Abstract
Objectives:
The aim was to identify the most important features of structural knee osteoarthritis (OA) progressors and classification using machine learning methods.
Methods:
Participants, features and outcomes were from the Osteoarthritis Initiative. Features were from baseline (1107), including articular knee tissues (135) assessed by quantitative magnetic resonance imaging (MRI). OA progressors were ascertained by four outcomes: cartilage volume loss in medial plateau at 48 and 96 months (Prop_CV_48M, 96M), Kellgren–Lawrence (KL) grade ⩾ 2 and medial joint space narrowing (JSN) ⩾ 1 at 48 months. Six feature selection models were used to identify the common features in each outcome. Six classification methods were applied to measure the accuracy of the selected features in classifying the subjects into progressors and non-progressors. Classification of the best features was done using an automatic machine learning interface and the area under the curve (AUC). To prioritize the top five features, sparse partial least square (sPLS) method was used.
Results:
For the classification of the best common features in each outcome, Multi-Layer Perceptron (MLP) achieved the highest AUC in Prop_CV_96M, KL and JSN (0.80, 0.88, 0.95), and Gradient Boosting Machine for Prop_CV_48M (0.70). sPLS showed the baseline top five features to predict knee OA progressors are the joint space width, mean cartilage thickness of the medial tibial plateau and sub-regions and JSN.
Conclusion:
In this comprehensive study using a large number of features (n = 1107) and MRI outcomes in addition to radiological outcomes, we identified the best features and classification methods for knee OA structural progressors. Data revealed baseline X-ray and MRI-based features could predict early OA knee progressors and that MLP is the best classification method.
Keywords: classification, feature selection, joint space narrowing, machine learning, magnetic resonance imaging, osteoarthritis, prediction
Key messages
Early prediction of knee osteoarthritis (OA) structural progression can be done with high accuracy and based on only a few features.
Baseline X-ray and magnetic resonance imaging-based features could predict early knee OA progressors.
Joint space width (JSW), mean cartilage thickness of the medial tibial plateau and sub-regions, and joint space narrowing are the most important for predicting knee OA progressors.
Introduction
Knee osteoarthritis (OA), a leading cause of disability worldwide, can be difficult to define as its development is often insidious and involves different subgroups.1 This disease is one of the main causes of chronic invalidity and use of health care services, an impact expected to double from 2009 to 2020 and again by 2030.2 OA can be found in many joints, but occurs most frequently in the knee. Disease evolution can be slow and span many years. With the guidelines currently available to clinicians, patient diagnosis occurs late; however, for some individuals, the progression/evolution can be fast.3–5
Although research is being done regarding the classification of knee OA patients, there is a great need to develop models that will enable the analysis of data sets using patient demographics, clinical and risk factor information, imaging and other features together for an early prediction with high classification accuracy of an individual’s risk for disease progression. Current automatic classification of OA is primarily based on a patient’s personal and familial history, demographic and clinical features and radiographs.6–12 Unfortunately, the information acquired does not lead to robust predictions or prognosis, in addition to having a relatively large precision error and low sensitivity. As reported previously,11 there are also several limitations associated with these studies. For example, mostly conventional risk factors have been included in the models while other features, such as the patient’s history, environmental (living environment) factors and magnetic resonance imaging (MRI) data, among others, should be considered. Furthermore, most studies included a small number of participants.6–10,12 Finally, these models were mostly used for classification purposes of knee OA, and not for feature/outcome selection for OA progression. At the same time that this work was performed, a recent published study13 using the Foundation for the National Institute of Health (FNIH) OA cohort and machine learning methodologies also looked at the identification of variables which include, among others, MRI features that are associated with OA progressors defined by radiographic and symptoms (pain).
In this work, we used machine learning (ML) algorithms on a fairly large set of subjects and features to develop advanced prediction models that provide high classification and prediction performance for the identification of OA progressors.
The main aim of this study was to identify the most important feature predictors of knee OA progression. To this end, we used four outcomes based on incidence of cartilage volume loss in medial tibial plateau (Prop_CV), the Kellgren–Lawrence (KL) grade, medial joint space narrowing (JSN) and 1107 predictor features including 135 quantitative MRI assessments.
Methods
Subjects, outcomes, and subjects selection
Subjects were from the Osteoarthritis Initiative (OAI) cohort (https://data-archive.nimh.nih.gov/oai/) which included 4796 individuals (for OAI participants enrolment, refer to Supplemental Methods). Written informed consent was obtained from subjects at participating sites and the study was approved by the institutional review boards at each site.
Outcomes
Four binary outcomes were used to predict OA progressors: incidence of cartilage volume loss in medial tibial plateau at 96 months and 48 months (Prop_CV_96M or 48M), KL grade ⩾ 2 at 48 months and medial JSN ⩾ 1 at 48 months. Data of the Prop_CV were assessed from quantitative MRI and the other outcomes were from the OAI.
Prop_CV_96M and 48M
For the Prop_CV, the following methodology was applied. The cartilage volume was first measured as described14,15 and the percentage of the cartilage volume loss in medial plateaus was calculated at 96 and 48 months [(CV_Med_Plat_96M and 48M- CV_Med_Plat_baseline)/CV_Med_Plat_ baseline] × 100. In order to have two distinct populations, progressors and non-progressors, we stratified as ‘1’ for the progressors according to the maximum of the highest tertile of the medial tibial plateau cartilage volume loss ⩾12.1% for 96 months and ⩾8.6% for 48 months, and for the non-progressors ‘0’ as the minimum of the smallest tertile of the medial tibial plateau cartilage volume loss, ⩽1.9% and 1.8%, respectively.
