Abbreviations
- ACTL6A
actin like 6A
- ALK
anaplastic lymphoma kinase
- ARID1A
AT‐rich interaction domain 1A
- ARID1B
AT‐rich interaction domain 1B
- ARMS
amplification refractory mutation system
- AUC
area under the receiver operating characteristic curve
- BRAF
B‐Raf proto‐oncogene
- CI
confidence interval
- CN‐ FGA
fraction of the genome altered by copy number alterations
- EGFR
epidermal growth factor receptor
- HR
hazard ratio
- ICI
immune checkpoint inhibitor
- KRAS
KRAS proto‐oncogene
- NSCLC
non‐small cell lung cancer
- OS
overall survival
- PBRM1
polybromo 1
- PD‐L1
programmed cell death ligand 1
- PFS
progression‐free survival
- RGD
Rat Genome Database
- ROC
Receiver operating characteristic
- ROS1
ROS proto‐oncogene 1
- SMARCA4
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4
- STK11
serine/threonine kinase 11
- SWI/SNF
SWItch/sucrose nonfermentable
- TMB
tumor mutation burden
- TP53
tumor protein p53
Dear Editor:
Although immune checkpoint inhibitor (ICI) has become an innovative treatment regimen for non‐small cell lung cancer (NSCLC) in recent years, only a small proportion of patients benefit from this regimen [1]. Nowadays, several biomarkers have been used to predict the effectiveness of ICI treatment, such as programmed cell death ligand 1 (PD‐L1) expression level in tumor tissues [2], tumor mutation burden (TMB) [3], and so on. However, single biomarker has not been found to be efficient enough to precisely identify treatment‐responsive patients; thereby indicating that multiple biomarkers would be a better option. Hence, we made a comprehensive analysis of several ICI efficacy‐associated biomarkers and constructed a prognostic classifier model for NSCLC patients receiving ICI treatment. Our main study endpoint was progression‐free survival (PFS).
Some evidences had indicated the prognostic value of common gene mutations, such as epidermal growth factor receptor (EGFR) and tumor protein p53 (TP53), for NSCLC treatment using ICI [1]. Additionally, the human SWItch/sucrose nonfermentable (SWI/SNF) chromatin‐remodeling complex was encoded by multi‐gene families. Mutations in these genes occurred broadly in cancer [4]. Previous studies have shown that tumors harbored SWI/SNF‐mutants, such as renal clear cell carcinoma, were sensitive to ICIs [5]. Thus, some SWI/SNF‐mutant genes were also taken into account in this study.
First, we downloaded and integrated the publicly available profiles of an ICI‐treated advanced NSCLC cohort from two datasets through cBioPotal online database (https://www.cbioportal.org/). Data processing and analyzing procedures were demonstrated in Supplementary Figure S1. Pooled dataset containing 86 NSCLC samples with complete requisite information for subsequent analysis was utilized for study. Parameters taken into consideration included age, gender, smoking history, treatment type, treatment line, mutation rate, frequency of fraction of the genome altered by copy number alterations (CN‐FGA), TMB, PD‐L1 score, as well as several gene mutation status and fusion status. Frequent SWI/SNF submit gene mutations which were deemed to mediate chromatin remodeling pathway, including actin like 6A (ACTL6A), AT‐rich interaction domain 1A (ARID1A), AT‐rich interaction domain 1B (ARID1B), polybromo 1 (PBRM1), and SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4 (SMARCA4), were extracted. These genes were selected based on previously literatures and the Rat Genome Database (RGD; https://rgd.mcw.edu/rgdweb/ontology; reference gene sets: altered SWI/SNF family mediated chromatin remodeling pathway).
