaminidase activity and its members display a similar structural architecture, i.e. an N-terminal β-sheet domain and a (β/α)8 TIM-barrel catalytic domain containing GH20-like catalytic residues. Some GH84 members, such as GH84C (NagJ) from Clostridium perfringens29,30, contain a C-terminal helical bundle analogous to that observed in FjGH20 (Supplemental Fig. 1). As in FjGH20, a loop from the helical bundle in NagJ also lines one face of the substrate binding pocket resulting in a tyrosine side chain being positioned analogously to Tyr538 in FjGH20 and Trp685 in SmGH20. In NagJ, this helical domain appears to act as a bridging domain between the catalytic TIM-barrel and multiple additional domains found closer to the C-terminus. While only distantly related in sequence (8% sequence identity shared between the full-length FjGH20 and the homologous domains of NagJ), the presence of an analogous C-terminal domain in FjGH20 to GH84 enzymes may suggest a closer relationship between the two GH families than has been previously suggested and/or could be a remnant of their common ancestry.
Structure of FjChiB
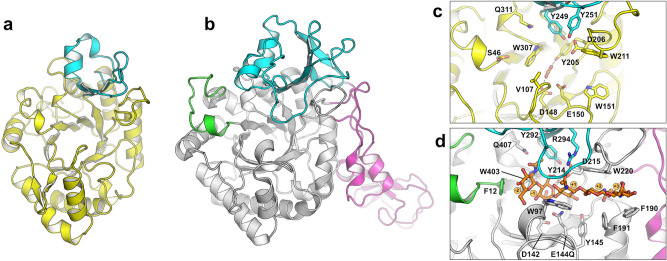
Although many GH18 chitinase structures have been determined to date (> 75 from distinct species), ChiB only shares up to 30% sequence identity to enzymes with solved structures and its structure could therefore provide novel insights into the GH18 family as a whole. To investigate these features, we pursued structural determination by X-ray crystallography and were able to solve the structure of FjChiB to 1.63 Å resolution. The asymmetric unit contained one protein molecule without contacts indicative of oligomerization. The overall FjChiB structure is similar to other structurally determined GH18 chitinases, having a (β/α)8 TIM-barrel fold and containing a CID between strand 7 and helix 7 of the (β/α)8-barrel (residues 249–286; Fig. 2). The electron density is well defined with only 13 residues from the N-terminus not possible to resolve in the final model. Notably, electron density for three residues of the C-terminal histidine tag used for affinity chromatography purification was resolved and modelled close to a symmetry related molecule’s substrate binding cleft.
Figure 2.
Structure of FjChiB. The overall structure of FjChiB (a) is shown next to SmChiB (b; PDB accession 1e6n). The chitinase insertion domain (CID) of each protein is colored cyan, and the -3-site capping loop and CBM family 5 domain of SmChiB are colored green and magenta, respectively. The active site architectures of FjChiB (c) and SmChiB (d) show the key residues lining the substrate binding clefts. The structure of SmChiB contained the catalytic residue substitution E144Q enabling the determination in complex with chitopentaose (orange sticks). Notably, the smaller CID and the lack of motifs equivalent to the capping loop of SmChiB lead to a more exposed substrate binding cleft in FjChiB.
Structural features of FjChiB and comparison to homologous structures
The catalytic motif (DxxDxDxE) common amongst GH18 family members, which supports a substrate-assisted catalytic mechanism2, is conserved in FjChiB (Asp146-Val147-Asp148-Leu149-Glu150; Fig. 2). Electron density consistent with a formate molecule, likely from the crystallization solution, was found in the active site positioned by hydrogen bonds with the hydroxyl moiety of Tyr205 and the carboxyl moiety of Glu150. The orientation and position of the formate molecule is similar to that of the acetyl group of a GlcNAc unit bound in the -1 subsite in several GH18 ligand complex structures. The overall architecture of the substrate-binding cleft of FjChiB is similar to that of other GH18 chitinases. Relative to SmChiB, a processive exo-acting enzyme that is amongst the best studied GH18 enzymes, the binding cleft of FjChiB has two distinct differences, described in more detail below, which leads to the cleft being more open and exposed to the bulk solvent.