KL grade ⩾ 2 at 48 months
For the KL grade outcome, KL grades 2, 3 and 4 were considered as progressors, and KL grade 0 and 1 as non-progressors. The baseline values were not considered in feature selection process as they were greatly affecting the selected features.
Medial JSN ⩾ 1 at 48 months
The assessment of JSN from the OAI was calculated according to the Osteoarthritis Research Society International (OARSI) scoring systems with grades 0–316 and subjects with JSN⩾1 were considered progressors as described.17
Subjects selection
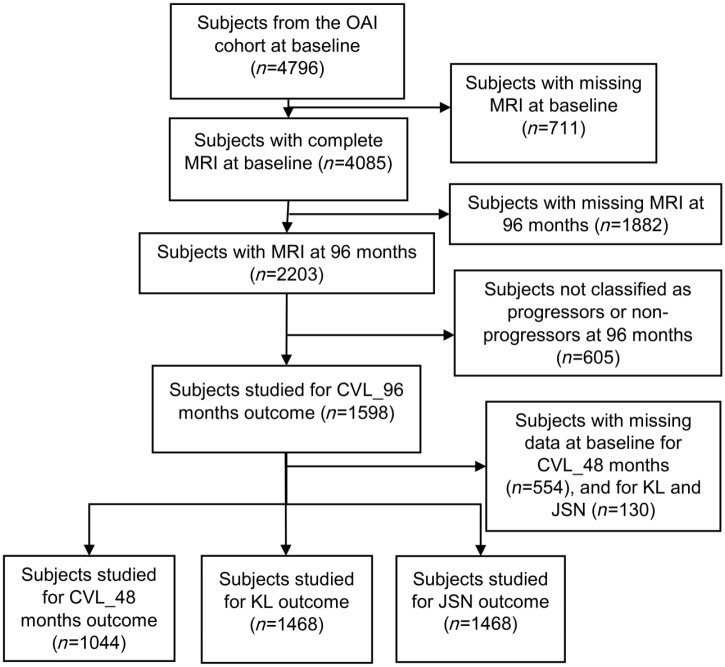
Flow chart of the subjects included/excluded is presented in Figure 1. Subjects were included if they have a MR image of the target knee (for definition, refer to Supplemental Methods) at baseline and at 96 months. Subjects (n = 1598) that were classified according to the outcome Prop_CV_96M (incidence of cartilage volume loss in medial tibial plateau at 96 months) as progressors or non-progressors (see description above) were included. In order to compare the results for all outcomes, we used the same individuals to study the other outcomes. Subjects with missing data at baseline for Prop_CV_48M, KL grade ⩾ 2 at 48 months and JSN ⩾ 1 at 48 months were excluded; as a result, 1044 were included for the Prop_CV_48M and 1468 for each KL grade ⩾ 2 at 48 months and JSN ⩾ 1 at 48 months outcomes.
Figure 1.
Flow chart of the subjects.
CVL_ 96 and 48 months, cartilage volume loss in medial tibial plateau at 96 and 48 months; JSN, joint space narrowing; KL, Kellgren–Lawrence; MRI, magnetic resonance imaging; n, number of individuals; OAI, Osteoarthritis Initiative.
Predictor features
The data and MR images are from the OAI database at the baseline visit. OAI contains 1187 features (AllClinical00 Dataset; https://nda.nih.gov/oai/full_downloads.html). The categories and subcategories of considered features are defined in Supplemental Table S1. We removed 215 features based on our experts’ opinions along with those including missing values greater than 20%. Features (1107) consist of 755 categorical variables and 352 numerical variables, in addition to 135 variables from quantitative MRI data. The MRI assessment included cartilage thickness, surface, volume14,15 and T2,18 meniscal extrusion,19,20 bone marrow lesions21 and synovial effusion size.22 All knee MRI acquisitions were acquired as described per the OAI protocol.23 (Details on the MRI methodology in Supplemental Methods, and a full list of features in Supplemental Tables S2 and S3).
Data analysis
To clean the data and impute the missing values (<20%), MICE (multivariate imputation by chained equation24) package in R was used. In order to combine the multiple imputed datasets, we used the pool() function in MICE package; this function combines the estimates from m repeated complete data analyses.
Machine learning algorithms applied for the feature selection in each outcome
Depending on how the ML algorithm learns the relationship between input features and outcomes, different ML algorithms may possibly end up using different features. Hence, the features that are selected and useful in a ML algorithm can turn out to be less useful and important in a regression-based model. We applied six methods to extract the most common and important features, including logistic regression with Least Absolute Shrinkage and Selection Operator (LASSO),25 logistic regression with Elastic Net regularization,26 Gradient Boosting machine (GBM),27 Random Forest, Information Gain (IG)28 and Multi-Layer Perceptron (MLP)29 to find the most important features as predictors for designing a more accurate prediction model. For the LASSO, glmnet30 package was used in R. In addition, for the IG method we applied WEKA data mining software (https://www.cs.waikato.ac.nz/ml/weka/). The rest of methods were performed using H2O31 and tuned using grid search in R. The areas under the receiving operating characteristic curves (AUC), sensitivity (also called true positive rate), and specificity (also called true negative rate) were calculated for all six methods. The AUC is a single number that can evaluate a model’s performance, regardless of the chosen decision boundary. A perfect ML model will have an AUC of 1.0, while a random prediction will have an AUC of 0.5.
