Abstract
The biological importance and varied metabolic capabilities of specific microbial strains have long been established in the scientific community. Strains have, in the past, been largely defined and characterized based on microbial isolates. However, the emergence of new technologies and techniques has enabled assessments of their ecology and phenotypes within microbial communities and the human microbiome. While it is now more obvious how pathogenic strain variants are detrimental to human health, the consequences of subtle genetic variation in the microbiome have only recently been exposed. Here, we review the operational definitions of strains (e.g., genetic and structural variants) as they can now be identified from microbial communities using different high-throughput, often culture-independent techniques. We summarize the distribution and diversity of strains across the human body and their emerging links to health maintenance, disease risk and progression, and biochemical responses to perturbations, such as diet or drugs. We list methods for identifying, quantifying, and tracking strains, utilizing high-throughput sequencing along with other molecular and “culturomics” technologies. Finally, we discuss implications of population studies in bridging experimental gaps and leading to a better understanding of the health effects of strains in the human microbiome.
Keywords: Microbial strains, Microbial communities, Microbiome, Metagenomics, Amplicons, 16S, Microbiome epidemiology
Background
The importance of phenotypes and physiology characteristic of specific microbial strains has been recognized as early as the nineteenth century. Robert Koch’s postulates, for example, differentiate between disease-causing “pathogens” and benign but closely related microbial variants [1]. While the surprising differences between otherwise similar microbial strains has thus been critical in infectious disease management and microbiology for centuries, it has only recently become accessible in the context of microbial communities and their ecology. It remains technically challenging to detect and differentiate among closely related microbial strains within communities, and we will discuss several high-throughput culture-independent and culture-based methods for doing so here. More importantly, though, the beginning of such work has shown strain variation in the human microbiome to be as important in the structure, function, immunology, and epidemiology of our “normal” microbial residents as it is in the definition of pathogenicity (Box 1).
Box 1.
Terminology for microbial community strain analysis
|
Strikingly, there is no universal definition of what constitutes a microbial strain (or, for that matter, species) [2, 3]. Many factors contribute to this difficulty, including the rapidity of microbial evolution, the plasticity of many microbial genomes, the prevalence of mobile elements and lateral transfers, the difficulty in differentiating between many microbial taxa or clades by non-molecular methods, and the overall natural history of microbiology and microbial systematics. This ambiguity has led to a field in which different microbial strains of the same species can differ by as much as 5% nucleotide identity, or 30% or more of their gene content [4]. As such, even apparently benign, phenotypically similar microbial strain variants can differ genomically more than most eukaryotic species, and most related terminology can be context-dependent or defined operationally: Species: microbial species have been variously defined based on (1) whole-genome or pangenome nucleotide or amino acid phylogenetic identity thresholds; (2) gross microbial physiology / morphology / phenotype; (3) phenotypes induced by a microbe on its host or environment (e.g., human pathogens); and (4) the host or environment of a microbe, e.g., a specific geographical or biochemical origin [5]. The more than 100-year history of microbial systematics must thus be constantly resolved against new, and emerging, molecular and phenotypic information, leading to operational definitions of microbial species in roughly the two categories of “clades defined as species at some previous point” versus “clades that meet specific quantitative phylogenetic criteria” [6]. These two definitions can be considered roughly equivalent if phylogeny (genotype) is considered to be a trait (i.e. phenotype) by which isolates or community members can be classified into self-similar groups. Species group or complex: a group of taxonomically defined species that are not well-differentiated based on genomic or other criteria [7]. These typically arise in microbial systematics due to multiple independent identifications of what later prove to be (essentially) the same organism. Conversely, individual taxonomically defined microbial species can later prove to represent implicit complexes, if they, e.g., are not initially differentiated by physiology but are later found to be molecularly distinct. Subspecies clade: in communities, an operationally defined group of related organisms or radius of phylogenetic divergence smaller than, and contained within, a parent species [8]. This allows microbial genotypes within communities to be manipulated independently of their potential systematics, since, e.g., some taxonomically defined species may unintentionally capture widely divergent genotypes (and are thus better described using multiple subspecies clades), while others may prove to be closely related or near-identical (and are thus better described as a single species complex). Historically, subspecies have also referred to phenotypically distinct groups within a species [5], which may or may not be monophyletic. Isolate: a presumed clonal strain grown, assayed, and manipulated (presumably) axenically (i.e., in monoculture), typically in vitro, after a process such as streaking and/or colony picking [9]. As per canonical references such as Bergey’s Manual [10], when not defined genomically, isolates have been commonly differentiated based on phenotypes such as morphology; medium specificity; serologic, phage, or bacteriocin sensitivity; biochemical reactions; pathogenicity; or other microbial physiology. Strain: Historically, this has meant a microbial isolate, although the definition is not well-suited to microbial community studies. In this context, the term is used variously to refer to a specific microbial genome or collection of clonally identical cells (i.e., a genotype); one or more colonies (believed to be) derived from the same progenitor cell; or most often, in practice, a collection of cells or genomes within a relatively small range of phylogenetic variation (i.e., a very narrow subspecies clade). |
Particularly within communities that are by definition collections of heterogeneous cells, it has proven to be technically challenging to detect and differentiate among cells containing such closely related but highly variable genomes. Indeed, it is not yet clear how clonally most microbial lineages remain within typical in vivo communities. This suggests both basic questions about the generation and maintenance of closely related genome variants in any microbial community, and also pressing translational questions regarding the personalization and health consequences of strains in the human microbiome. Because of the extensive genetic and genomic (i.e., functional) differences between even closely related microbial strains, work to date has only rarely been powered to associate “commensal” microbial strains with their health consequences [11–14]. Here, we thus review the ecology and effects known to date for microbial strain variants carried within the human microbiome, quantitative methods for their detection and epidemiology, and potential next steps including characterization of their surprisingly large pangenomic content of biochemical dark matter.
Unexpected microbial strain diversity in health and disease from population-scale investigations of the human microbiome
Culture-based comparative genetics of isolates has been a mainstay of microbial characterization for decades, and along with culture-independent techniques, it is increasingly important in an era of high-throughput “culturomics” and creative isolation methods [15, 16]. Especially for human pathogens that are both of clinical interest and relatively easily culturable, hundreds or thousands of genomes have been used in some cases to compare strains and their transmission, associate SNV and structural variation to microbial or host phenotype, and define the genetic and evolutionary architectures of species and other clades [17–19]. Metagenomic methods have the unique ability to extend these strain-specific investigations to almost any environment or microbe, while leveraging the insights already built up using isolate genomics. In particular, if a “strain” is considered to be a clonal genotype, it must correspond to a specific set of genes and resulting functionality. This functional perspective on strains has captured a wide range of operational architectures, since some processes are well-conserved across entire clades (e.g., butyrate production in Faecalibacterium prausnitzii [20, 21]). Others, conversely, are highly variable even within specific benign or pathogenic species—Escherichia coli in the gut being the most prominent example [22].
Strains in the human gut microbiome
The gut is the greatest reservoir of biomass in the human microbiome, the body’s largest immune exposure, the most well-studied contributor to microbiome-linked disease, and one of the most ecologically diverse human-associated microbial habitats [23]. It is also the source of several of the most canonical examples of radically different microbial physiology among closely related strains, such as the benign E. coli variants carried in most guts as compared to acute pathogens such as enterohemorrhagic E. coli (EHEC) O157:H7 [24], long-term risks such as colorectal cancer in association with colibactin production in pks + E. coli [25], or the probiotic E. coli Nissle 1917 [26]. Isolate cultures have identified other strain-specific characteristics associated with evolutionary advantages ranging from increased virulence [27], mobility [28], nutrient acquisition, antibiotic resistance [29], and defense [30].
Strains abundant in the infant gut are only rarely abundant in maternal microbiomes [31–34] and are often replaced within the first 1–2 years of life [35, 36]. Their similarity to maternal, familial, or generally environmental strains is also itself highly variable and species-specific [31, 32, 37], but even small structural variants may be crucial in immune programming during temporally specific developmental windows [38–41]. Like developmental variants of human gene products, such as hemoglobin forms [42], this dynamism in early life has functional consequences: Bifidobacterium longum, for example, is selected for human milk oligosaccharide (HMO) utilization [43] in breastfeeding infants, whereas closely related B. longum strains in the adult gut frequently possess the capacity to ferment carbohydrates, but not HMOs [44]. Strains abundant in the infant gut are only rarely abundant in maternal microbiomes [31–34] and are often replaced within the first 1–2 years of life [35, 45], but even small structural variants may be crucial in immune programming during temporally specific developmental windows [38–41]. Ultimately, microbial strain variants affect not only host and individual microbes’ physiology, but also the ecology and phylogenetics of the overall gut community: Helicobacter pylori is one of the best-known examples of resident microbial genetic variation paralleling that of human host populations [46], but this has recently been shown to be the case for multiple subsets of the gut microbiome, such as Prevotella copri [12] or Eubacterium rectale [47]. This leads to linkages between the evolution and diversification of gut microbial community strains and host migration, geography, and lifestyle [8, 48].
