Abstract
miR-206, miR-1a-1 and miR-1a-2 are induced during differentiation of skeletal myoblasts and promote myogenesis in vitro. miR-206 is required for skeletal muscle regeneration in vivo. Although this microRNA family is hypothesized to play an essential role in differentiation, a triple knockout of the three genes has not been done to test this hypothesis. We report that triple KO C2C12 myoblasts generated using CRISPR/Cas9 method differentiate despite the expected de-repression of the microRNA targets. Surprisingly, their mitochondrial function is diminished. Triple KO mice demonstrate partial embryonic lethality, most likely due to the role of miR-1a in cardiac muscle differentiation. Two triple KO mice survive and grow normally to adulthood with smaller myofiber diameter, diminished physical performance and an increase in PAX7 positive satellite cells. Thus, unlike other microRNAs important in other differentiation pathways, the miR-206 family is not absolutely essential for myogenesis and is instead a modulator of optimal differentiation of skeletal myoblasts.
Keywords: myomiRs, skeletal muscle differentiation, myogenesis, miR-1a-1, miR-1a-2
Introduction
The downregulation of pluripotency markers and activation of lineage-specific gene expression during differentiation allow for accurate development. Differentiation-induced microRNAs play a major role in this process by repressing their targets – genes responsible for self-renewal. Depletion of DGCR8 protein essential for biogenesis of microRNA in pluripotent cells decreases most active microRNA levels and inhibits differentiation (1). Since many microRNAs are induced during differentiation of specific tissue lineages, several of them have been tested for their importance in differentiation, particularly whether they act as a switch that is essential for differentiation, or as a modulator of differentiation. MicroRNAs regulate processes as early as gastrulation (2, 3), neural development (4–6), muscle development (7–11), bone formation (12, 13), skin development (14, 15) and hematopoiesis (16–18). Many of the studied microRNAs have been suggested to be essential, e.g. miR-206 and miR-1a for skeletal muscle myoblast differentiation (19–21), miR-144/451 for erythroid cells differentiation (22, 23), miR-17~92 during B lymphopoiesis and lung development (24), miR-15a-1 and miR-18a for development and function of inner ear hair cells in vertebrates (25), miR-219 for normal oligodendrocyte differentiation and myelination (26), miR-204 for differentiation of the retinal pigmented epithelium (27) and miR-375 for human spinal motor neuron development (28).
Myogenesis is a process of muscular tissue formation, which first occurs in vertebrate embryonic development (29), but also happens in adult muscle regeneration (30). The skeletal muscle satellite cells are the myogenic stem cells of adult muscles residing between the sarcolemma and basal lamina of muscle fibers (31). In normal conditions these tissue specific stem cells stay in a quiescent G0 state (32). Upon activation by injury or disease, they re-enter the cell cycle to establish a population of skeletal muscle progenitors (myoblasts), which differentiate further and fuse to produce myotubes. The major regulator of this process is Pax7 transcription factor (33, 34). The satellite cells express the transcription factor Pax7 in G0 and when activated, coexpress Myod1. Downregulation of Pax7 leads to differentiation into myotubes, whereas downregulation of Myod1 leads to a return to quiescence. An important player in Pax7 downregulation and differentiation induction is miR-206 (19, 20). Other microRNAs are also known to play important roles in skeletal muscle differentiation. A conditional skeletal muscle specific knockout of Dicer in mice leads to global loss of miRNAs in developing skeletal muscle, resulting in widespread apoptosis and abnormal myofiber morphology (35). Over the last 14 years miR-206, miR-1a-1 and miR-1a-2 have been hypothesized to not only be very important for muscle differentiation, but also essential for this process (7, 8, 19–21, 36–52). miR-206, miR-1a-1 and miR-1a-2 are members of the myomir family and are expressed from bicistronic loci. Interestingly, miR-206 and −1a have an 18/21 base match in sequence with each other and complete identity in the first eight nucleotides that constitute the seed sequence for target recognition. Even though all three are expressed in skeletal muscles, miR-1a-1 and miR-1a-2 are also expressed in cardiac muscle, where miR-206 is not expressed (39, 53). All three are upregulated during murine skeletal myoblast differentiation (39). Overexpression of miR-206 induces C2C12 differentiation, whereas simultaneous knockdown of miR-206 and miR-1a results in diminished differentiation (39). Based on this it was hypothesized that the three microRNAs collectively are essential for skeletal muscle differentiation.
Knockout of the miR-206 gene produced viable mice with no defect in skeletal muscle development, although there was a defect in skeletal muscle regeneration after extensive muscle injury (54). The loss of miR-206 in mice modeling amyotrophic lateral sclerosis leads to faster disease progression and lack of regeneration of neuromuscular synapses, again suggesting a role of the microRNA in response to tissue injury. In contrast, there was high embryonic lethality of miR-1a-2 knockout mice caused by various cardiac problems (41). Although miR-1a-1 knockout showed embryonic lethality in the 129 genetic background animals, knockout pups were obtained at the expected frequency in a mixed genetic background, proving that the miR-1a-1 knockouts can produce viable mice (50). Although the double KO of miR-1a-1 and miR-1a-2 was found to have high embryonic lethality, some mice survived with normal skeletal muscle development. By 10th postnatal day, however, all double knockout animals were dead because of serious cardiac defects (50). Moreover, knockout of two bicistronic loci (miR-1a-1/133a-2 and miR-1a-2/133a-1 dKO) showed complete embryonic lethality at embryonic stage E11.5 (49). The miR-1a/133a skeletal muscle specific dKO mice were alive, but had metabolic problems caused by incorrect functioning of mitochondria (51).
Overall, all animals had relatively normal skeletal muscle development, and this could be explained because at least one myomir remained intact in all these animals. A complete knockout of all three microRNAs, miR-206, miR-1a-1 and miR-1a-2, tests whether an essential role of these myomiRs in myogenesis was masked by the redundant function of the microRNA left in the cells.
Materials and Methods
Cell lines generation
Stable overexpression of inducible Cas9 in C2C12 cells
pCW-Cas9 (addgene #50661) vector was packed in the virus using psPAX2 (addgene #12260) and pMD2.G (addgene #12259) in 293T cells. C2C12 cells were transduced with the filtered supernatant containing virus. After 24 hours cells were treated with puromycin (C=2ug/ml; Sigma, Cat# P9620) and resistant cells were seeded to 96-well plates using single cell dilution method. Growing clones were examined for Cas9 expression by immmunoblotting after treatment with doxycycline (C=1ug/ml; Sigma, Cat# D9891) for 48h. Clones with high expression upon induction and low leakage level without doxycycline were next tested for differentiation efficiency by q-RT-PCR for Myod1, Myogenin and Myh3, and immunoblotting for MYOD1, MYOGENIN and MHC. The clone, which was the most similar to wild-type cells, was chosen for further experiments.
Knockout cell line generation
CRISPR protocol (55) with minor changes was followed to achieve deletion of miR-206, miR-1a-1 and miR-1a-2 genes. Briefly, sgRNAs were designed using CRISPR DESIGN tool: http://crispr.mit.edu/. Cas9 expression in C2C12 cells was induced 24h before sgRNAs transfection using doxycycline (C=1ug/ml). Cells were co-transfected with 6 different sgRNAs cloned into gRNA_GFP-T2 (addgene #41820), and a spiking vector coding for a resistance gene using Lipofectamine 3000 (Life Technologies, Cat# L3000015). After 24–48 hours cells were treated with hygromycin (C=300ug/ml; Life technologies, Cat# 10687–010), and resistant cells were seeded to 96-well plates using single cell dilution method. Growing clones were examined for desired deletion by PCR on extracted genomic DNA (Quick Extract DNA Extraction Solution, Lucigen., Cat# QE09050), and candidates with complete loss of all three WT PCR products (homozygous deletions) were confirmed by Sanger sequencing and q-RT-PCR for miR-206, miR-1a-1 and miR-1a-2. Oligonucleotides sequences are listed in Table S1.
Cell culture, differentiation assay
C2C12 cells were cultured in DMEM-high glucose medium (GE Healthcare Life Sciences co., Cat# SH30022.FS) with 10% FBS (Gibco, Cat# 10437–028), when differentiating, serum was switched to 2% horse serum (GE Healthcare Life Sciences co., Cat# SH30074.03) supplemented with (1) 1x ITS (Insulin-Transferrin-Selenium; Fisher, Cat# 41400045) and 5mM LiCl (Sigma; Cat# 203637) for PAX7 negative cells or (2) 1x ITS for PAX7 positive cells. PAX7 negative and positive cell lines were independently purchased.