Common features between the six feature selection methods
A feature is considered ‘common’ when it is selected in at least three out of six models. To find the importance of common features in each outcome, we calculated, in an initiative way, the average priority of each common feature. At first, we identified the priority numbers for the selected features using the coefficients obtained in each method, in which the highest coefficient has the priority number 1. The average priority of each common feature was calculated by dividing the sum of the priority number for each model into the number of models that have selected each common feature. Note that we considered LASSO and Elastic Net as one method (logistic regression) in this step. It should be mentioned that it is possible to have the same priority numbers for some features, as the averages are calculated using deterministic values. In common features, the top five priority numbers were considered as the most predictive features for each studied outcome.
Identification of the best classification model
This step identifies the best classification model using the common features. To classify the participants into two classes, progressors and non-progressors, and to ascertain the accuracy of the selected common variables, we performed automatic machine learning (AutoML) Interface for each outcome by tuning various sets of hyper-parameters (as described in Supplemental Methods). In this step, we used all of the common features, as well as the top five, to verify if the accuracy will change by reducing the number of features. This was done with six classifiers existing in H2O (Supplemental Table S4).
To assess the predictive power of the tested classification models, we partitioned the data into 80% training and 20% testing subsets. In addition, to ensure the generalizability of our prediction models, a repeated 10-fold cross-validation with 10 repeats on the 80% training data was performed, and the model tested on the holdout 20% of data. The averaged AUCs, sensitivity and specificity were calculated on the holdout test sets.
Prioritization of the features from the four outcomes as well as the outcome using Sparse Partial Least Square
To prioritize the major predictors among the features and outcomes, we applied the sparse partial least square (sPLS) regression method [mixOmix package (http://mixomics.org/) in R] enabling simultaneous feature and outcome selection in the several data sets (outcomes) as described.32 For the input features, we considered all of the common features for all of the outcomes except for Prop_CV_48M; if we considered those, we would have to drop 424 participants from all other outcomes. The reasons for not including Prop_CV_48M were that sPLS uses the outcomes and the features jointly in a single model, this outcome has a smaller number of participants than the others (Figure 1), and that the majority of the selected common features in this outcome were also selected in the other outcomes.
sPLS unravels the correlation structure between different data sets including same observations. In addition, it reduces dimensions of possibly correlated input features by summarizing the data into a set of values of linearly uncorrelated features called components.33 Each component consists of the most informative uncorrelated input features. sPLS searches for the largest covariance between orthogonal components (linear combination of features and outcomes). Regarding the number of components, sPLS selects the number of components that should be considered; there is a tuning process in sPLS for finding the right number of components. The objective is to determine the minimum number of components that account for most of the variation in data. Of note, the first component has the largest possible variance, meaning that the features selected in this component are the most important ones (for details refer to Supplemental Methods).
Results
Subjects
When comparing the four outcomes, the baseline characteristics of the included subjects were similar and no significant difference was found for age, bone mass index (BMI), gender and KL grade (Table 1).
Table 1.
Baseline characteristics of the included subjects.
| Outcomes | Prop_CV_96M (n = 1598) |
Prop_CV_48M (n = 1044) |
KL grade and JSN at 48M (n = 1468) |
|---|---|---|---|
| Age, years | 59.5 (8.5) | 60.5 (8.7) | 59.6 (8.6) |
| BMI, kg/m2 | 28.0 (4.6) | 28.5 (4.7) | 28.0 (4.6) |
| Gender, male | 725 (45.4%) | 468 (44.8%) | 644 (43.9%) |
| KL grade | |||
| 1 | 266 (25.2%) | 175 (24.6%) | 245 (25.1%) |
| 2 | 458 (43.5%) | 294 (41.4%) | 430 (44.1%) |
| 3 | 259 (24.6%) | 190 (26.7%) | 236 (24.2%) |
| 4 | 71 (6.7%) | 52 (7.3%) | 66 (6.8%) |
Results are shown as mean (standard deviation), and for gender and KL (Kellgren–Lawrence) as number of individuals (percent).
BMI, bone mass index; JSN, joint space narrowing; n, number of participants; Prop_CV_ 96 and 48 months, cartilage volume loss in medial tibial plateau at 96 and 48 months.
Statistical analysis was done with chi-square test, p value ⩽ 0.05 was considered significant. Data showed no statistical difference between groups.
The number of individuals classified as progressors for Prop_CV_96M and 48M, KL grade ⩾ 2 at 48 months and JSN at 48 months outcomes were 795 (49.7%), 514 (49.2%), 811 (55.2%) and 620 (42.2%), respectively.
Important features for prediction of OA knee progressors
For an initial prediction of knee OA progressors, we generated, from each outcome, the common features between the six methods. Data revealed that the top five common features are generally X-ray and MRI features (Table 2). The first 20 features for each outcome, the common features between the ML (highlighted in grey), and their priority numbers are in Supplemental Tables S5–S8.