One of the most crucial environmental factors related to this in the gut is diet, both acutely and over evolutionary time scales. However, the specifics of this relationship have been difficult to tease apart in human populations, due to the challenges of measuring diverse human diets, the confounding of long-term diet with other environmental factors, and the complexity of diet-microbial biochemical interactions. Indeed, diet represents only one aspect of gut microbial interaction with our biochemical environment, with several examples identified to date of strain-specific metabolism of drugs such as digoxin [49], metformin [50], acetaminophen [51], and potentially many others [52]. With respect to diet itself, De Filippis et al. [53], for example, found a greater abundance of P. copri among participants more closely adhering to a Mediterranean-style diet enriched with olive oil, fish, fruits, and vegetables. In contrast, Kovatcheva-Datchary et al. [54] observed that even on the same barley-rich diet, Prevotella was only enriched among select participants, potentially in a strain-specific manner. De Filippis et al. [55] later found similar heterogeneity among individuals on low-fat diets. Other examples include strains of short-chain fatty acid (SCFA)-producing bacteria with differential responses to fiber-enriched diets [56, 57]. Perhaps one of the most extreme examples of diet-linked strain specificity in the gut are among probiotic organisms such as Lactobacillus and Bifidobacterium, for which strains characteristic of fermented foods are highly distinct from those more typically resident in the human gut [58]. The health consequences of probiotics can also be strain-specific dependent either on the strain context of the microbiome being entered [59], or on the strain of the probiotic organisms, e.g., the recently proposed ability of some bifidobacteria to facilitate cancer immunotherapy [60].
Gut microbiome strains as risk factors in gastrointestinal and systemic disease
While many studies have linked overall microbiome structure or microbial species enrichments to gastrointestinal (GI) or systemic disease, relatively few have identified strain-specific microbial variants associated with these diseases. The inflammatory bowel diseases (IBD) are among the best-studied chronic gastrointestinal conditions with respect to the microbiome, and in IBD, subspecies of E. coli and Ruminococcus gnavus have each been associated with disease severity [61, 62]. Hall et al. [13] noted a particular subpopulation of R. gnavus strains more abundant in the IBD gut, enriched for adaptations to oxidative stress response, adhesion, and the utilization of iron and mucus. Bacteroides fragilis strains exhibit divergent behaviors leading to differential IgA induction in mouse models of IBD [63] and have been associated with host immunomodulatory effects in monocolonization [64]. While there are decades of work demonstrating the effects of such variants during animal monocolonization, understanding their effects in the human gut remains challenging, since the equivalent of a human genome-wide association study for most microbial community genetic variants (i.e., those not of very high penetrance) would be challenging, given the degree of multiple hypothesis testing necessary to account for the underlying microbial genetic variability [65, 66].
Studies of systemic disease outside of the gastrointestinal tract have also suggested functional roles for specific gut microbial strains. New-onset rheumatoid arthritis patients appear to be enriched for P. copri in the gut in some populations, for example, with evidence that this P. copri subset may be functionally or phylogenetically distinct [67]. Obesity and type 2 diabetes (T2D) have shown relatively weak taxonomic or functional shifts in the gut microbiome overall, but again using mice to avoid challenges in human population structure, specific strains of Akkermansia muciniphila proved to be causal in alleviating these metabolic conditions [68]. In human subjects, at least one study found SNPs specific to Bacteroides coprocola subpopulations within a T2D patient group [69]. More broadly, strain-specific promotion of several SCFA producers, including Bifidobacterium spp., Eubacterium spp., and Lactobacillus spp., was selectively enriched by dietary fiber in a randomized clinical trial, improving T2D parameters [70].
One of the most complex conditions bridging the gut microbiome, gastrointestinal, and systemic health has proven to be cancer. Particularly in colorectal cancer (CRC), specific microbial strain functionality can be readily shown to be locally causal, such as DNA-damaging production of colibactin by pks + E. coli as introduced above [71] or B. fragilis toxin [72]. Other microbes such as CRC-specific lineages of Fusobacterium nucleatum have been identified more recently, with mechanisms such as Fap2-mediated binding to host Gal-GalNAc [73] or immunomodulation via TIGIT [74] mediating both their carcinogenicity and their differentiation from typical oral F. nucleatum strains. Other mechanisms of microbial influence on GI or systemic cancer remain less well-understood, with strong evidence of resident microbial effects on immunotherapy responsiveness [75–77], but as yet few strain-specific culprits. Likewise, limited studies have shown intratumoral bacteria within and outside of the colon to be capable of direct metabolism of chemotherapeutics such as gemcitabine [78], with potentially many more such microbe-chemical interactions waiting to be discovered.
Strain carriage and variation in the body-wide human microbiome
While the strain epidemiology of the gut microbiome is perhaps best developed, similar examples exist of the effects of “commensal” and pathogenic strains throughout the human body habitat. As with the gut, the most extreme examples are those of well-studied pathogens [79], such as resistant variants of Staphylococcus aureus in the skin and nasal microbiomes [80]. More recently, combinations of culture-independent and high-throughput culture-based methods have exposed within-subject pathogen evolution over the course of months to years [81]. In these cases, as with pks + E. coli, resistance functionality such as mecA can be attributed to just one or a few loci that are genetically variable among strains via mobile chromosomal or plasmid-encoded elements [82]. More unexpectedly, however, recent findings have pointed to correspondingly strain-specific interactions with non-pathogenic commensals, such as coporphyrin III production by some Cutibacterium (formerly Propionibacterium) strains inducing Staphylococcus biofilm formation [83]. Indeed, due to their biogeographical heterogeneity relative to the gut, exposed topographical surfaces such as the skin, nasopharynx, and lung are among the few body areas where detailed ecology and persistence of multiple competing strains within an individual has been directly observed [84–86], e.g., among S. epidermidis strains in psoriasis [87].
Conversely, deep differentiation of strains within an individual is technically more challenging in the vaginal microbiome. Instead, this environment has revealed extensive subspecies heterogeneity between hosts within the dominant Lactobacillus and other species of the vagina, again raising issues regarding the exact definition of strains and species among different microbial clades. Specifically, analysis of the intraspecific diversity of vaginally dominant lactobacilli such as L. jensenii, L. iners, L. gasserii, and L. crispatus is complicated by the systematics of the clade, which has been under scrutiny for reorganization based on both isolate and culture-independent genomics [88, 89]. Nevertheless, vaginal Lactobacillus and other strains can be reasonably stable within individuals over time [90], with particularly large environmental changes such as pregnancy inducing shifts over the course of gestation [91]. As in the gut, such genetic variation between strains can affect health, such as in the determinants of pathogenicity in E. coli causing urinary tract infections [92, 93]. In examples from even more acute infectious disease, strain-specific Lactobacillus bioactivity can itself contribute to risk of sexually transmitted infection acquisition such as HIV, both due to direct microbial biochemistry [94] and its effect on host immunity [95].
Finally, oral microbiology has historically provided some of the first and most striking examples of phenotypic heterogeneity between closely related microbial isolates [96–98], and this trend holds true in the era of culture-independent sequencing and whole-community studies as well. Indeed, some of the earliest large population-scale surveys of the microbiome found oral site tropism to be a strong driver of subspecies differentiation [99–101], with stable genetic differences among related microbial colonizers of different surfaces—including different teeth—within the same mouth. These potentially adaptive, highly niche-specific variants have begun to be explored at scale, remaining stable within individual up to hundreds of days within subjects [102], but revealing extensive long-term plasticity between members of clades such as the Neisseria [11]. While there is extensive ongoing work regarding the role of overall oral microbial ecology in conditions from periodontitis [103] to pancreatic cancer [104] and heart disease [105], the ecological and genomic diversity of the oral microbiota has led to limited strain-specific associations to date. Several have been suggested for, e.g., Streptococcus variants in caries [106] or F. nucleatum in association with oral cancer [107]—suggesting intriguing links with its role in CRC. These include sufficient detail to implicate microbial processes such as polyamine biosynthesis, motility and chemotaxis, and immunostimulation (e.g., LPS and flagellar components), but without yet a clear picture of the many possible strains across which these functions may be distributed in the complex oral environment.