In SmChiB, a small insertion between β-strand 1 and α-helix 1 leads to a capping of the cleft at the -3 site 14, which likely explains why this enzyme favors exo-binding (at the non-reducing end of a chitin chain), rather than endo-binding2,31. FjChiB lacks this insertion and instead shows more similarity to the GH18 ChtII from the insect pest Ostrinia furnacalis, which also lacks this insertion and has been shown to be able to bind longer oligosaccharides beyond the -3 site32. Like several GH18 chitinases, FjChiB has a CID inserted between strand 7 and helix 7 of the (β/α)8-barrel which folds into a distinct domain and builds up one face of the active site cleft. In SmChiB, this region is large and folds over one end of the cleft effectively forming a tunnel and shielding the − 1 and + 1 sites from the bulk solvent2,14. There is significant diversity amongst GH18 chitinase members in the CID region, both in length and sequence, and the CID in FjChiB, while similar in overall structure, is shorter than the one found in SmChiB, leading to a much more open cleft at the + 1 and + 2 sites (Fig. 2). A small or absent CID, leading to a more exposed binding cleft, has been observed in other GH18 chitinases, such as the O. furnacalis ChtII32, and is commonly associated with endo-acting activity. Previous work has shown FjChiB to be an endo-acting chitinase8 and the openness of the active site cleft is consistent with this activity.
Structural investigation of the multi-modular FjChiA
FjChiA is indispensable for the growth of F. johnsoniae on crystalline chitin. Between its two GH18 chitinase domains, the protein contains a middle domain (FjChiA_M) with carbohydrate-binding functionality, which lacks close similarity to any previously studied proteins8. Attempts to crystallize either the full-length protein or the middle domain of FjChiA were unsuccessful. However, thanks to high sequence similarity, reliable homology modeling of the FjChiA N- and C-terminal GH18 chitinase domains was possible using PHYRE233. The modelling resulted in high-confidence structure predictions of TIM-barrel proteins consistent with both FjChiA_N and FjChiA_C belonging to the GH18 family (Supplemental Fig. 2). Both domains contain the conserved DxxDxDxE catalytic motif. Of characterized chitinases FjChiA_N is most similar to ChiW from Paenibacillus sp. str. FPU-7, a chitinase from Bacillus circulans WL-12, and SmChiA, while FjChiA_C is most similar to chitinases from Bacillus cereus NCTU2, Chromobacterium violaceum, and SmChiC (Supplemental Fig. 3). Our previous functional characterization of the individual catalytic domains suggested that FjChiA_N and FjChiA_C were exo- and endo-acting chitinases, respectively8. As discussed above, in GH18 members the presence of a large CID, that partially covers the substrate-binding cleft, is associated with a higher degree of exo- and processive characteristics while a smaller or absent CID, leading to a more open binding cleft, is associated with endo-acting activities2. As illustrated in Supplemental Figs. 2 and 3, a large and extensive CID domain is present FjChiA_N whereas the domain is much smaller in FjChiA_C, consistent with the observed enzyme activities8.