Baseline clinical characteristics for the entire cohort of our study are shown in Supplementary Table S1. COX regression was adopted to screen out prognostic variables. Since the fusion of anaplastic lymphoma kinase (ALK) and ROS proto‐oncogene 1(ROS1), and actin like 6A (ACTL6A) mutation did not exist in the enrolled NSCLC samples, they could not be analyzed. As shown in Supplementary Table S2, univariate analysis revealed that smoking history (hazard ratio [HR] = 0.48; 95% confidence interval [CI] = 0.28‐0.83; P = 0.009), treatment type (HR = 0.38; 95% CI = 0.17‐0.81; P = 0.012), TMB (HR = 0.54; 95% CI = 0.32‐0.93; P = 0.026), PD‐L1 score (HR = 0.55; 95% CI = 0.34‐0.88; P = 0.013), TP53 status (HR = 0.51; 95% CI = 0.32‐0.82; P = 0.006), EGFR status (HR = 2.25; 95% CI = 0.11‐4.57; P = 0.023), and ARID1B status (HR = 0.23; 95% CI = 0.06‐0.93; P = 0.040) were statistically significant for predicting PFS. The Kaplan‐Meier curves for these variables are shown in Supplementary Figure S2. Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) status (P = 0.775), B‐Raf proto‐oncogene (BRAF) status (P = 0.153), serine/threonine kinase 11 (STK11) status (P = 0.336) failed to show statistical significance. These might be due to the small sample size (n = 76). Next multivariate COX regression analysis was performed for 7 mentioned above variables with P < 0.05, utilizing “forward conditional” stepwise method. Eventually, 5 variables with P < 0.05, including EGFR status, ARID1B status, smoke history, treatment type, PD‐L1 score were finally retained to generate our prognostic model while TP53 status and TMB lost statistically significance (P > 0.05). (Supplementary Table S2). Next, we calculated the “risk score” for each of the 86 patients according to these 5 variables and their regression coefficients using the following formula:
The receiver operating characteristic (ROC) curves were plotted for EGFR status, ARID1B status, smoking history, Treatment type, PD‐L1 score as well as “risk score” for predicting PFS. Predictive ability was evaluated through area under the receiver operating characteristic curve (AUC). As shown in Figure 1A, our integrated prognostic model showed excellent predictive performance in 6month (AUC = 0.75, 95% CI = 0.61‐0.89) and 12‐month (AUC = 0.86, 95% CI = 0.70‐1.00) PFS as the AUC of “risk score” was the largest. The cutoff point ‐0.96 represented the highest Youden index (sensitivity + specificity − 1). It was selected as the threshold value of “risk score” for categorizing patients into high‐or low tumor progression risk group (Figure 1B). Kaplan‐Meier analysis revealed that patients in the high‐risk group had shorter PFS time than those in low‐risk group (median PFS: 10.0 months vs. 2.7 months, P < 0.001) (Figure 1C). Subsequently, a prognostic nomogram for predicting the PFS probability at 2‐, 6‐ and 12‐month was constructed based on the incorporated 5 prognostic factors identified above (Figure 1D). A certain score of each variable can be shown when a perpendicular line between the point scale and each variable is drawn. By adding all the scores as a total score and drawing a perpendicular line between the total point scale and PFS scales, we can estimate the predicted PFS rate. Thus, with this nomogram, clinicians could easily predict the efficacy of ICI treatment for NSCLC patient.
FIGURE 1.
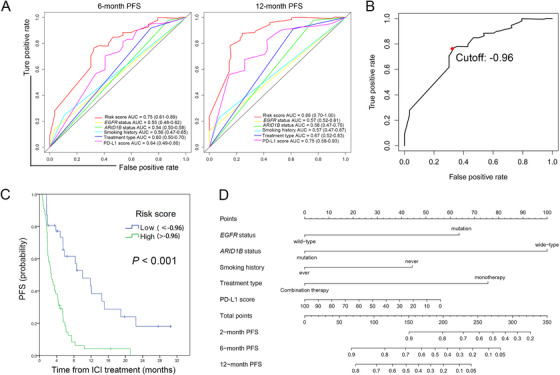
Predictive efficacy of variables finally selected for prognostic model generation and corresponding nomogram constructed. A. Time dependent ROC curves for evaluating 6‐and 12 ‐month PFS predictive efficacy. B. Cutoff point represented the highest Youden index (sensitivity + specificity − 1) in ROC curves was selected as the threshold value of “risk score” for categorizing patients into high‐or low risk group. C. Kaplan‐Meier curves demonstrated different PFS time for NSCLC patients in high or low‐risk group. D. Nomogram to predict the 2‐, 6‐, and 12‐month PFS probability of patients with NSCLC. Abbreviations: PFS, progression free survival; AUC, area under the receiver operating characteristic curve; EGFR, epidermal growth factor receptor; ARID1B, AT‐rich interaction domain 1B; ICI, immune checkpoint inhibitor; PD‐L1, programmed cell death ligand1
In terms of overall survival (OS), we also tried to explore potential prognostic factors aforementioned in 76 samples with survival data. Univariate COX regression found that only PD‐L1 score (HR = 0.35; 95% CI = 0.15‐0.83; P = 0.018) and treatment type (HR = 0.203; 95% CI = 0.048‐0.862; P = 0.031) demonstrated significant predictive value and their Kaplan‐Meier curves are shown in Supplementary Figure S3.
Next, we used tumor‐immune system interactions database (TISIDB) (http://cis.hku.hk/TISIDB/), an integrated repository portal publicly available for tumor‐immune system interactions to validate the ICI therapy efficacy of ARID1B mutation (Supplementary Figure S4). Our results indicated that ARID1B mutation was associated with ipilimumab treatment response for melanoma patients. Patients with ARID1B mutation demonstrated high response rate, with significant difference as compared to those with wide‐type (18.5% vs. 1.4% P = 0.002). As to NSCLC, a similar trend for a pembrolizumab treatment cohort was found. The response rate was 14.3% for ARID1B mutant patients versus 0% for ARID1B wild‐type patients, but probably because of the small sample size (34 in total), results did not reach statistical significance (P = 0.196).