RNA isolation and RT-PCR
RNA was isolated from cells and muscles homogenized in liquid nitrogen by TRIzol reagent (Life Technologies, Cat# 15596018) using Direct-zol RNA MiniPrep Plus Kit including DNase treatment (Zymo Research, Cat# R2052). cDNA synthesis for mRNA expression levels measurement was performed using Superscript III RT cDNA synthesis kit (Life Technologies co., Cat# 18080–051) with random hexamer and oligodT priming. After cDNA synthesis, qPCR was performed with Applied Biosystems 7500 Real-Time PCR Systems using SensiFAST™ SYBR® Hi-ROX Kit (BIOLINE, Cat# BIO-92020). miScript II RT (Qiagen, Cat# 218161) and miScript SYBR® Green PCR kits (Qiagen, Cat# 218075) were used for miRNA and pre-miRNA expression levels measurement. All primers used in this study are listed in Table S1.
Western blotting
Cells were lysed in IPH buffer (50mM Tris-Cl, 0.5% NP-40%, 50mM EDTA). TA muscles were homogenized in liquid nitrogen and lysed in ice cold buffer overnight (20mM Tris PH 8.0, 137mM NaCl, 2.7mM KCl, 1mM MgCl2, 1% Triton X-100, 10% glycerol, 1mM EDTA, 1mM DTT). Protein lysates were run on 10% polyacrylamide SDS-PAGE gel, transferred to nitrocellulose membranes. Membranes were blocked for 30 minutes in 5% milk containing PBST, and incubated overnight with primary antibody in 1% milk. Secondary antibody incubation was carried out for 1 hour after washing, and at 1:4000 dilution before washing and incubation with Millipore Immobilon HRP substrate. Primary antibodies were used as follows: mouse monoclonal MyoD (5.8A) (Santa Cruz co., Cat# sc-32758), mouse monoclonal Myogenin (F5D) (Santa Cruz co., Cat# sc-12732), rabbit polyclonal MYH3 (Proteintech, Cat# 22287–1-AP), mouse monoclonal HSP90 α/β (F-8) (Santa Cruz co., Cat# sc-13119), mouse monoclonal DNA Pol alpha (STK1) (Santa Cruz co., Cat# sc-13518), rabbit polyclonal NOTCH3 (M-134) (Santa Cruz co., Cat# sc-5593), rabbit polyclonal PAX7 (Sigma, Cat# SAB2107886), mouse monoclonal CCND1 (DCS6) (Cell Signaling, Cat# 2926), mouse monoclonal IGFBP5 (D-6) (Santa Cruz co., Cat# 515116). Secondary antibodies were used as follows: anti-rabbit IgG, HRP-linked Antibody (Cell Signaling, Cat# 7074S), anti-mouse IgG, HRP-linked Antibody (Cell Signaling, Cat# 7076S).
Immunofluorescence assay
Cells were plated on glass coverslips and collected in growth medium or after 5 days of differentiation. The coverslips were fixed with 4% paraformaldehyde in PBS for 15 min, permeabilized in 0.5% Triton X-100 in PBS, and blocked in 5% goat serum. The coverslips were incubated with primary rabbit polyclonal antibody MYH3 (Proteintech, Cat# 22287–1-AP) overnight at 4°C and then with goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Invitrogen, Cat# A-21244) for 1 h. Cells were stained with Hoechst 33342 (1 μg/mL; Life Technologies, Cat# H3570) for 2 minutes at room temperature, washed and then mounted with ProLong™ Gold Antifade Mountant (Life Technologies, Cat# P10144). The primary and secondary antibodies were diluted 1:400 and 1:1000, respectively. The fusion index was calculated by dividing the number of nuclei contained within multinucleated cells by the number of total nuclei in a field. Microscopy was performed using the Zeiss LSM 710 Confocal Microcopy and ImageJ Software for analysis (56).
Mitochondrial Stress Test
Oxygen Consumption Rate (OCR) for Mitochondrial Stress Test (MST) assay was measured using Seahorse XF24 Extracellular Flux Analyzer (Agilent). 20,000 cells per well were plated in growth medium in at least triplicate for each cell line and condition 48h before test. For DM1, 24h post seeding medium was switched to differentiation and MST assay was performed after 24h. One hour before MST assay medium was changed to MST medium (unbuffered, serum-free DMEM pH 7.4; Life Technologies, Cat# 12100046). For the MST assay, oligomycin (inhibits ATP synthase; Sigma, Cat# 75351), BAM15 (protonophore uncoupler, stimulates a maximum rate of mitochondrial respiration; Cayman, Cat# 17811), and Rotenone and Antimycin A (inhibits the transfer of electrons in complex I and III, respectively; Sigma, Cat# R8875, Cat# A8674) were injected to a final concentration of 2μM, 10μM, 1μM and 2μM, respectively over a 100-minute time course. At the end of each experiment, each assay was normalized to total protein count measured from a sister plate that was seeded concurrently with the experimental plate.
RNA-seq
RNA samples were isolated from proliferating or differentiating WT and tKO cells as described above. RNA-seq was performed by Beijing Novogene Co., Ltd. on poly(A) enriched RNA using the Illumina HiSeq instrument. We obtained >50 million paired-end 150 bp long reads for all conditions. RNA-Seq data was aligned to the reference genome - mouse assembly GRCm38/mm10 using STAR v2.5 (57) and quantified by HTSeq 0.6.1p1 (Python 2.7.5) (58). DESeq2 R package (59) was then applied to determine differentially expressed genes with a significant criterion padj <0.05. Gene Ontology was performed using clusterProfiler (60). GSEA (61) was used for gene set enrichment analysis. The list of miR-206/−1a targets was downloaded from TargetScan 7.1 (62). All RNA-Seq libraries data files are available under GEO accession number: GSE133260.
Short RNA-seq
RNA samples were isolated from proliferating or differentiating WT and tKO cells as described above. 0.5 ug of RNA was used for short RNA-seq library preparation with NEBNext Small RNA library kit (New England Biolabs, Cat# E7300). Shortly, 3’ preadenylated adaptors and then 5’ adaptors were ligated to RNA, followed by reverse transcription and PCR with indexes. Next, 8% TBE-PAGE gel was used for size selection (15–50nt). For sequencing we used Illumina NextSeq 500 sequencer with high-output, 75-cycles single-end mode at Genome Analysis and Technology Core (GATC) of University of Virginia, School of Medicine. Data trimmed by catadapt v1.15 (Python 2.7.5) (63) was aligned to the reference genome (gencode GRCm38.p5 Release M16, primary assembly) using STAR v2.5 with settings based on previous paper (64) and the total number of mapped reads were used for normalization. In general, mapped percentage is more than 95%. Unitas v.1.5.1 (with SeqMap v1.0.13) (65, 66) was used for miRNA abundance quantification with setting –species_miR_only – species mus_musculus to map the reads to mouse sequence of miRBase Release 22 (67). This setting (equivalent to –tail 2 –intmod 1 –mismatch 1 –insdel 0) will allow 2 non-templated 3’ nucleotides and 1 internal mismatch for miRNA mapping. For differential analysis, DESeq2 (59) was used on count matrix of mature microRNAs. All short RNA-Seq libraries data files are available under GEO accession number: GSE133255.
Mice
Mouse strains used in the study were obtained as follows: miR-206 KO mice provided by Eric Olson (68); miR-1a-1 and miR-1a-2 KO mice provided by Deepak Srivastava (41, 50). Double KO mice were generated by crossing single KO animals and then crossing their double heterozygous offspring. Triple KO mice were generated by crossing miR-206 miR-1a-1 double KO animals with miR-206 miR-1a-2 double KO animals and then crossing their offspring (miR-206 KO miR-1a-1 HET miR-1a-2 HET) as shown in Figure 4A. All animals were PCR-genotyped using gene-specific primers listed in Table S1. Work involving mice adhered to the guidelines of the University of Virginia Institutional Animal Care and Use Committee (IACUC), protocol number 3774.