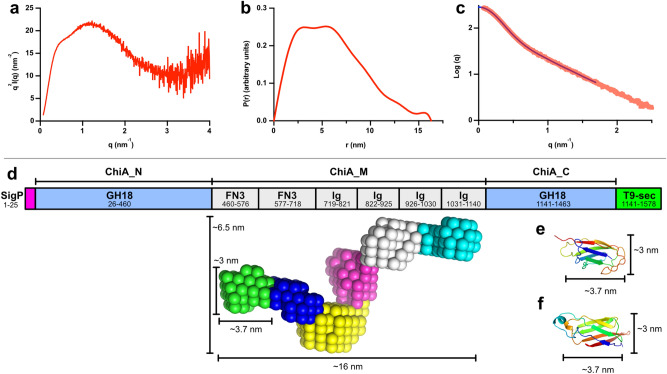
In the absence of atomic-level structural information, and to gain better insights into the overall structure of the multi-modular FjChiA, we utilized small angle X-ray scattering (SAXS) to determine a solution structure of FjChiA_M. Unfortunately, the full-length protein suffered from both aggregation and radiation damage issues, even when utilizing SAXS measurements coupled to size exclusion chromatography, and the data could not be utilized for analysis. The FjChiA_M protein, however, proved to be much more amenable to the technique and allowed for the generation of a low-resolution model of the domain (Table 1). Analysis of the data by a Kratky plot and the pair distance distribution function, P(r), indicated that the protein is modular and elongated with some degree of flexibility34 (Fig. 3). The ab initio calculation of the SAXS molecular envelopes of FjChiA_M consistently yielded an elongated protein comprised of 6 distinct modules each between 30 to 40 Å in length and ~ 30 Å wide (Fig. 3). The envelope of FjChiA_M is slightly compressed with a small rotation between the third and fourth modules.
Table 1.
SAXS data collection and analysis parameters.
| FjChiA_M | |
|---|---|
| Data collection | |
| Date | May 5, 2019 |
| Source | BL4-2 at SSRL |
| Wavelength (Å) | 1.127 |
| Sample to detector distance (m) | 1.8025 |
| Exposure time per frame (s) | 1.0 |
| Protein concentration (mg/mL) | 5.0 |
| Data analysis | |
| Rg (nm) [from Guinier approximation] | 4.72 |
| Rg (nm) [from P(r) function] | 4.75 |
| Dmax (nm) [from P(r) function] | 16.3 |
| Vporod (nm3) | 82.7 |
| SASBDB accession | SASDHU9 |
Figure 3.
Small angle X-ray scattering of FjChiA_M. The Kratky plot (a) indicates a structured protein with some degree of flexibility while the pair distance distribution function (b) is consistent with an elongated and modular protein. (c) The fit of the ab initio envelope is shown as a blue line (χ2 = 1.20; generated in ATSAS as described in the methods) alongside the experimental data shown in red. (d) The domain organization of FjChiA, including regions predicted for the signal peptide (SigP) and type-9 secretion signal (T9-sec), shown along with the FjChiA_M ab initio SAXS envelope with putatively defined discrete modules uniquely colored. The Fn3-like (e) and Ig (f) domains from the pilin protein BcpA from Bacillus cereus (PDB accession 3kpt) and an antifreeze protein from Marinomonas primoryensis (PDB accession 4p99), respectively, display between 20–30% sequence identity with the putatively annotated regions in FjChiA_M.
The FjChiA_M domain lacks significant sequence similarity to any previously characterized or structurally determined proteins, as determined by NCBI BLAST. However, protein structure predictions using PHYRE233 suggest that FjChiA_M is composed of six modules comprised of two Fn3-like domains (residues 471–577 and 578–718) followed by four immunoglobulin (Ig)-like domains (residues 719–821, 822–925, 926–1,030, and 1,031–1,140). Fn3-like and Ig-like domains are similar to each other, with both comprising seven to nine strands arranged into two β-sheets that pack onto each other35. However, Fn3-like domains tend to have shorter strands and longer intervening loop regions compared to Ig-like domains. The two FjChiA_M Fn3-like domains share 36% identity to each other and are both most closely related (20% and 22% sequence identity, respectively) to the Fn3-like domain of the pilin protein BcpA from Bacillus cereus (PDB accession 3kpt). The first two Ig-like domains share 75% sequence identity while the last two shares only 20 to 30% identity with each other and with the first two. Structure predictions of these last four domains are consistent with each comprising an Ig-like domain with each sharing between 20 to 30% sequence identity to Ig-like domains from the antifreeze protein (MpAFP) from Marinomonas primoryensis35. MpAFP is a large (1.5 MDa) protein comprised of > 100 tandem Ig-like domains that are proposed to extend and project the ice-binding domain of the protein away from the cell. Both Fn3-like and Ig-like domains are commonly found in extracellular carbohydrate-active enzymes and, while sometimes displaying weak carbohydrate-binding ability, it has been suggested that they can play a role in loosening and exfoliating chains from fibrous polysaccharides36. In SmChiA, an Fn3-like domain is connected to the catalytic domain in close proximity to the active site and may have a role in the interaction with polysaccharide substrates37,38, and it is possible that Ig-like modules could also have substrate interaction roles.