Strategies and approaches to identifying community strain diversity
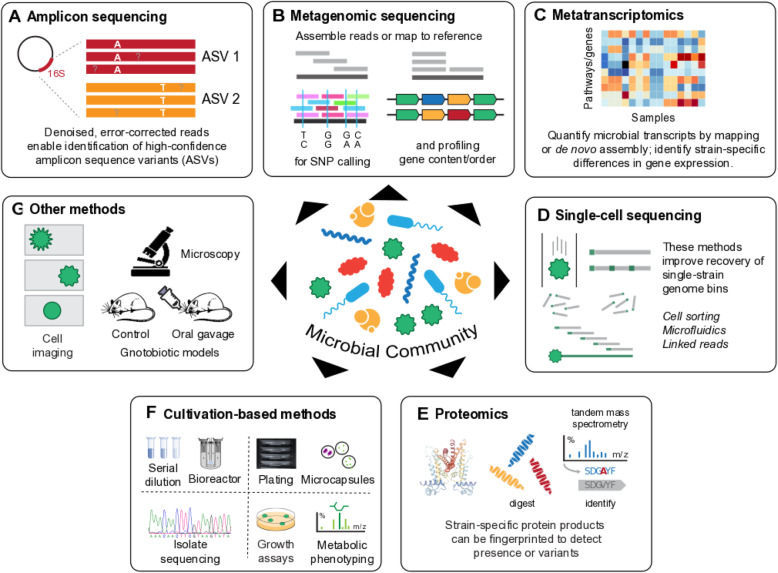
It is not our goal here to summarize the many methods that have been used to differentiate among microbial strains in culture over decades of microbiology [108, 109], so we will focus in this review mainly on culture-independent techniques, as well as some high-throughput culture-based methods appropriate for microbial communities (Fig. 1). In both of these categories, many strain definition methods rely on sequencing: assembly of culture-based isolates, or amplicon-based, shotgun metagenomic, or single-cell culture-independent approaches. Other molecular assays, particularly mass spectrometry (MS)-based proteomics, can be applied to strain-type either isolates or communities [110]. This is also true for MS- or NMR-based metabolomics or metabolic flux measurements [111]. Of course, microbial culture physiology and direct imaging has been used to differentiate among strains since the earliest microbiology, and in some cases, these time-tested methods can be applied to communities as well.
Fig. 1.
Strain identification approaches for microbial communities. This review summarizes a variety of high-throughput, often (but not always) culture-independent methods for strain identification within microbial communities. a Amplicon sequencing (e.g., 16S rRNA gene regions) can now be processed to near-strain-level fidelity, resulting in unique markers such as amplicon sequence variants (ASVs). b Shotgun metagenomic sequencing, either via assembly or using reference-based approaches, can identify strains broadly based on their single-nucleotide variants (SNVs) or structural variants (gene gain and loss events). c Whole-community transcriptomes can amplify the effects of gene gains or losses, or the effects of small variants that result in differential expression. d Single-cell methods can isolate individual microbial genomics directly from within communities, either via cell sorting and amplification, or through synthetic long-read/linked-read techniques. e High-throughput “culturomics” can be combined with rapid turnaround approaches such as peptide fingerprinting to strain-type isolates or microcolonies. f Relatedly, any combination of traditional isolation and high-throughput cultivation—batch, serial, or continuous—can be combined with growth, phenotypic, or molecular readouts for strain identification. g Finally, a variety of other approaches can be used with communities, ranging from flow- or high-content microscopic imaging to systems such as gnotobiotic animal model physiology and phenotyping
Strain identification from microbial community sequencing
The first breakthroughs in microbial strain identification from whole-community sequencing—like the first community-wide applications of sequencing generally—came from marker gene approaches relying on amplification of 16S rRNA gene variable regions (amplicon or “16S” sequencing, Table 1). In many cases, amplicon-based technologies struggle to differentiate closely related microbial strains, due both to technical (sequencing error, amplification noise, bioinformatics approximations) and biological (lack of nucleotide variants in the amplified regions) limitations [123, 124]. Once data generation platforms reached the fidelity necessary to preserve amplicon biological variation when present, however, several computational approaches emerged to classify such sequences in the most strain-specific manner possible. Oligotyping [125, 126] and Minimum Entropy Decomposition (MED) [114] rely on semi-supervised and unsupervised classification, respectively, of variant positions within otherwise-identical 16S amplicons that show statistically unusual distributions across sample sets (and are thus unlikely due to technical factors). Other types of sub-operational taxonomic unit (OTU) clustering [113] have subsequently extended this intuition to “exact” or “amplicon” sequence variants (ESVs or ASVs, respectively) using statistical error modeling (e.g., DADA2 [115]) or filtering before or after sequence identity clustering (e.g., Deblur [116] or UNOISE2 [117]). Strain-resolved 16S amplicons have been used with methods like these to very specifically link, e.g., Porphyromonas asaccharolytica ATCC 25260 and Parvimonas micra ATCC 33270 to CRC, for example [127], or to assess the temporal stability of strains in the gut [128]. With additional data generation efforts, they can also generally be extended to multiple -[129] or non-16S amplicons [130], such as the VaST system for identifying a minimum group of target loci for amplification [131]. While SNV diversity in sub-regions of the genome is typically highly correlated with that across the genome [8], the presence or absence of at least one reliably detected SNV within a single amplified 16S variable region can be so precise as to become highly clade- and protocol-specific [115].
Table 1.
Tools for strain identification in community amplicon and shotgun metagenomic sequencing. Methods and brief summaries of their algorithms for detecting and quantifying strains (by various definitions) from 16S rRNA gene amplicon or shotgun metagenomic sequencing. These are currently the two most prevalent assays for culture-independent strain detection within microbial communities. Note that we have excluded other experimental protocols from this summary, including single-cell, long-read, and synthetic long-read sequencing, since they generally require more than application of a specific software pipeline. These alternatives, and non-sequencing-based approaches, are described in more detail in the text
| Method | Platform | Authors’ description | Reference |
|---|---|---|---|
| Oligotyping | 16S rRNA gene amplicon | “oligotyping... Focus [es] on the variable sites revealed by the entropy analysis to identify highly refined taxonomic units” | [112] |
| Sub-OTU clustering | 16S rRNA gene amplicon | “we combine error-model-based denoising and systematic cross-sample comparisons to resolve the fine (sub-OTU) structure of moderate-to-high-abundance community members” | [113] |
| MED | 16S rRNA gene amplicon | “MED uses information uncertainty among sequence reads to iteratively decompose a dataset until the maximum entropy criterion is satisfied for each final unit” | [114] |
| DADA2 | 16S rRNA gene amplicon | “DADA2 implements a new quality-aware model of Illumina amplicon errors. Sample composition is inferred by dividing amplicon reads into partitions consistent with the error model.” | [115] |
| Deblur | 16S rRNA gene amplicon | “Deblur … compares sequence-to-sequence Hamming distances within a sample to an upper-bound error profile combined with a greedy algorithm to obtain single-nucleotide resolution.” | [116] |
| UNOISE2 | 16S rRNA gene amplicon | “UNOISE2... Cluster [s] the unique sequences in the reads. A cluster has a centroid sequence with higher abundance plus similar sequences having lower abundances.” | [117] |
| PathoScope | Shotgun metagenomic | “PathoID … reassign [s] ambiguously aligned sequencing reads and accurately estimate [s] read proportions from each genome in the sample.” | [118] |
| LSA | Shotgun metagenomic | “LSA... separates reads into biologically informed partitions and thereby enables assembly of individual genomes.” | [119] |
| PanPhlAn | Shotgun metagenomic | “PanPhlAn identifies which genes are present or absent within different strains of a species, based on the entire gene set of the species’ pangenome.” | [66] |
| MetaMLST | Shotgun metagenomic | “MetaMLST performs an in silico consensus sequence reconstruction of the allelic profile of the microbial strains in a metagenomics sample.” | [120] |
| MIDAS | Shotgun metagenomic | “MIDAS … is a computational pipeline that quantifies bacterial species abundance and intra-species genomic variation from shotgun metagenomes.” | [37] |
| ConStrains | Shotgun metagenomic | “ConStrains … exploits the polymorphism patterns in a set of universal bacterial and archaeal genes to infer strain-level structures in species populations.” | [121] |
| StrainPhlAn | Shotgun metagenomic | “StrainPhlAn … is based on reconstructing consensus sequence variants within species-specific marker genes and using them to estimate strain-level phylogenies.” | [8] |
| metaSNV | Shotgun metagenomic | “metaSNV … performs SNV calling for individual samples and across the whole data set, and generates various statistics for individual species” | [102] |
| DESMAN | Shotgun metagenomic | “DESMAN identifies variants in core genes and uses co-occurrence across samples to link variants into haplotypes and abundance profiles.” | [122] |
Notably, the earliest forms of full-length 16S rRNA gene sequencing avoided many of these issues by capturing biological variation across the entire locus with high fidelity [132], and this has recently become true again in higher throughput with the advancement of “long-read” technologies. Three main platforms can currently provide such long-reads: Pacific Biosciences, Oxford Nanopore, and linked-read analogs such as products from 10X Genomics and Loop Genomics. The extreme fidelity offered by Pacific Biosciences circular consensus sequencing (CCS) has been perhaps best-studied in this context, readily differentiating between single-nucleotide variants (SNVs, although sometimes not insertions or deletions) when they exist anywhere across the 16S rRNA gene locus between strains [133, 134]. Conversely, while Oxford Nanopore’s extremely cost-effective MinION can provide essentially full-length 16S rRNA gene reads, its error rates have restricted strain-specific applications to cases in which no other sequences highly homologous to microbes of interest are present in a community [135–137]. Finally, several protocols now exist facilitating “simulated” long- or linked-reads on a variety of platforms [138, 139], but those which have reached commercial viability are yet to be formally evaluated for amplicon profiling of microbial communities [140]. Similarly, these technologies can sometimes be applied to entire microbial genomes isolated from single cells (e.g., via sorting or microfluidics [48, 141]) or from cross-linked genome copies [138]. This abrogates the need for true metagenomic assembly or binning, as described below, although again with few quantitative studies of these emerging technologies in existence for whole-community profiling at the strain level.