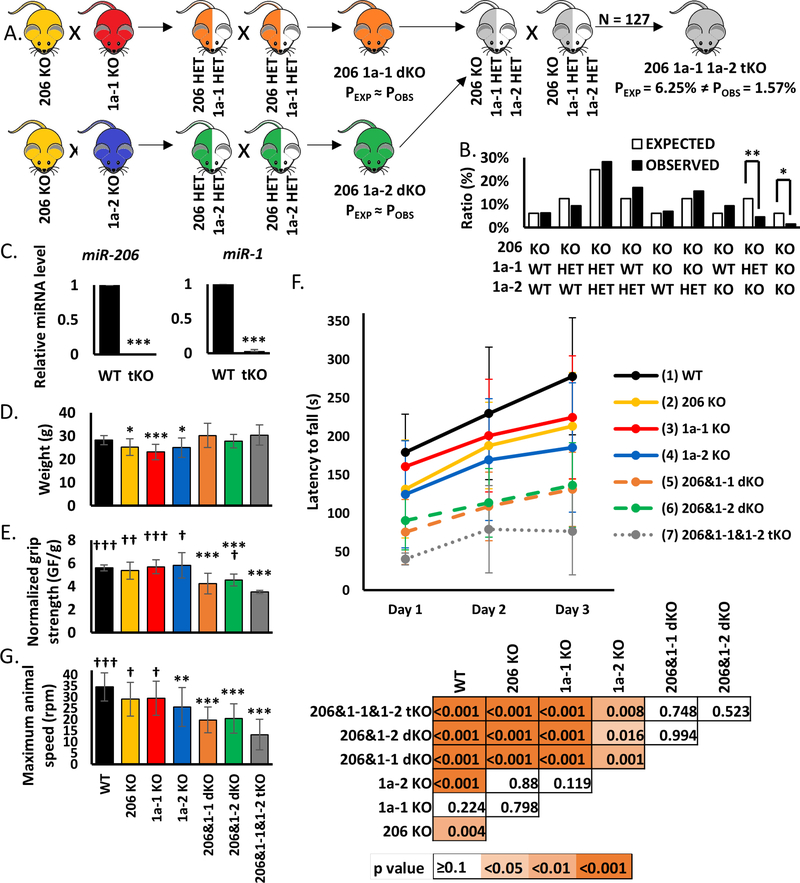
Figure 4.
Knockout of miR-206, miR-1a-1 and miR-1a-2 leads to partial embryonic lethality and diminishes adult mice physical potential
A) Scheme of animal crosses leading to generation of triple KO mice.
B) Genotypes of offspring generated from miR-206 KO miR-1a-1 HET miR-1a-2 intercrosses. In total 127 animals were genotyped. Percentage of expected and observed genotypes are given for weaning-age (3-week-old) pups. (**), (*) indicates p-value =< 0.01, =<0.05.
C) qRT-PCR analysis of microRNAs in TA skeletal; muscles from control wild-type (WT, n=3) and triple KO (tKO, n=2) animals. Levels of miRNAs were normalized to U6 snRNA. Levels are shown relative to WT. Values presented as mean +/− SD. Statistical significance was calculated using t-student test. (***) indicates p-value =< 0.001.
D) Animal weight used for physical performance tests. Statistical significance was calculated using t-student test. (***) indicates p-value =< 0.001. (*) indicates p-value =< 0.05. N = 13 for all genotypes, except for miR-206&1–1&1–2 tKO (N = 2).
E) Forelimb grip strength measured using grid normalized to respective body weights. Values presented as mean +/− SD. Statistical significance was calculated using t-student test. (***) indicates p-value =< 0.001 in comparison to WT mice. (†††), (††), (†) indicates p-value =< 0.001, =<0.01, =< 0.05 respectively in comparison to tKO mice. N = 13 for all genotypes, except for miR-206&1–1&1–2 tKO (N = 2).
F) Top: Latency to fall measured using rotating rod. Values presented as mean +/− SD. N = 13 for all genotypes, except for miR-206&1–1&1–2 tKO (N = 2). Bottom: Statistical significance heatmap calculated using one-way ANOVA test.
G) Maximum speed of rotation tolerated by the animals measured using rotating rod at the end of experiment (3rd day). Values presented as mean +/− SD. Statistical significance was calculated using t-student test. (***), (**) indicates p-value =< 0.001, =<0.01 respectively in comparison to WT mice. (†††), (†) indicates p-value =< 0.001, =< 0.05 respectively in comparison to tKO mice. N = 13 for all genotypes, except for miR-206&1–1&1–2 tKO (N = 2).
Physical endurance tests
Tests described below were performed on 12 weeks old mice, males and females (7+6) for all animals except tKO mice, where 2 males were examined.
Grip strength test
Mice were tested for their maximal grip strength with a grip strength meter (C.S.C. Force Measurement, Inc. AMETEK 2LBF Chatillon). The mice were placed on the force meter allowing forelimb to grip the grid. The mice were then slowly dragged backward until loss of grip. The bodies of the mice were kept in a horizontal position during the test. Three trials were repeated in 1-minute intervals. The average of the three trials was recorded as the maximum grip strength.
Accelerating rotarod test
Motor coordination and balance were assessed on an Economex accelerating rotarod (Columbus Instruments) that has the capacity to test five mice simultaneously. The testing procedure consists of three days, two runs on each day. The rod was adjusted to spin at a beginning speed of 4.0 r.p.m with an acceleration (0.12 r.p.m.) over 300s to final speed 40.0rpm. The maximum running time was 360s. Latency to fall from the rotarod and maximum animal speed for each run were recorded and averaged per each day.
Experiments involving mice adhered to the guidelines of the University of Virginia Institutional Animal Care and Use Committee (IACUC), protocol number 4064.
Muscle isolation, histological analyses and staining
The mice were sacrificed by carbon dioxide inhalation. The hearts, quadriceps and tibialis anterior muscles were dissected quickly, weighed, fixed with 4% paraformaldehyde or snap-frozen in liquid nitrogen and stored at −80 °C. All samples were collected at least 7 days after assessment of physical function from 3 males and 2 females for all animals except tKO mice (2 males).
All formalin-fixed paraffin-embedded (FFPE) sections, H&E staining and Masson’s trichrome staining were performed by Research Histology Core at University of Virginia (Charlottesville, USA). Fiber size was measured on H&E stained quadriceps and gastrocnemius muscles using ImageJ 1.50i (Java 1.6.0_24) (56) on three pictures from two animals per genotype (300 fibers per genotype in total). PAX7 immunohistochemistry was performed by Biorepository & Tissue Research Facility at University of Virginia on FFPE sections of quadriceps muscles. PAX7 positive cell number was counted per hundred myofibers using ImageJ 1.50i (Java 1.6.0_24) (56) on three pictures from two animals per genotype.
Immunofluorescence on FFPE sections was performed as previously described (69). In short, FFPE sections were deparaffinized and antigen-retrieved (by Biorepository & Tissue Research Facility at University of Virginia), washed, and blocked then incubated with anti-mitochondria antibody, clone 113–1, Cy3 conjugate (EMD Millipore, Cat# MAB1273C3) overnight. Slides were incubated with CuSO4 to reduce autofluorescence and mounted with Prolong Gold antifade reagent with DAPI. Images were taken with a Zeiss LSM 710 Multiphoton microscope with a 20× (NA 0.8) objective. Mitochondria content and nuclei per fiber count was performed using ImageJ 1.50i (Java 1.6.0_24) (56) on ten pictures per genotype (600 fibers per genotype in total).
Results
miR-206, miR-1a-1 and miR-1a-2 are not essential for C2C12 myoblasts differentiation
To determine whether miR-206 and miR-1a are necessary for murine myoblast proliferation and differentiation, we designed 6 different sgRNAs to delete all three genes at once (Figure 1A, Table S1). To increase knockout efficiency, we used C2C12 cell line with stable overexpression of Tet-inducible Cas9. miRNA gene deletions were confirmed by PCR on genomic DNA (Figure 1B) and Sanger sequencing of PCR products (Figure 1C). We didn’t detect any pre-miRNA or miRNA left in our differentiating triple KO (tKO) clones (Figure 1D and S1A). To ensure the reproducibility of our results we obtained triple KO clones independently from PAX7 negative (Figure 1A–D) and PAX7 positive (Figure S1) C2C12 myoblasts.
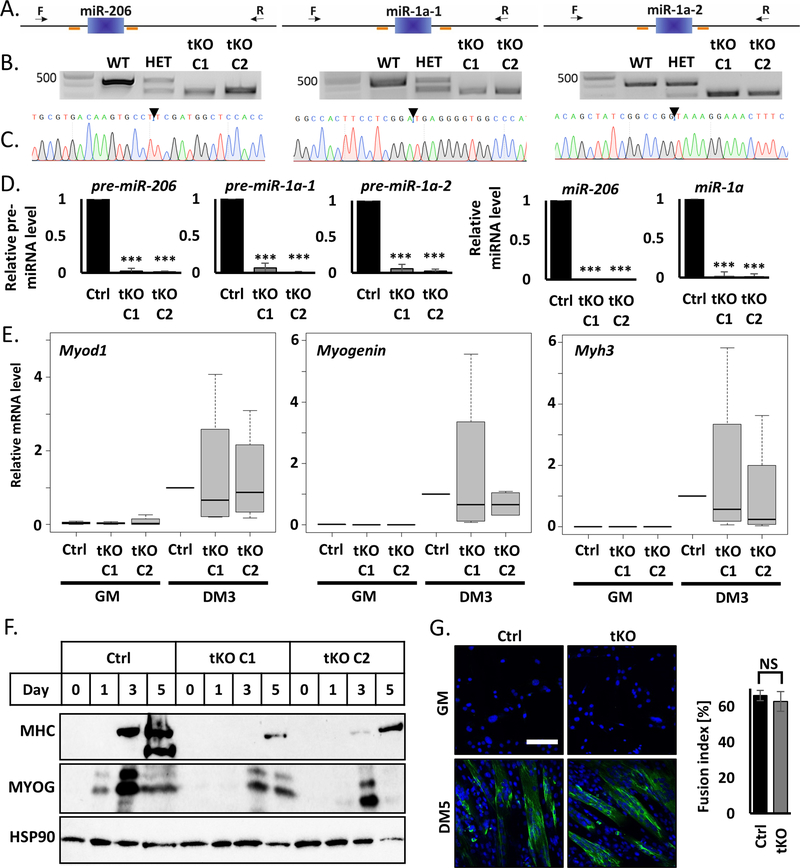
Figure 1.