Table 2.
The top five common features for each outcome.
| Outcome | Priority | Common features | Category |
|---|---|---|---|
| Prop_CV_96M | 1 | Cartilage thickness of the medial peripheral tibial plateau | MRI |
| 2 | Medial minimum joint space width | X-ray | |
| 3 | Mean cartilage thickness of the medial tibial plateau | MRI | |
| 4 | Kellgren-Lawrence grade 3 | X-ray | |
| 5 | Symptomatic osteoarthritis status in the right knee | X-ray | |
| Prop_CV_48M | 1 | Cartilage surface of the medial peripheral tibial plateau | MRI |
| 2 | Cartilage thickness of the medial peripheral tibial plateau | MRI | |
| 3 | Cartilage surface of the medial tibial plateau | MRI | |
| 4 | Medial minimum joint space width | X-ray | |
| 5 | Severe medial joint space narrowing in the right knee | X-ray | |
| KL_grade_48M | 1 | Radiographic osteoarthritis in the right knee | X-ray |
| 2 | Medial minimum joint space width | X-ray | |
| 3 | Osteophytes (definite) in the right knee | X-ray | |
| 4 | Moderate osteoarthritis in the right knee | X-ray | |
| 5 | Cartilage thickness of the posterior condyle | MRI | |
| JSN_48M | 1 | Kellgren-Lawrence grade 3 | X-ray |
| 2 | Medial minimum joint space width | X-ray | |
| 3 | Severe medial joint space narrowing in the right knee | X-ray | |
| 4 | Severe medial joint space narrowing in the left knee | X-ray | |
| 5 | Joint space narrowing in the lateral compartment in the right knee | X-ray |
JSN_48M, medial joint space narrowing ⩾1 at 48 months; KL_grade_48M, Kellgren–Lawrence grade ⩾2 at 48 months; MRI, magnetic resonance imaging; Prop_CV_96M, cartilage volume loss in medial tibial plateau at 96 months; Prop_CV_48M, cartilage volume loss in medial tibial plateau at 48 months; X-ray; radiography.
Classification of the best method for each outcome
Further, we looked at the best classification method using the AUC values for each outcome (Table 3) using the top five features and all of the common ones (Supplemental Tables S5–S8 and Table 2). AutoML in H2O package (Supplemental Table S4) was used to automatically discriminate the best classifier.
Table 3.
Classification results applied for each outcome.
| Outcome | Best classification method | All common features |
Top five common features |
||||
|---|---|---|---|---|---|---|---|
| AUC | SEN | SPE | AUC | SEN | SPE | ||
| Prop_CV_96M | Multi-Layer Perceptron | 0.80 | 0.86 | 0.69 | 0.73 | 0.95 | 0.63 |
| Prop_CV_48M | Gradient Boosting Machine | 0.70 | 0.79 | 0.55 | 0.70 | 0.79 | 0.55 |
| KL_grade_48M | Multi-Layer Perceptron | 0.88 | 0.86 | 0.77 | 0.87 | 0.80 | 0.77 |
| JSN_48M | Multi-Layer Perceptron | 0.95 | 0.87 | 0.90 | 0.92 | 0.86 | 0.90 |
AUC, area under the curve; JSN_48M, medial joint space narrowing ⩾1 at 48 months; KL_grade_48M, Kellgren–Lawrence grade ⩾2 at 48 months; Prop_CV_48M, cartilage volume loss in medial tibial plateau at 48 months; Prop_CV_96M, cartilage volume loss in medial tibial plateau at 96 months; SEN, sensitivity; SPE, specificity.
Data demonstrated (Table 3) that MLP classifier has achieved the highest AUC with moderate to high sensitivity and specificity in three out of the four outcomes (Prop_CV_96M, KL grade ⩾ 2 at 48 months grade and JSN ⩾ 1 at 48 months). Interestingly, compared with all of the common features, when considering the top five common features, there is a worsening in the AUC metric by 3–7%, with the most significant one for the Prop_CV_96M outcome. For the Prop_CV_48M, GBM was selected as the best classifier with the AUC of 0.70, which remained the same with the top 5 common features. The AUC for KL grade ⩾ 2 outcome was 0.88 while with the top five common features, there was only 1% worsening. Similarly, for the JSN ⩾ 1 at 48 months outcome, there was a 3% worsening in AUC value when the top 5 common features were studied (AUC = 0.92). This suggests that having more features increases the accuracy of all the outcomes, except for the Prop_CV_48M.