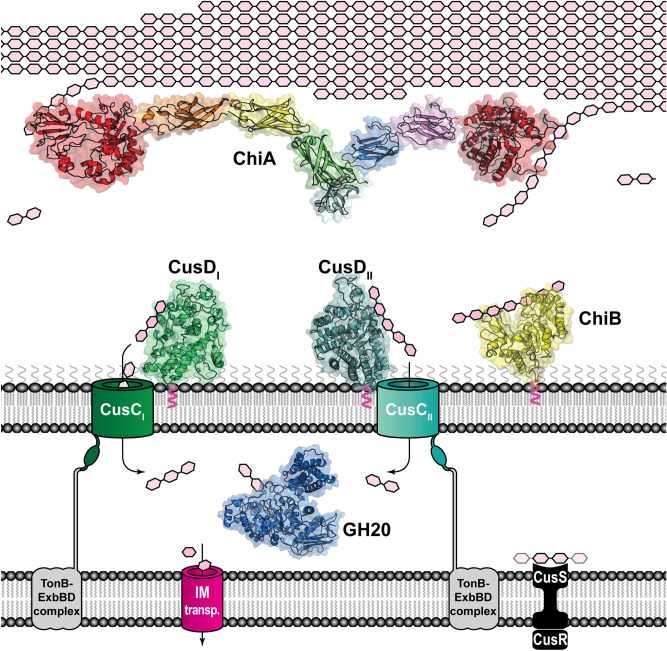
Collectively, the solution scattering results support the bioinformatic prediction that the FjChiA_M domain is composed of distinct modules which likely fold similarly to Fn3-like and Ig-like domains. Further, the protein is observed as elongated in solution in a fashion similar to “beads on a string” where each bead may sit on crystalline chitin and exfoliate chains for the two terminal chitinase domains. Our previous biochemical characterization8 showed that the FjChiA_M domain adheres to several insoluble and crystalline polysaccharides including α- and β-chitin and cellulose. The elongated conformation of the protein may be a feature that is important not only for adhesion/exfoliation but also for tethering FjChiA_N and FjChiA_C together physically for increased cooperativity between the domains. A tentative model of the FjChiA solution structure together with the entire ChiUL machinery is presented in Fig. 4.
Figure 4.
Proposed model of the ChiUL. FjCusDI (PDB accession 5j90), FjCusDII (PDB accession 5j5u), and FjChiB are outer membrane-bound lipoproteins where the former two bind oligosaccharides and facilitate import into the periplasm and the latter is an endo-acting chitinase. FjGH20 is a periplasmic chitobiase that cleaves imported oligosaccharides into GlcNAc for further metabolism. A modelled structure of ChiA based on homology models of the terminal GH18 chitinases, FjChiA_N and FjChiA_C, and spanned by homology models of the Fn3- and Ig-like domains of FjChiA_M fitted into the modelled SAXS envelope is visualized bound to, and possibly exfoliating, polysaccharides from insoluble chitin crystals.
Conclusions
Together with our previous characterization, the data presented here represent a holistic structural view of the chitin-interacting proteins of the F. johnsoniae ChiUL, including both carbohydrate-binding proteins and enzymes (Fig. 4). Our results in addition provide new structural information for both the GH18 and GH20 families. The FjGH20 structure reveals novel features previously not seen in structures of GH20 members, and the results suggest a stronger connection between families GH20 and GH84. FjChiA is an exceptionally powerful multi-modular chitinolytic enzyme and our SAXS model of its internal domain, FjChiA_M, showcases how F. johnsoniae has evolved this multidomain chitinase to form a complex enzyme with the terminal chitinase domains separated by an extended protein ‘spacer’ that also has carbohydrate-binding abilities. The exact mechanism of substrate-binding for the Fn3- and Ig-like domains of FjChiA_M remains elusive, but the current model provides a useful template for future studies. Further insights into the structure, dynamics and functional abilities of multi-catalytic molecular “machines” such as FjChiA may have implications not only for understanding chitin deconstruction, but also for understanding, and eventually designing and optimizing, enzymatic deconstruction of other recalcitrant polysaccharides.