Simultaneous knockout of miR-206, miR-1a-1 and miR-1a-2 in C2C12 murine myoblast does not block cell differentiation
A) Scheme of CRISPR/Cas9 experiment design. Blue blocks represent genes, orange lines sgRNAs sequences and black arrow genotyping primers localization. Left: miR-206, middle: miR-1a-1, right: miR-1a-2.
B) Representative picture of PCR genotyping results of triple KO C2C12 cells. Left: miR-206, middle: miR-1a-1, right: miR-1a-2. Top band – wild-type size, bottom band – KO size.
C) Representative picture of Sanger sequencing confirmation of the genotyping PCR product in tKO C1 C2C12 clone.
D) qRT-PCR analysis of differentiating (DM3) control cells (Ctrl) and triple KO clones (tKO C1 and tKO C2). Levels of pre-miRNAs were normalized to Gapdh and miRNAs – to U6 snRNA. Levels are shown relative to control cells (Ctrl DM3). Values represent three biological replicates, presented as mean +/− SEM. Statistical significance was calculated using t-student test. (***) indicates p-value =< 0.001.
E) qRT-PCR analysis of proliferating (GM) and differentiating (DM3) control cells (Ctrl) and triple KO clones (tKO C1 and tKO C2). Levels of Myod1, Myogenin and Myh3 mRNAs were normalized to Gapdh and shown relative to control cells (Ctrl DM3) in box and whiskers plots. Values represent four biological replicates, black line represents median. Statistical significance was calculated using t-student test.
F) Representative Western blot of time course of differentiation (GM, DM1, DM3, DM5) of control cells (Ctrl) and triple KO clones (tKO C1 and tKO C2). Protein levels for MYOGENIN and MHC were measured. HSP90 serves as a loading control.
G) Immunofluorescence analysis of fixed cells 5 days after differentiation (DM5). Cells were immunostained with antibodies against MHC. Hoechst 33342 was used to visualize nuclei. Quantification of the fusion index is presented on the right side. White line = 200μm.
C2C12 clones lacking miR-206 and miR-1a differentiate upon serum starvation producing similar levels of promyogenic mRNAs: Myod1, Myogenin and Myh3 as control cells (Figure 1E). The tKO clones also produce the corresponding proteins, which include markers of early (MYOGENIN) and late myogenesis (MHC), though their levels are lower in the PAX7 negative tKO cells (Figure 1F). Importantly, MYOGENIN level peaks at DM3 similarly in control and tKO cells, suggesting this is not delay in normal levels of promyogenic protein expression, but rather a decrease at the peak. In PAX7 positive tKO clones the Myod1, Myogenin and Myh3 mRNAs are induced normally during differentiation (Figure S1B), but we observe similar levels of MYOGENIN protein and slightly decreased MHC protein (Figure S1C). Moreover, the fusion index counted in the PAX7 positive cells (Figure 1G) shows there is no morphological differences upon differentiation between the control and tKO cells. A time course experiment at days 1, 3, 5 and 7 of C2C12 differentation shows no difference in myotube formation between PAX7 positive control C2C12 cells and tKO cells (Figure S1D).
Although triple KO C2C12 clones differentiate they are impaired in metabolic performance
By RNA-seq the triple KO clones differentiated efficiently as confirmed by hierarchical clustering which shows the differentiating tKO cells cluster with the differentiating wild type cells, separate from proliferating cells (Figure 2A). Nevertheless, 531 and 412 genes were differentially expressed in tKO in comparison to control cells in proliferating and differentiating conditions, respectively (Figure 2B). Among 232 genes upregulated during differentiation in the tKO cells are several involved in retinoic acid regulation (from the top 10: Aldh1a1, Aldh1a7, Brinp3 and Tceal5), which was previously described as important factor for myogenesis (70–72). Aldehyde dehydrogenases (Aldh) are also known to have promyogenic potential (73–75).
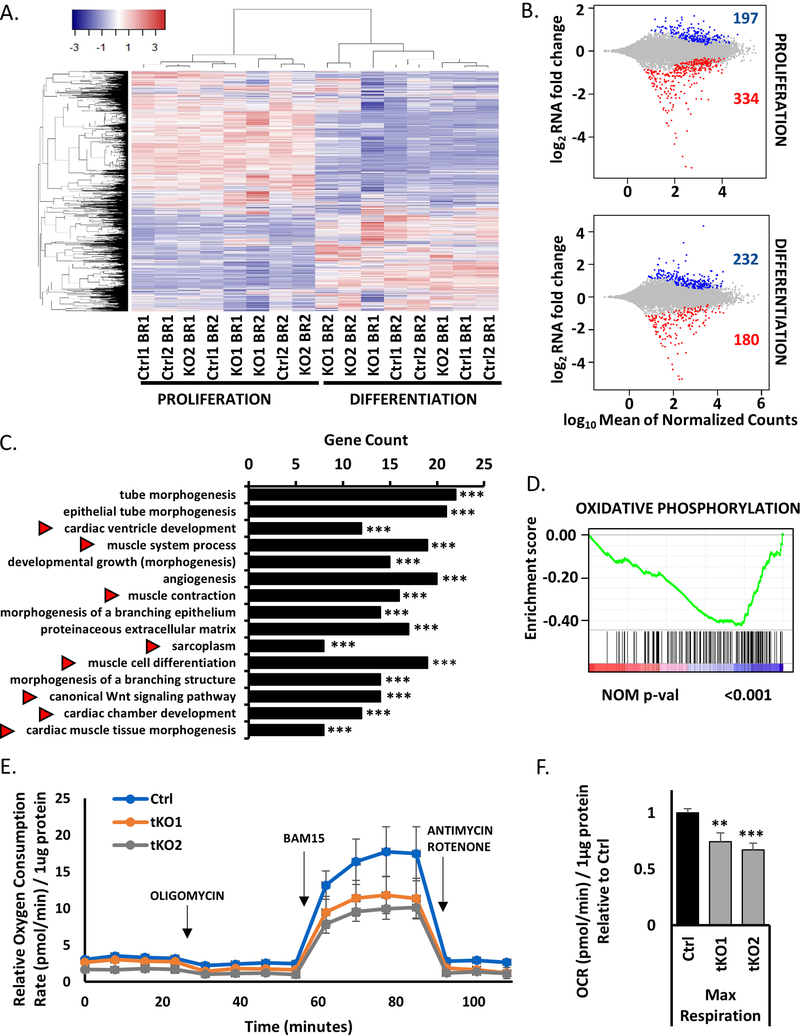
Figure 2.
RNAseq analysis confirms the tKO cells differentiate, but they have lower mitochondrial respiratory capacity than control cells during differentiation
A) Heatmap showing clustering of all RNA-seq samples and conditions based on expression (FPKM≥1) in proliferating conditions (left) and in differentiating conditions (right) in control cells (Ctrl1 and Ctrl 2) and triple KO clones (KO1 and KO2). There are two biological replicates for each condition (BR1 and BR2).
B) MA plots representing differentially expressed genes between control cells and triple KO clones in proliferation (top) and differentiation conditions (bottom). Upregulated genes are presented in blue and downregulated in red (padj<0.01).
C) Top 15 significant Gene Ontology terms enriched in downregulated genes in differentiating triple KO clones (tKO) in comparison to differentiating control cells (Ctrl). Red arrows show gene terms related to muscle development and regeneration. (***) indicates padj =< 0.001
D) Enrichment plot from gene set enrichment analysis (GSEA) showing the gene set involved in oxidative phosphorylation is enriched among differentially downregulated genes in differentiating tKO clones versus differentiating control cells.
E) Oxygen Consumption Rate (OCR) of differentiating (DM1) tKO C2C12 clones and control C2C12 cells measured at basal conditions and after the administration of the indicated compounds. Values represent five biological replicates, presented as mean +/− SEM. Levels are shown relative to the total protein content.