Overall, shotgun metagenomic approaches provide a richer profile of microbial communities’ genetic compositions, as they can in principle identify structural or SNVs anywhere within any microbe’s genome (Table 1). Two broad classes of analyses are currently able to identify microbial strains, the first based on the alignment of metagenomic nucleotides (typically unassembled) to a reference set of genes or genomes. This is generally efficient and sensitive, but of course only possible when sufficiently similar reference genomes (or prior metagenomic assemblies [142–144]) exist to permit direct mapping of metagenomic reads. Notably, “sufficiently similar” references need not be particularly high-identity with respect to a target metagenome. Instead, they must simply permit sufficient genome-wide mapping to identify SNVs or structural variants unique to strains in the community, which can be successful at up to several tens of percent overall nucleotide divergence.
Broadly speaking, four classes of reference-based community strain identification algorithms currently exist. The first identifies the one or more reference genotypes closest to those in a given community, with quantification based on some algorithm for ambiguity-resolved read mapping (e.g., PathoScope [118], Sigma [145]). The second identifies the dominant, potentially novel genotype (strain) per species; these include StrainPhlAn [8], MetaMLST [120], MetaSNV [146], and others [37]. These generally require deeper sequencing (up to 10× or more coverage of the strains to be targeted) and differ in their choice of which reference sequences to map against (e.g., complete genomes vs. universal core genes vs. species-specific marker genes) and the method and stringency of SNV identification. A third class of reference-based methods will further attempt to identify multiple strains per species within a metagenome, such as ConStrains [121] or DESMAN [122], requiring even deeper coverage and more stringent noise removal to prevent false positives. Finally, fourth, methods that rely on structural rather than SNV variants are generally more sensitive (appropriate for community members as rare as ~1× or lower coverage) and include PanPhlan [66] (which can be combined with gene-targeted functional profilers such as HUMAnN [147]), MIDAS [37], and others [4, 65].
Alternatively, when sufficiently similar reference genomes are not available, metagenomic assembly [142–144] can be used for highly novel strain discovery [148]. There is an inherent tension in assembly-based metagenomic strain profiling, as most assemblers seek to identify a single consensus sequence for each contig and require > 1× coverage of an entire genome (or region) to do so. This is appropriate when a single strain dominates its nearby phylogenetic space within a community, in which case less-common strains can be found by mapping metagenomic reads back to, e.g., a binned assembly [149–151] and identifying nucleotide or structural variants roughly as one would within complete genomes [8]. However, in the presence of too many closely related strains within a community, such a consensus sequence is not achievable in the first place, and most assemblers will not be able to provide a contig appropriate for mapping [152, 153]. Even when possible, this process can be further complicated by the high ecological and technical variability of microbial community assemblies, resulting in diverse coverage and confidence (dependent on sequencing depth and population strain admixture) and benefitting from manual inspection of putative variants [154, 155]. Algorithms facilitating this process include Latent Strain Analysis (LSA), which can refine strain-level taxonomy using covariant clusters across multiple related (e.g., longitudinal) samples [132]. Similarly, DESMAN uses statistical models not unlike those for ASV calling in amplicon data to identify variant genotypes well-supported across multiple samples’ co-assembly [122]. In a very few cases to date, strain variants within microbial communities have been identified via analogous differences in metatranscriptomic gene expression quantification, such as strain-specific variation in Eggerthella lenta metabolism of the cardiac drug digoxin [49].
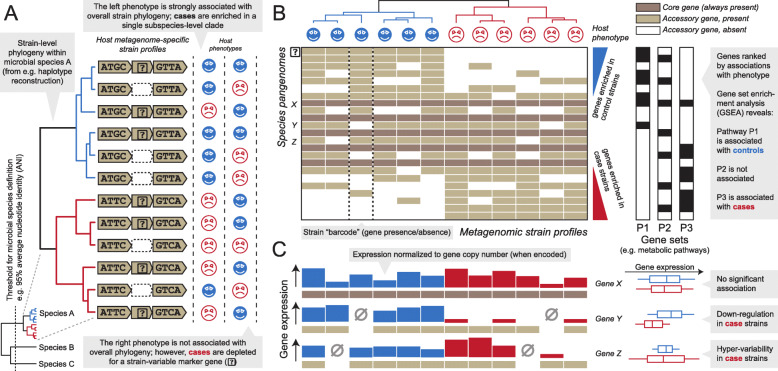
Whether from reference sequences or assemblies, SNV versus structural approaches are often complementary and can provide unique information regarding the same underlying community: SNVs (when detectable) identify finer-grained phylogenetic and evolutionary differences, but can be difficult to interpret functionally, whereas structural variants (i.e., gain or loss of full genes or genomic regions) have a lower limit of detection within communities and can speak directly to the biochemical roles of the affected genes (when known, Fig. 2). Unsurprisingly, each approach can provide different strengths and weaknesses. Structural variation can be captured well by reference-based approaches, which are sensitive to unique gene (non-)detection. However, it is very difficult to identify rearrangements (rather than gains or losses) using such techniques, and these are better identified by assembly-based methods instead (when they can be reliably differentiated from, e.g., chimeric assembly errors [157]). Conversely, SNV variation can be well-captured by either reference- or assembly-based approaches—the former more sensitively for organisms with representative isolates, the latter less sensitively but for novel organisms—and by either pangenome or whole-genome mapping approaches, depending where the most uniquely identifying polymorphisms occur. Finally, both structural variation and, to a lesser extent, nucleotide variation are particularly driven in microbial communities by mechanisms of genetic mobility, including all forms of lateral transfer, gene gain/loss, mobile elements, plasmids, and phage integration.
Fig. 2.
Microbial SNV, structural, and metatranscriptomic variants as features for genetic epidemiology in the human microbiome. Statistical approaches can link subspecies microbial features to human health phenotypes in several ways. a When microbial strains are identified using SNV genotypes (whether from genome bins, marker genes, core genes, etc.), any individual microbial SNV—or overall genotype—is typically of low prevalence and high variability. This means that it is extremely difficult to power significant associations with individual SNVs in reasonably sized human population studies. Instead, significant assortment of a host phenotype with strain phylogeny can be assessed, e.g., by PERMANOVA on per-species genetic distances [8] or by aggregating SNVs to genes or larger loci. b An extreme of this type of association test directly assesses the nonrandom assortment of genes’ presence or absence among microbial strain pangenomes in association with a phenotype of interest [66], since a gene loss (or gain) is essentially the “sum” of variants at every nucleotide within the gene. c Alternatively, even when no differences in genomic SNVs or structural variants are detectable at a study’s level of power, the transcriptional regulatory effects of these variants can be amplified, resulting in strain-specific differences in locus expression in association with a phenotype [156]
Other high-throughput molecular methods for strain identification in microbial communities
Other molecular technologies for microbial strain typing in communities are often limited to microbes that can be cultured or otherwise isolated, although advances in (semi-)automated anaerobic culture and nanoculture have made this feasible in high throughput as well. Particularly in clinical microbiology, near-strain variant typing via mass spectrometry peptide fingerprinting is commonplace for pathogen isolates [110, 158], due to its rapid turnaround time and low cost per individual sample relative to sequencing. The technology has some of the same caveats as ASV identification from sequence amplicons introduced above, however: amino acid variants must exist between the strains of interest in the profiled proteins, at a level detectable above experimental noise, and must be classifiable to a taxon of origin in a reference database or by clustering [159, 160]. While in principle the same types of strain-level protein variants could be detected using MALDI-TOF MS technologies in culture-independent community extracts, such applications remain extremely challenging, and instead, community proteomics are currently more commonly analyzed in a gene- or taxon-centric way [161].
Conversely, microbial imaging—arguably the first method for differentiating strains—has made the high-throughput leap to whole communities in several culture-independent forms that are, under appropriate circumstances, able to provide strain-level identification. In some cases, this can mean literally direct microscopy of microfluidically separated (or nanocultured) cells, using automated cell isolation and image analysis [162]. More molecular techniques include spectral or combinatorial fluorescent in situ hybridization (Combinatorial Labeling and Spectral Imaging or CLASI-FISH), which can currently identify over a dozen microbes within a community while maintaining spatial structure [163, 164]. Along with related techniques such as multilabel FISH (MiL-FISH) [165], this relies on the presence of sufficient genetic variants at the FISH-probed loci (often 16S rRNA gene regions) to be differentially bound by spectrally distinct probes, but can in some cases be extended to living bacteria [166]. This is also true for other microbial probe imaging methods such as flow cytometry [167] or light sheet microscopy [168], which can retain viable cells, but require probes or genetically manipulated microbes with loci capable of distinguishing between closely related strains.
