Abstract
There are no commercially available Food and Drug Administration–cleared rapid tests for Neisseria gonorrhoeae antimicrobial susceptibility testing. This study evaluated the performance of a laboratory-developed real-time polymerase chain reaction assay for genotyping the gyrA gene to determine antimicrobial susceptibility to ciprofloxacin. Validation and clinical performance of the gyrA assay were evaluated across 3 geographic locations (Los Angeles, San Francisco, Philadelphia). Following validation, clinical specimens were collected in Aptima Combo2® CT/NG transport medium from asymptomatic persons who tested positive for Neisseria gonorrhoeae and evaluated for assay percent reportable (i.e., proportion of N. gonorrhoeae–positive specimens that yielded a gyrA genotype). The percentage of gyrA genotyping results differed by laboratory and specimen type. The proportion of specimens that were reportable was best for urine/genital specimens (genotyped = 76.4% (95% confidence interval, 69.9–82%)) followed by rectal (genotyped = 67.2% (95% confidence interval, 63.4–70.6%)) and then pharyngeal specimens (genotyped = 36.1%, (95% confidence interval, 31.9–40.5%)). Overall, asymptomatic patients with N. gonorrhoeae yielded an interpretable genotype 57.2% (784/1370) of the time, of which 480 were wild-type gyrA, resulting in 61% (480/784) being potentially treatable with ciprofloxacin.
Keywords: Antibiotic resistance, Gonorrhea, Antimicrobial stewardship, Molecular surveillance
1. Introduction
Gonococcal infections continue to pose a significant risk to global public health, resulting in an estimated 78 million cases per year (Newman et al., 2015). The U.S. Centers for Disease Control and Prevention (CDC) recognize infection with multidrug resistant Neisseria gonorrhoeae as 1 of the 3 most urgent antimicrobial resistance threats in the United States (U.S.) (CDC, 2013). Gonococcal infections are the second most commonly reported notifiable sexually transmitted infection (STI) in the U.S., second only to Chlamydia trachomatis (CDC, 2015). Genome plasticity and acquired antimicrobial resistance, combined with the absence of antimicrobial susceptibility tests that provide results in a clinically relevant time frame for treatment of gonococcal infections, make antimicrobial resistance in this organism especially difficult to control (Buono et al., 2015).
In an effort to reduce the spread of antimicrobial resistant gonococci, the World Health Organization and the CDC regularly update the guidelines for the treatment of gonococcal infections. The most up-to-date guidance recommends dual therapy consisting of a single dose of 250 mg of intramuscular ceftriaxone and 1 g of oral azithromycin (Workowski et al., 2015). The first reported N. gonorrhoeae case with dual therapy failure in the United Kingdom occurred in 2016 (Fifer et al., 2016). N. gonorrhoeae with decreased susceptibility to ceftriaxone and/or azithromycin is most prevalent in Europe and Western Asia; however, cases have been reported in the United States and Canada, as well as some countries in South America, South Africa, and Australia (Wi et al., 2017). Fluoroquinolones are well-tolerated oral antimicrobials used historically for the successful treatment of gonococcal infections. The CDC eliminated that drug class as an empirical treatment choice for N. gonorrhoeae infections in 2007 due to the spread of fluoroquinolone-resistant N. gonorrhoeae (CDC, 2007. The most recent U.S. data indicate 19.2% of isolates of N. gonorrhoeae are resistant to ciprofloxacin nationwide (Kirkcaldy et al., 2016). As such, the majority of gonococcal infections could theoretically be treated with oral ciprofloxacin if the clinician knew the organism's ciprofloxacin susceptibility at the time of diagnosis.
Resistance to ciprofloxacin is the result of a highly conserved mutation in the serine 91 codon of the gyrase A (gyrA) gene (Tanaka et al., 1996). Multiple studies have previously demonstrated that targeting codon 91 in gyrA is a highly sensitive and specific method for rapid detection of ciprofloxacin susceptibility, directly from clinical specimens positive for N. gonorrhoeae (Allan-Blitz et al., 2017; Hemarajata et al., 2016a, b; Siedner et al., 2007; Tanaka et al., 1996). Herein, we describe the implementation and analytical performance of this gyrA genotyping assay at multiple clinical and public health laboratories. This assay has been utilized as part of a larger clinical trial (NCT02961751) evaluating the efficacy of treatment using ciprofloxacin for patients infected with strains of N. gonorrhoeae with a wild-type (WT) gyrA genotype, which are predicted to be ciprofloxacin susceptible (Allan-Blitz et al., 2017; Hemarajata et al., 2016a, b; Siedner et al., 2007).