F) Maximal Respiration of control C2C12 cells and tKO clones in differentiation (DM1). Values represent five biological replicates, presented relative to control cells as mean +/− SEM. (**), (***) indicates padj < 0.01 and 0.001 respectively. Levels are shown relative to the total protein content and control cells.
Gene Ontology (GO) analysis of differentiating triple KO clones reveals downregulation of genes involved in skeletal and cardiac muscle development pathways (Figure 2C). GO also shows that genes involved in mitochondria biogenesis and function are induced during differentiation of WT control cells, but not in the tKO cells (Figure S2A, B). The latter is supported by Gene Set Enrichment Analysis (GSEA), which shows only one significant category – an enrichment of genes related to oxidative phosphorylation among the genes downregulated in differentiated tKO cells vs. differentiated control cells (Figure 2D).
To assess whether tKO perturbed the mitochondrial metabolism of C2C12 cells, we analyzed oxygen consumption rate (OCR) as a measure of aerobic respiration using the Seahorse Bioscience XF24. The tKO clones had a lower OCR even at baseline than the control cells in differentiation conditions (Figure 2E). Furthermore, treatment of these clones with the mitochondrial uncoupling agent BAM15, which produces maximal oxygen consumption, revealed an even greater difference in OCR between control and tKO clones (Figure 2F). These results suggest that these three microRNA are required to maintain mitochondrial respiratory capacity, and are consistent with the findings from miR-1a−/−133a−/− skeletal muscle specific double KO animals that showed impairment of mitochondrial function (51). Thus, although the tKO C2C12 clones differentiate, their differentiation is slightly impaired compared to the WT cells and the cells suffer a downregulation of mitochondria function.
miR-206 and miR-1a targets are specifically upregulated during differentiation of triple KO C2C12 cells
The absence of miR-206 and miR-1a could be compensated by some other mechanism, in which case the targets of miR-206 and miR-1a will be unchanged in the tKO cells. We therefore analyzed the expression levels of the target mRNAs of these microRNAs (as specified in TargetScan 7.1; Table S2) in comparison to all other genes. The Cumulative Distribution Function (CDF) plots show that these targets are not changed in proliferation condition, when the three microRNA are not elevated (Figure 3A), however they are specifically upregulated in the tKO cells during differentiation, when the three microRNAs are normally induced (Figure 3B). When the ratio of target : non-target genes were examined in bins of genes distributed from the most-repressed to most-induced in the tKO, the targets were not enriched in any bin in proliferating myoblasts, but were enriched in the bins with induced genes under differentiating condition (bottom plots in Figure 3A and 3B). Thus, the shift for miR-206/−1a targets to the right in the CDF plot is due to the upregulation of the target genes in differentiating cells (Figure 3B). Interestingly, in C2C12 PAX7 positive cells we observe significant induction of Pax7 mRNA in proliferating cells (Figure S3A), but this induction is not significant in differentiation condition (Figure S3B). Other miR-206/−1a target mRNAs are not changed or mildly upregulated in tKO cells. At the protein level we detect mild upregulation of DNA Pol alpha (both in GM and DM3), NOTCH3 (GM), PAX7 (DM3), CCND1 (GM) and IGFBP5 (GM) (Figure S3C). Therefore, in tKO C2C12 cells the microRNA targets are de-repressed, both at RNA and protein level, suggesting that an unknown regulatory mechanism has not been brought into effect to substitute for the missing microRNAs.
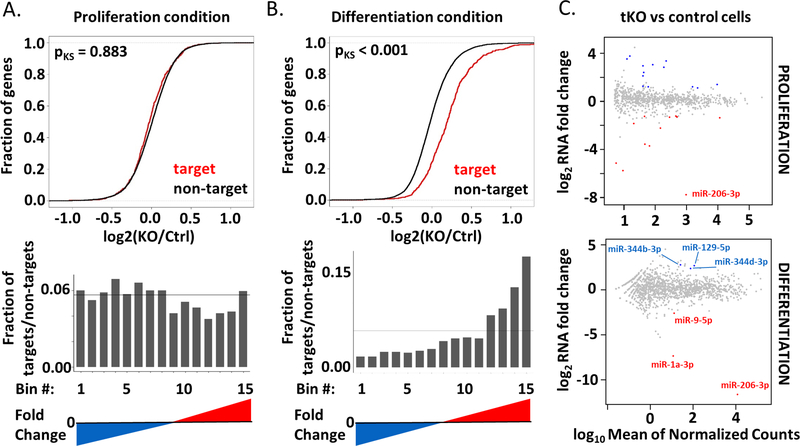
Figure 3.
Targets of miR-206/−1a are specifically upregulated in differentiating triple KO clones and are not affected by differentially expressed miRNAs.
A) Top: Cumulative distribution function (CDF) plot showing no change of the miR-206/−1 targets in proliferating triple KO clones (KO) versus control cells (Ctrl). The curves for the microRNA targets and non-targets are virtually superimposed. Bottom: Histogram of fraction of the miR-206/−1 targets, among all bins arranged from the most repressed to most induced in tKO vs Ctrl cells in growth medium. The genes are ranked from the most downregulated (blue) to the most upregulated (red). The horizontal line depicts the uniform distribution expected under the null hypothesis. Targets are based on match to 7mer sequence.
B) Top: Cumulative distribution function (CDF) plot showing upregulation of miR-206/−1 targets in differentiating triple KO clones (KO) versus differentiating control cells (Ctrl). Bottom: Histogram similar to that in Figure 3A, but in differentiating medium.
C) MA plots to identify differentially expressed miRNAs between control cells and triple KO clones in proliferation (top) and differentiation conditions (bottom). The microRNA abundance was measured by small RNA-seq. Upregulated microRNAs are presented in blue and downregulated in red (padj<0.1).
To address the hypothesis that other miRNAs could be upregulated in triple KO cells to compensate for the lack of miR-206/−1a we performed small RNA-seq in the tKO C2C12 cells. We detect 13 up- and 11 downregulated miRNAs in proliferating tKO cells in comparison to control cells (Figure 3C, S3D, E), and 3 up- and 1 downregulated miRNAs in differentiated tKO in comparison to control cells (Figure 3C, S3F). Nevertheless, the differentially expressed (particularly induced) miRNAs in tKO differentiating cells do not share seed sequence homology with miR-206/−1a, and as shown in Figure 3B, do not repress the miR-206/−1a targets. Thus it is unlikely that increase in other microRNAs in the tKO cells repress miR-206/1 targets to compensate for the lack of the latter, though we cannot rule out that another microRNA is more active in the tKO myoblasts to compensate for the lack of the three myo-miRs. We also found the variable change of the linked miR-133 gene product (2–4 fold increase or decrease) which does not meet the FDR < 0.1 threshold (Table S3).
Lack of miR-206, miR-1a-1 and miR-1a-2 leads to partial embryonic lethality
In order to generate triple KO animals, we first bred single KO of miR-206, miR-1a-1 or miR-1a-2, and then their double heterozygous offspring to obtain double KO animals (Figure 4A). Next, we crossed double KO of miR-206 and miR-1a-1 animals with double KO of miR-206 and miR-1a-2 animals, and subsequently their offspring (miR-206 KO miR-1a-1 HET miR-1a-2 HET) (Figure 4A). The genotypes were confirmed by PCR. Out of 127 genotyped animals we obtained 2 triple KO males. The expected probability of tKO mice was 6.25% and the observed one was 1.57% (Figure 4B), which means the lack of these three miRNAs leads to partial embryonic lethality. This partial embryonic lethality is probably due to the absence of miR-1a in the heart, based on previously reported lethality of miR-1a-1 miR-1a-2 double KO animals (49, 50) and the embryonic lethality we observe for miR-1a-1 and miR-1a-2 double KO animals. Lack of miR-206 and miR-1a expression in the skeletal muscle of the tKO animals was confirmed by q-RT-PCR (Figure 4C).
Adult triple KO animals have worse physical performance than control mice
Adult triple KO (tKO) mice had the same body weight as the control mice (Figure 4D). To test skeletal muscle function of the tKO animals we measured their grip strength in comparison to control, single KO and double KO mice. Even though the weights of the animals are very similar, there are differences in the force generated. The triple KOs are the weakest animals and this difference is significant in comparison to control, all single KOs and miR-206 miR-1a-2 double KO animals (Figure 4E). miR-206 miR-1a-1 double KO and miR-206 miR-1a-2 double KO were also significantly weaker than control animals (Figure 4E). The Rotarod experiments reveals that the tKO animals fall off the rotarod at an earlier acceleration than the other animal groups. They not only perform significantly worse in this test than control and all single KO animals, but also do not improve between Day 2 and Day 3 as do the other animals (Figure 4F). The tKO mice also have the lowest maximal speed on the third day of experiment of all animals tested (Figure 4G). The progressive decrease in performance from the single KO animals to the double KO animals and further decrease in the triple KO animals confirms that the three microRNAs do compensate for each other and are functionally redundant in skeletal muscle.