While many of these methods are in part or whole culture-independent, it is difficult to understate the importance of the “culturomics” renaissance in separating and characterizing microbial strain isolates from communities including the human microbiome [15, 16, 169]. While pathogen epidemiology has long relied on comparative genomics among up to tens of thousands of isolates, it has only recently become efficient to carry out large-scale isolation of commensal organisms from human populations or individuals [170, 171]. Doing so, however, opens up the ability to identify strain-level differences among isolates of the same species among individuals [12, 13, 172, 173], within an individual microbiome at different spatial locations [81, 174], or over time [170, 175]. Once isolated, of course, such microbial strains can be characterized by any number of standard methods, including differences among growth curves or media, chemical (e.g., antimicrobial) resistance, metabolic flux profiling, or amplicon or shotgun sequencing. Alternatively, whole-community culture via chemostat bioreactors [176] provides an intermediate environment in which strains that are rare in situ can sweep to dominance, or be perturbed in a controlled manner, to amplify differential phenotypes or sequences that may otherwise remain below the limit of detection. Finally, culture-based and culture-independent strain identification techniques blur in the areas of single-cell microbial isolation [177, 178] and microcolony growth [179, 180] from communities. Microfluidic techniques in this vein include gel microdroplets (GMDs) for single-cell amplification [181] or phenotyping [182], as well as microfluidic streak plates (MSPs) [183] that combine the specificity of single cells with the biomass of streaked colonies (if desired).
Particularly when considering culture-based and ex vivo/in vitro/model system assays, the combination of culture-independent high-throughput epidemiology with subsequent strain isolation or manipulation opens up a world of possibilities for characterizing novel health-relevant strains in the human microbiome. This review has taken an essentially “top down” perspective, akin to forward genetics, in which strain-specific features of interest (SNVs, gene cassettes, metabolism, etc.) are identified by various means from human population studies [184]. Such an approach leads naturally to the subsequent biochemical characterization of these variants, either via isolation from primary samples [15, 170] or by in silico retrieval of homologous sequences or related strains from databases or repositories (e.g., ATCC, BEI, DSMZ) [185]. Primary samples can be characterized as an entire community via gnotobiotics [186, 187] or continuous culture [188, 189], or individual isolate strains grown, characterized, or (when possible) genetically manipulated [15, 190, 191]. Such approaches dovetail nicely with “bottom up” approaches (analogous to reverse genetics) that identify and characterize health-relevant strains by directly beginning with isolates and assessing their phenotypes in gnotobiotic mono- or combinatorial colonization [192–197] or, when possible, human feeding [198–200] or microbiota transplant clinical trials [201–205].
Perspectives and future directions
As introduced above, the precise definition of “strain” is somewhat fluid throughout biology, let alone in microbiology [3] or microbial community biology [206]. While it has most often referred to a single colony isolate culture in the past, the introduction of technologies and tools for precisely resolved genetic variant identification within microbial communities has led to increased broadening of the term. It is now used with some frequency to mean a subspecies or intraspecific clade with relatively low genetic diversity, defined by core or pangenomic identity, nucleotide identity within an amplicon such as the 16S rRNA gene, or the other genotyping or phenotypic similarities described above. As has increasingly been discussed in the literature for microbial systematics overall [8, 207], this suggests the need for a more quantitative definition of strains or subspecies clades, particularly within naturally variant microbial communities. In the absence of a single consensus definition, it is extremely useful for individual studies to define their use of “strain” up front when describing culture-based or (especially) culture-independent microbial community research [174].
Regardless of their precise definition, several emerging technologies offer exciting new approaches for identifying, isolating, and characterizing health-relevant strains in the human microbiome. Historically, microbial genetic variants not associated with an overt, acute phenotype have gone largely undetected, until the relatively recent availability of whole-community profiling techniques by which they can be efficiently captured. Truly single-cell approaches reliant on individual microbial separation have been so far difficult to apply to human epidemiology, with methods for eukaryotic cells not transferring well at scale to the heterogeneity of microbial cell wall biochemistry [208] and methods from environmental community profiling difficult to apply to matrices as diverse as human stool or skin [209]. In addition to bioengineering for cell separation and lysis, advances in low-input, low-noise DNA isolation, amplification, and sequencing will help to address this challenge [210], as will nanoculture approaches that inherently amplify genomes in vivo [180]. Such methods for capturing strains from the human microbiome go hand-in-hand with additional technologies for characterizing them at scale, including cheaper experimental systems such as gut-on-chip [211, 212] or organoid variants [213, 214] that sit in between single isolate culture and rich gnotobiotic models. Ultimately, understanding human microbiome biology will require not just the detection of specific microbial genetic variants in communities, but their introduction and manipulation, including the theoretical ability to genetically perturb any microbial strain either after or even before isolation from its host community [173, 190].
Even in the absence of such technology, extensive work remains to be done to characterize the microbial strain diversity in the human microbiome that has already been uncovered. Of the tens of millions of gene families identified within the human microbiome [23, 99, 215], some ~ 75% are not biochemically characterized by anything more than (in some cases remote) homology to reference sequences, and ~ 25% are not closely homologous to any isolate open reading frames [216]. This astounding pool of biochemical dark matter may be unsurprising to microbial bioprospectors, who have mined primarily environmental communities for novel enzymatic and antimicrobial function for decades [217]. As such, it represents a remarkable potential for new bioactive discovery in human health as well, since human-associated microbes could easily be enriched for protein and metabolite products that modulate host responses [218]. In many of the examples described above, successful associations of SNV or structural variants in the microbiome with human phenotypes or environmental factors have led to genes of unknown function [13, 65, 66]. Strain-level epidemiology in the human microbiome can thus help to prioritize the daunting task of identifying and characterizing the “most interesting” novel microbial variants and products of greatest relevance to health.
Finally, the ways in which better techniques for strain characterization in the microbiome can benefit human health are themselves diverse. Cheap, rapid, and reproducible methods to quantify microbiome SNVs and genetic variants across human populations will allow the identification of precise microbial risk factors, much as did the standardization of human genetics platforms for genome-wide association studies (GWAS) [219]. Also analogously to GWAS, microbial strains can thus provide prognostic or diagnostic biomarkers for disease risk or diagnosis, or hints as to their underlying molecular mechanisms [220–222]. This has been the case for decades in for comparative genetics microbial isolates, and as the number and depth of metagenomes continues to increase, it will undoubtedly become practical in microbial communities as well [223, 224]. Conversely, features of strains found to be bioactive can be used to develop novel interventions for health maintenance or therapy. These can range from better targeting of existing fecal microbiota transplant (FMT) technologies based on donor or recipient strain content [225], to the rational design of synthetic FMTs [226], treatment response prediction for FMTs or prebiotics [227–230], or the eventual administration of genetically modified organisms or communities [231–234]. Recent work in strain-level epidemiology of microbial communities and the human microbiome is thus one of many important, ongoing efforts to realize the microbiome’s substantial translational potential.
Acknowledgements
We would like to thank Lea Wang, Siyuan Ma, and Nicole Levesque for their thoughtful input and assistance with the manuscript.
Abbreviations
- ASV
Amplicon sequence variant
- CCS
Circular consensus sequencing
- CLASI-FISH
Combinatorial Labeling and Spectral Imaging
- CRC
Colorectal cancer
- EHEC
Enterohemorrhagic E. coli
- ESV
Exact sequence variant
- FMT
Fecal microbiota transplant
- GI
Gastrointestinal
- GMDs
Gel microdroplets
- GWAS
Genome-wide association studies
- HMO
Human milk oligosaccharide
- IBD
Inflammatory bowel diseases
- LPS
Lipopolysaccharides
- MED
Minimum Entropy Decomposition
- MS
Mass spectrometry
- MSPs
Microfluidic streak plates
- OTU
Operational taxonomic unit
- SCFA
Short-chain fatty acid
- SNP
Single nucleotide polymorphism
- SNV
Single-nucleotide variant
- T2D
Type 2 diabetes
Authors’ contributions
Y.Y., L.H.N. and C.H. prepared the manuscript. Y.Y. and E.A.F. designed the figures. The author(s) read and approved the final manuscript.
Funding
This work was funded in part by Cancer Research UK Grand Challenge Initiative C10674/A27140 (Wendy S. Garrett) and by NIH NIDDK R24DK110499 (CH).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Falkow S. Molecular Koch's postulates applied to bacterial pathogenicity—a personal recollection 15 years later. Nat Rev Microbiol. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- 2.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond Ser B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dijkshoorn L, Ursing BM, Ursing JB. Strain, clone and species: comments on three basic concepts of bacteriology. J Med Microbiol. 2000;49:397–401. doi: 10.1099/0022-1317-49-5-397. [DOI] [PubMed] [Google Scholar]
- 4.Zhu A, Sunagawa S, Mende DR, Bork P. Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 2015;16:82. doi: 10.1186/s13059-015-0646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L. G. Wayne DJB, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr and H. G. Truper: Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. 1987.