2. Materials and methods
2.1. Overview of the study
Three testing facilities participated in this study: UCLA Clinical Microbiology Laboratory (UCLA), San Francisco Public Health Laboratory (SFPHL), and the Philadelphia Public Health Laboratory (PPHL). UCLA served as the study coordinator. UCLA generated seeded samples for the assay validation and prepared and pretested lots of primers, probes, and DNA controls for the testing facilities. The study was conducted in 3 phases. Phase 1 consisted of validation of the gyrA assay across the testing facilities. Phase 2 consisted of clinical testing of asymptomatic patients at the facilities. Phase 3 consisted of troubleshooting experiments performed at UCLA to attempt to determine the root cause of indeterminate results.
2.1.1. Assay description
For all 3 phases, the gyrA genotype assay was used as has been described elsewhere (Hemarajata et al., 2016a, b; Siedner et al., 2007). This assay was performed using specimens that were previously determined to be N. gonorrhoeae positive by commercially available nucleic acid amplification testing (NAAT) and were reflexed for gyrA genotyping to determine ciprofloxacin susceptibility. The following modifications were performed in this study design: At UCLA, DNA extraction was performed using a MagNA Pure 2.0 (Roche Diagnostics, Indianapolis, IN) and the MagNA Pure DNA large-volume kit. The other 2 facilities (SFPHL and PPHL) performed DNA extraction on a QIAcube (Qiagen, Valencia, CA) using the Qiagen QIAmp DNA Mini DNA extraction kit. In either case, 200 μL of specimen was extracted and 100 μL DNA eluted. PCR (gyrA genotype assay) was performed using 5 μL DNA template and 15 μL FastStart® DNA Master HybProbe mix (Hemarajata et al., 2016a, b) using a Roche LightCycler 480 (UCLA and PPHL) or a Roche LightCycler 2.0 (SFPHL).
Positive controls were made by extracting 200 μL of a 3.0 McFarland suspension of an MT (FQ4) and WT (FQ1) isolate on a MagNA Pure and eluting 100 μL template DNA. The negative control consisted of Neisseria meningitidis DNA. This species is known to cross-react with the gyrA primers but not probes. A collection of other non–N. gonorrhoeae isolates (Neisseria meningitidis, Neisseria sicca, Neisseria subflava, Neisseria mucosa, Neisseria cinerea, and Neisseria elongata) was previously evaluated and determined to not cross-react with the assay (Hemarajata et al., 2016a, b).
Data from all facilities were analyzed using the Meltcurve Genotyping Module in the Multi Color HybProbe Detection Format of the Lightcycler software (Hemarajata et al., 2016a, b). The melt temperature corresponded to the value obtained at the peak of curves, which is generated after taking the negative value of the first derivative of fluorescence generated per unit time according to Siedner et al. (2007). Samples with melt temperatures of 56 °C ± 1.5 °C were designated as mutant (MT), and samples with melt temperatures of 66 °C ± 1.5 °C were designated as wild type (WT).
2.2. Phase I: validation of gyrA genotype assay
2.2.1. Validation specimens
Accuracy, precision, and limit of detection (LOD) were evaluated at all 3 facilities using panels of contrived specimens, which were prepared at UCLA and shipped to the participating facilities. SFPHL and PPHL were blinded to the expected results for each panel. Contrived specimens were generated by first pooling N. gonorrhoeae–negative, deidentified Aptima Combo2® CT/NG NAAT remnants by specimen type (urine, rectal, pharyngeal, and genital). Twenty-two different isolates of N. gonorrhoeae were plated for isolation on Chocolate Agar plates (Remel, Lenexa, KS) and incubated for 24 h at 37 °C in a 5% CO2 environment. A suspension equivalent to a 3.0 McFarland was made from the colony growth in 0.85% saline (Hardy Diagnostics, Santa Ana, CA). Next, 4.5-mL aliquots of the pooled specimens were seeded with 500 μL of the individual bacterial suspensions. Ten-fold serial dilutions were prepared in pooled sample remnants to achieve expected concentrations of 102–105 CFU/mL N. gonorrhoeae for LOD studies. The accuracy was defined as the percent of seeded samples in the accuracy panel that were correctly identified as WT vs. MT by the gyrA assay based on knowledge of the genotype (at UCLA) for the isolate that was seeded into the remnant samples.