Skeletal muscles and hearts of triple KO animals reveal morphological abnormalities
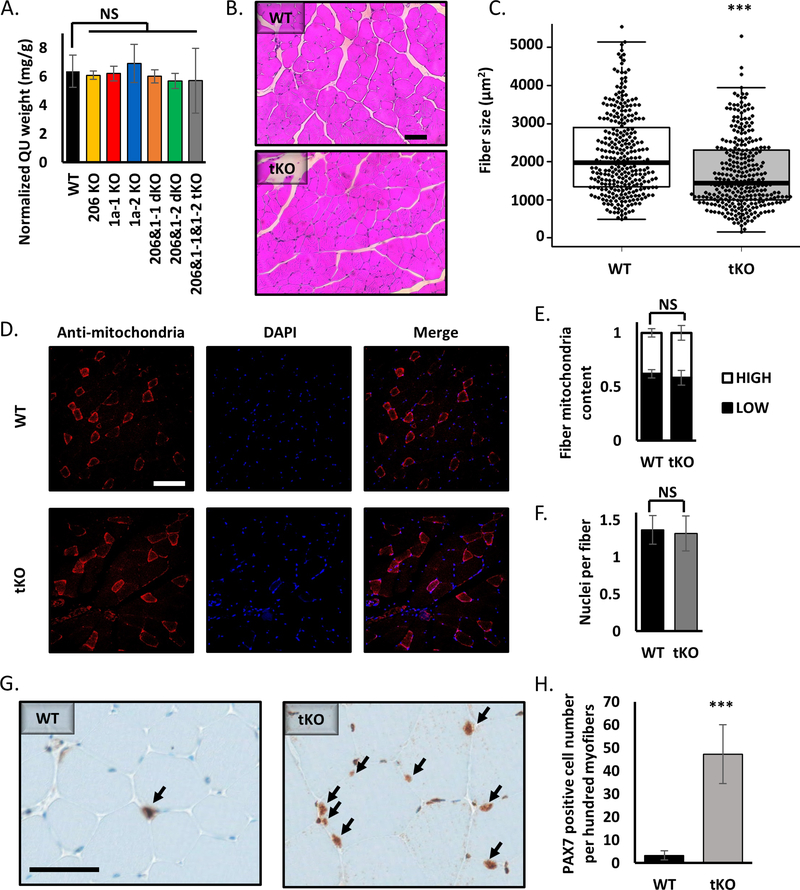
Even though the quadriceps size is very similar in all experimental mice (Figure 5A), the H&E staining shows that the average fiber cross-sectional area in the tKO animals lacking miR-206 and miR-1a is significantly smaller than in wild type mice (Figure 5B, 5C). Similar difference can be seen in the gastrocnemius (Figure S4A). Based on previous literature report (51) we decided to check the mitochondria content and organization in muscle fibers. Staining with anti-mitochondria antibody (Figure 5D) does not reveal any differences in fiber type content (Figure 5E). We also did not see a change in number of nuclei visible per fiber cross-section (Figure 5F). Moreover, we do not observe mitochondrial aggregates in the center of fibers or granulate-like patterns in tKO skeletal muscle fibers, phenotypes that were seen in miR-1a/−133a knockouts (51). PAX7 immunohistochemistry of quadriceps muscle reveals a 15x increase in PAX7 positive satellite cell number in the tKO animals (Figure 5G, 5H). Western blot analysis on TA muscles clearly shows upregulation of miR-206/−1a targets proteins: NOTCH3, PAX7 and CCND1 (Figure S4B). The size of heart in the triple KO animals is comparable to that in wild type (Figure S4A). Masson’s trichrome staining shows accumulation of fibrotic tissue in tKO, but not miR-206 miR-1a-1 dKO or miR-206 miR-1a-2 dKO hearts (Figure S4B).
Figure 5.
Knockout of miR-206, miR-1a-1 and miR-1a-2 leads to smaller muscle fiber diameter, but does not alter the content of mitochondria or nuclei in skeletal muscles.
A) Quadriceps muscle (QU) weight from animals used in this study. Statistical significance was calculated using t-student test. N = 5 for all group, except miR-206&1–1&1–2 tKO, where N=2.
B) Representative picture of hematoxylin and eosin (H&E) staining of quadriceps muscle cross-section from WT and tKO mice. Black line = 100μm.
C) Quantification of average fiber size based on H&E staining. Statistical significance was calculated using t-student test. (***) indicates p-value =< 0.001. N = 300 fibers per group.
D) Representative picture of anti-mitochondria staining of quadriceps muscle cross-section from WT and tKO mice. White line = 200μm.
E) Quantification of fiber mitochondria content based on anti-mitochondria staining. High mitochondria content fibers represent type I and IIa, low – type IIb and IId. Statistical significance was calculated using t-student test. N = 600 fibers per group.
F) Quantification of nuclei per fiber number based on DAPI (nuclei) and anti-mitochondria (fibers) staining. Statistical significance was calculated using t-student test. N = 600 fibers per group.
G) Representative picture of PAX7 immunohistochemistry of quadriceps muscle cross-section from WT and tKO mice. Black arrows point to the PAX7 positive cells. Black line = 50μm.
H) Quantification of PAX7 positive cell number based on IHC. Statistical significance was calculated using t-student test. (***) indicates p-value =< 0.001.
Discussion
Over the last decade many studies showed that myomiRs are important for muscle development and function (76–78). miR-206, miR-1a-1 and miR-1a-2 are genes encoding mature miR-206 and miR-1a, muscle specific miRNAs (myomiRs), which not only have identical seed sequence, suggesting functional redundancy, but also are highly expressed in cardiac and skeletal muscle along with miR-133a and miR-133b. miR-206, miR1a-1 and miR-1a-2 were suggested to be a critical factor for skeletal muscle differentiation (7, 8, 19, 20, 36–52). However, there was no model where all three microRNA genes are knocked out to prove the essentiality of these myomiRs in myogenesis.
Our study argues against an essential role of miR-206, miR-1a-1 and miR-1a-2 in skeletal muscle differentiation, but supports their functional importance in optimal skeletal muscle differentiation and performance. C2C12 myoblasts lacking miR-206, miR-1a-1 and miR-1a-2 are still able to differentiate, producing promyogenic mRNAs and proteins, and forming normal myotubes. These findings are true both for PAX7 negative and PAX7 positive murine myoblasts. Although differentiation was observed, in our RNA-seq there was a decrease in expression of genes in mitochondria- and skeletal muscle - related pathways in the triple KO C2C12 clones’ during differentiation. RNA-seq based screens and Western blot analyses revealed that the targets of these microRNAs remain de-repressed. The co-clustering of RNA-seq data from Ctrl and tKO cells in Figure 2A emphasize the similarities between differentiating Ctrl and tKO cells, despite the de-repression of the miR-206/1 targets, prove that the tKO C2C12 cells are able to differentiate well. Thus the simultaneous knockout of the three microRNAs has the expected de-repression of target genes and yet they are not essential for myogenesis. Loss of miR-1a/−133a leads to induction of Mef2a expression and further induction of Dlk1-Dio3 mega gene cluster (51). In our miR-206 miR-1a-1 miR-1a-2 triple KO model despite the decrease of miR-1a, Mef2a level is not changed in comparison to control cells, and the Dlk1-Dio3 gene cluster miRNA are not induced. This suggests that the changes seen in Wüst et al. (51) must be due to the loss of miR-133a.
miR-1 was previously reported as a mitochondrial translation enhancer during muscle differentiation (79). It enters the mitochondria and together with Ago2 but not GW182 stimulates the translation of specific mitochondrial genome-encoded transcripts. The triple KO C2C12 cells show a decrease in the maximal oxygen consumption rate suggesting mitochondrial dysfunction. The mitochondrial respiratory capacity in tKO C2C12 clones is significantly lower than in control C2C12 myoblasts and myotubes. Interestingly, Wüst et al. (51) observed mitochondrial aggregation and accumulation in the skeletal muscle fibers of miR-1a/−133a Pax7+-Cre mice, whereas we do not see changes in mitochondria structure or localization in our triple KO cells/muscles, suggesting that this phenotype may be caused by lack of miR-133a. Therefore, the miR-206 family (in this report) and miR-133a (51) are independently required for normal mitochondrial function.