Major features for prediction of OA knee progressors
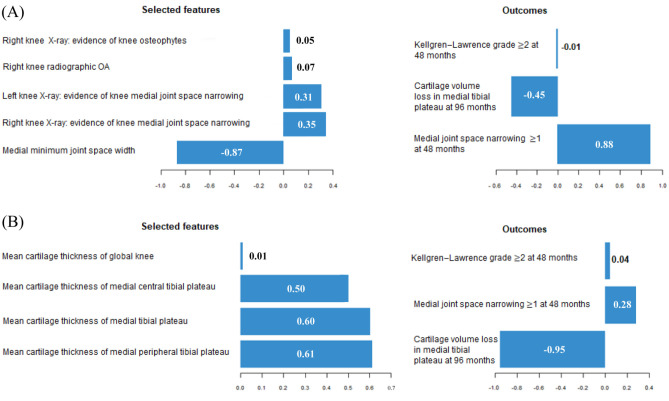
To draw a conclusion of the major features for predicting OA knee progressors, the sPLS method was applied. sPLS selected two components and were named component 1 and 2 (Figure 2). Data revealed that for the first component of sPLS (Figure 2A left side) only five X-ray features (including one negative and four positive coefficients) were selected. For example, the negative coefficient (–0.87) for a medial minimum joint space width (JSW) means that the probability of JSN at 48 months becomes more likely. From the outcomes (Figure 2A right side), the JSN is the most important one for identifying the abovementioned five features. For the second component, the most important features are the MRI-based (Figure 2B left side) mean cartilage thickness of the medial tibial plateau and sub-regions, and the most influential outcome (Figure 2B, right side) for selecting these features is the MRI-based Prop_CV_96M (–0.95). The positive sign of cartilage thickness features should be interpreted based on the negative sign of Prop_CV_96M outcome, that is, low baseline mean cartilage thickness is a predictor of a progressor. Altogether, data demonstrated that the most important features to discriminate the OA knee progressors are baseline medial minimum JSW, followed by MRI-based mean cartilage thickness of peripheral, medial, and central tibial plateau, medial JSN, and for the outcomes, JSN ⩾ 1 at 48 months and Prop_CV_96M.
Figure 2.
Coefficients for selected features and outcomes of (A) component 1 and (B) component 2. X-axis shows the coefficients of selected features and outcomes.
OA, osteoarthritis.
Discussion
This comprehensive study was carried out using state-of-the-art ML technologies by considering 1107 features from different categories at the same time and identifying the most important baseline features and the outcomes in classification of knee OA progressors. We have, in addition to X-ray features and outcome mostly used in such OA prediction studies, employed MRI as 135 features and as two outcomes to evaluate their potential in predicting the OA structural progressors. Although very few studies have included MRI evaluation as features, they were limited by including mostly cartilage alterations as a scoring system instead of quantitative assessment.10,12,34
To the best of our knowledge, this is the first study that has considered MRI-based outcomes in defining progressors and non-progressors for feature selection and classification of knee OA individuals. The majority of the previous studies on predictive models for OA progression have used the symptoms and/or X-ray as outcomes. Moreover, we chose for the outcomes to look at the knee structure instead of symptoms because it is believed that identification of disease progressors could be better modelled using the former, as although there is differing data in the literature, generally, symptoms do not correlate well with OA structural progression.
Data demonstrated that the most important features of the OA knee progressors, with a very high accuracy, are X-ray and MRI-based features and include baseline medial minimum JSW, followed by mean cartilage thickness of peripheral, medial, and central tibial plateau and medial JSN. Concerning the classification method for the features for each outcome, MLP outperforms in three out of four outcomes.
ML methods are gaining attention in medicine, however, there has not been any comprehensive and deep investigation using these methods in knee OA to identify the best features for an early prediction of the risk for OA disease progression. To prevent the bias that could happen for a regular feature selection with only one outcome, we used sPLS, which considered several outcomes at the same time for selecting the top features.
Our work was also strengthened by the use of six feature selection and classification methods in each outcome, and identification of the common features and best classifiers between these models, instead of one approach as usually performed. Hence, using different models could avoid the problem that a single ML method might select features that might not be important for another ML method. By reviewing the literature, comparable models were found only for KL grade and JSN outcomes (Supplemental Table S9) and in a study10 with AUCs of 0.79 and 0.55 respectively which illustrates the priority of our classification models [AUC = 0.88 and 0.95 respectively with MLP (Table 3)].
Data revealing that the mean cartilage thickness is a highly influential feature agrees with those showing that the low baseline cartilage thickness is the strongest predictor of OA progression.35,36 Moreover, the medial tibial plateau being selected could reflect that this region is within the first to be altered in OA patients.14,37,38 However, one could not disregard that it could also be due, at least in part, to a bias by indication, as the selection for OA progressors for the Prop_CV was the maximum of the highest tertile cartilage volume loss in this knee region. For the features, in addition to some MRI-based knee region, the JSW and JSN, as determined as a scoring method,39 were also selected and are in accordance with the MRI findings as their measurements are taken in the medial compartment. The X-ray findings support the reported significant association between the femorotibial cartilage changes and medial JSW changes.40 The relative superior importance of medial minimum JSW over MRI-based features could be a bias due to the fact that two of the outcomes are X-ray-based while only one MRI-based outcome is considered.