2.2.2. LOD studies
The LOD was defined for each specimen type (urine, rectal, and genital) as the lowest concentration of N. gonorrhoeae with a ≥90% detection rate for gyrA, which yielded an interpretable genotype based on melt-curve analysis, across 10 replicates. The LOD was predetermined this way at UCLA by testing a broad range of concentrations of MT and WT N. gonorrhoeae of each specimen type. The UCLA-defined LOD was then confirmed at each facility by testing 3 of each specimen type seeded with MT or WT N. gonorrhoeae at the UCLA-determined LOD, 1.0 log above the LOD, or 2.0 log above the LOD for a total of 18 specimens. Because each participating laboratory used different equipment, the LOD was not assumed to be the same across the different facilities and was reported as the lowest concentration at which 90% of samples were correctly genotyped for each specimen type at each participating laboratory.
2.2.3. Accuracy studies
Accuracy panels were prepared to range from 1 to 4 log above LOD. A total of 125 contrived specimens were included in the accuracy panel for each facility and included all 22 isolates of N. gonorrhoeae, which were randomly selected by UCLA. Each of the secondary laboratories (PPHL and SFPHL) was required to produce 95% concordant results to those obtained at the primary facility (UCLA) from the accuracy panel in order to satisfy the validation criteria.
2.2.4. Precision testing
The precision panel consisted of 15 contrived specimens seeded with a gyrA WT isolate in pooled specimen remnants (n = 5 each urine, rectal, and genital) and 15 contrived specimens seeded with a gyrA MT isolate in pooled specimen remnants (n = 5 each urine, rectal, and genital). N. gonorrhoeae was seeded at the LOD, 1 log above the LOD, or 2 log above the LOD. For intra-assay precision, each laboratory tested each specimen 3 times within a single run. For interassay precision, each laboratory tested the panel of specimens across 3 days. Results for the precision testing were considered satisfactory if all replicates on each day tested resulted in proper identification of MT or WT via melt-curve analysis.
2.3. Phase II: clinical testing
Clinical samples were collected as part of a larger clinical trial to determine the efficacy of ciprofloxacin for the treatment of asymptomatic N. gonorrhoeae infections with gyrA serine 91 genotype (NCT02961751). The study had several inclusion and exclusion criteria including but not limited to informed consent, age ≥ 18 years, and infection with a gyrA serine 91 WT genotype N. gonorrhoeae at ≥1 body site that is nonpharyngeal. Specimens were run at each laboratory using the validated assays to determine gyrA genotype.
2.4. Phase III: evaluation of indeterminate specimens
Some of the clinical specimens did not yield a genotype. Modifiable factors that could contribute to indeterminate results were further evaluated at 1 laboratory (UCLA). Total DNA added to the gyrA reaction was evaluated by maximizing the volume of specimen extracted (400 μLvs. 200 μL used in original protocol). The same 100-μL volume of product was eluted during this larger volume extraction. For those specimens, the volume of DNA added to each RT-PCR was also increased (20 μL in a 100-μL RT-PCR vs. 5 μL in a 20-μL RT-PCR). Sixty-five specimens (19 pharyngeal, 21 rectal, and 25 urine/genital) that initially yielded an indeterminate result were reevaluated using these modifications.
The potential for PCR inhibition from the Aptima Combo2® CT/NG assay specimen transport medium was also evaluated by pelleting cellular material from 40 of these 65 Aptima specimens (n = 16 rectal, n = 15 pharyngeal, and n = 9 urogenital) at 10,000×g for 10 min. Pellets were then resuspended in 200 μL PCR-grade water and subsequently extracted on the MagNA Pure (Roche) in parallel with 200 μL of the original specimen. These specimens were then retested using the original 20-μL gyrA genotyping assay.
2.5. Statistical analysis
2.5.1. Percent reportable
Prevalence calculations and confidence intervals for percent reportable genotype were calculated using the “Clinical Research Calculator 1” module at VasserStats.net.