Methods
Structure determination of FjGH20
The protein was produced and purified as previously reported8. Size exclusion chromatography for oligomerization analysis was performed on a HiLoad Superdex 200 16/60 column and an ÄKTA Explorer (GE Healthcare) using an isocratic gradient of 50 mM Tris pH 8.0 with 250 mM NaCl. Molecular weights were determined by standard curve using the Gel Filtration Markers Kit for Protein Molecular Weights 12–200 kDa (Sigma) as standards. Initial crystallization conditions were screened for using a Phoenix crystallization robot (Art Robbins Instruments) by the sitting-drop vapor-diffusion method. About 400 in-house made conditions were screened by using MRC plates with a 60 µl reservoir solution per well, and drop solutions were prepared by mixing 0.25 µl well solution and 0.25 µl protein solution at 20 mg/ml. Further optimization in Hampton 24-well hanging drop plates using 500 µl reservoirs and 1 µl + 1 µl drops, yielded crystals from 14% PEGMME 5 K and 0.1 M Na-Malonate pH 7.5. Diffraction data was collected at BL14.1 at Bessy (2016–04-16), and was integrated and scaled using XDS39 and AIMLESS40,41, respectively. The structure was solved by molecular replacement with Auto-Rickshaw42,43, with MoRDa44 identifying and using PDB accession 1now, human lysosomal beta-hexosaminidase45, as the template. An initial model was built using autobuilding in ARP/wARP46–49. Inspection of electron density maps was done in Coot50 with positional refinement in REFMAC51. The data collection, processing, and refinement statistics for all of the datasets can be found in Table 2.
Table 2.
Summary of crystallographic statistics.
| FjGH20 | FjChiB | |
|---|---|---|
| Data collection | ||
| Date | April 14, 2016 | May 3, 2017 |
| Source | BL 14.1 at Bessy | ID30A-3 at ESRF |
| Wavelength (Å) | 0.918409 | 0.9677 |
| Space group | P 21 21 21 | P 1 21 1 |
| Cell dimensions | ||
| a, b, c (Å) | 75.99, 124.55, 151.59 | 43.91, 66.44, 53.44 |
| α, β, γ (°) | 90, 90, 90 | 90, 98.31, 90 |
| No. of measured reflections | 1,033,044 (35,652) | 163,627 (14,163) |
| No. of independent reflections | 158,136 (7,637) | 34,484 (2,962) |
| Resolution (Å) | 49.1–1.7 (1.73–1.70) | 36.4–1.6 (1.69–1.63) |
| Rmerge (%) | 7.10 (39.4) | 4.30 (20.7) |
| CC1/2 (%) | 99.9 (86.8) | 99.8 (98.0) |
| Mean I/σI | 17.2 (3.2) | 19.7 (6.3) |
| Completeness (%) | 99.9 (98.4) | 90.7 (77.9) |
| Redundancy | 6.5 (4.7) | 4.8 (4.7) |
| Refinement | ||
| Rwork/Rfree | 0.142/0.175 | 0.157/0.189 |
| No. atoms | ||
| Protein | 10,214 | 2,523 |
| Ligand/ions | 22 | 15 |
| Water | 1544 | 338 |
| B-factors | ||
| Protein | 14.7 | 23.2 |
| Water | 23.9 | 34.2 |
| RMSD | ||
| Bond length (Å) | 0.020 | 0.006 |
| Bond angles (°) | 1.95 | 0.77 |
| PDB accession | 6YHH | 6XYZ |
Structure determination of FjChiB
FjChiB was produced and purified as previously reported8 and stored in 50 mM Tris buffer at pH 8.0 containing 50 mM NaCl. Crystallization conditions were screened for with a Mosquito robot (SPT Labtech) using the JCSG + screening kit (Molecular Dimensions) in MRC sitting drop plates. A condition which yielded a crystal hit was optimized in sitting drop plates with a reservoir volume of 40 µl and protein mixed with reservoir solution in a 1:1 ratio in 0.6 µl drop sizes using a protein stock solution at 20 mg/mL. The optimized formulation contained 0.15 M magnesium formate with 15% PEG3550 and yielded cuboid crystals within a week. An initial dataset diffracting to 1.85 Å was collected at the ESRF id23-2 (2014–12-02) which was integrated and scaled using