An in vivo model was generated by breeding single knockout animals in order to obtain animals lacking two or three miRNA genes. We obtained miR-206 miR-1a-1 and miR-206 miR-1a-2 double knockout animals with ratios similar to expected Mendelian ratios. miR-1a-1 miR-1a-2 dKO animals were embryonically lethal, most likely due to an essential function of these microRNAs in cardiac differentiation, as published previously (49, 50). Unexpectedly, we successfully generated triple knockout animals, clearly showing that these miRNAs are collectively not essential for skeletal muscle formation. The Mendelian ratio of the triple knockout mice is one-fourth of the expected rate and the decrease is statistically significant, suggesting that the viability of the tKO mice was not routine. We hypothesize that tKO could be generated in a mixed genetic background, which may decrease embryonic lethality, as reported for miR-1a-1 single KO animals by Heidersbach (50). The tKO males themselves did not yield any progeny despite multiple attempts at breeding. We hypothesize the triple KO males were infertile as even though breeding was observed, none of the females got pregnant. Interestingly, the mice lacking miR-206, miR-1a-1 and miR-1a-2 had worse physical performance than wild type, single miRNA knockout and double miRNAs knockout animals. The triple KO mice have weaker muscles and perform worse at rotarod test than wild type control animals. Although the rotarod results may be affected by cardiac defects of KO animals, the decrease in grip strength and decrease in skeletal myofiber diameter clearly show that skeletal muscle function is impaired. We have also found a striking (~15-fold) increase in number of PAX7 positive satellite cells in triple KO quadriceps muscles, not only confirming the knockout of the miR-206 family, but also opening the field for new questions about the role of these microRNAs in satellite cell proliferation and activation. We did not investigate adult muscle regeneration in the tKO mice, because regeneration studies require a larger experimental group than the two tKO mice we obtained. Since the miR-206 KO mice show a defect in regeneration after cardiotoxin injury (54), we are certain that regeneration after acute injury will also be impaired in the TKO mice. Note that the cardiotoxin injury experiments demand extensive and rapid myogenesis, much more than is necessary during normal development, and do not think that a requirement of the microRNAs during such regeneration dilutes our conclusion that these three genes, miR-206, miR-1a-1 and miR-1a-2 (and miR-1b) are not essential for normal skeletal muscle development and differentiation. Taken together, these findings emphasize that even though miR-206, miR-1a-1 and miR-1a-2 are not essential for the skeletal muscle formation, they are required for maintenance of its full physical capability. We also found that miR-206 KO or miR-1a-2 KO animals perform worse than WT mice in rotarod test, which, to our knowledge, is the first demonstration that even single microRNA gene deletion decreases the neuromuscular and cardiac performance. This effect increases with each additional microRNA gene deletion.
Extensive cooperation between several miRNAs, mRNAs and proteins leads to effective differentiation. Our study shows that lack of three specific myomiRs miR-206, miR-1a-1 and miR-1a-2 does not prevent myogenesis, but may lead to functional impairment of skeletal muscles, emphasizing the complex and non-binary nature of skeletal muscle differentiation. Although the overexpression of miR-206 promotes the in vitro differentiation (39), the effects of microRNA overexpression could be due to non-physiological levels of the microRNAs. The CRISPR-Cas9 derived knock out cell lines and knock out mice that we have described in this paper are the more precise approaches to test the role of these microRNAs in physiological differentiation, because we simply deplete the physiological levels of specific microRNAs. Combinatorial knockout of additional known myomiRs may be necessary to fully answer the question if any of these miRNAs are essential for myogenesis. Moreover, this study underscores the importance of joint in vitro and in vivo full knockout studies by showing that even small alterations in differentiation at the cellular level, may have a significant impact on physical activity with the adult skeletal muscle.
There is an interesting comparison to be made between the functional redundancy of the three myomiRs studied here and of the MyoD, Myf5 and Mrf4 trascription factor family. Just as overexpression of miR-206 pushes myoblasts towards differentiation (39), overexpression of Myod1 induces myogenic conversion of fibroblasts (80, 81) and ectopic Myod1 expression drives terminal differentiation of pluripotent stem cells into skeletal myotubes upon chemical treatment (82). Myod1 knockout in vitro inhibits the myoblast differentiation completely (83), but in vivo Myod1 knockout mice develop normal skeletal muscles (84). In a similar vein, knockout of individual microRNA genes, miR-206, miR-1a-1 or miR1a-2, did not interfere with differentiation in vivo (41, 49, 50, 68), but affected the regeneration of skeletal muscle after acute or chronic injury (54, 68). However, Myod1 is dispensable for skeletal muscle development in mice because of the functional redundancy between Myod1 and Myf5, so that mice with double knockout of the two genes do not show skeletal myogenesis (85). In a different Myod1 and Myf5 double KO model, where Mrf4 expression was not compromised, the embryonic myogenesis was rescued (86). Thus the myogenic transcription factors are essential for differentiation, with the essentiality masked by redundancy in actions of the three related transcription factors. This is not the case for the three microRNA genes, where even after removal of all three functionally redundant genes, skeletal muscle differentiation still occurs in vitro and in vivo: the three microRNAs studied here are not essential for differentiation.
However, the three microRNAs are clearly important for optimal skeletal muscle differentiation, judging by the defects in mitochondrial function, myofiber diameters and skeletal muscle performance that we report in this paper. We also demonstrate that it is possible to genetically separate the functional role of miRNAs coming from bicistronic loci like miR-1a and miR-133a and to genetically examine functional redundancy of as many as three microRNAs miR-206, miR-1a-1 and miR-1a-2.
Supplementary Material
Acknowledgments
We thank all members of the Dutta lab for helpful discussion, especially Dr. Etsuko Shibata for technical guidance regarding CRISPR/Cas9 method. We thank Research Histology Core, Genome Analysis and Technology Core, and Biorepository & Tissue Research Facility at University of Virginia (Charlottesville, USA). We thank Dr. Scott Zeitlin and Elise Braatz for help with grip strength measurement, and Ashley Bolte for help with rotarod experiment. This work was supported by a grant from the NIH (RO1 AR067712) (AD). RKP was supported by a Predoctoral Fellowship from the American Heart Association (18PRE33990261). The authors declare no competing financial interests.
Nonstandard abbreviations
- miRNA
microRNA (i.e. miR-206, miR-1a-1, miR-1a-2)
- mRNA
messenger RNA
- GM
growth medium
- DM
differentiation medium (i.e. DM3 – 3rd day of differentiating medium)
- WT
wild-type
- KO
knock-out
- dKO
double knock-out
- tKO
triple knock-out
- FDR
False Discovery Rate
- GO
Gene Ontology
- GSEA
Gene Set Enrichment Analysis
- CDF
Cumulative Distribution Function
- OCR
Oxygen Consumption Rate
- SD
standard deviation
- SEM
standard error of the mean
References
- 1.Wang Y, Medvid R, Melton C, Jaenisch R, and Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39, 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi WY, Giraldez AJ, and Schier AF (2007) Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274 [DOI] [PubMed] [Google Scholar]
- 3.Rosa A, Spagnoli FM, and Brivanlou AH (2009) The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16, 517–527 [DOI] [PubMed] [Google Scholar]
- 4.Krichevsky AM, Sonntag KC, Isacson O, and Kosik KS (2006) Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao C, Sun G, Li S, and Shi Y (2009) A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 16, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, and Gao FB (2010) MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Samal E, and Srivastava D (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220 [DOI] [PubMed] [Google Scholar]
- 8.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, and Wang DZ (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, and Srivastava D (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar S, Dey BK, and Dutta A (2010) MiR-322/424 and −503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell 21, 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey BK, Gagan J, Yan Z, and Dutta A (2012) miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev 26, 2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, and Stein GS (2008) A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A 105, 13906–13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, Xu K, Sheng ZF, Zhou HD, Wu XP, and Luo XH (2009) A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest 119, 3666–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson SJ, Zhang Z, Feng D, Flagg M, O’Loughlin E, Wang D, Stokes N, Fuchs E, and Yi R (2013) Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development 140, 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Zhang Z, O’Loughlin E, Wang L, Fan X, Lai EC, and Yi R (2013) MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol 15, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CZ, Li L, Lodish HF, and Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, and Croce CM (2008) MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol 15, 352–358 [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y, Wang D, Wang F, Li T, Dong L, Liu H, Ma Y, Jiang F, Yin H, Yan W, Luo M, Tang Z, Zhang G, Wang Q, Zhang J, Zhou J, and Yu J (2013) A comprehensive analysis of GATA-1-regulated miRNAs reveals miR-23a to be a positive modulator of erythropoiesis. Nucleic Acids Res 41, 4129–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, and Wang DZ (2010) microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol 190, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey BK, Gagan J, and Dutta A (2011) miR-206 and −486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 31, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang A, Dong C, Li B, Zhang Z, Chen Y, Ning C, Wu W, and Liu H (2019) MicroRNA-206 regulates cell proliferation by targeting G6PD in skeletal muscle. FASEB J, fj201900502RRRR [DOI] [PubMed] [Google Scholar]