2.5.2. Chi-square and Fisher's exact testing
Chi-square and Fisher's exact testing was used to evaluate the proportional differences for indeterminate results between the categorical variables laboratory (UCLA, SFPHL, and PPHL) and specimen type (urine, rectal, and pharyngeal) (socscistatistics.com).
3. Results
3.1. Results from validation of gyrA assay
3.1.1. Accuracy panel
All 3 laboratories successfully genotyped all 125 seeded clinical specimens in the accuracy panel, resulting in 100% concordance. No very major errors (i.e., false WT) or major errors (i.e., false MT) were observed (see Supplemental Data).
3.1.2. Precision panel
Intra- and interassay precision studies yielded results 100% concordant with expected.
3.1.3. LOD panel
All 3 of the laboratories were able to genotype specimens down to 9 × 102 CFU/mL with ≥90% detection of the gyrA results and 100% accurate genotype for all specimens genotyped.
3.2. Application of gyrA assay to clinical samples
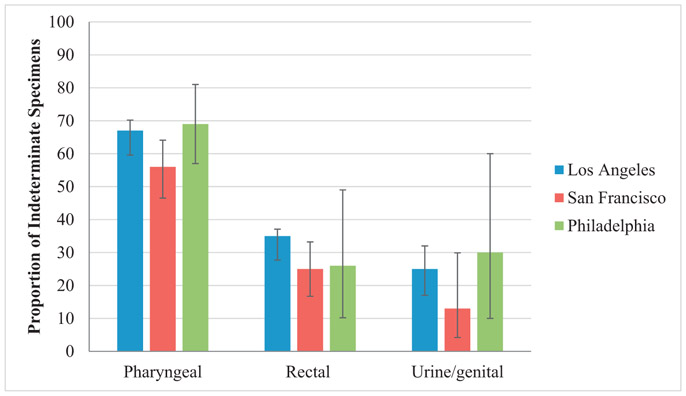
Launch of the assay at the 3 facilities was staggered, with 48 weeks of data available for UCLA, 37 for SFPHL, and 20 for PPHL. A total of 1370 prospective N. gonorrhoeae–positive specimens were tested across the 3 laboratories, including 49% (667/1370) rectal, 36% (499/1370) pharyngeal, and 15% (204/1370) urine/genital specimens. The percentage of N. gonorrhoeae–positive specimens that yielded a successful genotype varied considerably by specimen type (Fig. 1). Specimens that did not successfully yield a genotype result were considered “indeterminate.”
Fig. 1.
Frequency of indeterminate results for genotyping the gyrA gene from clinical specimen types. Error bars represent 95% confidence intervals.
As observed in previous studies, the indeterminate rate from pharyngeal specimens was high, ranging from 56% to 69% of all pharyngeal specimens tested by laboratories (Fig. 1) (Hemarajata et al. poster, 2016a, b). An unexpectedly high proportion of rectal swab specimens were also indeterminate, ranging from 24% to 35% of rectal specimens tested by laboratories. Of the urine/genital specimens tested, 13–30% were indeterminate. Therefore, the percentage of reportable specimens was the highest for urine/genital specimens, which were genotyped = 76.4% of the time (95% confidence interval, 69.9–82%).
Of the specimens that could be genotyped at UCLA, 57% (n = 319/ 557) were gyrA WT. At SFPHL, 72% (n = 134/185) were WT, and at PPHL, 64% (27/42) were WT (Table 1).
Table 1.
Proportion ofWT gyrA genotype results by study facility and specimen type.
| Specimen type | UCLA | SFPHL | PPHL |
|---|---|---|---|
| Pharyngeal | 64/110 (58%) | 44/65 (67%) | 3/5 (60%) |
| Rectal swab | 190/331 (57%) | 66/94 (70%) | 15/23 (65%) |
| Urine & genital swab | 65/116 (56%) | 24/26 (92%) | 9/14 (64%) |
| Column total | 319/557 (57%) | 134/185 (72%) | 27/42 (64%) |
The proportion of WT N. gonorrhoeae did not significantly vary by testing laboratory or anatomical source (P = 0.551, χ2 = 3.0425). The proportion of reportable results (WT and MT combined) also did not vary when compared by testing laboratory or anatomical source (P = 0.694, χ2 = 2.23). Therefore, post hoc pairwise statistical testing was not performed for reportable specimens combined.