- 6.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 7.Almeida LA, Araujo R. Highlights on molecular identification of closely related species. Infect Genet Evol. 2013;13:67–75. doi: 10.1016/j.meegid.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Brenner D, Staley J, Krieg N. Bergey’s manual of systematic bacteriology. New York: Springer; 2000. [Google Scholar]
- 11.Donati C, Zolfo M, Albanese D, Tin Truong D, Asnicar F, Iebba V, Cavalieri D, Jousson O, De Filippo C, Huttenhower C, Segata N. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nat Microbiol. 2016;1:16070. doi: 10.1038/nmicrobiol.2016.70. [DOI] [PubMed] [Google Scholar]
- 12.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, Armanini F, Manghi P, Bonham K, Zolfo M, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26:666–679. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain JM, Fournier PE, Raoult D. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 17.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Buso L, Golparian D, Corander J, Grad YH, Ohnishi M, Flemming R, Parkhill J, Bentley SD, Unemo M, Harris SR. The impact of antimicrobials on gonococcal evolution. Nat Microbiol. 2019;4:1941–1950. doi: 10.1038/s41564-019-0501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 21.Zou Y, Xue W, Luo G, Deng Z, Qin P, Guo R, Sun H, Xia Y, Liang S, Dai Y, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37:179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pena-Gonzalez A, Soto-Giron MJ, Smith S, Sistrunk J, Montero L, Paez M, Ortega E, Hatt JK, Cevallos W, Trueba G, et al. Metagenomic signatures of gut infections caused by different Escherichia coli Pathotypes. Appl Environ Microbiol. 2019;85. [DOI] [PMC free article] [PubMed]
- 23.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figler HM, Dudley EG. The interplay of Escherichia coli O157:H7 and commensal E. coli: the importance of strain-level identification. Expert Rev Gastroenterol Hepatol. 2016;10:415–417. doi: 10.1586/17474124.2016.1155449. [DOI] [PubMed] [Google Scholar]
- 25.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonnenborn U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol Lett. 2016;363. [DOI] [PubMed]
- 27.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira PH, Touchon M, Rocha EP. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kronheim S, Daniel-Ivad M, Duan Z, Hwang S, Wong AI, Mantel I, Nodwell JR, Maxwell KL. A chemical defence against phage infection. Nature. 2018;564:283–286. doi: 10.1038/s41586-018-0767-x. [DOI] [PubMed] [Google Scholar]
- 31.Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyoty H, Virtanen SM, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. 2018;24:146–154. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–145. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, Segata N, Bork P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28:561–568. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark A, Hagopian WA, Rewers MJ, She JX, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, A DL, Wu F, Perez-Perez GI, Chen Y, et al: Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016, 8:343ra382. [DOI] [PMC free article] [PubMed]
- 41.Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 42.Thom CS, Dickson CF, Gell DA, Weiss MJ. Hemoglobin variants: biochemical properties and clinical correlates. Cold Spring Harb Perspect Med. 2013;3:a011858. doi: 10.1101/cshperspect.a011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- 45.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Microbes and health Sackler colloquium: succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2010. [DOI] [PMC free article] [PubMed]
- 46.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 47.Karcher N, Pasolli E, Asnicar F, Huang K, Tett A, Manara S, Armanini F, Bain D, Duncan SH, Louis P, et al: Analysis of 1,321 Eubacterium rectale genomes from metagenomes uncovers complex phylogeographic population structures and subspecies functional adaptations. in review. [DOI] [PMC free article] [PubMed]
- 48.Brito IL, Yilmaz S, Huang K, Xu L, Jupiter SD, Jenkins AP, Naisilisili W, Tamminen M, Smillie CS, Wortman JR, et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 54.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Bjorck I, Backhed F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 55.De Filippis F, Pasolli E, Tett A, Tarallo S, Naccarati A, De Angelis M, Neviani E, Cocolin L, Gobbetti M, Segata N, Ercolini D. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25:444–453. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Wu G, Zhang C, Wu H, Wang R, Shen J, Wang L, Zhao Y, Pang X, Zhang X, Zhao L, Zhang M: Genomic Microdiversity of Bifidobacterium pseudocatenulatum Underlying Differential Strain-Level Responses to Dietary Carbohydrate Intervention. mBio 2017; 8:e02348-16. [DOI] [PMC free article] [PubMed]
- 57.Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, Zhang M, Wang L, Hou Y, Ouyang H, et al. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine. 2015;2:968–984. doi: 10.1016/j.ebiom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bottacini F, Medini D, Pavesi A, Turroni F, Foroni E, Riley D, Giubellini V, Tettelin H, van Sinderen D, Ventura M. Comparative genomics of the genus Bifidobacterium. Microbiology. 2010;156:3243–3254. doi: 10.1099/mic.0.039545-0. [DOI] [PubMed] [Google Scholar]
- 59.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 60.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang X, Monk JM, Nurk S, Akseshina M, Zhu Q, Gemmell C, Gianetto-Hill C, Leung N, Szubin R, Sanders J, et al. Metagenomics-based, strain-level analysis of Escherichia coli from a time-series of microbiome samples from a Crohn's disease patient. Front Microbiol. 2018;9:2559. doi: 10.3389/fmicb.2018.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 63.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeevi D, Korem T, Godneva A, Bar N, Kurilshikov A, Lotan-Pompan M, Weinberger A, Fu J, Wijmenga C, Zhernakova A, Segal E. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–48. doi: 10.1038/s41586-019-1065-y. [DOI] [PubMed] [Google Scholar]
- 66.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F, Truong DT, Tett A, Morrow AL, Segata N. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 67.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Li Z, Hu S, Zhang J, Wu J, Shao N, Bo X, Ni M, Ying X. Gut metagenomes of type 2 diabetic patients have characteristic single-nucleotide polymorphism distribution in Bacteroides coprocola. Microbiome. 2017;5:15. doi: 10.1186/s40168-017-0232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 71.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 77.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 2016;14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Miller RR, Godwin H, Knox K, Everitt RG, et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A. 2012;109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, Kishony R. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet. 2014;46:82–87. doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31. [DOI] [PMC free article] [PubMed]
- 83.Wollenberg MS, Claesen J, Escapa IF, Aldridge KL, Fischbach MA, Lemon KP. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio. 2014;5:e01286–e01214. doi: 10.1128/mBio.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, Program NCS, Belkaid Y, Segre JA, Kong HH. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9:eaal4651. [DOI] [PMC free article] [PubMed]
- 85.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tett A, Pasolli E, Farina S, Truong DT, Asnicar F, Zolfo M, Beghini F, Armanini F, Jousson O, De Sanctis V, et al. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes. 2017;3:14. doi: 10.1038/s41522-017-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wittouck S, Wuyts S, Meehan CJ, van Noort V, Lebeer S: A genome-based species taxonomy of the lactobacillus genus complex. mSystems 2019;4:e00264-19. [DOI] [PMC free article] [PubMed]
- 89.Salvetti E, Harris HMB, Felis GE, O'Toole PW. Comparative genomics of the genus Lactobacillus reveals robust Phylogroups that provide the basis for reclassification. Appl Environ Microbiol. 2018;84:e00993-18. [DOI] [PMC free article] [PubMed]
- 90.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJ, Huttenhower C. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A. 2015;112:E2930–E2938. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goltsman DSA, Sun CL, Proctor DM, DiGiulio DB, Robaczewska A, Thomas BC, Shaw GM, Stevenson DK, Holmes SP, Banfield JF, Relman DA. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 2018;28:1467–1480. doi: 10.1101/gr.236000.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, Gordon JI. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci Transl Med. 2013;5:184ra160. doi: 10.1126/scitranslmed.3005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nielsen KL, Stegger M, Kiil K, Godfrey PA, Feldgarden M, Lilje B, Andersen PS, Frimodt-Moller N. Whole-genome comparison of urinary pathogenic Escherichia coli and faecal isolates of UTI patients and healthy controls. Int J Med Microbiol. 2017;307:497–507. doi: 10.1016/j.ijmm.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nahui Palomino RA, Zicari S, Vanpouille C, Vitali B, Margolis L. Vaginal Lactobacillus inhibits HIV-1 replication in human tissues ex vivo. Front Microbiol. 2017;8:906. doi: 10.3389/fmicb.2017.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29–37. doi: 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, Pizza M, Dougan G, Rappuoli R, Moxon ER, Soriani M, Donati C. Genome sequencing of disease and carriage isolates of nontypeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci U S A. 2014;111:5439–5444. doi: 10.1073/pnas.1403353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 98.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Structure, function and diversity of the healthy human microbiome. Nature 2012, 486:207–214. [DOI] [PMC free article] [PubMed]
- 100.Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci U S A. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Costea PI, Munch R, Coelho LP, Paoli L, Sunagawa S. Bork P: metaSNV: a tool for metagenomic strain level analysis. PLoS One. 2017;12:e0182392. doi: 10.1371/journal.pone.0182392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038. doi: 10.1038/nrdp.2017.38. [DOI] [PubMed] [Google Scholar]
- 104.Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120–127. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goh CE, Trinh P, Colombo PC, Genkinger JM, Mathema B, Uhlemann AC, LeDuc C, Leibel R, Rosenbaum M, Paster BJ, et al. Association between nitrate-reducing oral bacteria and cardiometabolic outcomes: results from ORIGINS. J Am Heart Assoc. 2019;8:e013324. doi: 10.1161/JAHA.119.013324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Al-Hebshi NN, Baraniya D, Chen T, Hill J, Puri S, Tellez M, Hasan NA, Colwell RR, Ismail A. Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J Oral Microbiol. 2019;11:1557986. doi: 10.1080/20002297.2018.1557986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Al-Hebshi NN, Nasher AT, Maryoud MY, Homeida HE, Chen T, Idris AM, Johnson NW. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep. 2017;7:1834. doi: 10.1038/s41598-017-02079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol. 2004;70:4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sandrin TR, Goldstein JE, Schumaker S. MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom Rev. 2013;32:188–217. doi: 10.1002/mas.21359. [DOI] [PubMed] [Google Scholar]
- 111.Thommes M, Wang T, Zhao Q, Paschalidis IC, Segre D: Designing Metabolic Division of Labor in Microbial Communities. mSystems 2019;4:e00263-18. [DOI] [PMC free article] [PubMed]
- 112.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4:1111-9. [DOI] [PMC free article] [PubMed]
- 113.Tikhonov M, Leach RW, Wingreen NS. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2015;9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R: Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2017;2:e00191-16. [DOI] [PMC free article] [PubMed]
- 117.Edgar RC: UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016. 10.1101/081257.