- 22.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, and Weiss MJ (2008) A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A 105, 3333–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, and O’Carroll D (2010) The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207, 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, and Jacks T (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, and Avraham KB (2009) MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A 106, 7915–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, and Barres BA (2010) Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohana R, Weiman-Kelman B, Raviv S, Tamm ER, Pasmanik-Chor M, Rinon A, Netanely D, Shamir R, Solomon AS, and Ashery-Padan R (2015) MicroRNAs are essential for differentiation of the retinal pigmented epithelium and maturation of adjacent photoreceptors. Development 142, 2487–2498 [DOI] [PubMed] [Google Scholar]
- 28.Bhinge A, Namboori SC, Bithell A, Soldati C, Buckley NJ, and Stanton LW (2016) MiR-375 is Essential for Human Spinal Motor Neuron Development and May Be Involved in Motor Neuron Degeneration. Stem Cells 34, 124–134 [DOI] [PubMed] [Google Scholar]
- 29.Parker MH, Seale P, and Rudnicki MA (2003) Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet 4, 497–507 [DOI] [PubMed] [Google Scholar]
- 30.Chargé SB, and Rudnicki MA (2004) Cellular and molecular regulation of muscle regeneration. Physiol Rev 84, 209–238 [DOI] [PubMed] [Google Scholar]
- 31.MAURO A (1961) Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9, 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung TH, and Rando TA (2013) Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olguín HC, and Pisconti A (2012) Marking the tempo for myogenesis: Pax7 and the regulation of muscle stem cell fate decisions. J Cell Mol Med 16, 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, and Beauchamp JR (2006) Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119, 1824–1832 [DOI] [PubMed] [Google Scholar]
- 35.O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, and Harfe BD (2007) Essential role for Dicer during skeletal muscle development. Dev Biol 311, 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokol NS, and Ambros V (2005) Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev 19, 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon C, Han Z, Olson EN, and Srivastava D (2005) MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A 102, 18986–18991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, and Lodish HF (2006) Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A 103, 8721–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HK, Lee YS, Sivaprasad U, Malhotra A, and Dutta A (2006) Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson C, Catoe H, and Werner R (2006) MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 34, 5863–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, and Srivastava D (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell 129, 303–317 [DOI] [PubMed] [Google Scholar]
- 42.Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, and Münsterberg A (2008) Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol 321, 491–499 [DOI] [PubMed] [Google Scholar]
- 43.Yuasa K, Hagiwara Y, Ando M, Nakamura A, Takeda S, and Hijikata T (2008) MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct 33, 163–169 [DOI] [PubMed] [Google Scholar]
- 44.Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, and Giraldez AJ (2009) Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev 23, 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, and Asakura A (2010) MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol 191, 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, and Phylactou LA (2011) Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol 11, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goljanek-Whysall K, Pais H, Rathjen T, Sweetman D, Dalmay T, and Münsterberg A (2012) Regulation of multiple target genes by miR-1 and miR-206 is pivotal for C2C12 myoblast differentiation. J Cell Sci 125, 3590–3600 [DOI] [PubMed] [Google Scholar]
- 48.Gagan J, Dey BK, Layer R, Yan Z, and Dutta A (2012) Notch3 and Mef2c Proteins Are Mutually Antagonistic via Mkp1 Protein and miR-1/206 MicroRNAs in Differentiating Myoblasts. J Biol Chem 287, 40360–40370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wystub K, Besser J, Bachmann A, Boettger T, and Braun T (2013) miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet 9, e1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heidersbach A, Saxby C, Carver-Moore K, Huang Y, Ang YS, de Jong PJ, Ivey KN, and Srivastava D (2013) microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. Elife 2, e01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wüst S, Dröse S, Heidler J, Wittig I, Klockner I, Franko A, Bonke E, Günther S, Gärtner U, Boettger T, and Braun T (2018) Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by miR-1/133a-Mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster. Cell Metab 27, 1026–1039.e1026 [DOI] [PubMed] [Google Scholar]
- 52.Vergara HM, Ramirez J, Rosing T, Nave C, Blandino R, Saw D, Saraf P, Piexoto G, Coombes C, Adams M, and Domingo CR (2018) miR-206 is required for changes in cell adhesion that drive muscle cell morphogenesis in Xenopus laevis. Dev Biol 438, 94–110 [DOI] [PubMed] [Google Scholar]
- 53.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, and Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, and Olson EN (2012) microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122, 2054–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, and Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, and Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G, Wang LG, Han Y, and He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal V, Bell GW, Nam JW, and Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin M (2011) Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. pp. 10–12, EMBnet.journal [Google Scholar]
- 64.Faridani OR, Abdullayev I, Hagemann-Jensen M, Schell JP, Lanner F, and Sandberg R (2016) Single-cell sequencing of the small-RNA transcriptome. Nat Biotechnol 34, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 65.Gebert D, Hewel C, and Rosenkranz D (2017) unitas: the universal tool for annotation of small RNAs. BMC Genomics 18, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang H, and Wong WH (2008) SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24, 2395–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kozomara A, Birgaoanu M, and Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47, D155–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, and Olson EN (2009) MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang CC, Bajikar SS, Jamal L, Atkins KA, and Janes KA (2014) A time- and matrix-dependent TGFBR3-JUND-KRT5 regulatory circuit in single breast epithelial cells and basal-like premalignancies. Nat Cell Biol 16, 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halevy O, and Lerman O (1993) Retinoic acid induces adult muscle cell differentiation mediated by the retinoic acid receptor-alpha. J Cell Physiol 154, 566–572 [DOI] [PubMed] [Google Scholar]
- 71.Lamarche É, Lala-Tabbert N, Gunanayagam A, St-Louis C, and Wiper-Bergeron N (2015) Retinoic acid promotes myogenesis in myoblasts by antagonizing transforming growth factor-beta signaling via C/EBPβ. Skelet Muscle 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.El Haddad M, Notarnicola C, Evano B, El Khatib N, Blaquière M, Bonnieu A, Tajbakhsh S, Hugon G, Vernus B, Mercier J, and Carnac G (2017) Retinoic acid maintains human skeletal muscle progenitor cells in an immature state. Cell Mol Life Sci 74, 1923–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vauchez K, Marolleau JP, Schmid M, Khattar P, Chapel A, Catelain C, Lecourt S, Larghéro J, Fiszman M, and Vilquin JT (2009) Aldehyde dehydrogenase activity identifies a population of human skeletal muscle cells with high myogenic capacities. Mol Ther 17, 1948–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jean E, Laoudj-Chenivesse D, Notarnicola C, Rouger K, Serratrice N, Bonnieu A, Gay S, Bacou F, Duret C, and Carnac G (2011) Aldehyde dehydrogenase activity promotes survival of human muscle precursor cells. J Cell Mol Med 15, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vella JB, Thompson SD, Bucsek MJ, Song M, and Huard J (2011) Murine and human myogenic cells identified by elevated aldehyde dehydrogenase activity: implications for muscle regeneration and repair. PLoS One 6, e29226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Callis TE, Chen JF, and Wang DZ (2007) MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol 26, 219–225 [DOI] [PubMed] [Google Scholar]
- 77.Townley-Tilson WH, Callis TE, and Wang D (2010) MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol 42, 1252–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarthy JJ (2011) The MyomiR network in skeletal muscle plasticity. Exerc Sport Sci Rev 39, 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, and Fu XD (2014) MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, and Lassar AB (1988) MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411 [DOI] [PubMed] [Google Scholar]
- 81.Lattanzi L, Salvatori G, Coletta M, Sonnino C, Cusella De Angelis MG, Gioglio L, Murry CE, Kelly R, Ferrari G, Molinaro M, Crescenzi M, Mavilio F, and Cossu G (1998) High efficiency myogenic conversion of human fibroblasts by adenoviral vector-mediated MyoD gene transfer. An alternative strategy for ex vivo gene therapy of primary myopathies. J Clin Invest 101, 2119–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Genovese NJ, Domeier TL, Telugu BP, and Roberts RM (2017) Enhanced Development of Skeletal Myotubes from Porcine Induced Pluripotent Stem Cells. Sci Rep 7, 41833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cichewicz MA, Kiran M, Przanowska RK, Sobierajska E, Shibata Y, and Dutta A (2018) MUNC, an Enhancer RNA Upstream from the. Mol Cell Biol 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rudnicki MA, Braun T, Hinuma S, and Jaenisch R (1992) Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71, 383–390 [DOI] [PubMed] [Google Scholar]
- 85.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, and Jaenisch R (1993) MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 86.Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, and Tajbakhsh S (2004) Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466–471 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.