As in all studies, there are limitations. First, it would be interesting if the selected features could be applied in other cohorts for the model to be generalized and applied in clinical practice. Second, many of the MR images of the individuals at 96 months were unavailable, which greatly reduces the number of individuals included in our study. Third, although the definition of progressors/non-progressors could have affected the selected features, for JSN and KL grade the definitions were the same as the one used in the literature looking to identify the knee OA incidence.10 For MRI, as it is the first attempt to use such an outcome in modelling with ML to classify OA progressors, no true prior information exists of which region will be the best predictor. We elected for the medial plateau, as this region is among the first demonstrating cartilage loss in knee OA patients. Another region, the medial condyle, is also well-known to be one of the first to be involved in this tissue degradation; however, in some OA patients, the amount of cartilage left in this region is often very low (at the detection limit) and could bias the value of the cartilage volume loss over time. Moreover, the stratification of the progressors selected according to the maximum of the highest tertile of the medial plateau cartilage volume loss, ⩾12.1% for 96 months and ⩾8.6% for 48 months, was found to be most relevant based on previous published data,41 which reported a linear relationship between the rate of tibial plateau cartilage volume loss and incidence of total knee replacement, a hard outcome of the disease progression, in which for every 1% per year increase in the rate of tibial plateau cartilage loss, there was a 20% increased risk of undergoing a knee replacement. This value is about the same as the one obtained by our stratification. In addition, for the non-progressors, the threshold <1.9% and <1.8% for 96 and 48 months, respectively, of the medial tibial plateau cartilage volume loss translated into an annual loss of about 0.2–0.4%, which essentially represents no reasonable progression as previously reported.5
Another limitation could be that we did a preselection of the progressors and non-progressors instead of using the whole population. However, from a machine learning perspective, some subjects, particularly those lying on the boundary of the two categories, would be difficult to classify (label). By removing them, we aimed to avoid wrong labelling of patients for the binary feature selection process that could lead to bias and wrong classifications/predictions at the end. From a clinical perspective, we wanted the selected features to be sufficiently robust to guide to the clinical decision-making process for a given patient, as we sought to avoid tainting either group. It is our opinion that such a discrimination (avoiding individuals that may negatively impact with having stringent features) made more clinical sense. Moreover, this is not unusual and in a recently published article,42 the authors working on a multimodal machine learning based OA progression prediction model also preselected the individuals for their analysis.
Finally, further experimentation using variance inflation factor analysis showed the existence of collinearity between some MRI or X-ray features which could lead to a bias of the results. However, no known technique can truly ‘solve’ the problem of multicollinearity.43 Then, we employed multiple advanced feature selection methods plus sPLS by cross validating and tuning the hyperparameters to help identify the underlying structure within the predictor data set.
We acknowledge that not all the patients are evaluated with MRI. However, literature suggests that a combination of imaging techniques including X-ray and MRI provides the most comprehensive and effective assessment of the knee structure progression in OA44 and most of the recent research in this area are about developing prediction models based on these imaging data. In this line of thought, a recent report13 also confirmed that MRI-based features are comprised as predictors for knee OA progression in addition to a biomarker variable.
Results from this study are translational and could have high clinical relevance. The next step will be to develop a predictive tool for OA progressors based on our findings, which could be used to guide clinical decision-making allowing specific therapeutic interventions to be assigned. Moreover, such a prediction model could assist in the design of clinical trials in knee OA, as it may be useful in stratification of patients who will progress, as well as assist in discriminating the potential responders from non-responders for a given therapeutic approach.
In conclusion, this comprehensive study was carried out using state-of-the-art ML methods and revealed that early prediction of knee OA progression can be done with high accuracy and based on only a few features. This study identifies the features JSW, mean cartilage thickness of the medial tibial plateau and sub-regions and JSN as the most important for predicting knee OA progressors. These results could be used for the development of a tool enabling prediction of knee OA progressors.
Supplemental Material
Supplemental material, Suppl_Methods_TAB-20-02-034_Revision_1 for Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods by Afshin Jamshidi, Mickael Leclercq, Aurelie Labbe, Jean-Pierre Pelletier, François Abram, Arnaud Droit and Johanne Martel-Pelletier in Therapeutic Advances in Musculoskeletal Disease
Supplemental material, Suppl_Tables_TAB-20-02-034_Revision_1 for Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods by Afshin Jamshidi, Mickael Leclercq, Aurelie Labbe, Jean-Pierre Pelletier, François Abram, Arnaud Droit and Johanne Martel-Pelletier in Therapeutic Advances in Musculoskeletal Disease
Acknowledgments
The authors would like to thank the OAI participants and Coordinating Center for their work in generating the clinical and radiological data of the OAI cohort and for making them publicly available. The authors are grateful to ArthroLab Inc. for having provided the MRI data. None of the authors were part of the OAI investigator team. They also thank Julien Saint-Pierre, MSc, for his involvement in the first part of the statistical analysis and Jacqueline Brunet for preparing the manuscript.
Footnotes
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by the Osteoarthritis Research Unit and the Chair in Osteoarthritis, University of Montreal, Montréal, Québec, Canada; Computational Biology Laboratory, CHU de Québec Research Center - Université Laval; and Canada First Research Excellence Fund through the TransMedTech Institute, Montreal, Canada.
Conflict of interest statement: J-PP and JM-P are shareholders in ArthroLab Inc. and François Abram is an employee of ArthroLab Inc. The other authors have no conflicts of interest for this study.
ORCID iD: Johanne Martel-Pelletier  https://orcid.org/0000-0003-2618-383X
https://orcid.org/0000-0003-2618-383X
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Afshin Jamshidi, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), Montreal, Quebec, Canada; Laval University Hospital Research Centre, Quebec, Canada.
Mickael Leclercq, CHU de Québec Research Center - Université Laval, Quebec, Canada.
Aurelie Labbe, Department of Decision Sciences, HEC Montreal, Montreal, Quebec, Canada.
Jean-Pierre Pelletier, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), Montreal, Quebec, Canada.