- 118.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, Crandall KA, Johnson WE. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cleary B, Brito IL, Huang K, Gevers D, Shea T, Young S, Alm EJ. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat Biotechnol. 2015;33:1053–1060. doi: 10.1038/nbt.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zolfo M, Tett A, Jousson O, Donati C, Segata N. MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res. 2017;45:e7. doi: 10.1093/nar/gkw837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D. ConStrains identifies microbial strains in metagenomic datasets. Nat Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Quince C, Delmont TO. Raguideau S, Alneberg J, Darling AE, Collins G, Eren AM. DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol. 2017;18:181. doi: 10.1186/s13059-017-1309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Soergel DA, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012;6:1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. 2016;34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 125.Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One. 2011;6:e26732. doi: 10.1371/journal.pone.0026732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci U S A. 2014;111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A, Yamal JM, Hollister EB. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67:882–891. doi: 10.1136/gutjnl-2016-313189. [DOI] [PubMed] [Google Scholar]
- 128.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fuks G, Elgart M, Amir A, Zeisel A, Turnbaugh PJ, Soen Y, Shental N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome. 2018;6:17. doi: 10.1186/s40168-017-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang JY, Brooks S, Meyer JA, Blakesley RR, Zelazny AM, Segre JA, Snitkin ES. Pan-PCR, a computational method for designing bacterium-typing assays based on whole-genome sequence data. J Clin Microbiol. 2013;51:752–758. doi: 10.1128/JCM.02671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Furstenau TN, Cocking JH, Sahl JW, Fofanov VY. Variant site strain typer (VaST): efficient strain typing using a minimal number of variant genomic sites. BMC Bioinformatics. 2018;19:222. doi: 10.1186/s12859-018-2225-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, Gulati AS, McGill SK, Dougherty MK. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47:e103. doi: 10.1093/nar/gkz569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kai S, Matsuo Y, Nakagawa S, Kryukov K, Matsukawa S, Tanaka H, Iwai T, Imanishi T, Hirota K. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION nanopore sequencer. FEBS Open Bio. 2019;9:548–557. doi: 10.1002/2211-5463.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kerkhof LJ, Dillon KP, Haggblom MM, McGuinness LR. Profiling bacterial communities by MinION sequencing of ribosomal operons. Microbiome. 2017;5:116. doi: 10.1186/s40168-017-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Benitez-Paez A, Sanz Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinION portable nanopore sequencer. Gigascience. 2017;6:1–12. doi: 10.1093/gigascience/gix043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burke CM, Darling AE. A method for high precision sequencing of near full-length 16S rRNA genes on an Illumina MiSeq. PeerJ. 2016;4:e2492. doi: 10.7717/peerj.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Karst SM, Dueholm MS, McIlroy SJ, Kirkegaard RH, Nielsen PH, Albertsen M. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat Biotechnol. 2018;36:190–195. doi: 10.1038/nbt.4045. [DOI] [PubMed] [Google Scholar]
- 140.Wu I, Kim HS, Ben-Yehezkel T: A single-molecule long-read survey of human transcriptomes using LoopSeq synthetic long read sequencing. bioRxiv 2019. 10.1101/532135.
- 141.Woyke T, Doud DFR, Schulz F. The trajectory of microbial single-cell sequencing. Nat Methods. 2017;14:1045–1054. doi: 10.1038/nmeth.4469. [DOI] [PubMed] [Google Scholar]
- 142.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568:505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ahn TH, Chai J, Pan C. Sigma: strain-level inference of genomes from metagenomic analysis for biosurveillance. Bioinformatics. 2015;31:170–177. doi: 10.1093/bioinformatics/btu641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sahl JW, Schupp JM, Rasko DA, Colman RE, Foster JT, Keim P. Phylogenetically typing bacterial strains from partial SNP genotypes observed from direct sequencing of clinical specimen metagenomic data. Genome Med. 2015;7:52. doi: 10.1186/s13073-015-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 149.Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ. 2014;2:e603. doi: 10.7717/peerj.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 151.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guo J, Quensen JF, Sun Y, Wang Q, Brown CT, Cole JR, Tiedje JM. Review, evaluation, and directions for gene-targeted assembly for ecological analyses of metagenomes. Front Genet. 2019;10:957. doi: 10.3389/fgene.2019.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ghurye J, Pop M. Modern technologies and algorithms for scaffolding assembled genomes. PLoS Comput Biol. 2019;15:e1006994. doi: 10.1371/journal.pcbi.1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO Anvi'o: an advanced analysis and visualization platform for 'omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23:111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A. 2014;111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ayling M, Clark MD, Leggett RM. New approaches for metagenome assembly with short reads. Brief Bioinform. 2020;21:584–594. doi: 10.1093/bib/bbz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang L, Smart S, Sandrin TR. Biomarker- and similarity coefficient-based approaches to bacterial mixture characterization using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) Sci Rep. 2015;5:15834. doi: 10.1038/srep15834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Blanco-Miguez A, Meier-Kolthoff JP, Gutierrez-Jacome A, Goker M, Fdez-Riverola F, Sanchez B, Lourenco A. Improving phylogeny reconstruction at the strain level using peptidome datasets. PLoS Comput Biol. 2016;12:e1005271. doi: 10.1371/journal.pcbi.1005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rahi P, Vaishampayan P. Editorial: MALDI-TOF MS application in microbial ecology studies. Front Microbiol. 2019;10:2954. doi: 10.3389/fmicb.2019.02954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shi H, Colavin A, Lee TK, Huang KC. Strain library imaging protocol for high-throughput, automated single-cell microscopy of large bacterial collections arrayed on multiwell plates. Nat Protoc. 2017;12:429–438. doi: 10.1038/nprot.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Valm AM, Mark Welch JL, Borisy GG. CLASI-FISH: principles of combinatorial labeling and spectral imaging. Syst Appl Microbiol. 2012;35:496–502. doi: 10.1016/j.syapm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schimak MP, Kleiner M, Wetzel S, Liebeke M, Dubilier N, Fuchs BM. MiL-FISH: multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells. Appl Environ Microbiol. 2016;82:62–70. doi: 10.1128/AEM.02776-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Batani G, Bayer K, Boge J, Hentschel U, Thomas T. Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci Rep. 2019;9:18618. doi: 10.1038/s41598-019-55049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu Z, Cichocki N, Bonk F, Gunther S, Schattenberg F, Harms H, Centler F, Muller S: Ecological stability properties of microbial communities assessed by flow cytometry. mSphere 2018;3:e00564-17. [DOI] [PMC free article] [PubMed]
- 168.Wiles TJ, Wall ES, Schlomann BH, Hay EA, Parthasarathy R, Guillemin K: Modernized tools for streamlined genetic manipulation and comparative study of wild and diverse proteobacterial lineages. mBio 2018;9:e01877-18. [DOI] [PMC free article] [PubMed]
- 169.Ahern PP, Faith JJ, Gordon JI. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 2014;40:815–823. doi: 10.1016/j.immuni.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Poyet M, Groussin M, Gibbons SM, Avila-Pacheco J, Jiang X, Kearney SM, Perrotta AR, Berdy B, Zhao S, Lieberman TD, et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med. 2019;25:1442–1452. doi: 10.1038/s41591-019-0559-3. [DOI] [PubMed] [Google Scholar]
- 171.Lieberman TD: Seven billion microcosms: evolution within human microbiomes. mSystems 2018;3:e00171-17. [DOI] [PMC free article] [PubMed]
- 172.Hsu T, Gemmell MR, Franzosa EA, Berry S, Mukhopadhya I, Hansen R, Michaud M, Nielsen H, Miller WG, Nielsen H, et al. Comparative genomics and genome biology of Campylobacter showae. Emerg Microbes Infect. 2019;8:827–840. doi: 10.1080/22221751.2019.1622455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Garcia-Bayona L, Comstock LE: Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio 2019, 10. [DOI] [PMC free article] [PubMed]
- 174.Zhou W, Spoto M, Hardy R, Guan C, Fleming E, Larson PJ, Brown JS, Oh J. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell. 2020:454-70. [DOI] [PMC free article] [PubMed]