Consistent with the previous studies, EGFR mutation was a crucial unfavorable factor for ICI treatment efficacy, while PD‐L1 score, smoking history, treatment type (combination therapy) were favorable factors [1, 6]. Nowadays, TMB still remains a controversial biomarker for ICI therapy [7]. In this current study, although TMB demonstrated predictive value in univariate COX regression, it was not an independent factor in multivariate COX regression, which suggested that its predictive ability was relatively smaller when compared with other variables finally screened out for model construction. Previous studies suggested that patients harboring SWI/SNF gene mutations, like renal cell cancer patients with PBRM1 mutation, possess a higher capacity to respond to ICI treatment [4, 5]. In our current study, we found that ARID1B mutation conferred longer PFS time for NSCLC patients receiving ICI treatment, which had not been reported before. As one of the members of the human SWI/SNF chromatin remodeling complex, ARID1B mutation may alter the SWI/SNF chromatin remodeling pathway, resulting in significant changes in chromatin accessibility and contributing to overall genomic instability [8]. Su K et al. [9] revealed ARID1B mutation in multiple lung lesions from a single patient and suspected that ARID1B may play a significant role in lung cancer formation. Naito T et al. [10] discovered a higher proportion of PD‐L1‐positive and TMB level in NSCLC cases with loss of expression of one or more subunits of the SWI/SNF complex including ARID1B. However, whether ARID1B is a key cancer driver in lung cancer is still remained unknown.
The present study had several limitations. As a retrospective study based on public datasets, some information such as performance status, the therapeutic schedule before ICI treatment and the salvage treatments received after disease progression was not available, which might cause certain bias. The small available sample size might reduce the statistical efficiency, especially for infrequent mutations. We set PFS as the primary outcome rather than OS because PFS was relatively less affected by confounding factors; as mentioned above, thus might better represent the ICI efficacy. In our study, mutation status for genes were detected through targeted next generation sequencing, whether it could be alternatively detected by other methods widely used in clinical practice like amplification refractory mutation system (ARMS) method needed to be further elucidated. Moreover, other potential biomarkers such as tumor infiltrating lymphocytes and inflammatory cytokines [1] were not available for analysis in this study. This might to some extent impact the prediction accuracy of our model.
Our results indicated that ICI combination therapy conferred better PFS than monotherapy, but it may arise greater risk of adverse effects. Nowadays, the combination of ICI with chemotherapy and even targeted therapies are also wildly used in the real‐world clinical practice, therapeutic synergy effect may need to be considered, thus the conditions for predicting ICI treatment efficacy will be more complicated.
In summary, we constructed a comprehensive predictive classifier model for evaluating the efficacy of ICI therapy in NSCLC patients, supporting a personalized approach for clinical decision making. Patients with low risk scores can be considered as appropriate candidates for ICI treatment. A SWI/SNF‐mutant gene, ARID1B, serves as a novel molecular biomarker for predicting ICI treatment, which is worthy of in‐depth research. External cohorts, especially prospective cohorts with large sample size, are needed to validate and optimize our model and further discover more potential biomarkers associated with ICI therapy efficacy.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The DNA sequencing data and corresponding clinical information were downloaded from the cBioPotal online database (https://www.cbioportal.org/). The datasets analyzed during the current study are available from the corresponding author on reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by Medical Scientific Research Foundation of Guangdong Province, China (Grant No. A2020153).
AUTHORS' CONTRIBUTIONS
ZL and LZ contributed to the conception and design of the study. ZL and YZ contributed to the data acquisition and statistical analysis. All authors wrote and approved the final manuscript
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
Not applicable.
REFERENCES
- 1. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Larsen TV, Hussmann D, Nielsen AL. PD‐L1 and PD‐L2 expression correlated genes in non‐small‐cell lung cancer. Cancer Commun. 2019;39(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Folprecht G. Tumor mutational burden as a new biomarker for PD‐1 antibody treatment in gastric cancer. Cancer Commun. 2019;39(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang L, Zhao D, Qin K, Rehman FU, Zhang X. Effect and biomarker of Nivolumab for non‐small‐cell lung cancer. Biomed Pharmacother. 2019;117:109199. [DOI] [PubMed] [Google Scholar]
- 7. Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer. 2018;6(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin J. ARID1B: From the Garden of Eden to the Sahara. J Thorac Cardiovasc Surg. 2018;155(6):e193‐e4. [DOI] [PubMed] [Google Scholar]
- 9. Su K, Gao S, Ying J, Zou S, He J. Sequencing a super multiple synchronous lung cancer reveals a novel variant in driver gene ARID1B. J Thorac Cardiovasc Surg. 2018;155(6):e185‐e91. [DOI] [PubMed] [Google Scholar]
- 10. Naito T, Udagawa H, Umemura S, Sakai T, Zenke Y, Kirita K et al. Non‐small cell lung cancer with loss of expression of the SWI/SNF complex is associated with aggressive clinicopathological features, PD‐L1‐positive status, and high tumor mutation burden. Lung Cancer. 2019;138:35‐42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
The DNA sequencing data and corresponding clinical information were downloaded from the cBioPotal online database (https://www.cbioportal.org/). The datasets analyzed during the current study are available from the corresponding author on reasonable request.