François Abram, Medical Imaging Research and Development, ArthroLab Inc., Montreal, Quebec, Canada.
Arnaud Droit, CHU de Québec Research Center - Université Laval, Quebec, Canada.
Johanne Martel-Pelletier, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), 900 Saint-Denis, Suite R11.412, Montreal, Quebec H2X 0A9, Canada.
References
- 1. Cross M, Smith E, Hoy D, et al. The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis 2014; 73: 1323–1330. [DOI] [PubMed] [Google Scholar]
- 2. Bitton R. The economic burden of osteoarthritis. Am J Manag Care 2009; 15: S230–S235. [PubMed] [Google Scholar]
- 3. Jacobs CA, Vranceanu AM, Thompson KL, Lattermann C. Rapid Progression of Knee Pain and Osteoarthritis Biomarkers Greatest for Patients with Combined Obesity and Depression: Data from the Osteoarthritis Initiative. Cartilage 2020;11(1):38–46. doi: 10.1177/1947603518777577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Lange-Brokaar BJ, Bijsterbosch J, Kornaat PR, et al. Radiographic progression of knee osteoarthritis is associated with MRI abnormalities in both the patellofemoral and tibiofemoral joint. Osteoarthritis Cartilage 2016; 24: 473–479. [DOI] [PubMed] [Google Scholar]
- 5. Raynauld JP, Martel-Pelletier J, Berthiaume MJ, et al. Quantitative magnetic resonance imaging evaluation of knee osteoarthritis progression over two years and correlation with clinical symptoms and radiologic changes. Arthritis Rheum 2004; 50: 476–487. [DOI] [PubMed] [Google Scholar]
- 6. Yoo TK, Kim DW, Choi SB, et al. Simple scoring system and artificial neural network for knee osteoarthritis risk prediction: a cross-sectional study. PLoS One 2016; 11: e0148724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swan AL, Stekel DJ, Hodgman C, et al. A machine learning heuristic to identify biologically relevant and minimal biomarker panels from omics data. BMC Genomics 2015; 16(Suppl. 1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Losina E, Klara K, Michl GL, et al. Development and feasibility of a personalized, interactive risk calculator for knee osteoarthritis. BMC Musculoskelet Disord 2015; 16: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Long MJ, Papi E, Duffell LD, et al. Predicting knee osteoarthritis risk in injured populations. Clin Biomech (Bristol, Avon) 2017; 47: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lazzarini N, Runhaar J, Bay-Jensen AC, et al. A machine learning approach for the identification of new biomarkers for knee osteoarthritis development in overweight and obese women. Osteoarthritis Cartilage 2017; 25: 2014–2021. [DOI] [PubMed] [Google Scholar]
- 11. Jamshidi A, Pelletier JP, Martel-Pelletier J, et al. Machine-learning-based patient-specific prediction models for knee osteoarthritis. Nat Rev Rheumatol 2019; 15: 49–60. [DOI] [PubMed] [Google Scholar]
- 12. Ashinsky BG, Bouhrara M, Coletta CE, et al. Predicting early symptomatic osteoarthritis in the human knee using machine learning classification of magnetic resonance images from the osteoarthritis initiative. J Orthop Res 2017; 35: 2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nelson AE, Fang F, Arbeeva L, et al. A machine learning approach to knee osteoarthritis phenotyping: data from the FNIH biomarkers consortium. Osteoarthritis Cartilage 2019; 27: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martel-Pelletier J, Roubille C, Abram F, et al. First-line analysis of the effects of treatment on progression of structural changes in knee osteoarthritis over 24 months: data from the osteoarthritis initiative progression cohort. Ann Rheum Dis 2015; 74: 547–556. [DOI] [PubMed] [Google Scholar]
- 15. Dodin P, Pelletier JP, Martel-Pelletier J, et al. Automatic human knee cartilage segmentation from 3D magnetic resonance images. IEEE Trans Biomed Eng 2010; 57: 2699–2711. [DOI] [PubMed] [Google Scholar]
- 16. Culvenor AG, Engen CN, Oiestad BE, et al. Defining the presence of radiographic knee osteoarthritis: a comparison between the Kellgren and Lawrence system and OARSI atlas criteria. Knee Surg Sports Traumatol Arthrosc 2015; 23: 3532–3539. [DOI] [PubMed] [Google Scholar]
- 17. Hellio Le, Graverand MP, Mazzuca S, Duryea J, et al. Radiographic-based grading methods and radiographic measurement of joint space width in osteoarthritis. Radiol Clin North Am 2009; 47: 567–579. [DOI] [PubMed] [Google Scholar]
- 18. Raya JG, Dietrich O, Horng A, et al. T2 measurement in articular cartilage: impact of the fitting method on accuracy and precision at low SNR. Magn Reson Med 2010; 63: 181–193. [DOI] [PubMed] [Google Scholar]
- 19. Berthiaume MJ, Raynauld JP, Martel-Pelletier J, et al. Meniscal tear and extrusion are strongly associated with progression of symptomatic knee osteoarthritis as assessed by quantitative magnetic resonance imaging. Ann Rheum Dis 2005; 64: 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raynauld JP, Martel-Pelletier J, Berthiaume MJ, et al. Long term evaluation of disease progression through the quantitative magnetic resonance imaging of symptomatic knee osteoarthritis patients: correlation with clinical symptoms and radiographic changes. Arthritis Res Ther 2006; 8: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dodin P, Abram F, Pelletier JP, et al. A fully automated system for quantification of knee bone marrow lesions using MRI and the osteoarthritis initiative cohort. J Biomed Graph Comput 2013; 3: 51–65. [Google Scholar]