- 175.Zhao S, Lieberman TD, Poyet M, Kauffman KM, Gibbons SM, Groussin M, Xavier RJ, Alm EJ. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe. 2019;25:656–667. doi: 10.1016/j.chom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McDonald JA, Fuentes S, Schroeter K, Heikamp-deJong I, Khursigara CM, de Vos WM, Allen-Vercoe E. Simulating distal gut mucosal and luminal communities using packed-column biofilm reactors and an in vitro chemostat model. J Microbiol Methods. 2015;108:36–44. doi: 10.1016/j.mimet.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 177.Singer E, Wagner M, Woyke T. Capturing the genetic makeup of the active microbiome in situ. ISME J. 2017;11:1949–1963. doi: 10.1038/ismej.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bowers RM, Lee J, Woyke T. Sequencing of genomes from environmental single cells. Methods Mol Biol. 2018;1712:97–111. doi: 10.1007/978-1-4939-7514-3_8. [DOI] [PubMed] [Google Scholar]
- 179.Gao W, Navarroli D, Naimark J, Zhang W, Chao SH, Meldrum DR. Microbe observation and cultivation array (MOCA) for cultivating and analyzing environmental microbiota. Microbiome. 2013;1:4. doi: 10.1186/2049-2618-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Niepa TH, Hou L, Jiang H, Goulian M, Koo H, Stebe KJ, Lee D. Microbial nanoculture as an artificial microniche. Sci Rep. 2016;6:30578. doi: 10.1038/srep30578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fitzsimons MS, Novotny M, Lo CC, Dichosa AE, Yee-Greenbaum JL, Snook JP, Gu W, Chertkov O, Davenport KW, McMurry K, et al. Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome. Genome Res. 2013;23:878–888. doi: 10.1101/gr.142208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dong L, Chen DW, Liu SJ, Du W. Automated chemotactic sorting and single-cell cultivation of microbes using droplet microfluidics. Sci Rep. 2016;6:24192. doi: 10.1038/srep24192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jiang CY, Dong L, Zhao JK, Hu X, Shen C, Qiao Y, Zhang X, Wang Y, Ismagilov RF, Liu SJ, Du W. High-throughput single-cell cultivation on microfluidic streak plates. Appl Environ Microbiol. 2016;82:2210–2218. doi: 10.1128/AEM.03588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mallick H, Ma S, Franzosa EA, Vatanen T, Morgan XC, Huttenhower C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18:228. doi: 10.1186/s13059-017-1359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, et al. Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bess EN, Bisanz JE, Yarza F, Bustion A, Rich BE, Li X, Kitamura S, Waligurski E, Ang QY, Alba DL, et al. Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria. Nat Microbiol. 2020;5:56–66. doi: 10.1038/s41564-019-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oliphant K, Cochrane K, Schroeter K, Daigneault MC, Yen S, Verdu EF, Allen-Vercoe E: Effects of Antibiotic Pretreatment of an Ulcerative Colitis-Derived Fecal Microbial Community on the Integration of Therapeutic Bacteria In Vitro. mSystems 2020, 5. [DOI] [PMC free article] [PubMed]
- 189.Auchtung JM, Robinson CD, Britton RA. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs) Microbiome. 2015;3:42. doi: 10.1186/s40168-015-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bencivenga-Barry NA, Lim B, Herrera CM, Trent MS, Goodman AL. Genetic manipulation of wild human gut Bacteroides. J Bacteriol. 2020;202. [DOI] [PMC free article] [PubMed]
- 191.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Elzinga J, van der Oost J, de Vos WM, Smidt H. The use of defined microbial communities to model host-microbe interactions in the human gut. Microbiol Mol Biol Rev. 2019;83. [DOI] [PMC free article] [PubMed]
- 193.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358:359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Burns AR, Guillemin K. The scales of the zebrafish: host-microbiota interactions from proteins to populations. Curr Opin Microbiol. 2017;38:137–141. doi: 10.1016/j.mib.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lengfelder I, Sava IG, Hansen JJ, Kleigrewe K, Herzog J, Neuhaus K, Hofmann T, Sartor RB, Haller D. Complex bacterial consortia reprogram the volitogenic activity of enterococcus faecalis in a gnotobiotic mouse model of chronic immune-mediated colitis. Front Immunol. 2019;10:1420. doi: 10.3389/fimmu.2019.01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Piening BD, Zhou W, Contrepois K, Rost H, Gu Urban GJ, Mishra T, Hanson BM, Bautista EJ, Leopold S, Yeh CY, et al. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 2018;6:157–170. doi: 10.1016/j.cels.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 202.Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Taur Y, Coyte K, Schluter J, Robilotti E, Figueroa C, Gjonbalaj M, Littmann ER, Ling L, Miller L, Gyaltshen Y, et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med. 2018;10. [DOI] [PMC free article] [PubMed]
- 204.Bajaj JS, Kakiyama G, Savidge T, Takei H, Kassam ZA, Fagan A, Gavis EA, Pandak WM, Nittono H, Hylemon PB, et al. Antibiotic-associated disruption of microbiota composition and function in cirrhosis is restored by fecal transplant. Hepatology. 2018;68:1549–1558. doi: 10.1002/hep.30037. [DOI] [PubMed] [Google Scholar]
- 205.Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, Chang HJ, Coward S, Goodman KJ, Xu H, et al. Effect of oral capsule- vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA. 2017;318:1985–1993. doi: 10.1001/jama.2017.17077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marx V. Microbiology: the road to strain-level identification. Nat Methods. 2016;13:401–404. doi: 10.1038/nmeth.3837. [DOI] [PubMed] [Google Scholar]
- 207.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2019:1925-7. [DOI] [PMC free article] [PubMed]
- 208.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 209.Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, Dmitrieff E, Malmstrom R, Stepanauskas R, Woyke T. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc. 2014;9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- 210.Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 211.Poceviciute R, Ismagilov RF. Human-gut-microbiome on a chip. Nat Biomed Eng. 2019;3:500–501. doi: 10.1038/s41551-019-0425-0. [DOI] [PubMed] [Google Scholar]
- 212.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113:E7–15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 214.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 216.Joice R, Yasuda K, Shafquat A, Morgan XC, Huttenhower C. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 2014;20:731–741. doi: 10.1016/j.cmet.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Madhavan A, Sindhu R, Parameswaran B, Sukumaran RK, Pandey A. Metagenome analysis: a powerful tool for enzyme bioprospecting. Appl Biochem Biotechnol. 2017;183:636–651. doi: 10.1007/s12010-017-2568-3. [DOI] [PubMed] [Google Scholar]
- 218.Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wellcome Trust Case Control C Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25:667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–498. doi: 10.1038/nrmicro1157. [DOI] [PubMed] [Google Scholar]
- 224.Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet. 2017;18:41–50. doi: 10.1038/nrg.2016.132. [DOI] [PubMed] [Google Scholar]
- 225.Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 2018;23:229–240. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hudson LE, Anderson SE, Corbett AH, Lamb TJ. Gleaning insights from fecal microbiota transplantation and probiotic studies for the rational design of combination microbial therapies. Clin Microbiol Rev. 2017;30:191–231. doi: 10.1128/CMR.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Staley C, Kaiser T, Vaughn BP, Graiziger CT, Hamilton MJ, Rehman TU, Song K, Khoruts A, Sadowsky MJ. Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation. Microbiome. 2018;6:166. doi: 10.1186/s40168-018-0549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wilson BC, Vatanen T, Cutfield WS, O'Sullivan JM. The super-donor phenomenon in fecal microbiota transplantation. Front Cell Infect Microbiol. 2019;9:2. doi: 10.3389/fcimb.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Biesiekierski JR, Jalanka J, Staudacher HM. Can gut microbiota composition predict response to dietary treatments? Nutrients. 2019;11. [DOI] [PMC free article] [PubMed]
- 230.Hughes RL, Marco ML, Hughes JP, Keim NL, Kable ME. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models-part I: overview of current methods. Adv Nutr. 2019;10:953–978. doi: 10.1093/advances/nmz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Whitfill T, Oh J. Recoding the metagenome: microbiome engineering in situ. Curr Opin Microbiol. 2019;50:28–34. doi: 10.1016/j.mib.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 232.Pedrolli DB, Ribeiro NV, Squizato PN, de Jesus VN, Cozetto DA, Team AQAUai Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol. 2019;37:100–115. doi: 10.1016/j.tibtech.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 233.Bober JR, Beisel CL, Nair NU. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu Rev Biomed Eng. 2018;20:277–300. doi: 10.1146/annurev-bioeng-062117-121019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sonnenburg JL. Microbiome engineering. Nature. 2015;518:S10. doi: 10.1038/518S10a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.