Overall, the proportion of indeterminate N. gonorrhoeae varied significantly by testing laboratory and anatomical source (P < 0.00001, χ2 = 28.7). Therefore, post hoc pairwise Fisher's exact statistical testing was performed. There was a statistically significant number of indeterminate rectal and pharyngeal specimens between UCLA and SFPHL for (P < 0.05, P = 0.0016) as well as indeterminate urine and pharyngeal (P < 0.05, P = 0.02). The number of indeterminate pharyngeal and urine and indeterminate rectal and urine specimens also varied significantly between SFPHL and PPHL, respectively (P < 0.05, P = 0.0013 and P< 0.05, P = 0.0221). (See Table 2.)
Table 2.
Contingency table for χ2 analysis of indeterminate gyrA by laboratory and specimen type.
| Specimen type | UCLA | SFPHL | PPHL row total |
|---|---|---|---|
| Pharyngeal | 227 | 81 | 308 |
| Rectal swab | 180 | 31 | 211 |
| Urine & genital swab | 38 | 4 | 42 |
| Column total | 445 | 116 | 561 |
3.3. Further evaluation of indeterminate specimens at UCLA
The larger-volume (400 μL) DNA extraction and 100-μl gyrA PCR yielded a gyrA genotype for 10 of the 65 specimens (15%) tested by this method. Of 40 indeterminate specimens evaluated by pelleting and resuspension, 2 rectal specimens yielded a genotype (5%).
4. Discussion
Drug-resistant Neisseria gonorrhoeae continues to pose a threat to public health. The development of rapid molecular-based antimicrobial susceptibility testing can address this issue and potentially decrease resistance to antibiotics by offering more specific treatment options to physicians (Buono et al., 2015). The CDC recommend use of nucleic acid tests for the detection of N. gonorrhoeae infection (CDC, 2015). While exquisitely sensitive for N. gonorrhoeae, these assays do not provide data on antimicrobial susceptibility. As such, susceptibility data are currently only available for N. gonorrhoeae if a culture is performed, which is typically only conducted in the context of a suspected treatment failure. In this study, we successfully adapted a gyrA genotyping assay for prediction of ciprofloxacin susceptibility for use with DNA extracted from N. gonorrhoeae–positive specimens across 3 clinical laboratories. This study demonstrates that a second-step, genotype-based approach is feasible for real-time N. gonorrhoeae susceptibility testing in the U.S.
Important modifications of the assay as compared to our previously reported method include extraction of DNA from specimens in Aptima Combo2® CT/NG assay specimen transport medium. Hologic holds the majority market share (> 60%) for N. gonorrhoeae screening in the U.S. (Hologic, 2017), and as such, demonstration of the assay's feasibility with this specimen type is critical to potential downstream adoption by laboratories in the U.S. Additionally, we demonstrate that the test can be performed equally with 2 different extraction platforms: the QIAcube and MagNAPure. Specimens collected in Aptima Combo2® CT/NG assay specimen transport medium are stable at room temperature for 30 days, which may allow for specimen referral to local reference laboratories that can perform the gyrA assay. Indeed, this approach was used in Los Angeles where clinical specimens tested at the Los Angeles Public Health Laboratory were couriered to UCLA for reflex gyrA testing. This central-laboratory approach may increase the economic viability of performing a second-step gyrA genotype test, in particular for asymptomatic patients who may not have been treated empirically for N. gonnorhoeae infection (Alexander, 2009; Allan-Blitz et al., 2018).
The indeterminate frequency for the gyrA genotype assay was higher than initially expected (Hemarajata et al., 2016a, b) (Fig. 1). Nonetheless, results still conferred valuable information to clinicians about treatment options in 57.2% (784/1370) of the specimens tested. This low rate of positivity may relate to the fact that we only tested specimens from asymptomatic patients, as symptomatic patients were treated empirically with CDC-recommended therapy and excluded from this study. In contrast, in our previous work, all-comers were tested by the gyrA genotype assay (Hemarajata et al., 2016a, b). Because symptomatic patients may have higher N. gonorrheae load, adapting the method to a point-of-care environment may improve clinical sensitivity. Of 1370 prospective specimens tested in this study to date, a result of WT was obtained for 35% (480/1370). This suggests that treatment with ciprofloxacin might be appropriate for over a third of patients who would have been otherwise treated with dual therapy.