XDS39 and AIMLESS40,41, after which the structure was determined by molecular replacement with Auto-Rickshaw42,43, with MoRDa44 identifying and using PDB accession 3fnd, a putative chitinase from Bacteroides thetaiotaomicron, as the search template. An initial model was built using autobuilding in ARP/wARP46–49. A subsequent data set diffracting to 1.63 Å was collected at the ESRF id30a3 (2017-05-03), processed with XDS39, and the solution defined by rigid body refinement using Phenix Refine52 and the previously determined FjChiB structure. Since the new data set provided an improvement in resolution, only this dataset was pursued for further refinement and deposition. Coot50 and Phenix Refine52 were used in iterative cycles of manual and computational refinement. The data collection, processing, and refinement statistics for all of the datasets can be found in Table 2.
Small-angle X-ray scattering (SAXS) of FjChiA_M
The protein was produced and purified as previously reported8. X-ray scattering data were obtained at BL4-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) with a Pilatus3 X 1 M detector (Dectris) operated at 11.0 keV. Full-length ChiA and ChiA_M were buffer exchanged into 50 mM Tris pH 8.0 with 250 mM NaCl and 250 μM DTT, using a HiPrep 26/10 Desalting column and an ÄKTA Explorer (GE Healthcare). The proteins were concentrated to 20 mg/mL by ultrafiltration using a Vivaspin (GE Healthcare) 10 kDa molecular weight cut-off polyethersulfone spin column and a protein dilution series was created using the ultrafiltration filtrate. 10 images with 1 s exposure, taken at a distance of 1802.5 mm, were averaged and, after background subtraction of the buffer, were utilized for analysis in the ATSAS suite version 3.053. PRIMUS54 and GNOM55 were utilized to assess the data. The data for the full-length ChiA showed significant indications of aggregation and radiation damage and were not pursued further. Images of ChiA_M at concentrations of 1.25, 2.5, and 5.0 mg/mL yielded Guinier Rg estimates within 3% of each other with no indication of radiation damage and the data collected at 5 mg/mL were chosen for further analysis. DAMMIF56 was used to generate 100 models that were subsequently clustered by DAMCLUST57 and the top cluster was averaged by DAMAVER58 and then refined by DAMMIN59. The SAXS data collection and analysis parameters can be found in Table 1.
Supplementary information
Acknowledgements
The work performed at Chalmers University of Technology was supported by the Knut and Alice Wallenberg Foundation through the Wallenberg Wood Science Center. The Research Council of Norway program Syncnøyt (project # 247732) is gratefully acknowledged for financial support. The authors are grateful to the Join Berlin MX-Laboratory at BESSY II, Berlin, for beam time and assistance with data collection. The authors would like to thank Kjersti Johannessen and Kjersti Lian for setting up crystallization trials and Thomas Weiss, Ivan Rajkovic, and Tsutomu Matsui from SSRL for the SAXS data collection of FjChiA_M. The authors would also like to thank the Structural Biology group at the ESRF for assistance with both the BioSAXS and crystallography data collections. Work at NMBU, in Ås, Norway, was supported by a fellowship from the Norwegian University of Life Sciences (to PBP), Grants 214042 and 221568 from the Research Council of Norway (to PBP and VGHE, respectively), and by the European Research Council through Grant 336355 (“MicroDE”) to PBP. Open access funding provided by Chalmers University of Technology.
