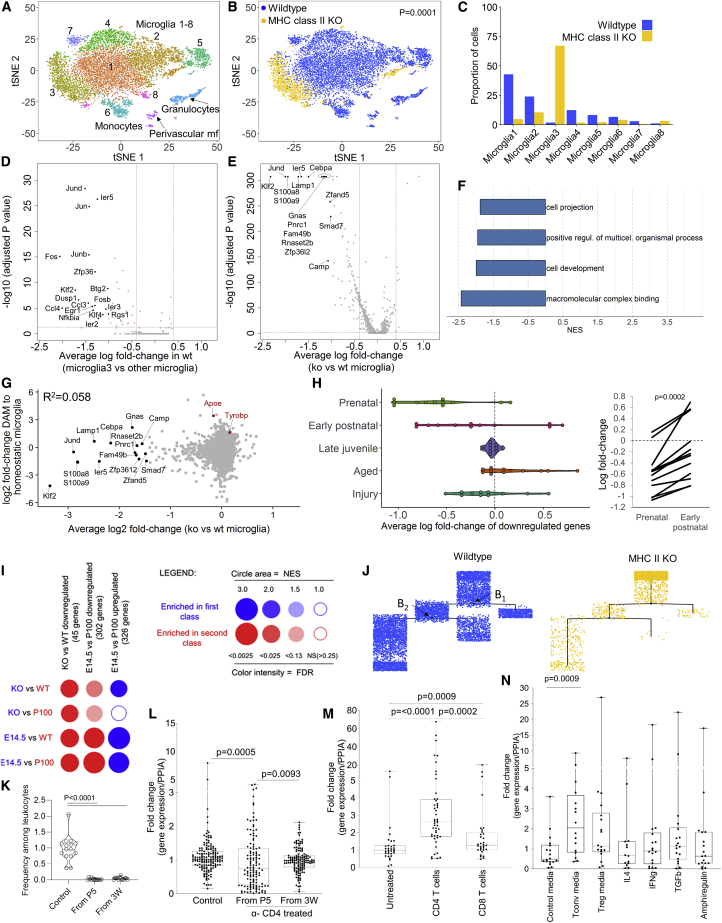
Figure 6.
CD4 T Cell Deficiency Traps Microglia in a Fetal-like Transcriptional State
10× single-cell sequencing was performed on 9,404 CD11b+ cells from the wild-type adult mouse brain and 2,278 CD11b+ cells from the MHC II KO adult mouse brain.
(A) t-SNE visualizing cell clusters built on the combined population.
(B and C) t-SNE visualizing cells originating from the (B) wild-type and MHC II KO condition with (C) relative proportions.
(D) Volcano plot of differential expression between cluster 3 microglia and non-cluster 3 microglia in the wild-type mouse.
(E and F) Volcano plot of differential expression between wild-type microglia and microglia from MHC II KO mice (E). High fold-change genes are labeled. Indicated cut-offs (signature genes) are used for (F) pathway analysis by GSEA.
(G) Fold-change of all expressed genes between wild-type and MHC II KO mice plotted against the fold-change of the same gene set in a reference dataset comparing healthy and damage-associated microglia (Keren-Shaul et al., 2017).
(H) Differentially expressed genes between wild-type and MHC II KO microglia plotted against a reference dataset of microglia spanning development, healthy adult status, aging, and neuroinjury (Hammond et al., 2019). Left: comparative expression in total microglia at each stage. Right: gene-level expression changes between the pre-natal and early post-natal period, paired t test.
(I) BubbleGUM analysis using gene sets from differential expression in wild-type versus MHC II KO microglia (this manuscript) and E14.5 versus day 100 microglia (Hammond et al., 2019). Red for wild-type and day 100 microglia, blue for MHC II KO and E14.5 microglia.
(J) The combined wild-type and MHC II KO microglia population were assessed for pseudotime trajectory, plotted separately for each genotype and showing branch points.
(K) Brain CD4 T cell numbers were assessed in control wild-type mice, or wild-type mice treated with anti-CD4 depleting antibody from day 5 or week 3 of age (n = 15,14,13).
(L) Microglia were sorted from control wild-type mice or anti-CD4 depleted mice. Expression of signature genes were assessed by qPCR, with data points reflecting 16 genes in 9–10 biological replicates.
(M) Neonatal brain slices were left untreated or exposed to CD4 or CD8 T cells for 14 days prior to microglia sorting. Expression of signature genes was assessed by qPCR, with data points reflecting 16 genes in 2–3 biological replicates.
(N) Neonatal microglia were cultured with control media, media from stimulated conventional or Tregs, or selected cytokines. Expression of signature genes were assessed by qPCR, with data points reflecting 10 genes in 2 biological replicates.
See also Figure S6.