Of the specimens that were successfully genotyped, the assay performance was better for urine/genital specimens (77.3%; 133/172) compared to rectal (65.4%; 342/523) and pharyngeal specimens (34.4%; 142/413). The poor performance for pharyngeal specimens is similar to that observed previously: 63.6% pharyngeal specimens that tested positive for N. gonnorheae in a previous study were found to be indeterminate by gyrA assay (Hemarajata et al., 2016a, b). The reason for the poor performance for pharyngeal samples remains unclear. This may result because lower levels of N. gonorrhoeae are present in the pharynx (Alexander, 2009) or because of cross-reactivity/interference due to the presence of organisms like N. meningitidis or other Neisseria commensal species (Alexander, 2009; Low and Unemo, 2016). Probit analysis was previously used to evaluate the sensitivity of the gyrA assay. The Probit module 90 on XLSTAT (Addinsoft, New York, NY) determined that the COBAS® 4800 CT/NT assay crossing point (Cp) value of ≤28.15 was associated with ≥95% detection of a genotype by gyrA genotyping assay for 100 seeded and clinical samples (Hemarajata et al., 2016a, b). Thus, only specimens with a low COBAS® crossing point (i.e., higher bacterial load) could be genotyped by the gyrA assay (Hemarajata et al., 2016a, b). Similarly, a Cp of ≤24.6 for pharyngeal swabs, ≤29.1 for urine, and ≤38.5 for rectal swabs was calculated for ≥95% sensitivity (Hemarajata et al. poster, 2016a, b). While the COBAS® assay is not quantitative, the lack of correlation between Cp and successful amplification of gyrA for pharyngeal specimens may point to an inhibition rather than an LOD issue.
In contrast, we previously observed only 6.7% indeterminate results using remnant DNA from the COBAS system for rectal swabs, whereas in the present study, the overall indeterminate rate for rectal swabs was much higher (25–35%, Fig. 1). It is possible that this discrepancy between the current data and our prior observations is due to the superior sensitivity of the Hologic® transcription-mediated amplification chemistry, which targets high–copy number 16S rRNA from N. gonorrhoeae (Buckley et al., 2016, Chernesky, 2002), as compared to the RT-PCR chemistry targeting DNA used by the COBAS® system. Neither assay is FDA-cleared for pharyngeal swabs; as such, LOD data are not available for that specimen type. However, comparison of the LOD for urine and genital specimens on the Aptima®, as compared to what is observed for the gyrA assay, demonstrates the expected differences of amplifying a low- versus high-copy target. The analytical sensitivity of the Aptima® assay is 250 CFU/mL in urine and 362 CFU per genital swab (Hologic® package insert, 2017). Our assay has an LOD of 900 CFU/mL for these 2 specimen types. As such, it is not surprising that our genotype detection rate was lower. While we observed limited success in maximizing the total amount of DNA added to reactions, this modification resulted in a more costly test due to increased reagent volumes in the RT-PCR and increased consumables.
Overall, of all genotyped specimens, 61% (480/784) were WT, which reflect the most current CDC data from GISP on ciprofloxacin susceptibility rates for N. gonorrhoeae in these regions (Elizabeth Torrone, personal communication to JD Klausner). Specifically, ciprofloxacin susceptibility rates as evaluated by the GISP program in 2016 were 70.3%, 77.3%, and 54.9%, respectively, for San Francisco, Philadelphia, and Los Angeles (Kirkcaldy et al., 2016). These data nicely reflect the 72%, 64%, and 57% WT gyrA genotypes initially observed in this study (Table 1). At the very least, the gyrA method, along with our previously published penA genotyping assay (Wong et al., 2017), could be used at these public health laboratories to monitor N. gonorrhoeae susceptibility in real time.
5. Conclusions
In summary, we demonstrate the introduction and use of a laboratory-developed, genotype-based assay for the detection of N. gonorrhoeae ciprofloxacin susceptibility across 3 laboratories. While detection rates were somewhat lower than expected, the assay yielded a genotype for a majority of specimens tested, potentially allowing more targeted therapy for more than half of these patients. Further developments of novel strategies to detect susceptibility are paramount to the health of both the public and individuals affected by gonococcal disease. Future data from this trial will evaluate the treatment outcomes for patients with WT gyrA results that were treated with ciprofloxacin.