Abbreviations
- PUL
Polysaccharide utilization locus
- ChiUL
Chitin utilization locus
- LPMO
Lytic polysaccharide monooxygenase
- Sus
Starch utilization system
- T9SS
Type IX secretion system
- TCS
Two-component sensor/regulator system
- Cus
Chitin utilization system
- GH
Glycoside hydrolase family
- CHO
Chitooligosaccharide
- ITC
Isothermal titration calorimetry
- GlcNAc
N-Acetylglucosamine
- PDB
Protein data bank
- RMSD
Root mean square deviation
- CBM
Carbohydrate binding module
- SAXS
Small angle X-ray scattering
Author contributions
S.M., V.G.H.E., P.B.P., G.B., and J.L. conceived the study. S.M. and J.L. produced and purified proteins. RH solved the FjGH20 structure, S.M., A.M., G.B., and J.L. solved the FjChiB structure. S.M. performed the SAXS experiments, assisted by G.B.. S.M. and J.L. drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-70749-w.
References
- 1.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaaje-Kolstad G, Horn SJ, Sorlie M, Eijsink VGH. The chitinolytic machinery of Serratia marcescens - a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 2013;280:3028–3049. doi: 10.1111/febs.12181. [DOI] [PubMed] [Google Scholar]
- 3.Grondin JM, Tamura K, Dejean G, Abbott DW, Brumer H. Polysaccharide Utilization Loci: fueling microbial communities. J Bacteriol. 2017;199:e00860–e1816. doi: 10.1128/jb.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsbrink J, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogowski A, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naas, A. E. et al. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? mBio5, 6. 10.1128/mBio.01401-14 (2014). [DOI] [PMC free article] [PubMed]
- 8.Larsbrink J, et al. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol. Biofuels. 2016;9:260. doi: 10.1186/s13068-016-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride MJ. Bacteroidetes gliding motility and the type IX secretion system. Microbiol. Spectr. 2019;7:1–9. doi: 10.1128/microbiolspec.PSIB-0002-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kharade SS, McBride MJ. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J. Bacteriol. 2014;196:961–970. doi: 10.1128/jb.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsbrink J, McKee LS. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 2020;110:63–98. doi: 10.1016/bs.aambs.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res42, D490–495. 10.1093/nar/gkt1178 (2014). [DOI] [PMC free article] [PubMed]
- 13.Perrakis, A. et al. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure2, 1169–1180. 10.1016/s0969-2126(94)00119-7 (1994). [DOI] [PubMed]
- 14.van Aalten, D. M. et al. Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9-Å resolution. Proc Natl Acad Sci U S A97, 5842–5847. 10.1073/pnas.97.11.5842 (2000). [DOI] [PMC free article] [PubMed]
- 15.Watanabe T, et al. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 1994;176:4465–4472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slámová K, Bojarová P, Petrásková L, Kren V. β-N-acetylhexosaminidase: What's in a name…? Biotechnol. Adv. 2010;28:682–693. doi: 10.1016/j.biotechadv.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Kerrigan JE, et al. Modeling and biochemical analysis of the activity of antibiofilm agent Dispersin B. Acta Biol. Hung. 2008;59:439–451. doi: 10.1556/ABiol.59.2008.4.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JK, Kim WJ, Park YI. Purification and characterization of an exo-type β-N-acetylglucosaminidase from Pseudomonas fluorescens JK-0412. J. Appl. Microbiol. 2011;110:277–286. doi: 10.1111/j.1365-2672.2010.04879.x. [DOI] [PubMed] [Google Scholar]
- 19.Mark BL, et al. Structural and functional characterization of Streptomyces plicatus beta-N-acetylhexosaminidase by comparative molecular modeling and site-directed mutagenesis. J. Biol. Chem. 1998;273:19618–19624. doi: 10.1074/jbc.273.31.19618. [DOI] [PubMed] [Google Scholar]
- 20.Tews I, et al. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 1996;3:638–648. doi: 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- 21.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drouillard S, Armand S, Davies GJ, Vorgias CE, Henrissat B. Serratia marcescens chitobiase is a retaining glycosidase utilizing substrate acetamido group participation. Biochem. J. 1997;328(Pt 3):945–949. doi: 10.1042/bj3280945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knapp S, et al. NAG-thiazoline, an N-acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 1996;118:6804–6805. doi: 10.1021/ja960826u. [DOI] [Google Scholar]