- 22. Li W, Abram F, Pelletier JP, et al. Fully automated system for the quantification of human osteoarthritic knee joint effusion volume using magnetic resonance imaging. Arthritis Res Ther 2010; 12: R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peterfy CG, Schneider E, Nevitt M, et al. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 2008; 16: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Azur MJ, Stuart EA, Frangakis C, et al. Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res 2011; 20: 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tibshirani R, et al. Regression shrinkage and selection via the lasso. J R Stat Soc 1996; 58: 267–288. [Google Scholar]
- 26. Zou H, Hastie T, et al. Regularization and variable selection via the elastic net. J R Statist Soc B 2005; 67: 301–320. [Google Scholar]
- 27. Natekin A, Knoll A, et al. Gradient boosting machines, a tutorial. Front Neurorobot 2013; 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zelic I, Kononenko I, Lavrac N, et al. Induction of decision trees and Bayesian classification applied to diagnosis of sport injuries. J Med Syst 1997; 21: 429–444. [DOI] [PubMed] [Google Scholar]
- 29. Souzaa FAA, Araújo R, Matias T, et al. A multilayer-perceptron based method for variable selection in soft sensor design. J Process Control 2013; 23: 1371–1378. [Google Scholar]
- 30. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- 31. H2O.ai. R Interface for H2O, R package version 3.10.0.8. 2016. [Google Scholar]
- 32. Le Cao KA, Rossouw D, Robert-Granie C, et al. A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Mol Biol 2008; 7: 35. [DOI] [PubMed] [Google Scholar]
- 33. González I, Cao KAL, Davis MJ, et al. Visualising associations between paired ‘omics’ data sets. BioData Mining 2012; 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma L, Hochberg M, Nevitt M, et al. Knee tissue lesions and prediction of incident knee osteoarthritis over 7 years in a cohort of persons at higher risk. Osteoarthritis Cartilage 2017; 25: 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eckstein F, Kwoh CK, Boudreau RM, et al. Quantitative MRI measures of cartilage predict knee replacement: a case-control study from the osteoarthritis initiative. Ann Rheum Dis 2013; 72: 707–714. [DOI] [PubMed] [Google Scholar]
- 36. Eckstein F, Le Graverand MP, Charles HC, et al. Clinical, radiographic, molecular and MRI-based predictors of cartilage loss in knee osteoarthritis. Ann Rheum Dis 2011; 70: 1223–1230. [DOI] [PubMed] [Google Scholar]
- 37. Everhart JS, Abouljoud MM, Poland SG, Flanigan DC. Medial compartment defects progress at a more rapid rate than lateral cartilage defects in older adults with minimal to moderate knee osteoarthritis (OA): data from the OA initiative. Knee Surg Sports Traumatol Arthrosc 2019; 27(8): 2401–2409. doi:10.1007/s00167-018-5202-1 [DOI] [PubMed] [Google Scholar]
- 38. Raynauld JP, Martel-Pelletier J, Bias P, et al. Protective effects of licofelone, a 5-lipoxygenase and cyclo-oxygenase inhibitor, versus naproxen on cartilage loss in knee osteoarthritis: a first multicentre clinical trial using quantitative MRI. Ann Rheum Dis 2009; 68: 938–947. [DOI] [PubMed] [Google Scholar]
- 39. Altman RD, Gold GE, et al. Atlas of individual radiographic features in osteoarthritis, revised. Osteoarthritis Cartilage 2007; 15(Suppl. A): A1–A56. [DOI] [PubMed] [Google Scholar]
- 40. Pelletier JP, Raynauld JP, Berthiaume MJ, et al. Risk factors associated with the loss of cartilage volume on weight-bearing areas in knee osteoarthritis patients assessed by quantitative magnetic resonance imaging: a longitudinal study. Arthritis Res Ther 2007; 9: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cicuttini FM, Jones G, Forbes A, et al. Rate of cartilage loss at two years predicts subsequent total knee arthroplasty: a prospective study. Ann Rheum Dis 2004; 63: 1124–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tiulpin A, Klein S, Bierma-Zeinstra SMA, et al. Multimodal machine learning-based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci Rep 2019; 9: 20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dormann CF, Elith J, Bacher S, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013; 36: 27–46. [Google Scholar]
- 44. Braun HJ, Gold GE. Diagnosis of osteoarthritis: imaging. Bone 2012; 51: 278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Suppl_Methods_TAB-20-02-034_Revision_1 for Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods by Afshin Jamshidi, Mickael Leclercq, Aurelie Labbe, Jean-Pierre Pelletier, François Abram, Arnaud Droit and Johanne Martel-Pelletier in Therapeutic Advances in Musculoskeletal Disease
Supplemental material, Suppl_Tables_TAB-20-02-034_Revision_1 for Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods by Afshin Jamshidi, Mickael Leclercq, Aurelie Labbe, Jean-Pierre Pelletier, François Abram, Arnaud Droit and Johanne Martel-Pelletier in Therapeutic Advances in Musculoskeletal Disease