Acknowledgments
We thank Hologic for providing specimen collection media free of charge. We are grateful to the following collaborators from the Neisseria gonorrhoeae gryA Study Group for their ardent participation, data collection, innovation, and commitment to providing exceptional participant care: Azza Abdin (PDPH); Sagar Amin (AHF); Onika Anglin (PDPH); Lenore Asbel (PDPH); Robert Bolan (LA LGBT); Degina Booker (MSDH); Claire Bristow (UCSD); Kerry Buchs (PDPH); Anthony Cristillo (SSS); Gregory Deye (DMID); Anne DeMeis (PDPH); Olivia Ellis (UCLA); Omai Garner (UCLA); Vicky Caruthers (UCLA); Tami Truong (UCLA); Stephanie Horiuchi (UCLA); Samuel Rosenthal (UCLA); Brian Bolan (UCLA); Mikkel Cheng (UCLA); Risa Flynn (LA LGBT); Elizabeth Formentini (DMID); Morgan Gapara (SSS); Elizabeth Hawkins (SSS); Amelia Herrera (SFDPH); Lei Huang (DMID); R. M. Humphries (Accelerate Diagnostics); Shelley Campeau (Accelerate Diagnostics); Peera Hemarajata (LACPHL); Nikki Green (LACPHL); Ruel Torres (LACPHL); Eduardo Ramos (LACPHL); Mwenda Kudumu (SSS); Gwendolyn Maddox (SSS); Godfred Masinde (SFDPH); Mark McGrath (AHF); Cassandra McQuaid (SFDPH); Sheldon Morris (UCSD); Mark Pandori (ACPHD); Susan Philip (SFDPH); Stephanie Piper (EMMES); Jonathan Powell (EMMES); Drew Ragland (SSS); Mindy Ratra (PDPH); Ali Talan (LA LGBT); Phoebe Lyman (LA LGBT); Stephanie Trammell (SFDPH); Daphne Ware (MSDH); Michael Wierzbicki (EMMES); Peter A. Wolff (DMID); Toni Waymer (SSS); Arick Wong (SFDPH); AIDS Healthcare Foundation (AHF); Alameda County Public Health Department (ACPHD); Division of Microbiology and Infectious Diseases, NIAID, NIH (DMID); The Emmes Corporation (EMMES); Los Angeles County Public Health Laboratory (LACPHL); Los Angeles LGBT Center (LA LGBT); Mississippi State Department of Health (MSDH); Philadelphia Department of Public Health (PDPH); San Francisco Department of Public Health (SFDPH); Social & Scientific Systems, Inc. (SSS); University of California, San Diego (UCSD).
Funding
Overall support for the STAR Sexually Transmitted Infections Clinical Trials Group (STAR STI CTG) and protocol DMID 15-0090 is provided by the National Institute of Allergy and Infectious Diseases Division of Microbiology and Infectious Diseases (HHSN272201300014I/HHSN27200006). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Declaration of Interest: Specimen collection media were provided free of charge by Hologic.