- 25.Prag G, et al. Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J. Mol. Biol. 2000;300:611–617. doi: 10.1006/jmbi.2000.3906. [DOI] [PubMed] [Google Scholar]
- 26.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997;321(Pt 2):557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier T, et al. The X-ray crystal structure of human beta-hexosaminidase B provides new insights into Sandhoff disease. J. Mol. Biol. 2003;328:669–681. doi: 10.1016/s0022-2836(03)00311-5. [DOI] [PubMed] [Google Scholar]
- 28.Briliūtė J, et al. Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat. Microbiol. 2019;4:1571–1581. doi: 10.1038/s41564-019-0466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao FV, et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ficko-Blean E, et al. Portrait of an enzyme, a complete structural analysis of a multimodular β-N-acetylglucosaminidase from Clostridium perfringens. J. Biol. Chem. 2009;284:9876–9884. doi: 10.1074/jbc.M808954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hult EL, Katouno F, Uchiyama T, Watanabe T, Sugiyama J. Molecular directionality in crystalline beta-chitin: Hydrolysis by chitinases A and B from Serratia marcescens 2170. Biochem. J. 2005;388:851–856. doi: 10.1042/BJ20050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, Qu M, Zhou Y, Yang Q. Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects. J. Biol. Chem. 2018;293:2652–2660. doi: 10.1074/jbc.RA117.000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernadó P, Svergun DI. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol. Biosyst. 2012;8:151–167. doi: 10.1039/c1mb05275f. [DOI] [PubMed] [Google Scholar]
- 35.Vance, T. D. et al. Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice. Biosci Rep34. 10.1042/bsr20140083 (2014). [DOI] [PMC free article] [PubMed]
- 36.Kataeva IA, et al. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 2002;68:4292–4300. doi: 10.1128/aem.68.9.4292-4300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrakis A, Ouzounis C, Wilson KS. Evolution of immunoglobulin-like modules in chitinases: Their structural flexibility and functional implications. Fold Des. 1997;2:291–294. doi: 10.1016/S1359-0278(97)00040-0. [DOI] [PubMed] [Google Scholar]
- 38.Uchiyama T, et al. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J. Biol. Chem. 2001;276:41343–41349. doi: 10.1074/jbc.M103610200. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 2005;61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 43.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1089–1097. doi: 10.1107/S0907444909029643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagin A, Lebedev A. MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr. Sect. A. 2015;71:s19. doi: 10.1107/S2053273315099672. [DOI] [Google Scholar]
- 45.Mark BL, et al. Crystal structure of human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease. J. Mol. Biol. 2003;327:1093–1109. doi: 10.1016/S0022-2836(03)00216-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamzin, V. S. & Wilson, K. S. in Methods in Enzymology Vol. 277 269–305 (Academic Press, London, 1997). [DOI] [PubMed]
- 47.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 48.48Morris, R. J., Perrakis, A. & Lamzin, V. S. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr.58, 968–975, 10.1107/S0907444902005462 (2002). [DOI] [PubMed]
- 49.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr.68, 352–367. 10.1107/S0907444912001308 (2012). [DOI] [PMC free article] [PubMed]
- 53.Franke, D. et al.ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr.50, 1212–1225. 10.1107/S1600576717007786 (2017). [DOI] [PMC free article] [PubMed]
- 54.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. doi: 10.1107/S0021889803012779. [DOI] [Google Scholar]
- 55.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- 56.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volkov, V. V. a. S., D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr.36, 860–864. 10.1107/S0021889803000268 (2003).
- 59.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.