References
- Alexander S The challenges of detecting gonorrhoea and chlamydia in rectal and pharyngeal sites: could we, should we, be doing more? Sex Transm Infect 2009;85: 159–60. [DOI] [PubMed] [Google Scholar]
- Allan-Blitz L, Hemarajata P, Humphries RM, Wynn A, Segura E, Klausner JD. A cost analysis of gyrase A testing and targeted ciprofloxacin therapy versus recommended 2-drug therapy for Neisseria gonorrhoeae infection. Sex Transm Dis 2018;45(2):87–91. 10.1097/OLQ.0000000000000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan-Blitz L-T, Humphries RM, Hemarajata P, Pandori MW, Siedner MJ, Klausner JD. Implementation of a rapid genotypic assay to promote targeted ciprofloxacin therapy of Neisseria gonorrhoeae in a large health system. Clin Infect Dis 2017;64:1268–70. 10.1093/cid/ciw864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C, Trembizki E, Donovan B, Chen M, Freeman K, Guy R, et al. A real-time PCR assay for direct characterization of the Neisseria gonorrhoeae GyrA 91 locus associated with ciprofloxacin susceptibility. J Antimicrob Chemother 2016;71(2):353–6. 10.1093/jac/dkv366. [DOI] [PubMed] [Google Scholar]
- Buono SA, Watson TD, Borenstein LA, Klausner JD, Pandori MW, Godwin HA. Stemming the tide of a drug-resistant Neisseria gonorrhoeae: the need for an individualized approach to treatment. J Antimicrob Chemother 2015;70:374–81. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control & Prevention (CDC). Update to CDC's sexually transmitted diseases treatment guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. Morb Mortal Wkly Rep 2007;56:332–6. [PubMed] [Google Scholar]
- Centers for Disease Control & Prevention (CDC). Antibiotic resistance threats in the United States. Atlanta, GA: Centers for Disease Control and Prevention; 2013. [Google Scholar]
- Centers for Disease Control & Prevention (CDC). Sexually transmitted disease surveillance 2014. Atlanta, GA: Centers for Disease Control and Prevention; 2015. [Google Scholar]
- Chernesky MA. Chlamydia trachomatis diagnostics. Sex Transm Infect 2002;78:232–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifer H, Natarajan U, Alexander S, Hughes G, Golparian D, Unemo M. Failure of dual antimicrobial therapy in treatment of gonorrhea. N Engl J Med 2016;374(25). 10.1056/NEJMc1512757. [DOI] [PubMed] [Google Scholar]
- Hemarajata P, Yang S, Humphries RM, Klausner JD. American Society for Microbiology. Poster: Clinical Sensitivity of a Real-Time PCR Assay Targeting gyrA Gene for Prediction of Ciprofloxacin Resistance in Neisseria gonorrhoeae; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P, Yang S, Soge OO, Humphries RM, Klausner JD. Performance and verification of a real-time PCR assay targeting the gyrA gene for prediction of ciprofloxacin resistance in Neisseria gonorrhoeae. J Clin Microbiol 2016;54:805–8. 10.1128/JCM.03032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hologic proxy statement. http://eproxymaterials.com/interactive/holx2017proxy, 2017. Hologic APTIMA Combo 2® assay package insert. http://www.hologic.com/sites/default/files/package%20inserts/502487-IFU-PI_001_01_1.pdf.
- Kirkcaldy RD, Harvey A, Papp JR, del Rio C, Olusegun SO, King HK, et al. Neisseria gonorrhoeae antimicrobial susceptibility surveillance – the gonococcal isolate surveillance project, 27 sites, United States, 2014. MMWR Surveill Summ 2016;65(SS-7):1–19. 10.15585/mmwr.ss6507a1. [DOI] [PubMed] [Google Scholar]
- Low N, Unemo M. Molecular tests for the detection of antimicrobial resistant Neisseria gonorrhoeae: when, where, and how to use? Curr Opin Infect Dis 2016;29(1 ):45–51. [DOI] [PubMed] [Google Scholar]
- Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, Low N, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE 2015;10 (12), e0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedner MJ, Pandori M, Castro L, Barry P, Whittington WL, Liska S, et al. Real-time PCR assay for detection of quinoloneresistant Neisseria gonorrhoeae in urine samples. J Clin Microbiol 2007;45:1250–4. 10.1128/JCM.01909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Otsuki M, Nishino T, Kobayashi I, Matsumoto T, Kumazawa J. Mutation in DNA gyrase of norfloxacin-resistant clinical isolates of Neisseria gonorrhoeae. Genitourin Med 1996;72(4):295–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi T, Lahra MM, Ndowa F, Bala M, Dillon J-AR, Ramon-Pardo P, et al. Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med 2017;14(7), e1002344 10.1371/journal.pmed.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LK, Henarajata P, Soge OO, Humphries RM, KlausnerJD. Real-time PCR targeting penA mosaic XXXIV type for prediction of extended-spectrum cephalosporins susceptibility in clinical Neisseria gonorrhoeae isolates. Antimicrob Agents Chemother 2017; 61(11), e01339–7. 10.1128/AAC.01339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workowski KA, Bolan GA, Centers for Disease Control and Prevention. Sexually transmitted diseases treatment guidelines. MMWR Recomm Rep 2015;64(RR-03):1–137. 10.15585/mmwr.rr6404a1. [DOI] [PMC free article] [PubMed] [Google Scholar]